Abstract
Purpose:
To develop a prostate tumor habitat risk scoring (HRS) system based on multiparametric MRI (mpMRI) referenced to prostatectomy Gleason Score (GS) for automatic delineation of Gross Tumor Volumes (GTVs). A workflow for integration of HRS into radiotherapy (RT) boost volume dose escalation was developed in the framework of a Phase II randomized clinical trial (BLaStM).
Materials and Methods:
An automated quantitative mpMRI based 10 point pixel by pixel method was optimized to prostatectomy GSs and volumes using referenced Dynamic Contrast Enhanced and Apparent Diffusion Coefficient sequences. The HRS contours were migrated to the planning CT for boost volume generation.
Results:
There were 51 regions of interest (ROIs) in 12 patients who underwent radical prostatectomy (RP) (26 with GS≥7 and 25 with GS6). The resultant heat maps showed inter- and intra-tumoral heterogeneity. The HRS6 level was significantly associated with RP ROIs (slope 1.09, r=0.767; p<.0001). For predicting the likelihood of cancer, GS≥7 and GS≥8, HRS6 AUCs were 0.718, 0.802 and 0.897, respectively. HRS was superior to the Prostate Imaging, Reporting and Diagnosis System 4/5 classification, wherein the AUCs were 0.62, 0.64 and 0.617, respectively (difference with HR6, p<.0001). HRS maps were created for the first 37 assessable patients on the BLaStM trial. There were an average of 1.38 habitat boost volumes per patient at a total boost volume average of 3.6 cc.
Conclusions:
An automated quantitative mpMRI based method was developed to objectively guide dose escalation to high risk habitat volumes based on prostatectomy GS.
Summary:
Prostate multiparamateric MRI has a high sensitivity and specificity for identifying tumor regions in the prostate; but, there is subjectivity in defining high risk 3D volumes that could be boosted, as opposed to entire prostate dose escalation. Registering to prostatectomy Gleason score, a habitat risk score based on pixel by pixel quantitative diffusion and perfusion was developed and then applied to guiding radiotherapy boost volumes in the background of a randomized Phase II clinical trial.
INTRODUCTION:
Radiation dose escalation improves the control of intermediate to high risk prostate cancer, with doses above 80 Gy improving outcomes(1). Limiting the highest radiation doses to the Gross Tumor Volume (GTV), as opposed to whole prostate, is hypothesized to result in equivalent tumor control without increasing side effects(2–4). A major obstacle is defining the GTV in a systematic and reproducible way. While the use of multiparametric MRI (mpMRI) for GTV identification is gaining momentum because of its diagnostic reliability for distinguishing intraprostatic tumors of Gleason Score (GS) 7 or above(5, 6), there is considerable variability in how boost volumes are defined. Currently, the Prostate Imaging, Reporting and Diagnosis System (PIRADS) is the standard of care for region of interest (ROI) identification and risk classification(7, 8). However, there is subjectivity in PIRADS, the system was not designed for 3D volume assessment, a wealth of quantitative information in mpMRI is ignored, and PIRADS does not accurately and reproducibly elucidate inter- and intra-lesional spatial heterogeneity.
In this report, a quantitative mpMRI analysis technique that combines perfusion (Dynamic Contrast Enhanced (DCE)) and diffusion mpMRI sequences to identify distinct pathophysiologic regions, or “Habitats”(9) is described. In two prior reports, we described: (i) DCE-Score(10), and (ii) Apparent Diffusion Coefficient (ADC) thresholds and volumes to describe risk based on Gleason score (GS). We describe herein a pixel by pixel habitat risk scoring (HRS) system that combines the quantitative DCE and ADC information by referencing to a prostatectomy dataset. The automatically generated heat maps were used prospectively to guide radiotherapy (RT) boost volumes in a randomized Phase II clinical trial, BLaStM, comparing two methods of increasing dose to the mpMRI-defined tumor habitat region(s).
MATERIALS AND METHODS:
Patients:
For the development of the prostate mpMRI analysis techniques, the Institutional Review Board (IRB) approved a protocol for retrospective review of patients who have undergone prostate mpMRI. Patients that underwent RP during 2016 and had mpMRI on 3T Discovery MR750 magnet (GE, Waukesha, WI) were identified. The IRB waived the need for informed consent for the prostatectomy cohort. The BLaStM clinical trial was open for accrual in February 2015 and the patients were consented prospectively.
