Abstract
Background
Foot-and-mouth disease (FMD) caused by foot-and-mouth disease virus (FMDV) is one of the most highly infectious diseases in livestock, and leads to huge economic losses. Early diagnosis and rapid differentiation of FMDV serotype is therefore integral to the prevention and control of FMD. In this study, a series of serotype-specific reverse transcription recombinase polymerase amplification assays combined with lateral flow dipstick (RPA-LFD) were establish to differentiate FMDV serotypes A, O or Asia 1, respectively.
Results
The serotype-specific primers and probes of RPA-LFD were designed to target conserved regions of the FMDV VP1 gene sequence, and three primer and probe sets of serotype-specific RPA-LFD were selected for amplification of FMDV serotypes A, O or Asia 1, respectively. Following incubation at 38 °C for 20 min, the RPA amplification products could be visualized by LFD. Analytical sensitivity of the RPA assay was then determined with ten-fold serial dilutions of RNA of VP1 gene and the recombinant vector respectively containing VP1 gene from FMDV serotypes A, O or Asia1, the detection limits of these assays were 3 copies of plasmid DNA or 50 copies of viral RNA per reaction. Moreover, the specificity of the assay was assessed, and there was no cross reactions with other viruses leading to bovine vesicular lesions. Furthermore, 126 clinical samples were respectively detected with RPA-LFD and real-time PCR (rPCR), there was 98.41% concordance between the two assays, and two samples were positive by RPA-LFD but negative in rPCR, these were confirmed as FMDV-positive through viral isolation in BHK-21 cells. It showed that RPA-LFD assay was more sensitive than the rPCR method in this study.
Conclusion
The development of serotype-specific RPA-LFD assay provides a rapid, sensitive, and specific method for differentiation of FMDV serotype A, O or Asia1, respectively. It is possible that the serotype-specific RPA-LFD assay may be used as a integral protocol for field detection of FMDV.
Keywords: FMDV, Serotype-specific, Recombinase polymerase amplification, Lateral flow dipstick
Background
Foot-and-mouth disease (FMD) is an acute, highly contagious disease that affects cloven-hoofed animals, often resulting in huge economical losses in terms of trade and animal productivity. Recent outbreaks of FMD in Taiwan, Japan, South Korea and the United Kingdom have directly caused the culling of millions of animals, compensated heavily by the government [1, 2]. The responsible virus, FMD virus (FMDV), was a single-stranded positive-sense RNA virus belonging to the Aphthovirus genus in the family Picornaviridae. There were seven serotypes including O, A, C, Asia 1, and South African Territories (SAT) 1, 2 and 3, which together manifest a distinct geographical distribution [3]. FMDV serotype A, C and O are widely distributed across the world while Asia 1 and SAT 1–3 mainly occur in Asia and Africa, respectively. Several outbreaks of FMD serotype Asia 1, O and A have been recorded in mainland provinces of Southern China during 1999–2013 [4–6]. Early diagnosis of FMDV is therefore essential to providing valuable epidemiological information, and initiating the appropriate prevention and control strategies.
FMDV can be detected from blood, esophageal-pharyngeal fluid, nasal fluid, saliva, and other excretions of FMDV infected animals before clinical symptoms [7, 8] start to show. Currently, there are three typical assays for FMDV diagnosis including virus isolation, antigen enzyme-linked immunosorbent assay (Ag-ELISA) and real-time RT-PCR (rRT-PCR) used in FMDV reference laboratories [9]. However, these diagnostic tests require special equipment and professionally trained personnel. Another alternative propose is to use isothermal assays for diagnosis of FMDV. To date, there are four isothermal assays to detect FMDV: reverse transcription loop-mediated isothermal amplification (RT-LAMP) [10, 11], reverse transcription recombinase polymerase amplification (RT-RPA) [12], and nucleic acid sequence based amplification [13, 14]. RT-LAMP and RT-RPA have also been used to distinguish various serotypes in clinical samples [11, 12]. However, LAMP assay needs more primers than PRA, leading to longer amplicons and difficult designs in cases of highly variable viruses.
The RPA method is probably the one promising direction capable of rapid diagnosis of many different pathogens [12, 15–18]. The amplification relies on recombinase, single stranded binding protein, and strand displacing DNA polymerase at a constant temperature. The RPA products could be analyzed with gel electrophoresis, fluorescence monitoring based on probes, or simple visualization with a lateral flow dipstick (LFD) [19–22].
In the present study, a reverse transcription serotype-specific RPA-LFD assay was established, and evaluated as a field method for diagnosis and typing of FMDV serotypes A, Asia 1 or O, respectively.
Results
Design and optimization of serotype-specific RPA primers and probe
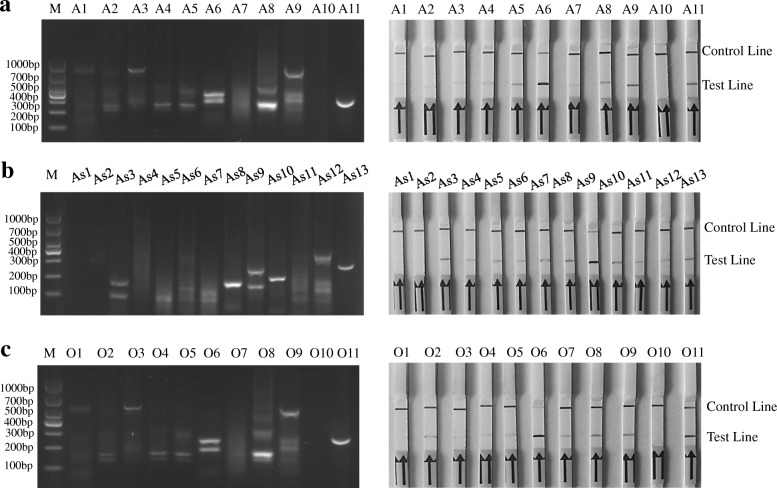
The TwistAmp nfo reactions were performed to screen the candidate primer/probes for the RPA-LFD assay, the products were analyzed on 2% agarose gel. A6, As9, O6 sets of the primer/probes were respectively screened as serotype-specific RPA primers and probe for FMDV serotypes A, Asia-1, or O. They respectively produced 346 bp and 334 bp, 286 bp and 182 bp, 231 bp and 190 bp amplification products and their RPA-LFD test line appeared faster and darker than other sets within 5 min (Table 1 and Fig. 1).
Table 1.
