Abstract
ChlA-F is a novel conformation-derivative of Cheliensisin A, styryl-lactone isolates that show potent anti-tumor potential in vivo and vitro. However, the anti-cancer activity and its potential mechanisms underlying ChlA-F action have never been explored. In the present study, we evaluated the potency of ChlA-F on autophagy-mediated anchorage-independent growth inhibition in human high-grade invasive bladder cancer (BC) cells. We found that ChlA-F treatment significantly inhibited anchorage-independent growth of human BC cells by inducing autophagy in a Sestrin-2 (SESN2)-dependent fashion. Our results revealed that ChlA-F treatment specifically induced SESN2 expression via increasing its transcription and mRNA stability. On one hand, ChlA-F treatment markedly attenuated Dicer protein abundance, in turn abolishing miR-27a maturation and further relieving miR-27a binding directly to SESN2 mRNA 3’UTR, thereby promoting SESN2 mRNA stabilization. On the other hand, ChlA-F treatment promoted Sp1 abundance and consequently mediated SESN2 transcription. These results demonstrate that its activation of the autophagic pathway through specifically promoting SESN2 expression mediates the anti-cancer effect of ChlA-F, which offers insights into the novel anti-cancer effect of ChlA-F on BC, as well as providing therapeutic alternatives against human BC.
Keywords: Anchorage-independent growth; Cheliensisine A-fluoride (ChlA-F); Macro-autophagy (Autophagy); miR-27a, Sestrin-2
1. Introduction
Bladder cancer (BC) is the most common malignant tumor in the urinary tract. It is the sixth most common cancer in the US with an estimated 74,000 new cases and 16,000 deaths, accounting for about 5% of all new cancers and about 3% of all deaths in the USA in the year 2015 [1]. Although various therapies have been developed to treat cancer patients, natural products have been a well-spring source of anticancer drugs and drug leads for decades [2–5]. Cheliensisin A (Chel A), a styryl-lactone isolated from Goniothalamus cheliensis Hu, has been shown to have anticancer activities both in vitro and in vivo [6]. However, its poor water solubility has precluded Chel A as a potential drug candidate [7, 8]. Given the pharmaceutical potential of Chel A and its unresolved water solubility problems in further drug development, a structure-activity study of Chel A has been carried out to search for druggable candidates for the development of a new ChleA derivative with strong anti-cancer potency. ChlA-F has been identified as being appropriate for this purpose [8]. Although ChlA-F possesses better water solubility and chemical stability, which makes it as a potential drug candidate [8], its anti-cancer activity and the molecular mechanisms underlying its biological effects have never been explored.
Autophagy (macro-autophagy) is an intercellular homeostatic process that is important for cellular digesting protein aggregates, nutrient deposits, dysfunctional organelles,eliminating toxic protein inclusion and regulating signal transduction machinery by selective degradation of damaged molecules [9]. Given that autophagy is a highly conserved self-degradative and catabolic process that targets cellular contents to the lysosomal compartment for degradation and possesses an essential function in cellular stress responses and survival [10], the emerging role of autophagy in cancer cells is one of a double-edged sword [11–16]. On one hand, autophagy enables tumor cells to tolerate stress including a hypoxic microenvironment, starvation, and probably some forms of therapy [17]. On the other hand, autophagy plays an important role in damage mitigation in response to stress that can limit tumorigenesis by either clearing away damaged proteins and organelles, or by maintaining energy homeostasis through intracellular recycling, which ultimately can prevent the genome damage that drives tumorigenesis [18–21]. Sestrin2/SESN2, also known as Hi95, SES2 or SEST2, includes a member of the sestrin family of PA26-related proteins and functions as a regulator of cell growth and survival in cell response to different stress conditions and senescence [22, 23]. Emerging evidence indicates that SESN2 is involved in autophagic responses because it can bind to p62 and promotes autophagic degradation of p62 [9, 24]. SESN2 has been reported to act as a novel p53-targeted gene that participates in autophagy induction [25].Our most recent study reveals that SESN2 is remarkably down-regulated in clinical human BC tissues in comparison to their paired adjacent normal bladder tissues and induction of SESN2 level in cancer cells mediate the anti-cancer activity of ISO in human BC cells, demonstrating that SESN2 acts as a tumor suppressor in human BC cells [26]. Our study also revealed that SESN2’s upregulation via JNK/c-Jun-dependent transcription is crucial for autophagic induction and the anti-cancer effect of the anti-cancer compound isorhapontigenin (ISO) [26]. The interesting observation made by Maiuri et al. also indicates that SESN2 acts as a new positive regulator of autophagy in p53-proficient but not in p53-deficient cells [25]. In our present studies, by using loss-of-function studies, we demonstrated that ChlA-F treatment resulted in SESN2 induction through both activation of Sp1-dependent transcription and inhibition of miR-27a-mediated mRNA degradation, and that the SESN2 upregulation led to an autophagic induction and anti-cancer activity of ChlA-F in human BC cells.