mpMRI acquisition:
mpMRIs of the prostate were acquired at 3T Discovery MR750 magnet (GE, Waukesha, WI) with 32-channel phased array pelvis coil. Axial T2 weighted-MRI (T2w): 2D Fast spinecho, 1.25×1.25×2.5mm resolution; acquisition matrix = 256×256 and 72 slices (no gap); repetition time (TR)/echo time (TE)/Number of Excitations (NEX)=5700/85/1; echo train length =16; flip angle 120°; (i) Diffusion Weighted Imaging (DWI): single-shot echo-planar imaging (EPI) utilizing the diffusion-module and fat-suppression pulses; 2.5×2.5×2.5 mm resolution; acquisition matrix 128×128 and 36 slices; TR/TE/NEX 9500/53/1; b-values 50, 500 and 1000 s/mm2. The ADC map was calculated on the GE MR console, using a mono-exponential fit. (ii) Twelve DCE series were acquired with the same spatial resolution and spacing as the T2w, using a dynamic 3D fast spoiled gradient echo T1 weighted (T1w) sequence utilizing a parallel imaging, acceleration factor of 2; temporal resolution: 30–34s, TR=3.77–4.05 ms, TE=1.69–1.78 ms following intravenous bolus injection of a paramagnetic gadolinium chelate - 0.1 mmol of gadobenate-glumine (Bracco Diagnostics Inc., Princeton, NJ) per kilogram of body weight. The contrast is administered with a power injector (Spectris, Medrad Inc., Warrendale, PA) at 2 mL/s and followed by a 20-mL saline flush.
Habitat Risk Score:
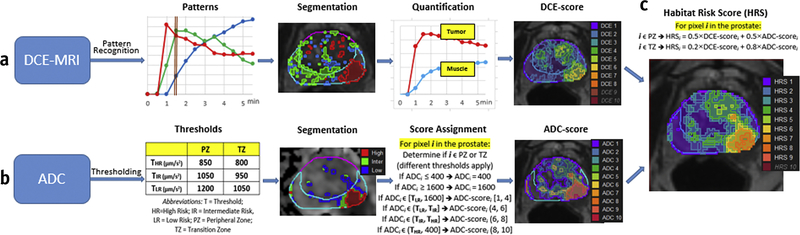
The HRS combines (i) DCE-scores, modified from Parra et al (10) in 71 radiotherapy patients, and (ii) ADC thresholds and volumes, modified from Tcshudi (11) in 137 radiotherapy patients. From these refinements in the DCE-scores and In this manuscript, the combination of DCE and ADC quantitative analyses into HRS was developed in the prostatectomy cohort. HRS is implemented in Java in a plug-in extension to MIM (MIM Cleveland, OH) (Suppl.Figure 1). The input consists of DCE-MRI and ADC series with contours of the prostate, peripheral zone (PZ) and a representative sample of the gluteus muscle (GM) (Suppl.Figure 1b); the transition zone (TZ) is derived from the PZ and prostate volumes. The flow chart in Figure 1 illustrates the steps for HRS calculation, as described below:
Figure 1. Flow-chart of Habitat Risk Score (HRS) construction.
(a) DCE-MRI analysis: </p>DCE-MRI data is baseline corrected by the average of δ = 2 pre-contrast points. Non Negative Matrix Factorization (NMF) is applied to the matrix D(X, t) with κ=3. D is represented as a product of κ basic temporal contrast signatures S(t) and their weights W(X), i.e. D ~W × S. The identified three basic temporal contrast signatures S(t) are shown in box, labelled Patterns; the well perfused pattern, Swp (depicted in red) is automatically selected as the pattern with maximum area under the curve (AUC) between 0 and 90 sec (brown bar). The segmented weights of the patterns W(t) are shown in corresponding colors in box, labelled Segmentation (α=60, β=10). The area in red depicts the region of interest (ROI) for the well-perfused pattern: ROIwp. Two curves are shown in box, labelled Quantification: the average DCE-curve from the ROIwp (Tumor) and Gluteus Maximus (Muscle). Semi-quantitative feature (Late AUC) is extracted from these two curves: Σi (Tumor) and Σim (Muscle). The ratio Σi/Σim is propagated for each pixel in the prostate, using Wwp. A value between 1 to 10 is assigned to each pixel of the prostate, using σ1=.5 and σ2=2.5 (the range is bound by the γth and (100-γ)th; γ=5). DCE-score is depicted as a heat map (Note that DCE9 and DCE10 are empty contours). (b) ADC analysis: Thresholds ΤHR, TIR and TLR for volumes at high risk, intermediate risk and low risk for cancer are identified separately for peripheral zone (PZ) and transition zone (TZ). The segmented volumes are shown in box, labelled Segmentation. The ADC is mapped on 10-point scale, using the rules in Score Assignment box and depicted as a heat map. (c) HRS is calculated for each pixel, using η1= 0.5 and η2= 0.5 in PZ and η1= 0.2 and η2 =0.8 in TZ. HRS is depicted as a heat map (Note that HRS10 is an empty contour).
Quantitative analysis of DCE-MRI:
The DCE analysis (Figure 1a) is described in detail in Parra et al(10). Briefly:
Pattern recognition: Non Negative Matrix Factorization (NMF) is applied to signal vs time curves of all pixels within the prostate(12). Let D be the data matrix, containing the individual pixel’s signal vs time curves in its rows (baseline corrected by the average of δ pre-contrast points). D is represented as a product of κ basic temporal contrast signatures S(t) and their weights W(X) in each pixel, i.e. D ~W × S under the constraint that all elements of W and S are non-negative; S(t) are normalized to one. κ is estimated by Principal Component Analysis (PCA) of D as the number of significant Principal Components(13).