The sequences of primers and probes designed for screening in the study
| Primer/probe set Name | Sequence(5′ → 3′) | The location on accession number | Product sizes (bp) | |
|---|---|---|---|---|
| F2 | ATGGAGCACCTGAGGCAGCACTGGACAACA | 3452–3481 | 221 | |
| A1 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 203 |
| R1 | [Biotin]CGTTGAGAAGGGCACAGTCGTATTGAAACA | 3643–3672 | ||
| F2 | ATGGAGCACCTGAGGCAGCACTGGACAACA | 3452–3481 | 258 | |
| A2 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 250 |
| R2 | [Biotin]GCACGAGGAGTTCTTGGATCTCCGTGGCTC | 3680–3709 | ||
| F2 | ATGGAGCACCTGAGGCAGCACTGGACAACA | 3452–3481 | 352 | |
| A3 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 229 |
| R3 | [Biotin]GCAGGGGCAATAATTTTCTGCTTGTGTCTG | 3774–3803 | ||
| F1 | CACCTGAGGCAGCACTGGACAACACGAGCAA | 3458–3488 | 215 | |
| A4 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 203 |
| R1 | [Biotin]GCCGTAGTTGAAGGAGGCAGGAAGCTGTGC | 3643–3672 | ||
| F1 | CACCTGAGGCAGCACTGGACAACACGAGCAA | 3458–3488 | 252 | |
| A5 | P1 | [FAM] CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 240 |
| R2 | [Biotin]GCACGAGGAGTTCTTGGATCTCCGTGGCTC | 3680–3709 | ||
| A6 (Optimal set) | F1 | CACCTGAGGCAGCACTGGACAACACGAGCAA | 3458–3488 | 346 |
| P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 334 | |
| R3 | [Biotin]GCAGGGGCAATAATTTTCTGCTTGTGTCTG | 3774–3803 | ||
| F3 | TGAGGCAGCACTGGACAACACGAGCAACCC | 3462–3461 | 211 | |
| A7 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 203 |
| R1 | [Biotin]GCCGTAGTTGAAGGAGGCAGGAAGCTGTGC | 3643–3672 | ||
| F3 | TGAGGCAGCACTGGACAACACGAGCAACCC | 3462–3461 | 248 | |
| A8 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 240 |
| R2 | [Biotin]GCACGAGGAGTTCTTGGATCTCCGTGGCTC | 3680–3709 | ||
| F3 | TGAGGCAGCACTGGACAACACGAGCAACCC | 3462–3461 | 342 | |
| A9 | P1 | [FAM]CACTGGACAACACGAGCAACCCCACTGCTTA[dSpacer] TATAAAGCACCGTTCACA[C3-spacer] | 3470–3519 | 334 |
| R3 | [Biotin]GCAGGGGCAATAATTTTCTGCTTGTGTCTG | 3774–3803 | ||
| F1 | AAAAGCAACCCATTACCCGCCTGGCACTCC | 3582–3611 | 285 | |
| As1 | P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3596–3645 | 271 |
| R2 | [Biotin]GACTCTTCCCCGTAGGTTGTCTTCCCGTTG | 3837–3866 | ||
| As2 | F1 | AAAAGCAACCCATTACCCGCCTGGCACTCC | 3582–3611 | 145 |
| P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3596–3645 | 131 | |
| R3 | [Biotin]GGGAGTGCCAGGCGGGTAATGGGTTGCTTT | 3697–3726 | ||
| F2 | AACCCAACCGCCTACCAAAAGCAACCCATT | 3566–3595 | 153 | |
| As3 | P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3596–3645 | 123 |
| R1 | [Biotin]CGGTGTAAGGGAGTGCCAGGCGGGTAATGG | 3689–3718 | ||
| F2 | AACCCAACCGCCTACCAAAAGCAACCCATT | 3566–3595 | 301 | |
| As4 | P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3596–3645 | 271 |
| R2 | [Biotin]GACTCTTCCCCGTAGGTTGTCTTCCCGTTG | 3837–3866 | ||
| F2 | AACCCAACCGCCTACCAAAAGCAACCCATT | 3566–3595 | 161 | |
| As5 | P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3596–3645 | 131 |
| R1 | [Biotin]CGGTGTAAGGGAGTGCCAGGCGGGTAATGG | 3689–3718 | ||
| F3 | CGAATCAGCAGACCCAGTTACCACCACAGT | 3274–3303 | 445 | |
| As6 | P2 | [FAM]TGAAACTCACACAGCTCAAGAACACCCAAACT[dSpacer] TTGATCTTATGCAAATC[C3-spacer] | 3378–3427 | 341 |
| R1 | [Biotin]CGGTGTAAGGGAGTGCCAGGCGGGTAATGG | 3689–3718 | ||
| F3 | CGAATCAGCAGACCCAGTTACCACCACAGT | 3274–3303 | 593 | |
| As7 | P2 | [FAM]TGAAACTCACACAGCTCAAGAACACCCAAACT[dSpacer] TTGATCTTATGCAAATC[C3-spacer] | 3378–3427 | 489 |
| R2 | [Biotin]GACTCTTCCCCGTAGGTTGTCTTCCCGTTG | 3837–3866 | ||
| F3 | CGAATCAGCAGACCCAGTTACCACCACAGT | 3274–3303 | 453 | |
| As8 | P2 | [FAM]TGAAACTCACACAGCTCAAGAACACCCAAACT[dSpacer] TTGATCTTATGCAAATC[C3-spacer] | 3378–3427 | 349 |
| R3 | [Biotin]GGGAGTGCCAGGCGGGTAATGGGTTGCTTT | 3697–3726 | ||
| As9 (Optimal set) | F3 | CGAATCAGCAGACCCAGTTACCACCACAGT | 3274–3303 | 286 |
| P2 | [FAM]TGAAACTCACACAGCTCAAGAACACCCAAACT[dSpacer] TTGATCTTATGCAAATC[C3-spacer] | 3378–3427 | 182 | |
| R4 | [Biotin]GAGAAGTAGTACGTCGCAGACCGAAGTAGCG | 3530–3559 | ||
| F3 | CGAATCAGCAGACCCAGTTACCACCACAGT | 3274–3303 | 445 | |
| As10 | P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3378–3427 | 341 |
| R1 | [Biotin]CGGTGTAAGGGAGTGCCAGGCGGGTAATGG | 3689–3718 | ||