2. Materials and Methods
2.1. Plasmids and Reagents.
GFP-LC3 and the c-Jun dominant negative mutant (PRC-CMV/TAM67) were described in our previous studies [26, 27]. shRNA constructs against human SESN2 (V2LHS-117405),Sp1 and ELAVL1 were purchased from Open Biosystems (Pittsburg, PA, USA). The miR-27a overexpression construct was obtained from Addgene (Cambridge, MA, USA). Plasmids were prepared by the Plasmid Preparation/Extraction Maxi kit from QIAGEN (Valencia, CA, USA). The antibodies specific against phospho-JUN Ser63 (2361S), total c-JUN (9165S), E2F1(3742S), BECN1 (3495S), total NFκB RELA (4764S), phospho-NFκB RELA S536 (3033S), LC3B(2775S), GAPDH(5174S), Atg5(12994S), Atg7(8558P), Dicer(5362P), were purchased from Cell Signaling Technology (Beverly, MA, USA); The antibodies specific for SESN2 (sc-292558), Sp1 (sc-H225) and ELAVL1(sc-5261), were bought from Santa Cruz Biotechnology (Santa Cruz, CA, USA), The antibody against HNRNPD/AUF1 (ARP40238_T100) was bought from Aviva (San Diego, CA, USA). The antibodies specific against ACTB (A5441) and NCL (N2662) were obtained from Sigma-Aldrich Corporation (St. Louis, MO, USA). Bafilomycin A1 (sc-201550) was purchased from Santa Cruz Biotechnology. Chloroquine (CQ) was acquired from Frontier Scientific (Logan, UT), while actinomycin D (Act D) (50–76-0) was bought from Fisher Scientific (Pittsburgh, PA, USA).
2.2. General information for chemical procedures.
ChlA-F with purity over 99% was prepared according to the developed protocol (see Patent ZL201310034985.5 and PCT/CN2014/071751). All the reactions were performed under argon atmosphere using flame-dried glassware unless otherwise noted. Dichloromethane (DCM) was treated as anhydrous solvents prior to use. All reagents were commercial available and used without further purification unless indicated otherwise. Flash Chromatographies were performed with 300–400 mesh silica gels. Thin layer chromatographies (TLC) were carried out on GF254 plates (0.25 mm layer thickness). Visualization of the developed chromatogram was performed by fluorescence quenching or by ceric ammonium molybdate. Yields reported were for isolated, spectroscopically pure compounds. High-resolution mass spectra (HRMS) were taken on a VG Auto Spec-3000 or on a Finnigan MAT 90 instrument. Optical rotations were measured with a Horiba SEPA-300 polarimeter. 1H–NMR and 13C–NMR experiments were performed on a Bruker AM-300 NMR spectrometer at ambient temperature. Chemical shifts were given in δ with TMS as internal reference.
Preparation of cheliensisin A:
Cheliensisin A was isolated as white crystal with purity more than 99.0% from Goniothalamus cheliensis according to the previously reported procedure [28].
Procedures for preparation of ChlA-F:
(7S,8R)-7- Hydroxyl −8-fluoro cheliesisin A
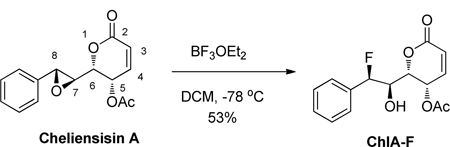
To a solution of cheliensisine A (548 mg, 2 mmol) in DCM (5 mL) at −78oC under N2 was added a solution of BF3·OEt2 (202 µL) in DCM (3 mL) dropwise. The mixture was stirred at this temperature for 0.5 hour until no starting material was detected. The mixture was quenched by a saturated aqueous NaHCO3 solution. The aqueous phase was extracted with dichloromethane (30 mL×3). The combined organic layers were washed with brine, dried over anhydrous Na2SO4 and concentrated. The crude product was purified by flash chromatography on silica gel (ethyl acetate/petrol ether = 1:1.5) to afford ChlA-F as a white foam (312 mg, 53%). High-resolution electrospray ionisation mass spectrometry (m/z): calculated for C15H15FO5Na [M+Na]+, 317.0801,found 317.0795.
For performing the in vitro experiments, ChlA-F was dissolved in Dimethyl sulfoxide (DMSO, Sigma-Aldrich Corporation, USA, 67–68-5) to make a stock concentration at 8 mM. It was the further diluted in DMEM/Ham’s F12 medium with final DMSO concentration at 0.1% (v/v) for cell culture experiments. The same amount of DMSO was used as a vehicle control in all experiments.