Patterns: The well-perfused (wp) temporal pattern, Swp, is automatically selected as the pattern with maximum area under the curve (AUC) between 0 and 90 sec (Figure 1a, brown bar). This time frame was chosen because it best discriminates the well-perfused pattern from the others (13).
Segmentation: The corresponding weights Wwp represent an intensity map of the distribution of well-perfused pixels in the data. To delineate the suspected tumor area ROIwp, several segmentation approaches are implemented in HRS. The parameter α, describes how ‘pure’ the pattern is in a pixel. The parameter β is related to the assignment of the segmented ROI in the PZ or transition zone (TZ). For instance, if α=60, ROIwp is defined by pixels for which Wwp is >60% of the total sum of the weights; similarly, if β=20, then if at least 20% of ROIwp is in PZ, the ROIwp is considered to be in PZ.
Quantitation: Six semi-quantitative features Σ1 to Σ6 related to the washin and washout components of the DCE-curves are implemented in HRS. These features are: early AUC/AUFC (sum of the raw/fitted(14) points @60, 90,120 s); late AUC/AUFC (sum of the last raw/fitted(14) 4 points); wash-in(14) and wash-out(15) were calculated using the average DCE signal in ROIwp. Corresponding features were calculated for Gluteus Maximus muscle (Σim).
DCE-Score: The estimate of a given Σi, normalized Σim in ROIwp is used to propagate this score to each pixel of the data, using Wwp. The range of Σi/Σim: σ1 and σ2, is determined in a series of patients (>50) and mapped to 1 to 10 scale, where the range is bound by the γth and (100-γ)th percentile of the feature distribution. Using Σi/Σim estimate in the ROIwp and Wwp, a value between 1 to 10 is assigned to each pixel of the prostate.
In summary, the free-parameters for DCE analysis are δ, κ, α, β, Σi, σ1, σ2, γ. These parameters were optimized in DCE-MRIs from as described previously(10) and used as default settings in HRS (Suppl.Figure 1c). The results of the DCE-MRI analysis for two features (early and last AUC) in correlation with GS are shown in Suppl.Figure 2. Following normalization with a gluteus muscle (GM) volume DCE signal, the quantitative features in the PZ were significantly correlated with GS.
Quantitative analysis of ADC:
The ADC analysis is illustrated in Figure 1b. A search algorithm was developed to span ADC values in order to determine optimal ADC thresholds (T) (ΤHR, TIR and TLR, THR<TIR<TLR) for the identification of volumes at high risk (HR), intermediate risk (IR) and low risk (LR) for cancer(11). Because of the differential diffusion characteristics of PZ and TZ, thresholds were determined separately. To exclude outliers, ADC values between τ1 and τ2 (e.g. 400 and 1600 μm2/s) are considered. Two sets of thresholds ΤHR, TIR and TLR (for PZ and TZ) and τ1 and τ2 are user-defined parameters for ADC analysis (Suppl.Figure 1c).
The thresholds in PZ/TZ were determined as: ΤHR=900/800; TIR=1100/850; TLR=1300/1050 μm2/s; τ1=400 and τ2=1600 (Suppl.Figure 1c). The size of the identified volumes at high, intermediate and low risk were significantly correlated with GS (Suppl.Figure 3).
Using these parameters as are the default settings in HRS, the ADC is mapped on 10-point scale, using the following rule: intensities between τ2 and TLR are mapped between 1 to 4; (TLR, TIR]➔(4,6]; (TIR, THR]➔(6,8]; [τ1,THR]➔(8,10] (Figure 1b).
Construction of HRS:
Both DCE- and ADC-scores are calculated at the resolution of the corresponding sequence. HRS is estimated for each pixel at the lower resolution (ADC) image as a weighted sum of DCE- and ADC-scores. These weights (η1 and η2) are set differently for PZ and TZ (Figure 1c). HRS volumes, smaller than ε (cc) are removed (Suppl.Figure 1c).
Evaluation of Habitat Risk Score performance:
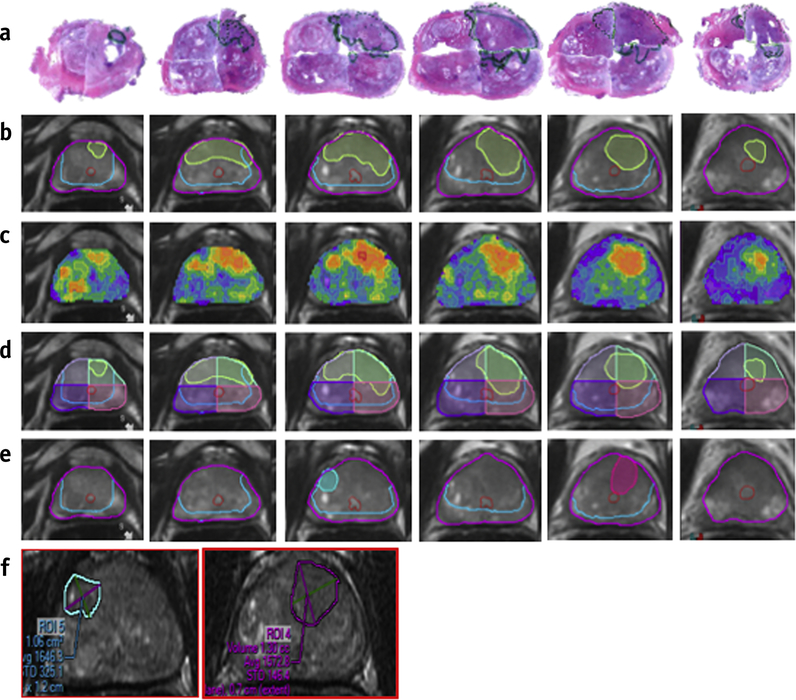
Radical prostatectomy (RP) specimens were used for HRS evaluation. RP specimens were handled in accordance with a well-established procedure (16). The prostate was cut in quadrants and tumor nodules contoured and graded by a urologic pathologist (ONK)(17, 18) (Suppl.Methods). The annotated slides were scanned and prostate quadrants were “stitched” into a pseudo whole-mount RP sample (Figure 2a).