| F3 | CGAATCAGCAGACCCAGTTACCACCACAGT | 3274–3303 | 593 | |
| As11 | P1 | [FAM]ACCCGCCTGGCACTCCCTTACACCGCTCCC[dSpacer]ACCGTGTGCTTGCAACAGT[C3-spacer] | 3596–3645 | 271 |
| R2 | [Biotin]GACTCTTCCCCGTAGGTTGTCTTCCCGTTG | 3837–3866 | ||
| F1 | CAACACCACCAACCCAACGGCGTACCATAA | 3570–3599 | 161 | |
| O1 | P1 | [FAM]CGTACCATAAGGCGCCGCTTACCCGGCTTA[dSpacer] ATTGCCCTACACGGCACCA[C3-spacer] | 3590–3639 | 141 |
| R1 | [Biotin]GAGCCAGCACTTGGAGATCGCCTCTCACGT | 3701–3730 | ||
| F1 | CAACACCACCAACCCAACGGCGTACCATAA | 3570–3599 | 343 | |
| O2 | P1 | [FAM]CGTACCATAAGGCGCCGCTTACCCGGCTTA[dSpacer] ATTGCCCTACACGGCACCA[C3-spacer] | 3590–3639 | 323 |
| R2 | [Biotin]CAAGGACTGCTTTACAGGTGCCACTATTTT | 3883–3912 | ||
| F1 | CAACACCACCAACCCAACGGCGTACCATAA | 3570–3599 | 210 | |
| O3 | P1 | [FAM]CGTACCATAAGGCGCCGCTTACCCGGCTTA[dSpacer] ATTGCCCTACACGGCACCA[C3-spacer] | 3590–3639 | 190 |
| R3 | [Biotin]TTGATGGCACCGTAGTTGAAAGAAGTAGGC | 3751–3779 | ||
| F2 | GGAGCACCTGAAGCAGCCTTGGACAACACC | 3549–3578 | 182 | |
| O4 | P1 | [FAM]CGTACCATAAGGCGCCGCTTACCCGGCTTA[dSpacer] ATTGCCCTACACGGCACCA[C3-spacer] | 3590–3639 | 141 |
| R1 | [Biotin]GAGCCAGCACTTGGAGATCGCCTCTCACGT | 3701–3730 | ||
| F2 | GGAGCACCTGAAGCAGCCTTGGACAACACC | 3549–3578 | 364 | |
| O5 | P1 | [FAM]CGTACCATAAGGCGCCGCTTACCCGGCTTA[dSpacer] ATTGCCCTACACGGCACCA[C3-spacer] | 3590–3639 | 323 |
| R2 | [Biotin]CAAGGACTGCTTTACAGGTGCCACTATTTT | 3883–3912 | ||
| O6 (Optimal set) | F2 | GGAGCACCTGAAGCAGCCTTGGACAACACC | 3549–3578 | 231 |
| P1 | [FAM]CGTACCATAAGGCGCCGCTTACCCGGCTTA[dSpacer] ATTGCCCTACACGGCACCA[C3-spacer] | 3590–3639 | 190 | |
| R3 | [Biotin]TTGATGGCACCGTAGTTGAAAGAAGTAGGC | 3751–3779 | ||
| F3 | GGGGACCTTACCTGGGTGCCAAATGGAGCA | 3524–3553 | 217 | |
| O7 | P2 | [FAM]CAAATGGAGCACCTGAAGCAGCCTTGGACAA[dSpacer]ACCACCAACCCAACGGCGTAC[C3-spacer] | 3542–3596 | 189 |
| R1 | [Biotin]GAGCCAGCACTTGGAGATCGCCTCTCACGT | 3701–3730 | ||
| F3 | GGGGACCTTACCTGGGTGCCAAATGGAGCA | 3524–3553 | 389 | |
| O8 | P2 | [FAM]CAAATGGAGCACCTGAAGCAGCCTTGGACAA[dSpacer]ACCACCAACCCAACGGCGTAC[C3-spacer] | 3542–3596 | 371 |
| R2 | [Biotin]CAAGGACTGCTTTACAGGTGCCACTATTTT | 3883–3912 | ||
| F3 | GGGGACCTTACCTGGGTGCCAAATGGAGCA | 3524–3553 | 256 | |
| O9 | P2 | [FAM]CAAATGGAGCACCTGAAGCAGCCTTGGACAA[dSpacer]ACCACCAACCCAACGGCGTAC[C3-spacer] | 3542–3596 | 238 |
| R3 | [Biotin]TTGATGGCACCGTAGTTGAAAGAAGTAGGC | 3751–3779 | ||
Note: F:forward primer, R reverse primer, P probe, FAM Carboxyfluorescein, dSpacer A tetrahydrofuran residue, C3-spacer 3’-block
Fig. 1.
Screening of the primers/probes for the FMDV serotype-specific RPA-LFD assay. a Agarose gel electrophoresis and LFD detection of RT-RPA products amplified with different primer and probe sets of FMDV serotype A. Lane M was DNA Marker DL1000. A1 to A9 were different primer and probe sets. A6: the optimal primer and probe set, and the estimated size of the RPA amplified fragment were 346 bp and 334 bp. A10: negative control, (DNase-free water). A11: positive control (supplied by Twist Amp nfo kit). b Agarose gel electrophoresis and LFD detection of RT-RPA products amplified with different primers/probe sets of FMDV serotype Asia 1. As2 to As10 were different primer and probe sets. As9: the optimal primer/probe set, and the estimated size of the RPA amplified fragment were 286 bp and 182 bp. As1: negative control (DNase-free water). As10: positive control (supplied by Twist Amp nfo kit). c Agarose gel electrophoresis and LFD detection of RT-RPA products amplified with different primers/probe sets of FMDV serotype O. O1 to O9 were different primer and probe sets. O6: the optimal primer/probe set, and the estimated size of the RPA amplified fragment were 231 bp and 190 bp. O10: negative control (DNase-free water). O11: positive control (supplied by Twist Amp nfo kit)
Besides that, different reaction conditions were optimized for the serotype-specific RPA assay. Results showed that a reaction temperature at 38 °C (Fig. 2a) and a incubation time of 20 min or longer (Fig. 2b) can best promote the amplification efficiency. Thus, the amplification reaction for the serotype-specific RPA assay should be carried out at 38 °C for 20 min.
Fig. 2.
