Abstract
Animals’ olfactory systems rely on proteins, olfactory receptors (ORs) and odorant-binding proteins (OBPs), as their native sensing units to detect odours. Recent advances demonstrate that these proteins can also be employed as molecular recognition units in gas-phase biosensors. In addition, the interactions between odorant molecules and ORs or OBPs are a source of inspiration for designing peptides with tunable odorant selectivity. We review recent progress in gas biosensors employing biological units (ORs, OBPs, and peptides) in light of future developments in artificial olfaction, emphasizing examples where biological components have been employed to detect gas-phase analytes.
Biosensors in Artificial Olfaction
The possibility of mimicking nature, building artificial intelligent systems to perform complex tasks, and dealing with large sets of data is gaining increasing relevance in all areas, including biological sciences [1]. The mysteries of olfaction, in particular human olfaction, still intrigue scientists and, not surprisingly, it is considered the least understood sense [2,3]. Artificial olfaction systems aim to mimic the sense of smell and typically consist of electronic nose devices (e-noses) (see Glossary) that include an array of gas sensors associated with signal-processing tools [4–6]. Classically, in an e-nose, a sample enters the system through an inlet that guides the gas molecules to a chamber where the sensing material is deposited. The interaction of the volatile organic compounds (VOCs) with the sensing material generates a signal (for example, electric, optical, or gravimetric) that is transduced and further processed by a signal-processing computer.
Glossary.
Biophotonic sensor: a sensor that detects the interaction of photons and biomaterials.
Carbon nanotubes (CNTs): carbon atoms arranged in a tube-shaped nanostructure.
Electrochemical impedance spectroscopy (EIS): a spectroscopic technique that measures the impedance of an electrochemical system to an applied potential.
Electronic nose devices (e-noses): electronic devices with the purpose of detecting odorants.
Film bulk acoustic resonator (FBAR): a device comprising a piezoelectric material inserted between two electrodes, acoustically isolated from the contiguous medium.
Field-effect transistor (FET): a semiconductor channel with electrodes at both ends, denoted as drain and source. The conductivity between the drain and source terminals is controlled by an electric field in the device.
Homology modeling: modeling a protein 3D structure by means of a known experimental structure of a homologous protein sequence.
Limit of detection (LoD): the lowest analyte concentration that can be reliably distinguished from a blank.
Lipocalin: proteins that transport hydrophobic molecules.
Microcantilever: a rigid microscale plate anchored at one end and free at the other end to allow structural fluctuations. It measures mechanical properties of materials, such as resonance frequency, amplitude of vibration, and bending.
Molecular docking: a method for predicting the right orientation of a molecule when bound to a receptor.
Nanodisc: soluble nanoscale phospholipid bilayers able to host membrane proteins.
Odorant-binding protein (OBP): soluble proteins secreted in the nasal mucus of vertebrates and in the lymph of insect chemosensory sensilla.
Olfactory receptor (OR): a membrane protein channel expressed in olfactory neurons, triggered by odorant binding.
Phage display: a combinatorial protein expression technique with bacteriophages linking proteins to their respective DNA.
Piezoelectric: the ability of a material to generate electricity upon mechanical stress.
Quartz crystal microbalance (QCM): a technique that measures mass variation per unit area by measuring the change in frequency of a quartz crystal resonator.
Resonator: a system that naturally oscillates at some frequencies, called resonant frequencies, with greater amplitude than at others.
Self-assembled monolayer (SAM): one molecule layer of material bound to a surface in an ordered way resulting from its physico-chemical characteristics.
Silicon nanowire (SiNW): a nanowire formed from a silicon precursor by etching of a solid or through catalyzed growth from a vapor or liquid phase. SiNW sensors transduce electrical signals.
Surface acoustic wave (SAW): a sound wave that travels parallel to the surface of an elastic material, with its displacement amplitude decaying into the material. SAW sensors are microelectromechanical systems that transduce an input electrical signal into a mechanical wave. This mechanical wave is influenced by physical phenomena, and the wave is transduced back into an electrical signal.
Volatile organic compounds (VOCs): organic compounds with high vapor pressure at room temperature, like odors, pheromones, and aromas.
Odors are typically composed of a set of VOCs. The detection of VOCs is of significant interest, not only to understand biological processes, but also in several scientific and technological areas. For instance, VOC sensors hold great promise in the early diagnosis of diseases [7–12], food quality control [13–15], security [16], agriculture [17], environmental monitoring [18], and insect supervision [19,20]. The broad range of application areas for enoses has been accelerating the progress of gas-sensing technologies, with an extensive research and market space for developing new sensing materials and devices [2]. Current gas-sensing materials, including those commercially available, include mostly metal oxide semiconductors and conductive polymers. The main drawbacks of these systems are the low stability and high promiscuity towards VOC molecules, resulting in low selectivity. As such, incorporating the sensing components of biological olfaction systems into gas-sensing materials can increase the VOC selectivity of the resultant gas sensors and bio-electronic noses (Figure 1) [4,6,19–22]. Still, most published works report biosensing using VOC analytes in solutions [23–27], in contrast to the real-life implementation of e-noses to analyse gaseous samples. Therefore, this review focuses only on recent research trends in gas-phase biosensing.
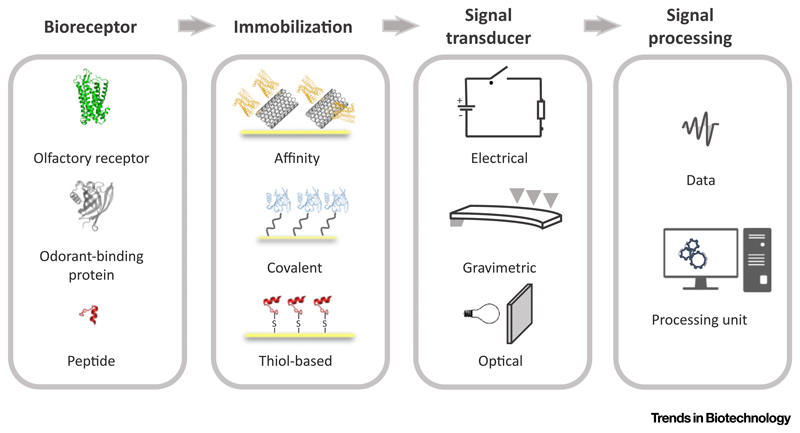
Figure 1. Scheme of Main Constituents of Protein and Peptide-Based Biosensors for Artificial Olfaction.
A bioreceptor known to bind to volatile organic compounds (VOCs) – olfactory receptors, odorant binding proteins or VOC affinity peptides – is selected for immobilization. Surface immobilization can be made through different methods including affinity, covalent spacers, and thiol-based chemistry. The biosensor is then applied in a signal transducing system which may be, for example, electrical, gravimetric, or optical. The collected data is finally processed to yield qualitative or quantitative information about target VOCs.
Olfactory Receptor Gas Biosensors
Olfactory receptors (ORs) are believed to recognize different odorants, so odorant recognition depends on the OR being activated and on the extent of its activation [28,29] (Box 1). Identifying which odorants specifically bind to a certain OR is called deorphanization [30,31]. The deorphanization of ORs is a crucial step to understand several aspects of the smelling sensory process, including the complete knowledge of an OR’s activity, the impact of the physicochemical properties of odorant molecules in an OR’s selectivity, and the way in which information is accurately processed to generate the recognized pattern that culminates in the final olfactory perception [31]. These studies must take into account the ability of an odorant to be recognized by multiple ORs, and the fact that a specific OR can recognize several odorants. Some ORs have already been isolated and applied to gas-sensing biosensors (Table 1).
Box 1. Differences between Proteins and Peptides Currently Used in VOC Biosensing.