2.3. Cell culture and Transfections
The normal urinary epithelial cell line UROtsa and the different genetic backgrounds human BC cell lines RT4 (p53 wild-type), T24T (p53 mutant) and UMUC3 (p53 mutant) were described in our previous studies[26, 29, 30]. The T24T cell was cultured at 37℃ in a 5% CO2 incubator in a 1:1 mixture of DMEM/Ham’s F12 medium supplemented with 5% FBS, 2 mmol/L L-glutamine and a mixture solution of 100 IU penicillin and 100 µg/ml streptomycin; and the UMUC3 cells were cultured in DMEM supplemented with 10% FBS, 2 mmol/L L-glutamine and a mix solution of 100 IU penicillin and 100 µg/ml streptomycin. UROtsa cells were cultured with 1640 supplemented with 10% FBS and RT4 cells were cultured with McCoy’s 5A supplemented with 10% FBS. The cell lines were regularly authenticated every 6–12 months to evaluate viability, recovery, growth, morphology and chemical response, as well as by testing STR loci and gender using the PowerPlex® 16 HS System provided by Genetica DNA Laboratories (Burlington, NC, USA) as stated in our previous studies [26, 30]. The transfections were carried out with specific plasmid constructs using PolyJet™ DNA in Vitro Transfection Reagent (SignaGen Laboratories, Gaithersburg, MD) according to the manufacturer’s instructions. The stable transfection selection of SESN2, Sp1, c-Jun, and miR-27a in T24T cells was subjected to puromycin selection for 4 to 6 weeks. The surviving stable transfectants were pooled as stable mass culture as described in our previous studies [26, 30].
2.4. Human bladder cancer tissues
Fourteen pairs of human BC samples and their matched adjacent normal bladder tissues were obtained from patients underwent radical cystectomy in the Department of Urology, Union Hospital of Tongji Medical College, Huazhong University of Science and Technology (Wuhan, China) from 2013 to 2015. All specimens were immediately snap-frozen in liquid nitrogen after surgical resection. Histological and pathological diagnoses were confirmed and the tissues were defined by a certified clinical pathologist according to the 2004 World Health Organization Consensus Classification and Staging System for bladder neoplasms (Table S1). All specimens were obtained with appropriate informed consent from the patients and a supportive grant obtained from the Medical Ethics Committee of China.
2.5. Western Blot.
Cells were extracted with cell lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 1% SDS, and 1 mmol/L Na3VO4), and protein concentrations were determined by NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). The cell extracts were subjected to SDS-PAGE, then transferred to polyvinylidene fluoride membranes (Bio-Rad, Hercules, CA, USA). The protein band specifically bound to the primary antibody was detected by Typhoon FLA 7000 (GE Healthcare, Chicago, IL, USA) using an alkaline phosphatase–linked secondary antibody and an enhanced chemifluorescence Western Blot system as described in our previous studies[30].
2.6. RT-PCR and Quantitative Real-Time PCR.
Total RNAs were extracted with TRIzol reagent (Invitrogen, USA) following ChlA-F treatment, according to the manufacturer’s instructions, and the cDNAs were synthesized with the Thermo-Script RT-PCR system (Invitrogen). The mRNA amount present in the cells was measured by semi-quantitative RT-PCR. The primers for human sestrin2 were 5’-CAA CTC TGG GGG CTT TGA GT-3’ (Forward) and 5’-ACC TTC TCT GAG TGG CGG AA-3’ (Reverse). The primers for human gapdh were 5’-AGA AGG CTG GGG CTC ATT TG-3’(Forward) and 5’-AGG GGC CAT CCA CAG TCT TC-3’ (Reverse). The PCR products were separated on 2% agarose gels, stained with Ethidium Bromide, and scanned for the images under UV light with Alpha Innotech SP Image system (Alpha Innotech Corporation, San Leandron, CA, USA).
Total microRNAs were extracted using the miRNeasy Mini Kit (QIAGEN, USA), reverse transcription was then performed using the miScript II RT Kit (QIAGEN), and quantitative PCR was performed using miScript PCR Starter Kit (QIAGEN) according to the manufacturer’s protocol. U6 was used as the endogenous normalizer. Cycle threshold (CT) values were determined, and the relative expression of microRNAs was calculated by using the values of 2-△△CT, as described in our publications [30].
2.7. Luciferase Reporter Assay.
The cells were transfected with the indicated luciferase reporter in combination with the pRL-TK vector (Promega, Fitchburg, WI, USA) as an internal control, and the luciferase activities were determined using a microplate luminometer, as described in previous studies[30].