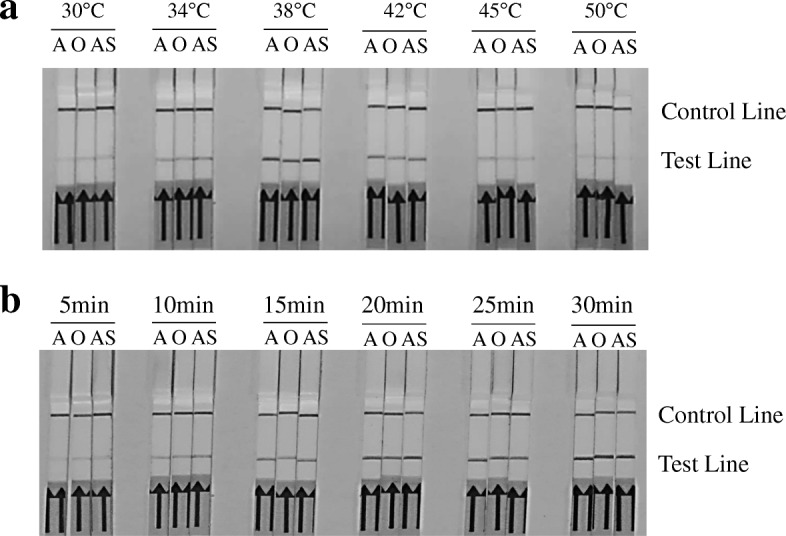
Optimization of reaction temperature and time for FMDV serotype-specific RPA-LFD assays. a The RPA-LFD performs effectively in a wide range of constant reaction temperatures. b The amplified products can be visible on the LFD at 5 min or longer
Sensitivity and specificity of the FMDV serotype-specific RPA reaction
To determine the sensitivity of RPA assay, positive vector and RNA standard of three FMDV serotypes were diluted from 3 × 106 to 3 × 100 copies/μL and 5 × 107 to 5 × 101 molecules/μL, respectively. RNA was reversely transcribed to cDNA. All reverse transcriptional cDNA and positive vectors of each dilution were respectively used as templates in the RPA reactions. All RPA reaction products were than respectively tested on LFD, and the limit of detection with the RPA-LFD was 3 × 100 dilution for positive vector (Fig. 3a) and 5 × 101 dilution for RNA standard (Fig. 3b).
Fig. 3.
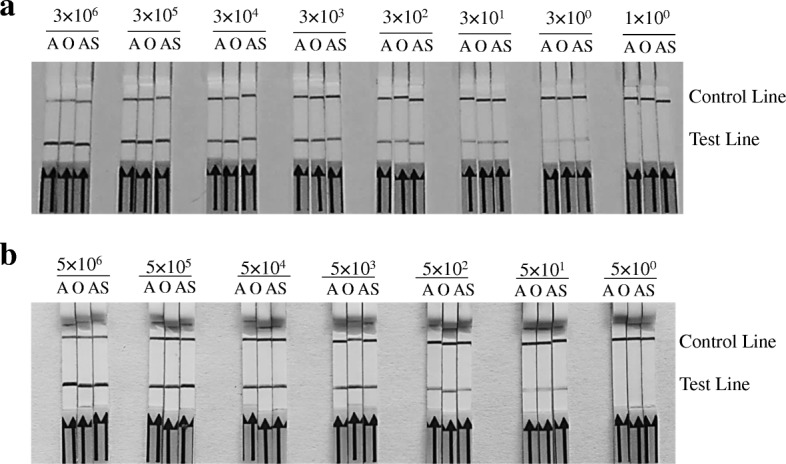
The sensitivity of FMDV serotype-specific RPA-LFD assays. a Sensitivity of the standard plasmids. Molecular sensitivity of RPA-LFD was determined using 10-fold serially diluted 3 × 106 to 3 × 100 copies and 100 copy of FMDV DNA standard plasmids per reaction as template. The minimum limits for virus detection of RPA-LFD were 3 × 100 copies. b Sensitivity of the RNA standard. The cDNA of reverse transcription using 10-fold serially diluted 5 × 106 to 5 × 100 RNA molecular was used in molecular sensitivity of RPA-LFD. The minimum limits detection of RPA-LFD were 5 × 100 RNA. A: primers/probe set of FMDV serotype A. AS: primers/probe sets of FMDV serotype Asia 1. O: primers/probe sets of FMDV serotype O. Samples were tested in triplicate with one reaction and independently repeated 3 times
The other viral pathogens with similar clinical signs, including BEV, BVDV, BEFV, VSV, IBRV, and SVDV, were used to assess the specificity of the assay. IBRV DNA and other viral cDNA were respectively detected with the RPA-LFD. There was no cross reaction with the other bovine viral pathogens with similar clinical signs (Fig. 4). The cDNA of other FMDV epidemic virus strains in China were used for detection of serotype-specific RPA-LFD. There was no cross-reactivity with different serotype strains of FMDV, so the specific primer and probe sets could differentiate the corresponding serotype virus.
Fig. 4.
The specificity of the FMDV RPA-LFD assays. Other bovine viral pathogens with similar clinic and etiologies were used to assess the specificity of the assays. There was no cross-reaction with BVDV, IBRV, BEV, BEFV, BVSV and SVDV. NC: negative control. A: primers/probe set of FMDV serotype A. AS: primers/probe set of FMDV serotype Asia 1. O: primers/probe set of FMDV serotype O. Samples were tested in triplicate with one reaction and three separate assays
Performance of FMDV serotype-specific RPA-LFD assay on clinical samples
To evaluate the diagnostic sensitivity of the FMDV RPA-LFD assay, cDNA obtained from each specimen of 126 clinical samples were detected with RPA-LFD and rPCR, respectively. RPA-LFD identified 41 negative and 85 positive samples (25 vesicular material, 14 saliva, 10 aerosol, 14 oesophageal-pharyngeal fluid, 9 blood and 13 nasal swabs) (Table 2). Of the 85 positive samples, 32 were serotyped as serotype A, 17 as serotype Asia 1, and 36 as serotype O. The concordance between FMDV RPA-LFD and rPCR was 98.41% (124/126).
Table 2.