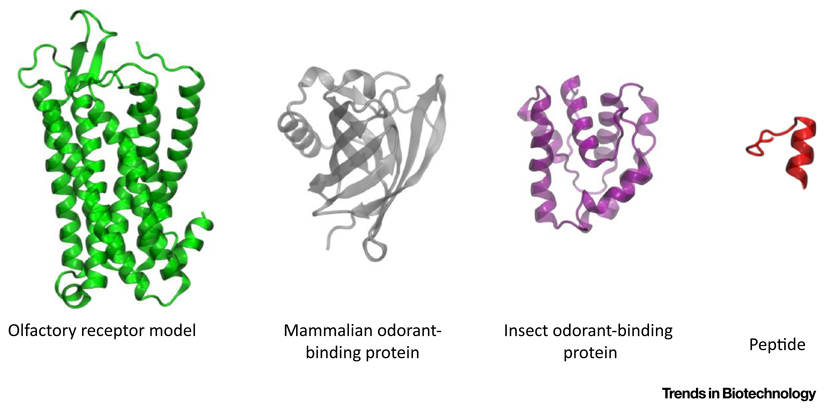
The biomolecules applied so far in gas biosensing include ORs, OBPs, and peptides, with remarkable structural differences highlighted in Figure I.
ORs are members of the G protein-coupled receptor family: they are structurally defined by seven trans-membrane α-helices, about 320 amino acid residues in length [29]. These proteins, upon volatile binding, generate a signal that is transmitted from the olfactory sensory neurons, where ORs are located, to the brain in a recognized pattern that culminates in olfactory perception.
Mammalian OBPs belong to the lipocalin superfamily of proteins, transport proteins [38,41,42] that are 150–160 residues in length [43] and have typical lipocalin-fold of eight anti-parallel β-sheets and a short α-helix close to the C terminal. The β-sheets form an antiparallel β-barrel shaping a central pocket for ligand binding [44]. Insect OBPs present well-conserved folding with protein chains of 130–150 amino acids [40]. The tertiary protein structure comprises a compact set of six α-helices with a hydrophobic cavity. Additionally, the folding is stabilized by the presence of three disulfide bonds [39,40]. These disulfide bonds are considered the fingerprint of insect OBPs. Notwithstanding amino acid variability, the overall structure of insect OBPs is highly conserved, even among members of different orders of insects [39,40,78].
Peptides represent a simple and low-cost option for biosensors. Ranging from approximately 5 to 15 protein residues, they can be synthetically or biologically produced. Their selectivity towards target VOCs is easier to tune due to their small size [20,56]. VOC-sensing peptides have been developed by different approaches (Box 2, Figure 2), and applied in several substrates and sensing devices, as summarized in Table 2.
Figure I. Structural Features of Odor-Sensing Proteins and Peptides.
Table 1. Protein Biosensors in Gas Sensing.
| Protein | VOCs | Support | Transducer | LoD/measureda | Source | Expression system | Refs | |
|---|---|---|---|---|---|---|---|---|
| Olfactory receptors | OR-10 | Diacetyl | Gold | QCM | 1 x 10–12 M | C. elegans | E. coli | [32] |
| SAW | 1.2 x 10–11 mM | MCF-7 cells | [34] | |||||
| hOR 17–40 hOR3A1 | Helional | CNT | Interdigitated microelectrode array, Current-voltage | 0.02 ppt | Homo sapiens | MC18 (Saccharomyces cerevisiae) HEK-293T cells | [36] | |
| mOR174-9 mOR203-1 mOR256-17 | Acetophenone, Others | CNT | CNT transistors, Current-gate Voltage | 1–10 μM | Mus musculus | S. cerevisiae | [35] | |
| Odorant-binding proteins | AaegOBP22 | N,N-diethyl-meta-toluamide (DEET) | Gold | ZnO film bulk acoustic resonators | – | A. aegypti | E. coli | [55] |
| pOBP | Ethanol; methanol | Si | Si-substrate with interdigitated electrodes (EIS) | 20 ppma; 10 ppma | Sus scrofa | Pig nasal tissue | [46] | |
| pOBP | (R)-(−)-1-octen-3-ol(octenol); (R)-(−)-carvone (carvone) | Gold | SAW | 0.48 ppm; 0.72 ppm | S. scrofa | Pig nasal tissue | [47] | |
| wtbOBP | (R)-(−)-1-octen-3-ol(octenol); (R)-(−)-carvone (carvone) | SAW | 0.2–0.23 ppm (carvone); 0.18–0.21 ppm (octenol) | Bos taurus | BL21-DE3 E. Coli | [49] | ||
| pOBP wtbOBP dmbOBP | (R)-(−)-1-octen-3-ol(octenol); (R)-(−)-carvone (carvone) | Gold | SAW | wtpOBP: 25.9 (octenol), 7.0 (carvone) Hz/ppm; wtbOBP: 3.5 (octenol), 5.4 (carvone) Hz/ppm; dmbOBP: 6.0 (octenol), 9.2 (carvone) Hz/ppm; | S. scrofa; B. Taurus | Pig nasal tissue; BL21-DE3 E. Coli | [51] | |
| pOBP wtbOBP dmbOBP | (R)-(−)-1-octen-3-ol(octenol); (R)-(−)-carvone (carvone) | Gold | SAW | 0.48 ppm (octenol); 0.74 ppm (carvone) | S. scrofa; B. Taurus | Pig nasal tissue; E. coli | [52] | |
| wtbOBP | (R)-(−)-1-octen-3-ol (octenol) | Gold | Solidly mounted resonator | 7 ppm | B. taurus | BL21-DE3 E. coli | [48] | |
| wtbOBP | (R)-(−)-1-octen-3-ol(octenol); (R)-(−)-carvone (carvone) | Gold | SAW | 0.18 ppm; 0.2 ppm | B. taurus | BL21-DE3 E. Coli | [49] | |
| wtbOBP | (R)-(−)-1-octen-3-ol (octenol) | Gold | SAW | 2 ppm | B. taurus | BL21-DE3 E. coli | [50] | |
| wtbOBP, dmbOBP | DMMP | Silicon nitrate | Photonic ring resonator | 6.8 ppb | B. taurus | BL21-DE3 E. coli | [53] |
Indicates when the detected concentration was not the LoD, but the “measured” in that paper.
The first development of gas-phase biosensors for VOC detection used the receptor OR-10 from Caenorhabditis elegans. This protein was expressed in Escherichia coli, and its membrane fraction containing OR-10 coated on a quartz crystal for quartz crystal microbalance (QCM) measurements. The biosensor responded to diacetyl with a detectable concentration as low as 10−12 M [32]. However, using a crude membrane extract on the sensor surface may have distorted the response of OR-10 to VOCs due to the presence of the lipidic fraction of the membrane, as phospholipids are also known to bind odorant-like molecules [33]. Still, these reports highlighted the potential of ORs in gas sensing and the hurdles of dealing with cellular membranes for OR stabilization and immobilization. A similar approach, expressing OR-10 in human breast cancer MCF-7 cells, assessed the influence of VOCs binding to phospholipids, using control sensors analysed in parallel [34]. The controls produced very low-frequency shifts in a surface acoustic wave (SAW) as a response to the tested VOCs (diacetyl, butanone, and 2,3-pentanedione). Moreover, the response to diacetyl was the most significant with a limit of detection (LoD) of 1.2 × 10−14 M [32]. The use of a control sensor elucidated that the contribution of the affinity of VOCs to phospholipid molecules is negligible compared with their affinity to ORs, consolidating a new strategy for VOCs biosensor development.
A different methodology for incorporating ORs in a gas sensor was explored using mouse ORs (mOR) in nanodiscs [35]. Nanodiscs with mORs were coupled to carbonnanotubes (CNTs) via His-tag interaction. In contrast with other examples where the ORs were in the membrane fraction [32–34], in this case the nanodiscs were deposited on the sensor surface. To certify the efficiency of the nanodisc sensors, the same mORs were incorporated in digitonin micelles, deposited in CNTs, and tested for VOC sensing (Table 1). The mORs–CNT biosensors revealed broad agreement of results for the micelle and nanodisc systems. The difference observed between mOR nanodisc and micelles sensors was in their stability over time. Micelle sensors remained active for 5 days in contrast with nanodisc sensors, which were stable for 10 weeks after an initial decrease in response. The longer integrity may be due to the higher stability of the nanodisc structure in contrast to the micelles [35].