Figure 2. Schema of approach for correlating histopathology with Habitat Risk Score (HRS) and PIRADS.
(a) stitched quantrantized prostatectomy H&E-stained histopathology sections. (b) Corresponding T2-weighted MRI transverse slices with contours of the prostate (magenta), urethra (red), peripheral zone (PZ) (light blue) and tumor nodules from histopathology, mapped in light green; (c) HRS represented as heat-map, overplayed on the T2w; (d) quadrants overlays (last row); (e) PIRADS areas are transferred from radiologists drawn contours (f).
The pathologist’s GS specific volume contours were transferred to the mpMRI in MIM using freehand ROIs (rpROIs)(19) (Figure 2b, Suppl.Methods). Independently, the HRS heat maps were generated (Figure 2c) and each MRI slice divided into four cross-sectional quadrants complimentary to the pathologic specimen (Figure 2d). PIRADS contours were also transferred to MIM (Figure 2e,f).
Logistic regression models were used to assess the association between the HRS and three histopathology outcomes in each tumor nodule: Cancer vs No Cancer, GS≥7 vs No Cancer/GS=6 and GS≥8 vs No cancer/GS=6,7. The same approach was used for association between PIRADS4/5 and the above histopathology parameter. Receiver operating characteristic (ROC) curves and areas under the ROC (AUCs) were obtained both for HRS and PIRADS4/5. For model validation, we used: (i) apparent, and (ii) bootstrap validation approaches. In the apparent validation, same data were used to build a model and calculate the AUCs. This approach usually gives an optimistic estimate of AUC. We also obtained optimism-corrected AUCs via bootstrap validation with 200 repetitions (20).
Implementation of HRS in Radiation Treatment of patients:
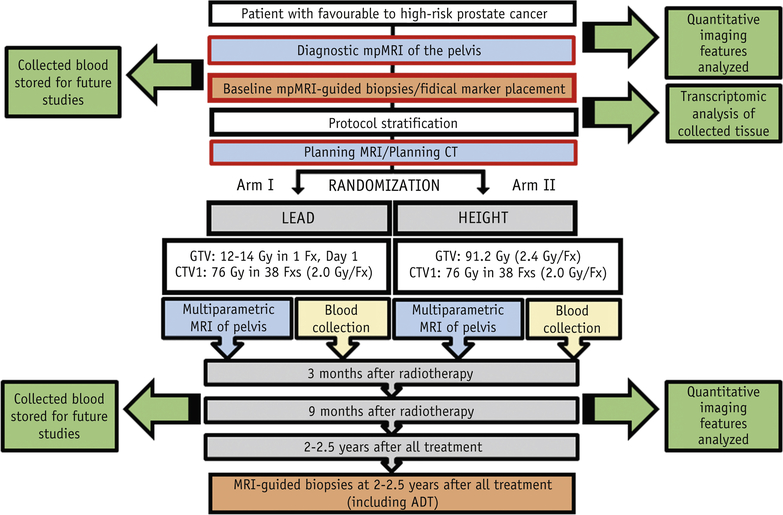
Aside from developing a heat map to direct prostate biopsies, a main goal was to automate and implement a workflow for the objective identification of tumor volumes for dose escalation as a means of potentially reducing the toxicity of dose escalating the entire gland, as has been the tradition(1). The implementation was facilitated through an institutional Phase II randomized clinical trial “MRI-Guided Prostate Boosts Via Initial Lattice Stereotactic vs Daily Moderately Hypofractionated Radiotherapy (BLaStM)”, http://clinicaltrials.gov: NCT02307058 (Figure 3). Patients are randomized into one of two treatment arms. In Arm I, a high RT dose (12–14 Gy) is given on day 1 to a portion near the center of the mpMRI delineated gross tumor volume(s) or GTV(s), followed by standard fractionation RT to the entire prostate and proximal seminal vesicles (SVs) to 76 Gy in 38 treatments. In Arm II, a daily simultaneous integrated boost (SIB) to the mpMRI delineated GTVs is at 2.4 Gy per fraction, while the remaining prostate and proximal SVs are treated at 2 Gy per fraction to 76 Gy. More details for BlaStM are provided in Suppl.Methods.