Comparative performance of serotype-specific RT-LFD-RPA and rRT-PCR assays for detection of suspected clinical specimens and serotyping of FMDV
| LFD-RPA | Real-time | (qPCR) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Samples name | A | Asia 1 | O | FMDV positive | FMDV Negative | A | Asia 1 | O | FMDV positive | FMDV Negative | |
| vesicular material | 7 | 3 | 15 | 25 | 5 | 7 | 3 | 15 | 25 | 5 | |
| nasal swab | 6 | 2 | 5 | 13 | 7 | 6 | 2 | 5 | 13 | 7 | |
| saliva | 5 | 3 | 6 | 14 | 15 | 5 | 3 | 6 | 14 | 15 | |
| oesophageal pharyngeal fluid | 6 | 4 | 4 | 14 | 3 | 6 | 4 | 4 | 14 | 3 | |
| blood | 4 | 2 | 3 | 9 | 7 | 4 | 2 | 3 | 9 | 7 | |
| aerosol | 4 | 3 | 3 | 10 | 4 | 3 | 3 | 2 | 8 | 6 | |
| Total | 32 | 17 | 36 | 85 | 41 | 31 | 17 | 35 | 83 | 43 | |
It is worth noting that 4 aerosol specimens of FMDV serotype A were designated as positive by the RPA-LFD assay, but only 3 of them were FMDV serotype A positive detected by the rPCR, and 3 aerosol specimens of FMDV serotype O were differentiated with the RPA-LFD assay, whereas 2 of them were FMDV serotype O positive using the rPCR. BHK-21 cells were used to isolate viruses and identify the two inconsistent aerosol specimens, and CPE of cells inoculated with two inconsistent samples appeared untill the third passage. CPE-positive cell and control cell culture were respectively harvested and detected for FMDV using rPCR and RPA-LFD. As expected, CPE-positive cells were respectively identified for FMDV serotype O and A, and the control cells were FMDV negative, it showed that the sensitivity of RPA-LFD assay was higher than the rPCR in this study.
Discussion
There are a series of methods used as detection of FMDV, such as Ag-ELISA, virus separation, and rRT-PCR. However, the above methods have been either too time-consuming or require high-precision instruments to meet practical needs [19, 20]. RPA isothermal amplification techniques can amplify nucleic acids and detect the products without a requirement of special instrument or complex operations [21–24]. It is worth mentioning that human body heat can indeed incubate RPA reactions under certain limit resources [25]. Moreover, it is not required to store the lyophilized RPA reagents with a cooling chain because they can actually be stably stored at room temperature for a longer time [26].
Although it has been proved that the pan-specific real-time RT-RPA (rRT-RPA) and RPA-LFD technique can provide rapid and accurate diagnosis of FMDV [12, 18], they are difficult to distinguish the serotypes of FMDV for the rRT-RPA assay. The VP1, a surface exposed-capsid protein, take a pivotal role in the antigenicity as a major viral antigen, and plays an important role in pathogenicity of FMDV as its binding to viral receptors of host cells. Because of heterogeneity, the nucleotide sequence encoding VP1 is widely used to determine genetic relationships between different strains and to trace the provenance and transmission route of epidemic FMDV strains [27–30]. In this study, primer and probe sets specific for serotypes O, A, or Asia-1 FMDV were designed based on the alignment of the nucleotide sequences of viral VP1 gene of the above serotypes strains circulating in Asia, respectively. The primer and probe sets A6, As9, and O6 screened for RPA in this study could perform effective and accurate detection of different FMDV serotype of A/China/5/99, Asia1/AF/72, and O/HNK/CHA/05 (see Fig. 1) as intended. Furthermore, the serotype-specific RPA-LFD assay successfully detected the epidemic strains of FMDV in China (Table 2), and provided a more robust assessment method regarding the serotype specificity.
FMDV is mainly transmitted by aerosol. Viral RNA was detected in aerosol samples from FMDV suspected farm at 1–3 days before infected cattle appeared clinical signs [31]. Based on the aerogenous characteristics of the FMDV, it’s proven to be a valuable technique that aerosol samples were used to detect viral RNA in infected farms [31, 32]. In our study, viruses in aerosol were also detected and their serotypes were respectively differentiated by the FMDV RPA-LFD assay. Therefore, it may be the potential integral monitoring strategies for prevention, control and eradication of FMD using this technique.
It is a crucial step for clinical detection with any molecular diagnostic assay that the nucleic acids are extracted from tissue and cell samples, the Punch-it™ kit can easily isolate nucleic acid from different samples via paper chromatography, and becomes one of the ideal tools for the extraction of DNA/RNA. Recent study showed that the DNA isolated with the Punch-it™ kit could be used in molecular assays [33]. In our study, nucleic acids of different samples, including vesicular material, blood, oesophageal-pharyngeal fluid, saliva, aerosol, and nasal swabs, were successfully extracted using the Punch-it™ kit. The method only takes 10 min to extract the nucleic acid without centrifugation, so it is relatively simple and rapid, and it is even more important that the extracted nucleic acids can directly serve as templates in RPA-LFD assays. In the present study, the extracted total RNA needed to be reverse transcribed into cDNA in the two-step RPA-LFD assay, whereas reverse transcription and RPA were performed in one reaction using the TwistAmp™ exo kit (TwistDx Limited, UK) and with the addition of reverse transcriptase in previously studied RT-RPA assays [12]. We would like to further simplify the test to make it more suitable for field use in future. Moreover, sophisticated instrumentations were required in RT-RPA [12, 15, 16], whereas RPA-LFD assay in this study only needs a thermos metal bath for incubation at 38 °C and amplified products can be direct visible on the LFD without requirements of instruments. Therefore, it may be an effective way to detect clinical samples in the field.
Conclusions
In the present study, the serotype-specific FMDV RPA-LFD assay was successfully developed, and will be helpful for detection of FMDV infection during FMD outbreaks. Because RPA-LFD assay is a simple, specific, rapid, and serotype-specific method, it is possible to be a general differentiated protocol for diagnostics of FMDV, especially for detection of clinical samples in the field.
Methods
Virus and clinical specimens
In this study, cDNA of three serotypes of FMDV reference strains including type O strain China/5/99, type A strain AF/72, and type Asia 1 strain HNK/CHA/05, which were provided by Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, were positive controls of various serotypes used to optimize primer and probe sets of RPA. The cDNA of other epidemic FMDV strains in China including type A (HuBWH/2009, Mya98, GSLX/2010, GDMM/2013), type Asia1 (ZB/58, HeB/05, YS/05, HN/06, BR/Myanmar/06, WHN/06), and type O (LY/2000, CC/03, GZ/2010, BY/2010, HKN/2011, GD/2013, GD/2015), were used to provide a more robust assessment of serotype-specific RPA. Other viral pathogens causing similar clinical vesicular signs, including bovine ephemeral fever virus (BEFV), vesicular stomatitis virus (VSV), bovine viral diarrhea virus (BVDV), bovine enterovirus (BEV), infectious bovine rhinotracheitis virus (IBRV),and swine vesicular disease virus (SVDV), were stored by the Ruminant Diseases Research Center, Shandong Normal University, and used for cross-reactivity testing. To compare the detection sensitivity between RPA-LFD reactions and rPCR, 126 clinical specimens (30 vesicular materials, 29 salivas, 14 aerosols, 16 bloods, 17 oesophageal-pharyngeal fluids, and 20 nasal swabs) were collected from suspected cases of FMD in the Chinese endemic region from 2013 to 2017.