A human OR (hOR)-based biosensor for VOC sensing has been coupled to carboxylated polypyrrole nanotubes [36]. Conductivity measurements revealed a very high sensitivity of this biosensor towards helional (0.02 ppt). Moreover, it presented selectivity towards helional when compared with analog VOCs like 3,4-methylenedioxy dihydrocinnamic acid, piperonal, safrole, and phenyl propanol. These analogs had at least 2/3 lower electrical resistance change upon binding to the hOR than helional in the reported biosensor measurements [36].
The difficult, expensive, and time-consuming handling of membrane proteins may have led to the pursuit of simpler and more robust biomolecules as recognition agents [37]. Therefore, gas-sensing biosensors based on the soluble odorant-binding proteins (OBPs) and small peptides have gained traction [20,38,39].
OBP Gas Biosensors
OBPs are an essential component of the olfactory system. They are in the first line of olfaction, located in the nasal epithelium of mammals (nose) and in the sensillar lymph of insects (e.g., antennae) [39,40]. Less soluble VOCs are believed to first bind to OBPs that carry them to the ORs in the olfactory sensory neurons. More hydrophilic VOCs are able to cross the mucosa or lymph to bind directly to ORs. Furthermore, OBPs are believed to act as scavengers when VOCs are present in high amounts. Vertebrate and insect classes of OBPs differ in their 3D structure (Box 1). The OBPs from vertebrates belong to the lipocalin superfamily of proteins, which are essentially transport proteins [38,41,42]. Insect OBPs are also stabilized by the presence of three disulfide bonds [39,40]. OBPs can withstand high temperatures before experiencing denaturation, and after unfolding they frequently refold, restoring their initial structure [42]. Interestingly, when binding high-affinity ligands, the stability of pig OBP (pOBP) increases at higher temperatures [45]. Insect OBPs are even more resistant, as the three interlocked disulfide bonds prevent proteolytic degradation and thermal denaturation [40,42].
Developments in OBP-based biosensors for gas sensing started to flourish in the last decade (Table 1). Initially, OBP biosensors were tested against volatiles and odor solutions, with measurements of analytes in gas phase only reported by the end of the 2000s. The first described OBP biosensor to detect ethanol and methanol in gas phase involved pOBP [46]. The protein immobilized on a silicon substrate for electrochemical impedance spectroscopy (EIS) measurements, and was able to detect ethanol and methanol vapors (Table 1). Further research revealed that pOBP can also bind octenol and carvone [47]. Octenol is an important VOC for food quality control, as it is correlated with the presence of fungi and molds. pOBP was immobilized onto the gold coating of a resonator, and the sensor was able to detect R-carvone (a spearmint-like flavor and odorant), but with a lower sensitivity than for octenol. These two VOCs have also been thoroughly tested with bovine OBP (bOBP) [48–50], and in sensor arrays of pOBP, bOBP both wild type (wt) and double-mutant (dm) with higher affinity to carvone [51,52]. The above-mentioned reports have shown that the LoDs for octenol and carvone of pOBP were 0.48 and 0.72 ppm [47], and of bOBP were 0.18 and 0.2 ppm, respectively [49]. For a different application, a biophotonic sensor using the same wtbOBP and dmbOBP as sensing elements was developed to detect dimethyl methylphosphonate (DMMP), a precursor of sarin gas [53]. The bOBPs were deposited onto the silicon nitride surface of a biophotonic resonator chip, resulting in a stable sensor that was responsive for over 3 months and could detect 6.8 ppb of DMMP in 15 minutes. Furthermore, a pOBP microcantilever biosensor was recently reported; although the system is in early development, the sensor successfully detected 2-isobutyl-3-methoxypyrazine in a gas chamber [54]. Taken together, these examples reveal some plasticity of OBP structures in identifying different chemical structures. It is in fact important to merge structural information on OBP–VOC interaction for protein design and engineering towards target VOCs of interest, which currently still remains a challenge due to the limited structural information available for a wide variety of VOC structures (Boxes 1 and 2; Figure 2, Key Figure).
Box 2. Protein and Peptide Engineering in VOC Sensing.
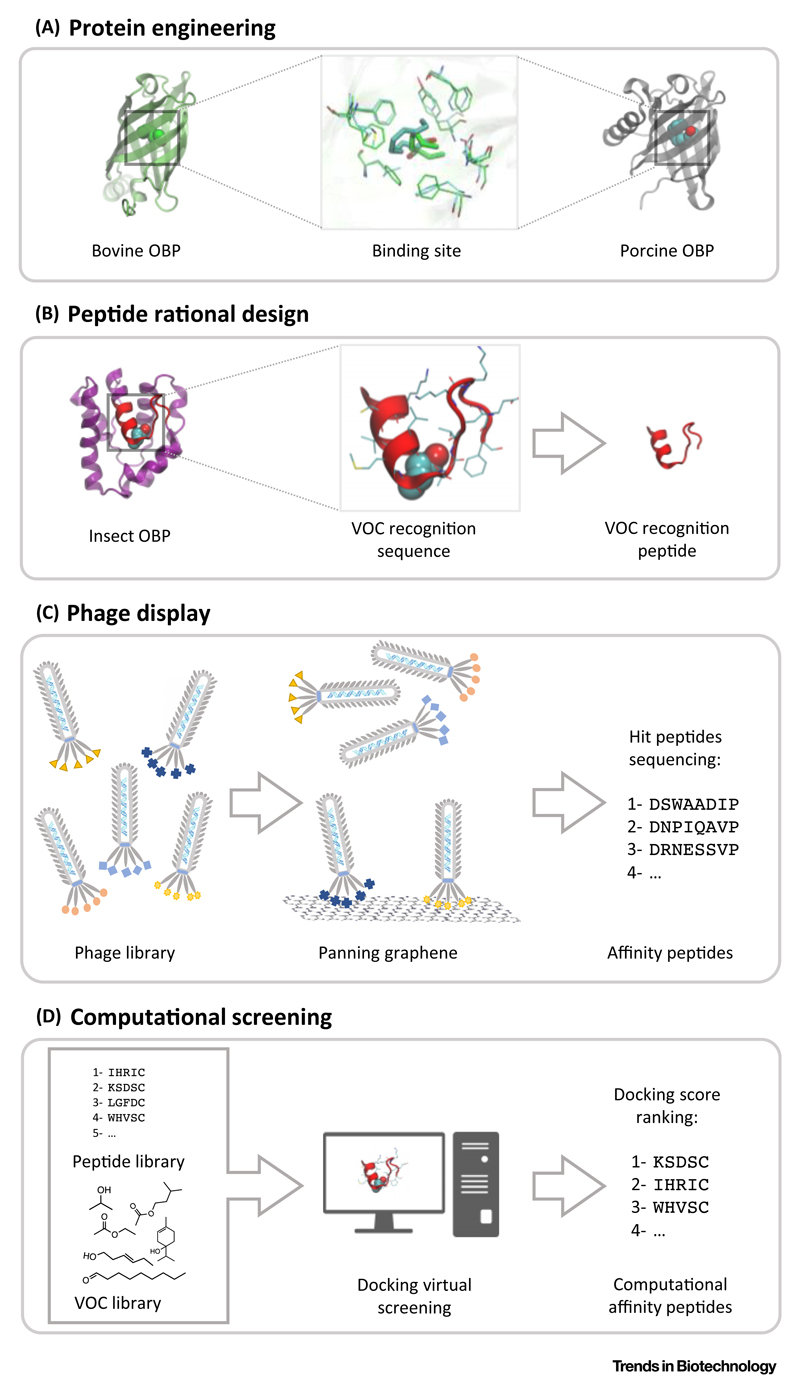
To tailor proteins and peptides for binding target VOCs, several protein engineering techniques may be applied (Figure 2). Predominant techniques involve site-specific protein mutations (Figure 2A), the design of peptides inspired by the VOC recognition sites of olfactory receptors and odorant-binding proteins (Figure 2B), and screening of peptide libraries by phage display and virtual screening approaches (Figure 2C,D).