Figure 3. Schema of the BLaStM Phase II randomized clinical trial.
“MRI-Guided Prostate Boosts Via Initial Lattice Stereotactic vs Daily Moderately Hypofractionated Radiotherapy (BLaStM), http://clinicaltrials.gov: NCT02307058.
The relevant HRS key steps are designated with red frames in Figure 3, which include registrations of the diagnostic mpMRI, to the CT- and MRI-simulation images that are done after fiducial marker placement (Suppl.Methods).
RESULTS:
Habitat Risk Score:
HRS was calculated in 12 prostatectomy patients with available mpMRI preoperatively, using η1=0.8 and η2=0.2 for TZ and η1=0.5 and η2=0.5 for PZ; ε =0.01 cc. The ability of HRS to discriminate between Cancer vs No Cancer, GS≥7 vs No Cancer or GS=6, and GS≥8 vs No Cancer or GS=6 or 7, was compared to PIRADS4/5 in 51 tumor nodules (26 GS ≥ 7 and 25 GS=6) (Suppl.Table 1). The HRS algorithm had a higher sensitivity than PIRADS4/5 at most levels, ranging from 100% for HRS4 to 45.58% for HRS9, compared to PIRADS4/5 of 25.2% for Cancer vs No Cancer, 31.4% for GS≥7 vs No Cancer or GS=6 and 33.7% for GS≥8 vs No Cancer or GS=6 or 7. However, the specificities for HRS were lower than for PIRADS4/5. In addition, the individual DCE and ADC-scores operating characteristics were calculated (Suppl.Table 1). HRS performed better than the DCE and ADC models alone. While in the majority of the cases ADC has better sensitivity and DCE better specificity, HRS has balanced sensitivity and specificity.
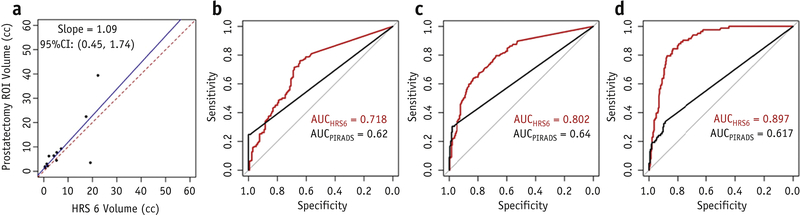
The lesion volumes of GS≥7 in the RP were in good agreement with HRS6 volumes within the prostate (Figure 4a). From the apparent validation, the corresponding AUCs of HRS6 are shown in Figure 4b-d, together with AUCs based on PIRADS4/5 only. HRS6 was chosen because it was the most consistent at achieving the main objective of detecting significant cancers of GS≥7 (AUC=0.802, 95%CI: 0.756–0.847). In contrast, PIRADS had an AUC=0.64 (95%CI: 0.602–0.678). The difference between the two AUCs was statistically significant (p<.0001). From the bootstrap validation (Suppl.Table 2), optimism-corrected AUC for HRS6 was 0.798 (95%CI: 0.68–0.924) for predicting the likelihood of GS≥7. In contrast, PIRADS had an AUC=0.635 (95%CI: 0.534–0.707).
Figure 4. HRS performance.
(a) Correlation of HRS6 volumes with prostatectomy ROI volumes with GS≥7; ROC curves of PIRADS and HRS6 in discriminating: (b) Cancer vs No Cancer; (c) GS≥7 vs No Cancer or GS=6; and (d) GS≥8 vs No cancer or GS=6,7.
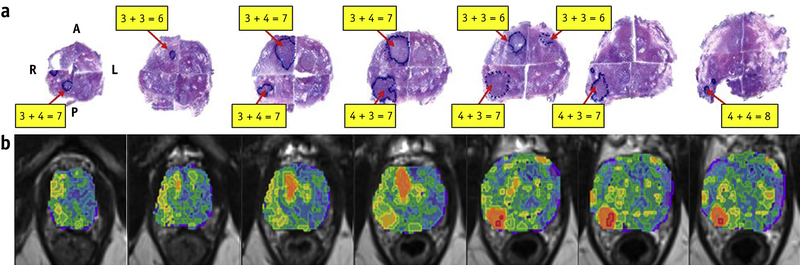
HRS mapping of inter- and intralesional heterogeneity is demonstrated in Figure 5. The GS of the right posterior nodule changes from 3+4=7 and 4+3=7 to 4+4=8; the GS of the right anterior nodule changes from 3+3=6 to 3+4=7 and back to 3+3=6. Note the correlation between the red color intensities of the HRS heat-map with the higher microscopic tumor grade.
Figure 5: Correlation of habitat risk score (HRS) maps with histopathology.