Isolation of viral RNA/DNA and synthesis of cDNA
Viral RNA and DNA were isolated using the MiniBEST Viral RNA/DNA Extraction Kit (TaKaRa, Dalian, China) following respective instructions. The amounts of viral RNA was measured using a Biophotometer plus (Eppendorf, USA). The extracted RNA was template used to synthesized cDNA with reverse transcription using random primers in a total volume of 10 μL according to the instructions of the PrimeScript™ RT Master Mix (Takara, Dalian, China). All viral DNA and cDNA were stored at − 70 °C for further employment.
Generation of DNA/RNA molecular standard
The viral VP1 gene recombinant vectors respectively containing RPA amplified region of FMDV serotype O, A, or Asia 1 (named pET32a-A-FMDV-VP1, pET32a-AS1-FMDV-VP1 and pET32a-O-FMDV-VP1) were constructed and used for the analytical sensitivity. The Positive plasmids were measured using a Biophotometer plus (Eppendorf, USA), respectively. The quantity of copies was calculated by the formula: DNA copy number (copies/μL) = (M × 6.02 × 1023 × 10− 9)/(n × 660), M: molecular weight, n:plasmid concentration (g/μL) measured at 260 nm.
RNA molecular standards were prepared as described in previous study [17] with certain modifications. The linearized recombinant vectors with SgrA I (New England Biolabs, USA) were purified with the MiniBEST DNA Fragment Purification Kit (Takara, Dalian, China), and then used as template for RNA transcription with the RiboMAX Large Scale RNA Production System-T7 (Promega, USA). Furthermore, the RNA was measured using the Quant-iTTM RiboGreen RNA Assay Kit (Thermo Fisher Scientific, Germany) according to the manufacturer’s instructions. The quantity of copies was calculated by the equation: Amount (copies/μL) = [RNA concentration (g/μL)/(transcript length in nucleotides× 340)] × 6.02 × 1023.
Design of serotype-specific RPA primers and probe
A multiple sequence alignment of FMDV strains of serotype A, Asia 1, or O was respectively performed to find highly conserved region of the FMDV VP1 gene. The following reference sequences of three serotypes found in GenBank database were respectively used: KT968663(A/HY/CHA/2013), FJ755082 (A/PAK/1/2006), KY322679 (A/TAI/4/2014), FJ755052 (A/IRN/51/2005), KY404935 (A/A01NL), EF149010 (Asia 1/HNK/CHA/05), EF614458 (Asia1/MOG/05), AY687334 (Asia1/IND 491/97), GU931682 (Asia1/YS/CHA/05), AY687333 (Asia1/IND 321/01), HQ009509 (O/China/5/99), LC149720 (O/JPN/2010–362/3), JN998086 (O/GZ/CHA/2010), AF095876(O/Taipei-150). RT-RPA primers and probes specific for serotypes O, A or Asia-1 of FMDV were designed against the consensus sequence for this region and were synthesized by Sangon Biotech, respectively. RPA primers/probes were synthesized and labeled as described in previous study [18]. Oligonucleotide sequences of RPA primers and probes of specific serotype A, Asia-1, or O of FMDV are listed in Table 1 (accession numbers KT968663, EF149010 and HQ009509, respectively).
FMDV serotype-specific RPA assays
Serotype-specific primers and probes were screened as described in previous study [25] with some modifications. In brief, RPA was performed using a TwistAmp™ nfo kit (TwistDx Limited, UK). The freeze-dried enzyme pellet was dissolved with 47.5 μLof solution containing 29.5 μL rehydration buffers, 2.1 μL forward and reverse primers (10 μM), 0.6 μL probe (10 μM), 11.2 μL of sterile water, 2 μL of cDNA of FMDV reference strains, and then 2.5 μL magnesium acetate (280 mM) was added. Assays were completed in a thermos metal bath at 38 °C for 20 min. The amplified products were then put through a 2% (w/v) agarose gel electrophoresis to screen the optimal and serotype-specific primer and probe sets. The optimal reaction conditions were determined by testing various reaction temperatures and incubation times.
LFD double label with anti-FAM gold conjugates and anti-Biotin antibodies (Milenia Biotec GmbH, Germany) were used to visualize the RPA amplified products. 1 μL of RPA products were diluted with 99 μL Dipstick Assay Buffer (Milenia Biotec GmbH, Germany), and then tested by LFD. FMDV serotype-specific positives are indicated by the visualization of both a test line and control line simultaneously perceptible on the LFDs after 5 min, while the negative reactions only generate a control line. The cDNA of other FMDV epidemic virus strains in China were used for evaluation of serotype-specific RPA-LFD.
Sensitivity and specificity of the RPA-LFD assay
To determine the DNA analytical sensitivity of the RPA-LFD assay, the recombinant plasmids pET32a-A-FMDV-VP1, pET32a-As1-FMDV-VP1, and pET32a-O-FMDV-VP1 were respectively the standard DNA template of FMDV serotype A, Asia 1 and O. The RPA-LFD assays were performed with ten-fold serial dilutions of the recombinant vector ranging from 3 × 106 to 3 × 100 copies per microliter for respective serotypes. To detect the RNA analytical sensitivity, RNA standards of three FMDV serotypes were diluted from 5 × 107 to 5 × 101 molecules/μL. 10 μL RNA was used as template to synthesize cDNA in 20 μL reverse transcription reaction system using PrimeScript™ RT Master Mix (Takara, Dalian, China) in accordance with the manufacturer’s instructions. 2 μL cDNA of each dilution was used as a template in the RPA reactions. DNA plasmid/RNA samples were detected with three separate assays, respectively.
The specificity of the method was assessed using other viral pathogens with similar clinical symptoms, including BEFV, VSV, BVDV, BEV, IBRV, and SVDV. The IBRV DNA was extracted and viral cDNA were reverse transcribed from isolated other viral RNA. Positive controls and negative controls for RPA were constructed using recombinant vectors and RNase free water.