A detailed analysis of the VOC-binding pocket from OBPs reveals a close similarity among the binding site of OBPs from different organisms, such as pig and bovine OBPs (Figure 2A), even though the target VOC is different. These properties led to the development of sensor arrays including OBPs from different organisms and mutated OBPs, in order to better discriminate detected VOCs [51,52]. An example mutation is the bovine double mutant described to increase the sensitivity of bOBP to octenol and carvone [51].
Another approach consists of a peptide that was rationally designed by extracting the sequences responsible for VOC recognition in ORs and OBPs. Due to their smaller sizes, peptides are usually easier to immobilize in an oriented and predefined way onto sensor surfaces. Peptides are also somewhat more stable than ORs and more resilient to reusability and shelf-life. Successful examples have been reported for peptides derived from human OR1E1 [58,61,79] and dog OR-P30955 [57,60]. Three different peptides derived from insect OBPs were successfully produced and implemented in gas sensors to detect 3-methyl-butanol [63,80], 1-hexanol [63], and trinitrotoluene [65] (Figure 2B).
Combinatorial screening techniques, such as phage display and virtual screening, can broaden peptide sequence variability and find sequences for specific VOCs. Phage display is implemented when a VOC analog is immobilized onto a surface or a VOC-analog surface is used to screen randomized peptides for binding [66–68,71] (Figure 2C). Computational virtual screening of peptide libraries is also used in the discovery of VOC-binding peptides [69,72,75] (Figure 2D). The top-performing peptides are usually synthetized and experimentally tested in gas sensing, with satisfactory correlation between in silico and experimental data [69,75].
Figure 2.
Key Figure
Protein and Peptide Discovery and Engineering for Gas Sensing
For a Figure360 author presentation of Figure 2, see the figure legend at https://doi.org/10.1016/j.tibtech.2018.07.004.
For a Figure360 author presentation of Figure 2, see the figure legend at https://doi.org/10.1016/j.tibtech.2018.07.004.
(A) Superimposition of the binding pockets of bovine and porcine odorant-binding proteins (OBPs) reveals how small differences in residue composition lead to different volatile organic compound (VOC) affinities [51]. (B) The extraction of volatile binding sequence from the fruit fly OBP LUSH, as a way to rationally develop peptides for VOC detection [62,63]. (C) Phage-display panning screens a multitude of peptide sequences against a graphitic surface mimicking benzene [68]. (D) Computational techniques predict the best peptide–VOC pairs to be experimentally tested [70].
The only reported biosensor based on an insect OBP for VOC detection in the gas phase used an OBP from the mosquito Aedes aegypti (AaegOBP22) [55]. The purified OBP was deposited in a ZnO thin film bulk acoustic resonator (FBAR), and a vapor flow of N,N-diethyl-meta-toluamide (DEET), the main component of mosquito repellents, was used for detection measurements. The AaegOBP22 biosensor detected the presence of DEET through a drop in the response frequency of the FBAR sensor, depending on a higher or lower amount of loaded OBP [55].
Peptide-Based Gas Biosensors
Peptides are promising alternatives to ORs and OBPs as molecular recognition units in VOC sensing. Due to their smaller size, peptides represent a simple and low-cost option for biosensors (Box 1). Additionally, the selectivity towards target VOCs is easier to tune [20,56]. VOC-sensing peptides have been developed by different approaches (Box 2, Figure 2), and applied in several substrates and sensing devices, as summarized in Table 2.
Table 2. Peptide Biosensors in Gas Sensing.
| Peptidea | VOCs | Support | Transducer | LDL/measuredb | Development | Refs |
|---|---|---|---|---|---|---|
| NQLSNLSFSDLC (dORp61) Dog OR – P30955 | Trimethylamine (TMA); ammonia; acetic acid; ethyl acetate; methanol | Gold Surface | Piezoelectric multiarray analyzer | – | OR-based design | [57] |
| ACSDAQVNE (dORp188) Dog OR – P30955 | TMA; ammonia; acetic acid; ethyl acetate; methanol; benzene | – | ||||
| FLSNLSFSDLC (hORp61) human OR1E1 | TMA | Gold surface | Piezoelectric multiarray analyzer | 6.7 Hz | OR-based design | [58] |
| FFLLFGC (hORp103) human OR1E1 | O-xylene | 5.1 Hz | ||||
| DLESFLC (hORp109) human OR1E1 | Ammonia | Gold surface/silicon (SiNW) | Piezoelectric multiarray analyzer/conductance | 11 Hz/100 ppmb | [58,59] | |
| RVNEWVIC (hORp193) human OR1E1 | Acetic acid | Gold surface/silicon (SiNW) | Piezoelectric multiarray analyzer/QCM/conductance | 28 Hz/3.1 ± 1.2 ppm/100 ppmb | [58,59,61] | |
| LHYTTIC (PAC1) dog OR – P30955 | Acetic acid; butyric acid | Gold surface | QCM (multiarray) | – | OR-based design | [60] |
| TIMSPKLC (PAC2) dog OR – P30955 | Acetic acid; butyric acid | |||||
| DLESC (PAM1) human OR1E1 | Dimethyl amine | |||||
| ELPLGCG (PAM2) | Acetic acid; dimethylamine | |||||
| SLMAGTVNKKGEFC (LUSH OBP) | 3-methyl-1-butanol; 1-hexanol | Gold surface | QCM | 1–3 ppm | OBP-based design | [62] |
| TKCVSLMAGTVNKKGEFFFF (LUSH OBP) | 3-methyl-1-butanol | CNT | Field-effect transistor | 1 fM | OBP-based design | [63] |
| HSSYWYAFNNKTGGGGWFVI (P1-ASP1C peptide) (honeybee antenna OBP) | Trinitrotoluene (TNT) | SWCNT | Field-effect transistor | 12 ppb | OBP-based design | [65] |
| WHYQRPLMPVSI (TNT-BP) | TNT | Gold surface | Thermal desorption GC-MS | – | Phage display | [66] |
| HPNFSKYILMPVSI (DNT-BP) | 2,4-dinitrotoluene (DNT) | |||||
| CH3ONH-KMHTASLSQPLMGC-CONH2 (DNT-BP1C) | DNT | Gold surface | QCM liquid phase/microcantilever | –/431 ppt | Phage display | [67,71] |
| DSWAADIP (GP1) | Benzene | Gold surface | Microcantilever | 121 ppb | Phage display | [68] |
| DNPIQAVP (GP2) | Toluene; xylene; hexane | 2.2 ppm (toluene); 28 ppm (xylene); 1 ppm (hexane) | ||||
| DRNESSVP (BP1) | Benzene; toluene | – | ||||
| AYSSGAPPMPPF (A3) | Acetonitrile; dichloromethane; methyl salicylate | Gold nanoparticles | Inductor–capacitor– resistor resonators | – | Phage display | [81] |
| NFQGI | 2,3.7,8-TCDD (2,3,7,8-tetrachlorinated dibenzo-p-dioxin | Gold Surfaces | QCM | 10–20 ppbb | Rational design (virtual screening) | [72] |
| NFGGQ | ||||||
| NFQGF | ||||||
| CG | 2-propanol; acetone; acetonitrile; butane-2,3-dione; ethanol; ethyl acetate; ethyl butanoate; ethyl octanoate; hex-3-en-1-ol; hexane; isopentylacetate; nonanal; octanal; terpin-4-ol | Gold nanoparticles | QCM | Sensor array (discriminates all tested VOCs) | Rational design (virtual screening) | [69]/[75] |
| ECG (glutathione) | ||||||
| CIHNP | ||||||
| CIQPV | ||||||
| CRQVF | ||||||
| IHRIC | Terpinen-4-ol > hexane > octanal > nonanal | Gold nanoparticles | QCM | Sensor array (discriminates all tested VOCs) | Rational design (virtual screening) | [70] |
| KSDSC | Small alcohols (2-propanol; ethanol) | |||||
| LGFDC | Ethanol, 2-propanol; acetone; butane-2,3-dione; ethyl acetate | |||||
| TGKFC | Alcohols (hex-3-en-1-ol; terpinen-4-ol) and aldehydes (nonanal; octanal) | |||||
| WHVSC | Esters (ethyl acetate; ethyl butanoate; ethyl octanoate; isopentyl acetate) and hexane | |||||
| IHRIC | Alcohols (1-butanol; 1-hexanol; 2-methyl-1-propanol; ethanol; hexen-3-en-1-ol) and esters (ethyl acetate; ethyl-methyl-2-butyrate; isopentyl acetate) | Zinc oxide nanoparticles | QCM | Sensor array (discriminates all tested VOCs) | Rational design (Virtual screening) | [76] |
| LAWHC | ||||||
| TGKFC | ||||||
| WHVSC |
Peptide sequences provided as one-letter codes.