(a) Pseudowhole mount H&E-stained histopathology sections. The consecutive axial slices are displayed from apex (left) to base (right). Tumor nodules are labeled with the corresponding Gleason Score (GS). The right posterior nodule displays heterogeneity with GS increasing from 3+4=7, to 4+3=7, and then to 4+4=8 (apex to base). The GS of the right anterior tumor nodule changes from 3+3=6 to 3+4=7 and back to 3+3=6; (b) Corresponding HRS maps, displayed on axial T2w slices. Image data was resampled at the same spacing (0.3 to 0.4 cm) as the histopathology specimens. The red color intensities of the heat-map are associated with the higher microscopic GS.
Abbreviations: A = anterior; L = left; P = posterior; R = right
The BLaStM Trial:
The BLaStM trial was used as a backdrop to the hypothesis that automated quantitative mpMRI could be used to guide the planning of dose escalation to high risk tumor habitat volumes in the prostate. The BLaStM trial is an investigator-initiated Phase II randomized protocol that is comparing two methods of delivering dose escalated boosts to targeted volumes (details in Suppl.Methods). As of November 2017, there were 57 assessable BLaStM patients. The clinical characteristics of the analyzed cohort are summarized in Suppl.Table 3. Twenty patients were excluded (37 assessable) from the analyses because Androgen Deprivation Therapy (ADT) was started prior to the diagnostic mpMRI.
HRS for GTV delineation in the BLaStM trial:
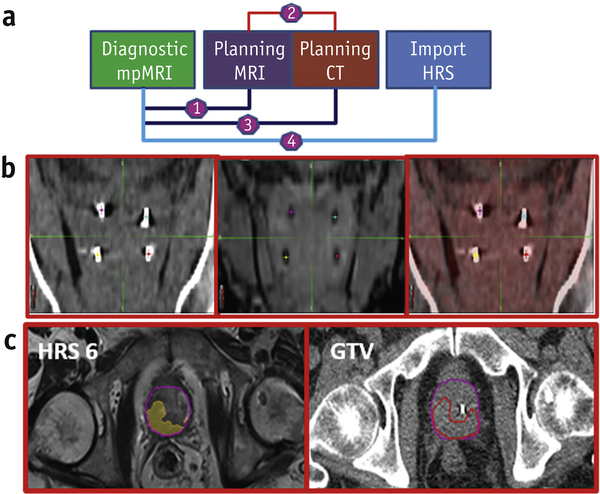
The implementation of HRS6 in the treatment planning of the patients in BLaStM is shown in Figure 6. The diagnostic mpMRI is rigidly registered to the planning MRI, by an experienced physicist (KRP), focusing on anatomical matching and specifically balancing alignment of the prostate, prostate/rectum interface and prostate/bladder interface. The planning MRI is rigidly registered to the planning CT utilizing the four gold fiducials that are visible on both acquisitions employing a commercially available point based alignment method based on linear least squares minimization. Finally, the diagnostic mpMRI generated HRS contours are registered to the planning CT. The HRS6 contours provide a recommended GTV; although all of the HRS5 through HRS10 contours are viewable, when present (not every patient has pixels at the higher HRS values). While the HRS6 contour is now being used prospectively, all 37 assessable patients were analyzed. The dosimetry for patients in treatment Arms I and II are shown in Suppl.Figure 4. The mean number of lesions were 1.38 per patient (Table 1). The mean lesion volume (+/−SD) was 3.36±3.74 cc. The median lesion volume was 1.48 (range 0.19–15.34) cc.
Figure 6: Workflow for incorporation of HRS in BLaStM patient RT planning.
(a) Schematic of the registrations utilized and the migration of the HRS structures: (1) Using prostate anatomical matching, the diagnostic mpMRI is registered with the planning MRI; (2) The planning MRI is fused to the planning CT, using fiducial matching; (3) Using (1) and (2), the diagnostic mpMRI generated HRS contours are migrated to the planning CT; (b) Registration of the planning CT (left) and planning MRI (center) using fiducial matching and the alignment (right); (c) The final result for one patient is displayed where the HRS 6 contour has been migrated to the planning CT using the methods described.
Table 1:
Summary of HRS analysis in BlaStM patients
| Patients | Prostate Volume (cc) | Mean No. habitats/Pt | Habitat Volume/Pt (mean ± SD cc) | Habitat Volume/lesion (mean ± SD cc) | PZ N (%) | TZ N (%) | Both N (%) |
|---|---|---|---|---|---|---|---|
| Total (n = 37) | 39.6 | 1.38 | 3.6 ± 3.74 | 2.83 ± 3.34 | 16 (43.2) | 10 (27.1) | 11 (29.7) |
| Intermediate risk (n = 21) | 42.1 | 1.48 | 3.37 ± 3.93 | 2.67 ± 3.34 | 9 (42.9) | 4 (19.0) | 8 (38.1) |
| High risk (n = 16) | 36.2 | 1.25 | 3.91 ± 3.68 | 3.76 ± 3.22 | 7 (43.8) | 6 (37.5) | 3 (18.7) |
Abbreviations: PZ = Peripheral Zone; TZ = Transition Zone
DISSCUSION:
Prostate cancer radiation dose escalation improves clinical outcomes and there is evidence that doses beyond 80 Gy result in substantive improvements(1, 21). However, when the whole prostate is dose escalated there is an increase in grade 3 complication risk(22). An alternative approach is to dose escalate beyond 80 Gy only to determinate tumor areas in the prostate. While multiple groups(2–4) have sought to direct radiation dose to such high risk tumor regions in lieu of escalating dose to the entire prostate, there has been considerable variability and subjectivity in defining boost volumes. In order to reproducibly identify prostate high risk habitat volumes in 3-dimensions, we describe an automated method using quantitative parameters extracted from standard mpMRI sequences.