Diagnosis of clinical specimens with FMDV serotype-specific RPA-LFD assays
To compare the diagnostic sensitivity between RPA-LFD and rPCR, 126 clinical specimens were collected from bovine farms suspected with infection of FMDV in China from 2013 to 2017 (details listed in Table 2). RNAs were isolated from the clinical samples using a Punch-it™ Kit (Nanohelix, Daejeon, South Korea) following manufacturers’ instructions. A 1 mm punched disk, containing the nucleic acids, was added with 10 μL of reverse transcript reaction system using PrimeScript™ RT Master Mix (Takara, Dalian, China), which contained 2 μL 5 × PrimeScript RT Master Mix and 8 μL RNase Free dH2O. Reverse transcription for each sample was completed in two tubes using a thermos metal bath at 37 °C for 15 min. 2 μL of cDNA was then used in both RPA-LFD and rPCR reactions. For the rPCR, serotype-specific primers and probes for serotypes O, A or Asia-1 FMDV were employed as previously described [23]. Reactions were performed with Premix Ex Taq™ Kit (Takara, Dalian, China) for respective serotypes.
Samples were positive in RPA-LFD, but negative in rPCR, were further tested for presence of FMDV. The virus was isolated using BHK-21 cells (provided by China Center for Type Culture Collection) as described in the previous study [18]. The cytopathic effect (CPE) was examined at 24 h, 48 h, and 72 h, respectively. If there was no CPE after 72 h, Cell cultures were passaged. CPE-positive, CPE-negative and control cell cultures were respectively harvested and examined again for FMDV using RPA-LFD and rPCR.
Acknowledgments
Yang He from Emory University in the United State revised the text of manuscript.
Funding
This study was partly supported by grants from National Natural Science Fund of China (31672556, 31872490, 31502064), Taishan Scholar and Distinguished Experts (H.HB), National Primary Research & Development Plan (2018YFD0501605–06), Primary Research & Development Plan of Shandong Province (2016GNC113006, 2018GNC113011).
Availability of data and materials
The data supporting the results of the study were included in the manuscript.
Abbreviations
- BEFV
Bovine ephemeral fever virus
- BEV
Bovine enterovirus
- BVDV
Bovine viral diarrhea virus
- ELISA
Immune-histopathology and enzyme-linked immunosorbent assay
- FMDV
Foot-and-mouth disease virus
- IBRV
Infectious bovine rhinotracheitis virus
- LAMP
Loop-mediated isothermal amplification
- LFD
Lateral flow dipstick
- PCR
Polymerase chain reaction
- RPA
Recombinase polymerase amplification
- rPCR
Real time PCR
- RT
Reverse transcription
- SAT
South African Territories
- SVDV
Swine Vesicular Disease Virus
- VSV
Vesicular stomatitis virus
Authors’ contributions
W.HM and H.PL performed the experiments and drafted manuscript, Z.GM analyzed the data, Y. L isolated virus, H.HB and G.YW designed and instructed the experiments. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Experimental protocols for collecting bovine clinical samples were carried out in strict accordance with the Animal Ethics Procedures and Guidelines of the People’s Republic of China, and the animal study proposal was approved by Shandong Normal University Animal Care and Use Committee. All cattle owners signed an informed consent before participation in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hongmei Wang, Email: hongmeiwang@sdnu.edu.cn.
Peili Hou, Email: apeilihou@163.com.
Guimin Zhao, Email: zgmnefu@163.com.
Li Yu, Email: 910115446@qq.com.
Yu-wei Gao, Email: gaoyuwei@gmail.com.
Hongbin He, Email: hongbinhe@sdnu.edu.cn.
References
- 1.Domingo E, Escarmís C, Baranowski E, Ruiz-Jarabo CM, Carrillo E, Núñez JI, Sobrino F. Evolution of foot-and-mouth disease virus. Virus Res. 2003;91(1):47–63. doi: 10.1016/S0168-1702(02)00259-9. [DOI] [PubMed] [Google Scholar]
- 2.Jamal SM, Belsham GJ. Foot-and-mouth disease: past present and future. Vet Res. 2013;44:116. doi: 10.1186/1297-9716-44-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brito BP, Rodriguez LL, Hammond JM, Pinto J, Perez AM. Review of the global distribution of foot-and-mouth disease virus from 2007 to 2014. Transbound Emerg Dis. 2017;64:316–332. doi: 10.1111/tbed.12373. [DOI] [PubMed] [Google Scholar]
- 4.Zheng H, He J, Guo J, Jin Y, Yang F, Lv L, Liu X. Genetic characterization of a new pandemic Southeast Asia topotype strain of serotype O foot-and-mouth disease virus isolated in China during 2010. Virus Genes. 2012;44(1):80–88. doi: 10.1007/s11262-011-0670-0. [DOI] [PubMed] [Google Scholar]
- 5.Yang X, Zhou YS, Wang HN, Zhang Y, Wei K, Wang T. Isolation, identification and complete genome sequence analysis of a strain of foot-and-mouth disease virus serotype Asia1 from pigs in southwest of China. Virol J. 2011;8:175. doi: 10.1186/1743-422X-8-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He CQ, Liu YX, Wang HM, Hou PL, He HB, Ding NZ. New genetic mechanism, origin and population dynamic of bovine ephemeral fever virus. Vet Microbiol. 2016;182:50–56. doi: 10.1016/j.vetmic.2015.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Charleston B, Bankowski BM, Gubbins S. Chase-topping ME, Schley D, Howey R, Barnett PV, Gibson D, Juleff ND, Woolhouse MEJ. Relationship between clinical signs and transmission of an infectious disease and the implications for control. Science. 2011;332:726–729. doi: 10.1126/science.1199884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranjan R, Biswal JK, Subramaniam S, Singh KP, Stenfeldt C, Rodriguez LL, Pattnaik B, Arzt J. Foot-and-mouth disease virus-associated abortion and vertical transmission following acute infection in cattle under natural conditions. PLoS One. 2016;11(12):e0167163. doi: 10.1371/journal.pone.0167163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight-Jones TJ, Robinson L, Charleston B, Rodriguez LL, Gay CG, Sumption KJ, Vosloo W. Global foot-and-mouth disease research update and gap analysis: 4-diagnostics. Transbound Emerg Dis. 2016;63(1):42–48. doi: 10.1111/tbed.12523. [DOI] [PubMed] [Google Scholar]