Indicates when the detected concentration was not the LoD, but the “measured” in that paper.
Peptide sequences for VOC sensing were developed in the early 2000s based on dog and hORs. The first OR-based peptide sequences were designed by maintaining the dog OR regions responsible for VOC binding. The selected peptide chains, ranging from 9 to 14 amino acid residues, were deposited on a gold surface of a piezoelectric multiarray analyzer. Two immobilized peptide sequences, orp61 (12-mer) and orp188 (9-mer), successfully detected trimethylamine (TMA) and ammonia, opening a new path for the development of peptide-based volatile biosensors [57]. In another work, a series of peptides were designed inspired by a homology model of the human OR1E1. Several VOCs were docked to each transmembrane sequence of hOR1E1 using molecular docking and the four most promising peptides were synthesized. The lead peptides, horp61 (12-mer), horp103 (7-mer), horp109 (8-mer), and horp193 (8-mer), were immobilized on silicon nanowires (SiNWs), and used to detect TMA, o-xylene, ammonia, and acetic acid [58,59]. Other QCM sensor arrays with peptides from dog OR (LHYTTIC, TIMSPKLC) and human OR1E1 (DLESC, ELPLGCG) have also been assembled. This system could discriminate more VOCs than previous examples, including acetic acid, butyric acid, ammonia, dimethyl amine, chlorobenzene, and benzene [60]. The affinity of the peptide horp193 towards acetic acid was further confirmed by QCM studies, revealing a LoD of 3.8–2 ppm. This work also reported that a lower amount of peptide in the sensing film originates better reproducibility and shelf-life, as opposed to higher peptide densities [61].
When utilizing OBPs as scaffolds to extract putative VOC-sensing peptide sequences, some of the problems associated with OR-inspired peptide design are solved. The existence of 3D structures of several OBPs means that their binding sites are generally well defined and the need to predict homology models becomes obsolete. With the aim of detecting food contamination by Salmonella, two peptides were extracted from the sequence of Drosophila melanogaster LUSH-OBP (Table 2) [62,63]. Knowing that the main VOC released from Salmonella-contaminated meat is 3-methyl-1-butanol, the LUSH-OBP was selected due to its known sensitivity and selectivity towards alcohols [62]. Hence, both selected sequences contained alcohol-binding residues and also different C terminal ends for their immobilization on gold surfaces for QCM analysis [62] and CNTs for field-effect transistor (FET) studies [63]. QCM measurements revealed that the immobilized peptide could bind to 3-methyl-1-butanol and 1-hexanol, with a LoD of 10 ppm, whereas the peptide immobilized on CNT demonstrated a very selective response for 3-methyl-1-butanol, with a LoD of 1 fM. The different reported LoDs are a consequence of not only the different substrates for peptide immobilization and resultant transducing systems, but also the experiment setup, which was performed with gases for the QCM measurements and in solution for the CNT assays. In the CNT assays, the diffusion effects of the VOCs in the sensing chamber were discarded, although this effect is known to be one of the main difficulties in VOCs biosensors [62,63].
Another interesting OBP-based peptide was developed to detect the explosive trinitrotoluene (TNT). In this case, a hybrid peptide was assembled, with an amino acid sequence presenting affinity towards the CNT surface (HSSYWYAFNNKT) [64], and a TNT recognition site based on the binding site of ASP1 (GGGGWFVI), an OBP from the antennae of Apis mellifera [65]. The CNT-hybrid peptide was then exposed to TNT in a saturated vapor chamber for FET analysis. The positive result for TNT binding reveals that the methodology of developing hybrid peptides with a specific group for immobilization on the substrate’s surface can be successful, as the VOC binding moiety of the peptide is completely available for VOC detection and not anchored to the surface [65].
The use of OBPs and ORs to develop peptide sequences for gas sensing is somehow limited to the diversity of VOCs known to bind these proteins. Therefore, methodologies like phage display [66–68], virtual screening [69], and combinatorial peptide libraries [70] broaden the range of peptide sequences with distinct physico-chemical properties, improving affinity and selectivity towards target VOCs or VOC classes.
The application of phage display has been very fruitful in unveiling new specific peptide sequences with an affinity towards certain target molecules (Figure 2). In this method, a phage library is panned against a solid surface resembling the VOC structure or against an immobilized target, which can be the target VOC itself or an analog handled in solution or in crystal phases. The choice of the target for screening in phage display is of utmost importance, as it will determine the affinity and selectivity of the selected peptide sequences. The first attempt to translate phage display panning methods to gas phase sensing was performed to search for peptides with an affinity for TNT and 2,4-dinitrotoluene (DNT) [66]. Two peptides, TNT-BP and DNT-BP, were discovered to selectively bind to TNT and DNT in liquid phase, when panning two commercial phage display libraries (linear 12-mer and constrained 7-mer) against crystal forms of TNT and DNT. However, after translating the system to the gas phase, the low vapor pressure of TNT did not allow gas-phase affinity testing for TNT-BP. DNT-BP successfully demonstrated affinity in binding DNT through thermal desorption gas chromatography-mass spectrometry (GC-MS) analysis [66]. Later, a linear 12-mer commercial phage display library was panned against a gold-immobilized DNT analog [4-(2,4-dinitrophenyl)butan-1-amine] [67]. A DNT affinity peptide was reported (DNT-BP1C) and gas phase measurements in a microcantilever system, revealed a high sensitivity of the peptide to DNT [71]. In a recent example, an 8-mer proprietary phage display peptide library was screened against a self-assembled monolayer (SAM) of benzene on gold, and against a graphite surface (consisting of hexagonal aromatic ring structures, here used as benzene mimetics), with the aim of selecting benzene binders able to discriminate single carbon analogs of benzene, namely toluene and xylene [68]. Three peptides were selected, one presenting affinity to the benzene SAM and two towards the graphite surface (Table 2). Upon peptide immobilization on surfaces with different functional groups, a microcantilever setup for gas testing showed the potential to differentiate the VOCs benzene, toluene, and xylene.
One of the major challenges in employing phage display technology to search for VOC binders is adapting the panning methodologies and translating the panning results from the liquid phase to gas-sensing devices. A critical factor is how to immobilize such small targets as VOCs onto the surfaces employed during panning, and how to reduce nonspecific interactions, as all these methods were developed mainly to look for binders in aqueous conditions against large targets, mainly proteins.
Computational virtual screening allows ‘in silico’ testing of VOC–peptide binding with fast and low-cost routines. This approach has been applied to the discovery of gas-sensing peptides (Figure 2) [69,72]. The first reported effort using a computational screening method focused on finding peptides to detect dioxins. A library of 11 pentapeptides was constructed based on a binding site model for dioxins (-NFQGR-) [73]. After calculating the theoretical binding energy, the three most promising pentapeptides were employed in a QCM-based gas biosensor. The three peptides (NFQGI, NFGGQ, NFQGF) identified the presence of 2,3,7,8-tetrachlorinated dibenzo-p-dioxin (2,3,7,8-TCDD) within a dioxin mixture containing 17 compounds [72].