The depiction of habitats in Figure 5 visually describes these areas. The current standard of care uses radiologist’s drawn contours that are outlined typically on one imaging slice. The significance of the presented techniques relative to PIRADS4/5 is appreciated in Figure 2 where a radiologist has contoured two areas for PIRADS assessment (Figure 2e,f), whereas the HRS maps (Figure 2c) depict the entire 3D volume of the tumor. While one could argue that the radiologist in this example could have done a better job of depicting the volume of interest, PIRADS was not designed for contouring, 3D volume contouring is labor-intensive (not practical for radiologists), and subjectivity in contouring would likely be considerable. Quantitative imaging analytic methods are sure to reduce subjectivity and variability in the depiction of high risk volumes of interest. Moreover, pixel by pixel risk assignment displays intratumoral spatial heterogeneity, when present; this degree of habitat resolution is not possible at the purely human level. HRS also holds the promise to detect some of the so-called “invisible” lesions that are not seen by the radiologists. About 50% of these lesions have genomic alterations, associated with adverse outcome (23).
The HRS classification system was arbitrarily assigned a 10 point scale per pixel. Our data suggest that this could be collapsed to fewer levels. The associations with GS were seen mainly at HRS5 and above, with HRS6 showing reasonable discrimination between cancer vs no cancer, GS≥7 vs other, and GS≥8 vs other (Figure 4, Suppl Table 1). The workflow developed imports the HRS 5–10 contours to the planning CT and during the boost planning on BLaStM attention is paid to the HRS6 contours. In the current HRS version, the weights for DCE- and ADC-scores were selected based on an ad-hoc approach. The rational for the under-weighting DCE (η2=0.2) in TZ is that TZ is better vascularized than PZ and therefore there are more false positives on DCE-MRI in TZ (24). This is also recognized in the current version of PIRADS.v2 definitions (8), where DCE is considered only in PZ. We used equal weights for ADC and DCEMRI and in PZ. HRS is undergoing further testing in larger RP dataset. This will allow for separate tests for lesions in the PZ and TZ and further refinement of the weights. The dissemination of the existing software will also provide a platform for tests and adjustments, carried out by different groups. The malleable software design allows for multiple parameter adjustments, while the ones used in the current version of HRS are set as defaults.
The DCE analysis considers the MR signal-vs-time curves rather than the contrast agent concentration changes over time. The calculated DCE features have limitations related to the fact that they do not accurately reflect contrast medium concentration in tissues and can be influenced by scanner settings (including gain and scaling factors). The normalization by the DCE signal-vs-time from the muscle mitigates these effects to certain extent. Another limitation of the study is that mpMRI data were obtained under the same conditions for all patients. The generalizability of the HRS parameters in other MRI sequences, vendors, magnetic field strengths and coils (endorectal vs body) should be investigated. Until then, it should be assumed that the parameter defaults, utilized here are valid for mpMRI data acquired under identical conditions. However, HRS may be calibrated by utilizing the large array of user-defined parameters. In addition, the application of the thresholds requires contours of the PZ and TZ, for which a prostate atlas would decrease significantly the need of manual contouring(25).
CONCLUSIONS:
An automated quantitative method was developed to identify habitats in the prostate determinate with the potential to be determinate of outcome. The HRS system assigns risk pixel by pixel in mpMRI sequences and was referenced to prostatectomy. The HRS6 contour was associated with GS risk and was then used to guide radiotherapy boost volumes.
Supplementary Material
Acknowledgments
Funding: This work was supported by National Institutes of Health [grant numbers R01CA189295 and R01CA190105].