- 10.Ferris NP, Nordengrahn A, Hutchings GH, Reid SM, King DP, Ebert K, Paton DJ, Kristersson T, Brocchi E, Grazioli S, Merza M. Development and laboratory validation of a lateral flow device for the detection of foot-and-mouth disease virus in clinical samples. J Virol Methods. 2009;155(1):10–17. doi: 10.1016/j.jviromet.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Waters RA, Fowler VL, Armson B, Nelson N, Gloster J, Paton DJ, King DP. Preliminary validation of direct detection of foot-and-mouth disease virus within clinical samples using reverse transcription loop-mediated isothermal amplification coupled with a simple lateral flow device for detection. PLoS One. 2014;9(8):e105630. doi: 10.1371/journal.pone.0105630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abd El Wahed A, El-Deeb A, El-Tholoth M, Abd El Kader H, Ahmed A, Hassan S, Hoffmann B, Haas B, Shalaby MA, Hufert FT. Weidmann M. a portable reverse transcription recombinase polymerase amplification assay for rapid detection of foot-and-mouth disease virus. PLoS One. 2013;8(8):e71642. doi: 10.1371/journal.pone.0071642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau LT, Reid SM, King DP, Lau AMF, Shaw AE, Ferris NP. Yu ACH. Detection of foot-and-mouth virus by nucleic acid sequence-based amplification (NASBA) Vet Microbiol. 2008;126(1–3):101–110. doi: 10.1016/j.vetmic.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, Peng Z, Xu S, Fu F, Yu J, Sun W, Xu S, Li J, Wang J. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg Dis. 2017;64(5):1337–1341. doi: 10.1111/tbed.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng Y, Wang J, Liu L, Lu Y, Tan K, Chang YZ. Development of real-time recombinase polymerase amplification assay for rapid and sensitive detection of canine parvovirus 2. BMC Vet Res. 2017;13(1):311. doi: 10.1186/s12917-017-1232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou P, Zhao G, He C, Wang H, He H. Biopanning of polypeptides binding to bovine ephemeral fever virus G1 protein from phage display peptide library. BMC Vet Res. 2018;14(1):3. doi: 10.1186/s12917-017-1315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Wang J, Li R, Liu L, Yuan W. Rapid and sensitive detection of canine distemper virus by real-time reverse transcription recombinase polymerase amplification. BMC Vet Res. 2017;13(1):241. doi: 10.1186/s12917-017-1180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang HM, Zhao GM, Hou PL, Yu L, He CQ, He HB. Rapid detection of foot-and-mouth disease virus using reverse transcription recombinase polymerase amplification combined with a lateral flow dipstick. J Virol Methods. 2018;261:46–50. doi: 10.1016/j.jviromet.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Ding NZ, Qi QR, Gu XW, Zuo RJ, Liu J, Yang ZM. De novo synthesis of sphingolipids is essential for decidualization in mice. Theriogenology. 2018;106(3):227–236. doi: 10.1016/j.theriogenology.2017.09.036. [DOI] [PubMed] [Google Scholar]
- 20.Jamal SM, Belsham GJ. Development and characterization of probe-based real time quantitative RT-PCR assays for detection and serotyping of foot-and-mouth disease viruses circulating in west eurasia. PLoS One. 2015;10(8):e0135559. doi: 10.1371/journal.pone.0135559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalaby MA, El-Deeb A, El-Tholoth M, Hoffmann D, Czerny CP, Hufert FT, Weidmann M, Abd El Wahed A. Recombinase polymerase amplification assay for rapid detection of lumpy skin disease virus. BMC Vet Res. 2016;12(1):244. doi: 10.1186/s12917-016-0875-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P, Wang H, Zhao G, He C, He H. Rapid detection of infectious bovine Rhinotracheitis virus using recombinase polymerase amplification assays. BMC Vet Res. 2017;13(1):386. doi: 10.1186/s12917-017-1284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Huang YH, Liu SZ, Zhang L, Li BT, Zhao XX, Fu Y, Liu JJ, Zhang XX. Pseudomonas reactans, a bacterial strain isolated from the intestinal flora of Blattella germanica with anti-Beauveria bassiana activity. Environ Entomol. 2013;42(3):453–459. doi: 10.1603/EN12347. [DOI] [PubMed] [Google Scholar]
- 24.Zhao G, Wang H, Hou P, He C, Huan Y, He H. Rapid and visual detection of Mycobacterium avium subsp. paratuberculosis by recombinase polymerase amplification combined with a lateral flow dipstick. J Vet Sci. 2018;19(2):242–250. doi: 10.4142/jvs.2018.19.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crannell ZA, Rohrman B, Richards-Kortum R. Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS One. 2014;9(11):e112146. doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lillis L, Siverson J, Lee A, Cantera J, Parker M, Piepenburg O, Lehman DA, Boyle DS. Factors influencing recombinase polymerase amplification (RPA) assay outcomes at point of care. Mol Cell Probes. 2016;30(2):74–78. doi: 10.1016/j.mcp.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuel AR, Knowles NJ. Foot-and-mouth disease type O viruses exhibit genetically and geographically distinct evolutionary lineages (topotypes) J Gen Virol. 2001;82:609–621. doi: 10.1099/0022-1317-82-3-609. [DOI] [PubMed] [Google Scholar]
- 28.Cottam EM, Thébaud G, Wadsworth J, Gloster J, Mansley L, Paton DJ, King DP, Haydon DT. Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus. Proc R Soc B. 2008;275:887–895. doi: 10.1098/rspb.2007.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Xie S, Zhou J. Use of animal models for the imaging and quantification of angiogenesis. Exp Anim. 2018;67(1):1–6. doi: 10.1538/expanim.17-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamal SM, Ferrari G, Ahmed S, Normann P, Curry S, Belsham GJ. Evolutionary analysis of serotype a foot-and-mouth disease viruses circulating in Pakistan and Afghanistan during 2002–2009. J Gen Virol. 2011;92:2849–2864. doi: 10.1099/vir.0.035626-0. [DOI] [PubMed] [Google Scholar]
- 31.Pacheco JM, Brito B, Hartwig E, Smoliga GR, Perez A, Arzt J, Rodriguez LL. Early detection of foot-and-mouth disease virus from infected cattle using a dry filter air sampling system. Transbound Emerg Dis. 2017;64(2):564–573. doi: 10.1111/tbed.12404. [DOI] [PubMed] [Google Scholar]
- 32.Nelson N, Paton DJ, Gubbins S, Colenutt C, Brown E, Hodgson S, Gonzales JL. Predicting the ability of preclinical diagnosis to improve control of farm-to-farm foot-and-mouth disease transmission in cattle. J Clin Microbiol. 2017;55(6):1671–1681. doi: 10.1128/JCM.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Wang HY, Kim S, Park SD, Yu K, Kim HY, Uh Y, Lee H. Evaluation of the punch-it™ NA-sample kit for detecting microbial DNA in blood culture bottles using PCR-reverse blot hybridization assay. J Microbiol Methods. 2016;128:24–30. doi: 10.1016/j.mimet.2016.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the results of the study were included in the manuscript.