Another study involved the virtual docking screening of 5 peptides (cysteinylglycine, glutathione, CIHNP, CIQPV, CRQVF) (Table 2), towards 14 distinct VOCs [69]. The results showed a 100% positive correspondence between computational and experimental data for ethyl acetate, ethyl butanoate, and hexane, and an overall matching trend of 60%. The same set of computationally discovered peptides [69,74] (Table 2), was used in the ‘Ten 2009’ electronic nose (Tor Vergata Sensors Group, Rome, Italy), equipped with an 8-QCM sensor array. Contemporaneously, another setup for the e-nose was assembled using only metallo-tetra-phenylporphyrin-coated chips. The peptide-based e-nose arrangement was able to discriminate between artificially contaminated and regular white, milk, and dark chocolate samples with a higher accuracy than the porphyrin sensor array [75].
Another study used a semi-combinatorial virtual screening approach to evaluate a peptide library of 8000 tripeptides for affinity towards 58 compounds from five distinct chemical classes (alcohols, aldehydes, esters, hydrocarbons, and ketones) [70]. After an automated docking procedure, a subset of 120 tripeptides was selected based on their ability to differentiate chemical classes. Then, a new tetrapeptide library was designed basedon these 120 tripeptidesand docked against the 58 volatiles. Five tetrapeptides (IHRI, KSDS, LGFD, TGKF, and WHVS) with different affinity to alcohols, aldehydes, esters, hydrocarbons, and ketones were selected for gas sensing (Table 2). Using a gas sensor QCM array, the peptides were immobilized on gold nanoparticles and tested for binding the target volatiles. In this work, the experimental results for four of the five peptides confirmed the predictions obtained by the computational calculations [70]. Although peptide-based sensors areableto successfullydetect different VOCs, waterproduces a shift in the signal in QCM measurements. A recent work overcame this obstacle by immobilizing four peptides (IHRIC, LAWHC, TGKFC, WHVSC) on zinc oxide nanoparticles. These four biosensors together were able to discriminate alcohols and esters (Table 2) and were further tested in juice samples. The different juice samples were discriminated, supporting the usage of zinc oxide as a support for peptides in VOC detection for high water content samples [76].
Concluding Remarks and Future Perspectives
Using biological recognition units in sensing devices improves their selectivity and sensitivity towards defined targets, with proven pronounced expression in samples analyzed in liquid states. Despite the extremely valuable knowledge derived from testing VOC biosensors in solution, an essential feature is sometimes overlooked: the behavior of these biosensors in testing analytes in the gas phase. Although several obstacles are currently being investigated, there are still challenges to inspire future developments. These include, for example, assessing humidity interference during VOC detection, reproducing gas-phase measurements obtained from protein-based biosensors, enhancing the stability of proteins and peptides upon immobilization onto surfaces and ensuring their compatibility with nanofabrication approaches, and improving the selectivity of VOC biosensors when analyzing extremely complex samples closer to real-life settings.
Some trends are evident for gas-phase protein biosensors. Over the last 5 years, there has been a decreased interest in OR-based and an increase in OBP- and peptide-based biosensors. Certainly, using membrane protein receptors as ORs is more challenging, expensive, and time consuming, when compared with soluble and robust proteins such as OBPs. Although OBPs are easier to express and to handle than ORs, they still lack the simplicity and ease to generate diversity when compared with peptides. This shortcoming justifies the burgeoning interest in peptide-based biosensors. They are easy to assemble through chemical and biological means, and their engineering profits from a variety of techniques (phage display, rational design, virtual screening). Overall, all three systems reported in this review should still be considered in the future, as the sensitivity and selectivity derived from ORs and OBPs may be translated to peptide and protein design and perhaps directly correlate amino acid sequences with VOC selectivity and sensitivity.
In artificial olfaction, though, the combination of biological recognition units has not yet reached its full potential, in some cases due to the difficulty in establishing VOC markers from complex mixtures [77]. However, adapting protein engineering, phage display, and computational tools to address the molecular recognition between biological binders and analytes in gas phases has made tremendous progress and has matured over the last 15 years. As such, the tools to develop VOC binders are in place, as well as the methodologies to interface the bioreceptors with gas-sensing devices (see Outstanding Questions) [2].
Outstanding Questions.
How can VOC-identifying analytical tools help to define VOC markers for relevant societal problems?
Will volatolomics, together with gas-sensing advances, contribute to advanced diagnostic tools?
What should be included in a ‘control VOC set’ for protein and peptide gas-sensing benchmarks?
Could de novo computational protein design be a useful tool to predict new VOC-binding proteins and peptides?
Will advances in VOC-biomolecular recognition promote smell implementation in artificial life systems?
An investment in the expansion of tested VOCs for the development of gas biosensors is required, as recent examples report a very limited number and chemical diversity of VOC targets. In almost all areas of VOC sensing, gas samples are rich in the variety of chemical entities, and it is essential to address molecular discrimination in current and future research. The increasing improvements in protein and peptide engineering may be very useful to increase the stability of biomolecules (for example, through the use of nanodiscs) or to expand the variability in generating binders against less common VOCs (for example, through the molecular modeling and in vitro evolution techniques).
With the developments in volatolomics, we expect that the implementation of gas-phase biosensors to address biological questions will become a reality in the near future. High-impact social and economic areas will be the central application of this technology. Early disease diagnostics and health monitoring, pollutant and fire hazards in the environment, spoilage and quality monitoring in food and agriculture, explosive and chemical/biological weaponry detection in safety, and odor identification in prosthetics are just some potential examples for gas biosensing. Overall, the development of protein and peptide VOC biosensors will be important to attain the desired sensitivity and selectivity in VOC detection, either in single-gas sensors for a defined analyte or in arrays of gas-sensing materials coupled to artificial intelligence tools for signal processing.
Supplementary Material
Supplemental information associated with this article can be found online at https://doi.org/10.1016/j.tibtech.2018.07.004.
Highlights.
Artificial olfaction is being implemented in key societal areas like early disease diagnostics, food safety hazards, air quality, and security.
Protein- and peptide-based VOC biosensors, coupled with cutting-edge transducers, increase the selectivity and sensitivity in detecting key VOCs with limits of detection in the order of ppb.
Gas-phase testing, rather than VOC solutions, is becoming the state of the art in biosensor validation.
Combinatorial techniques such as phage display and virtual screening are advancing the discovery of new VOC-binding peptides.
Progresses in biomolecule immobilization are steadily increasing the reusability and shelf-life of biosensors, while maintaining the desired selectivity.
Acknowledgements
This work was supported by the European Research Council through the grant reference SCENT-ERC-2014-STG-639123 (2015-2020), and by the Unidade de Ciências Biomoleculares Aplicadas-UCIBIO, which is financed by national funds from FCT/MEC (UID/Multi/04378/2013) and co-financed by the ERDF under the PT2020 Partnership Agreement (POCI-01-0145-FEDER-007728). A.J.M. Barbosa and A.R. Oliveira thank fellowships SFRH/BPD/112543/2015 and SFRH/BD/128687/2017 from FCT/MCTES, Portugal.