Footnotes
Conflict of Interest: None
Statistical analysis: Deukwoo Kwon (dkwon@med.miami.edu, 305–243-0181; Isildinha M.Reis (ireis@med.miami.edu, 305-243-5884)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Morris WJ, Tyldesley S, Rodda S, et al. Androgen Suppression Combined with Elective Nodal and Dose Escalated Radiation Therapy (the ASCENDE-RT Trial): An Analysis of Survival Endpoints for a Randomized Trial Comparing a Low-Dose-Rate Brachytherapy Boost to a Dose-Escalated External Beam Boost for High- and Intermediate-risk Prostate Cancer. Int J Radiat Oncol Biol Phys. 2017;98(2):275–85. [DOI] [PubMed] [Google Scholar]
- 2.Bauman G, Haider M, Van der Heide UA, Menard C. Boosting imaging defined dominant prostatic tumors: a systematic review. Radiother Oncol. 2013;107(3):274–81. [DOI] [PubMed] [Google Scholar]
- 3.Lips IM, van der Heide UA, Haustermans K, et al. Single blind randomized phase III trial to investigate the benefit of a focal lesion ablative microboost in prostate cancer (FLAME-trial): study protocol for a randomized controlled trial. Trials. 2011;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.von Eyben FE, Kiljunen T, Kangasmaki A, et al. Radiotherapy Boost for the Dominant Intraprostatic Cancer Lesion-A Systematic Review and Meta-Analysis. Clin Genitourin Cancer. 2016;14(3):189–97. [DOI] [PubMed] [Google Scholar]
- 5.Isebaert S, Van den Bergh L, Haustermans K, et al. Multiparametric MRI for prostate cancer localization in correlation to whole-mount histopathology. J Magn Reson Imaging. 2013;37(6):1392–401. [DOI] [PubMed] [Google Scholar]
- 6.Futterer JJ, Briganti A, De Visschere P, et al. Can Clinically Significant Prostate Cancer Be Detected with Multiparametric Magnetic Resonance Imaging? A Systematic Review of the Literature. Eur Urol. 2015. [DOI] [PubMed] [Google Scholar]
- 7.Barentsz JO, Richenberg J, Clements R, et al. ESUR prostate MR guidelines 2012. Eur Radiol. 2012;22(4):746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging - Reporting and Data System: 2015, Version 2. Eur Urol. 2016;69(1):16–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269(1):8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parra NA, Pollack A, Chinea FM, et al. Automatic Detection and Quantitative DCE-MRI Scoring of Prostate Cancer Aggressiveness. Front Oncol. 2017;7:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tschudi Y Association of Prostate Volumes with Restricted Diffusion and Prostate Cancer Aggressiveness. Medical Physics. 2016;43(6):3832-+. [Google Scholar]
- 12.Stoyanova R, Huang K, Sandler K, et al. Mapping Tumor Hypoxia In Vivo Using Pattern Recognition of Dynamic Contrast-enhanced MRI Data. Transl Oncol. 2012;5(6):437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang YC, Ackerstaff E, Tschudi Y, et al. Delineation of Tumor Habitats based on Dynamic Contrast Enhanced MRI. Sci Rep. 2017;7(1):9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huisman HJ, Engelbrecht MR, Barentsz JO. Accurate estimation of pharmacokinetic contrastenhanced dynamic MRI parameters of the prostate. J Magn Reson Imaging. 2001;13(4):607–14. [DOI] [PubMed] [Google Scholar]
- 15.Chen YJ, Chu WC, Pu YS, et al. Washout gradient in dynamic contrast-enhanced MRI is associated with tumor aggressiveness of prostate cancer. J Magn Reson Imaging. 2012;36(4):912–9. [DOI] [PubMed] [Google Scholar]
- 16.Tjionas GA, Epstein JI, Williamson SR, et al. Average Weight of Seminal Vesicles: An Adjustment Factor for Radical Prostatectomy Specimens Weighed With Seminal Vesicles. Int J Surg Pathol. 2015;23(8):617–22. [DOI] [PubMed] [Google Scholar]
- 17.Kryvenko ON, Epstein JI. Prostate Cancer Grading: A Decade After the 2005 Modified Gleason Grading System. Arch Pathol Lab Med. 2016;140(10):1140–52. [DOI] [PubMed] [Google Scholar]
- 18.Kryvenko ON, Epstein JI. Changes in prostate cancer grading: Including a new patient-centric grading system. Prostate. 2016;76(5):427–33. [DOI] [PubMed] [Google Scholar]
- 19.Wibmer A, Hricak H, Gondo T, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol. 2015;25(10):2840–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steyerberg EW. Clinical Prediction Models. New York: Springer; 2010. [Google Scholar]
- 21.Eade TN, Hanlon AL, Horwitz EM, et al. What dose of external-beam radiation is high enough for prostate cancer? Int J Radiat Oncol Biol Phys. 2007;68(3):682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodda S, Tyldesley S, Morris WJ, et al. ASCENDE-RT: An Analysis of Treatment-Related Morbidity for a Randomized Trial Comparing a Low-Dose-Rate Brachytherapy Boost with a Dose-Escalated External Beam Boost for High- and Intermediate-Risk Prostate Cancer. Int J Radiat Oncol Biol Phys. 2017;98(2):286–95. [DOI] [PubMed] [Google Scholar]
- 23.Salami S, Hovelson D, Udager A, et al. Targeted Next Generation Sequencing to Characterize Magnetic Resonance Imaging Visible and Invisible Prostate Cancer: Biological Insights and Therapeutic Implications. Journal of Urology. 2017;197(4):E595–E. [Google Scholar]
- 24.Noworolski SM, Vigneron DB, Chen AP, Kurhanewicz J. Dynamic contrast-enhanced MRI and MR diffusion imaging to distinguish between glandular and stromal prostatic tissues. Magn Reson Imaging. 2008;26(8):1071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson AS, Piper J, Curry K, et al. Evaluation of An Atlas-Based Segmentation Method for Prostate and Peripheral Zone Regions On MRI. Medical Physics. 2015;42(6):3606-. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.