References
- 1.Castelvecchi D. Can we open the black box of AI? Nature. 2016;538:20–23. doi: 10.1038/538020a. [DOI] [PubMed] [Google Scholar]
- 2.Wang P, et al. Bioinspired Smell and Taste Sensors. Springer; 2015. [Google Scholar]
- 3.McGann JP. Poor human olfaction is a 19th-century myth. Science. 2017;356:eaam7263. doi: 10.1126/science.aam7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitzgerald JE, et al. Artificial nose technology: status and prospects in diagnostics. Trends Biotechnol. 2017;35:33–42. doi: 10.1016/j.tibtech.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, et al. An overview of an artificial nose system. Talanta. 2018;184:93–102. doi: 10.1016/j.talanta.2018.02.113. [DOI] [PubMed] [Google Scholar]
- 6.Persaud KC. Towardsbionic noses. Sens Rev. 2017;37:165–171. [Google Scholar]
- 7.Broza YY, et al. Hybrid volatolomics and disease detection. Angew Chem Int Ed Engl. 2015;54:11036–11048. doi: 10.1002/anie.201500153. [DOI] [PubMed] [Google Scholar]
- 8.Krilaviciute A, et al. Detection of cancer through exhaled breath: a systematic review. Oncotarget. 2015;6:38643–38657. doi: 10.18632/oncotarget.5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vishinkin R, Haick H. Nanoscale sensor technologies for disease detection via volatolomics. Small. 2015;11:6142–6164. doi: 10.1002/smll.201501904. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald J, Fenniri H. Cutting edge methods for non-invasive disease diagnosis using e-tongue and e-nose devices. Biosensors. 2017;7:59. doi: 10.3390/bios7040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruz H, et al. Development of e-nose biosensors based on organic semiconductors towards low-cost health care diagnosis in gynecological diseases. Mater Today Proc. 2017;4:11544–11553. [Google Scholar]
- 12.Rocco G. Every breath you take: the value of the electronic nose (e-nose) technology in the early detection of lung cancer. J Thorac Cardiovasc Surg. 2018;155:2622–2625. doi: 10.1016/j.jtcvs.2017.12.155. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava AK, et al. Nanosensors and nanobiosensors in food and agriculture. Environ Chem Lett. 2018;16:161–182. [Google Scholar]
- 14.Loutfi A, et al. Electronic noses for food quality: a review. J Food Eng. 2015;144:103–111. [Google Scholar]
- 15.Zanardi E, et al. New insights to detect irradiated food: an overview. Food Anal Methods. 2018;11:224–235. [Google Scholar]
- 16.Giannoukos S, et al. Chemical sniffing instrumentation for security applications. Chem Rev. 2016;116:8146–8172. doi: 10.1021/acs.chemrev.6b00065. [DOI] [PubMed] [Google Scholar]
- 17.Cui S, et al. Plant pest detection using an artificial nose system: a review. Sensors. 2018;18:378. doi: 10.3390/s18020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szulczyński B, et al. Different ways to apply a measurement instrument of e-nose type to evaluate ambient air quality with respect to odour nuisance in a vicinity of municipal processing plants. Sensors (Switzerland) 2017;17:E2671. doi: 10.3390/s17112671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dung T, et al. Applications and advances in bioelectronic noses for odour sensing. Sensors. 2018;18:103. doi: 10.3390/s18010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wasilewski T, et al. Bioelectronic nose: current status and perspectives. Biosens Bioelectron. 2017;87:480–494. doi: 10.1016/j.bios.2016.08.080. [DOI] [PubMed] [Google Scholar]
- 21.Wasilewski T, et al. Advances in olfaction-inspired biomaterials applied to bioelectronic noses. Sens Actuators B Chem. 2018;257:511–537. [Google Scholar]
- 22.Son M, et al. Bioelectronic nose: an emerging tool for odor standardization. Trends Biotechnol. 2017;35:301–307. doi: 10.1016/j.tibtech.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Sanmartí-espinal M, et al. Quantification of interacting cognate odorants with olfactory receptors in nanovesicles. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-16997-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsuno H, et al. Novel cell-based odorant sensor elements based on insect odorant receptors. Biosens Bioelectron. 2015;65:287–294. doi: 10.1016/j.bios.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 25.Mulla MY, et al. Capacitance-modulated transistor detects odorant binding protein chiral interactions. Nat Commun. 2015;6:1–9. doi: 10.1038/ncomms7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotlowski C, et al. Fine discrimination of volatile compounds by graphene-immobilized odorant-binding proteins. Sens Actuators B Chem. 2018;256:564–572. [Google Scholar]
- 27.Larisika M, et al. Electronic olfactory sensor based on A. mellifera odorant-binding protein 14 on a reduced graphene oxide field-effect transistor. Angew Chem Int Ed Engl. 2015;54:13245–13248. doi: 10.1002/anie.201505712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antunes G, Simoes de Souza FM. Olfactory receptor signaling. Methods Cell Biol. 2016;132:127–145. doi: 10.1016/bs.mcb.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 29.Behrens M, et al. Structure–function relationships of olfactory and taste receptors. Chem Senses. 2018;43:81–87. doi: 10.1093/chemse/bjx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Fouchier A, et al. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat Commun. 2017;8 doi: 10.1038/ncomms15709. 15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva Teixeira CS, et al. Unravelling the olfactory sense: from the gene to odor perception. Chem Senses. 2016;41:105–121. doi: 10.1093/chemse/bjv075. [DOI] [PubMed] [Google Scholar]
- 32.Sung JH, et al. Piezoelectric biosensor using olfactory receptor protein expressed in Escherichia coli. Biosens Bioelectron. 2006;21:1981–1986. doi: 10.1016/j.bios.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Wu TZ. A piezoelectric biosensor as an olfactory receptor for odour detection: electronic nose. Biosens Bioelectron. 1999;14:9–18. doi: 10.1016/s0956-5663(98)00086-4. [DOI] [PubMed] [Google Scholar]
- 34.Wu C, et al. A biomimetic olfactory-based biosensor with high efficiency immobilization of molecular detectors. Biosens Bioelectron. 2012;31:44–48. doi: 10.1016/j.bios.2011.09.037. [DOI] [PubMed] [Google Scholar]
- 35.Goldsmith BR, et al. Biomimetic chemical sensors using nanoelectronic readout of olfactory receptor proteins. ACS Nano. 2011;5:5408–5416. doi: 10.1021/nn200489j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SH, et al. Mimicking the human smell sensing mechanism with an artificial nose platform. Biomaterials. 2012;33:1722–1729. doi: 10.1016/j.biomaterials.2011.11.044. [DOI] [PubMed] [Google Scholar]
- 37.Wu C, et al. Biomimetic sensors for the senses: towards better understanding of taste and odor sensation. Sensors (Basel) 2017;17:E2881. doi: 10.3390/s17122881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persaud KC, Tuccori E. Biosensors based on odorant binding proteins. In: Park TH, editor. Bioelectronic Nose. Springer; 2014. pp. 171–190. [Google Scholar]
- 39.Pelosi P, et al. Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol Rev. 2018;93:184–200. doi: 10.1111/brv.12339. [DOI] [PubMed] [Google Scholar]
- 40.Brito NF, et al. A look inside odorant-binding proteins in insect chemoreception. J Insect Physiol. 2016;95:51–65. doi: 10.1016/j.jinsphys.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Northey T, et al. Crystal structures and binding dynamics of odorant-binding protein 3 from two aphid species Megoura viciae and Nasonovia ribisnigri. Sci Rep. 2016;6 doi: 10.1038/srep24739. 24739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelosi P, et al. Structure and biotechnological applications of odorant-binding proteins. Appl Microbiol Biotechnol. 2014;98:61–70. doi: 10.1007/s00253-013-5383-y. [DOI] [PubMed] [Google Scholar]
- 43.Mastrogiacomo R, et al. An odorant-binding protein is abundantly expressed in the nose and in the seminal fluid of the rabbit. PLoS One. 2014;9:45–48. doi: 10.1371/journal.pone.0111932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiefner A, et al. Crystal structure of the human odorant binding protein, OBP IIa. Proteins. 2015;83:1180–1184. doi: 10.1002/prot.24797. [DOI] [PubMed] [Google Scholar]
- 45.Paolini S, et al. Porcine odorant-binding protein: structural stability and ligand affinities measured by fourier-transform infrared spectroscopy and fluorescence spectroscopy. Biochim Biophys Acta. 1999;1431:179–188. doi: 10.1016/s0167-4838(99)00037-0. [DOI] [PubMed] [Google Scholar]
- 46.Capone S, et al. Electrical characterization of a pig odorant binding protein by impedance spectroscopy. Proc IEEE Sens. 2009;2009:1758–1762. [Google Scholar]
- 47.Di Pietrantonio F, et al. Surface acoustic wave biosensor based on odorant binding proteins deposited by laser induced forward transfer. IEEE Int Ultrason Symp. 2013;2013:2144–2147. [Google Scholar]
- 48.Cannata D, et al. Odorant detection via solidly mounted resonator biosensor. IEEE Int Ultrason Symp. 2012;2012:1537–1540. [Google Scholar]
- 49.Di Pietrantonio F, et al. Tailoring odorant-binding protein coatings characteristics for surface acoustic wave biosensor development. Appl Surf Sci. 2014;302:250–255. [Google Scholar]
- 50.Palla-Papavlu A, et al. Preparation of surface acoustic wave odor sensors by laser-induced forward transfer. Sens Actuators B Chem. 2014;192:369–377. [Google Scholar]
- 51.Di Pietrantonio F, et al. Detection of odorant molecules via surface acoustic wave biosensor array based on odorant-binding proteins. Biosens Bioelectron. 2013;41:328–334. doi: 10.1016/j.bios.2012.08.046. [DOI] [PubMed] [Google Scholar]
- 52.Di Pietrantonio F, et al. A surface acoustic wave bioelectronic nose for detection of volatile odorant molecules. Biosens Bioelectron. 2015;67:516–523. doi: 10.1016/j.bios.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 53.Bonnot K, et al. Biophotonic ring resonator for ultrasensitive detection of DMMP as a simulant for organophosphorus agents. Anal Chem. 2014;86:5125–5130. doi: 10.1021/ac500903s. [DOI] [PubMed] [Google Scholar]
- 54.Possas-Abreu M, et al. Biomimetic diamond MEMS sensors based on odorant-binding proteins: sensors validation through an autonomous electronic system. ISOCS/IEEE Int Symp Olfaction Electron Nose; 2017. [Published online May 2017]. [DOI] [Google Scholar]
- 55.Zhao X, et al. Protein functionalized ZnO thin film bulk acoustic resonator as an odorant biosensor. Sens. Actuators B Chem. 2012;163:242–246. [Google Scholar]
- 56.Sankaran S, et al. Biology and applications of olfactory sensing system: a review. Sens Actuators B Chem. 2012;171–172:1–17. [Google Scholar]
- 57.Wu TZ, Lo YR. Synthetic peptide mimicking of binding sites on olfactory receptor protein for use in “electronic nose”. J Biotechnol. 2000;80:63–73. doi: 10.1016/s0168-1656(00)00228-5. [DOI] [PubMed] [Google Scholar]
- 58.Wu TZ, et al. Exploring the recognized bio-mimicry materials for gas sensing. Biosens Bioelectron. 2001;16:945–953. doi: 10.1016/s0956-5663(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 59.McAlpine MC, et al. Peptide-nanowire hybrid materials for selective sensing of small molecules. J Am Chem Soc. 2008;130:9583–9589. doi: 10.1021/ja802506d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu HH, et al. Direct characterization and quantification of volatile organic compounds by piezoelectric module chips sensor. Sens Actuators B Chem. 2009;137:741–746. [Google Scholar]
- 61.Panigrahi S, et al. Olfactory receptor-based polypeptide sensor for acetic acid VOC detection. Mater Sci Eng C Mater Biol Appl. 2012;32:1307–1313. doi: 10.1016/j.msec.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 62.Sankaran S, et al. Odorant binding protein based biomimetic sensors for detection of alcohols associated with Salmonella contamination in packaged beef. Biosens Bioelectron. 2011;26:3103–3109. doi: 10.1016/j.bios.2010.07.122. [DOI] [PubMed] [Google Scholar]
- 63.Son M, et al. Bioelectronic nose using odorant binding protein-derived peptide and carbon nanotube field-effect transistor for the assessment of Salmonella contamination in food. Anal Chem. 2016;88:11283–11287. doi: 10.1021/acs.analchem.6b03284. [DOI] [PubMed] [Google Scholar]
- 64.Pender MJ, et al. Peptide-mediated formation of single-wall carbon nanotube composites. Nano Lett. 2016;6:40–44. doi: 10.1021/nl051899r. [DOI] [PubMed] [Google Scholar]
- 65.Kuang Z, et al. Biomimetic chemosensor: designing peptide recognition elements for surface functionalization of carbon nanotube field effect transistors. ACS Nano. 2009;4:452–458. doi: 10.1021/nn901365g. [DOI] [PubMed] [Google Scholar]
- 66.Jaworski JW, et al. Evolutionary screening of biomimetic coatings for selective detection of explosives. Langmuir. 2008;24:4938–4943. doi: 10.1021/la7035289. [DOI] [PubMed] [Google Scholar]
- 67.Jang H-J, et al. Identification of dinitrotoluene selective peptides by phage display cloning. Bull Korean Chem Soc. 2010;31:3703–3706. [Google Scholar]
- 68.Ju S, et al. Single-carbon discrimination by selected peptides for individual detection of volatile organic compounds. Sci Rep. 2015;5:9196. doi: 10.1038/srep09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pizzoni D, et al. Selection of peptide ligands for piezoelectric peptide based gas sensors arrays using a virtual screening approach. Biosens Bioelectron. 2014;52:247–254. doi: 10.1016/j.bios.2013.08.044. [DOI] [PubMed] [Google Scholar]
- 70.Mascini M, et al. Tailoring gas sensor arrays via the design of short peptides sequences as binding elements. Biosens Bioelectron. 2017;93:161–169. doi: 10.1016/j.bios.2016.09.028. [DOI] [PubMed] [Google Scholar]
- 71.Hwang KS, et al. Peptide receptor-based selective dinitrotoluene detection using a microcantilever sensor. Biosens Bioelectron. 2011;30:249–254. doi: 10.1016/j.bios.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 72.Mascini M, et al. Piezoelectric sensors for dioxins: a biomimetic approach. Biosens Bioelectron. 2004;20:1203–1210. doi: 10.1016/j.bios.2004.06.048. [DOI] [PubMed] [Google Scholar]
- 73.Kobayashi S, et al. A theoretical investigation of the conformation changing of dioxins in the binding site of dioxin receptor model; role of absolute hardness–electronegativity activity diagrams for biological activity. J Mol Struct. 1999;475:203–217. [Google Scholar]
- 74.Compagnone D, et al. Gold nanoparticles-peptide based gas sensor arrays for the detection of food aromas. Biosens Bioelectron. 2013;42:618–625. doi: 10.1016/j.bios.2012.10.096. [DOI] [PubMed] [Google Scholar]
- 75.Compagnone D, et al. Sensors and actuators B: chemical quartz crystal microbalance gas sensor arrays for the quality control of chocolate. Sens Actuators B Chem. 2015;207:1114–1120. [Google Scholar]
- 76.Mascini M, et al. Peptide modified ZnO nanoparticles as gas sensors array for volatile organic compounds (VOCs) Front Chem. 2018;6:105. doi: 10.3389/fchem.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palma SICJ, et al. Machine learning for the meta-analyses of microbial pathogens’ volatile signatures. Sci Rep. 2018;8:3360. doi: 10.1038/s41598-018-21544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carraher C, et al. Towards an understanding of the structural basis for insect olfaction by odorant receptors. Insect Biochem Mol Biol. 2015;66:31–41. doi: 10.1016/j.ibmb.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 79.Cui Y, et al. Biomimetic peptide nanosensors. Acc Chem Res. 2012;45:696–704. doi: 10.1021/ar2002057. [DOI] [PubMed] [Google Scholar]
- 80.Sankaran S, et al. Olfactory receptor based piezoelectric biosensors for detection of alcohols related to food safety applications. Sens Actuators B Chem. 2011;155:8–18. [Google Scholar]
- 81.Nagraj N, et al. Selective sensing of vapors of similar dielectric constants using peptide-capped gold nanoparticles on individual multivariable transducers. Analyst. 2013;138:4334–4339. doi: 10.1039/c3an00088e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.