2.8. Anchorage-independent growth assay.
Anchorage-independent growth in soft agar (soft-agar assay) was performed, as described in our earlier studies [26, 30]. Briefly, the 1×104 cells mixed with ChlA-F at a final concentration of 8 µmol/L or vehicle control in 2% FBS Basal Medium Eagle (BME) containing 0.33% agar were seeded over the basal layer containing 0.5% agar and 2% FBS/BME in each well of 6-well plates. The plates were incubated in 5% CO2 in an incubator at 37℃ for 3 weeks. Colonies were captured under a microscope, and only colonies with over 32 cells were counted. The results are presented as Mean ± SD obtained from three independent experiments.
2.9. Confocal laser scanning microscopy.
T24T GFP-LC3 cells were seeded on chambers(1 × 104/well)in a 1:1 mixture of DMEM/Ham’s F12 medium supplemented with 5% FBS and cultured at 37℃with 5% CO2 incubator. When the cell density reaches to the optimal 70~80% confluency,the cells were treated with 8 µM ChlA-F for the time points as indicated. The cells were washed with pre-warmed 1xPBS once, then subjected to 4% paraformaldehyde for 30min at room temperature. Then the cells were washed with pre-warmed 1xPBS twice and the chamber membrane was permeabilized with 1xPBS containing 0.2% Triton X-100 for 15 min at room temperature, then stained with 0.1 mg/ml DAPI (Sigma-Aldrich Corporation, #9542) for 30 min. The slides were washed 3 times with 1x PBS and mounted with antifade reagent (Molecular Probes, P36930, Eugene, OR, USA). All cellular images were captured using an inverted Leica fluorescence microscope (Wetzlar, Germany). Cells with 5 or more intense GFP-LC3 puncta were considered autophagic cells, whereas those with diffuse cytoplasmic GFP-LC3 staining were considered non-autophagic cells. The percentages of the GFP-LC3-positive cell were calculated based on at least 200 counted cells. The number of GFP-LC3 puncta per cell was counted for at least 50 cells.
2.10. Live-cell imaging.
Live-cell imaging was performed using the Lysosome Staining Kit (AAT Bioquest) according to the manufacturer’s instructions. Briefly, T24T(RFP-GFP-LC3B) cells were seeded into a 35mM glass bottom culture dish (MatTek Corporation) and cultured in the incubator with 5% CO2 at 37℃. When the cell density reaches the 70% confluence, the cells were washed once with pre-warmed 1x PBS and then incubated in 1.5 ml of lysosome blue dye-working solution (dilute 3µl LysoBriteTM Blue into 1.5 ml of Live Cell Staining Buffer according to the manufacture’s protocol) containing 2% FBS in an incubator with 5% CO2 at 37℃for 2 hours. The culture dishes were washed twice with pre-warmed 1x HBSS (Gibco) and were then filled with 2 ml CO2-Independent medium containing 5% FBS and 8µM of ChlA-F. Cells were imaged overnight every 5 min using the inverted Leica fluorescence microscope (Wetzlar, Germany) at three channels with laser 405,488 and 594.
2.11. Chromatin Immunoprecipitation (ChIP) Assay.
ChIP was performed using the EZ-CHIP kit (Millipore Technologies) according to the manufacturer’s instructions and as described in our previous publication [31]. Briefly, T24T cells genomic DNA and the proteins were cross-linked with 1% formaldehyde. The cross-linked cells were pelleted, resuspended in cell lysis buffer, and sonicated to generate 200–500bp chromatin DNA fragments. After centrifugation, the supernatants were diluted 10-fold, then incubated overnight with either anti-Sp1 antibody or the control rabbit IgG at 4℃. The immune complex was captured by protein G-agarose saturated with salmon sperm DNA, then eluted with the elution buffer. Cross-linked DNA-protein was reversed by heating overnight at 65℃. DNA was purified by PCR. To specifically amplify the region containing the putative responsive elements on the human SESN2 promoter, PCR was performed with the following two pairs of primers: 5’-CATTCACCCGAGGCGGACTA-3’ (Forward),5’-CGGTCCAGCCAATCAGAGGT-3’ (Reverse) and 5’-ACAGACCTCTGATTGGCT-3’ (Forward),5-’CTCTGACACCAGCAGTT-3’ (Reverse). The PCR products were separated on 2% agarose gels and stained with ethidium bromide, the images were scanned under a UV light with Alpha Innotech SP Image system (Alpha Innotech Corporation, San Leandron, CA, USA).
2.12. Statistical analysis
The student’s T-test was used to determine significant differences and p<0.05 was considered as a significant difference between the compared groups.
3. Results
3.1. ChlA-F induced autophagy concurrent with the inhibition of Anchorage-independent Growth in human BC cells.
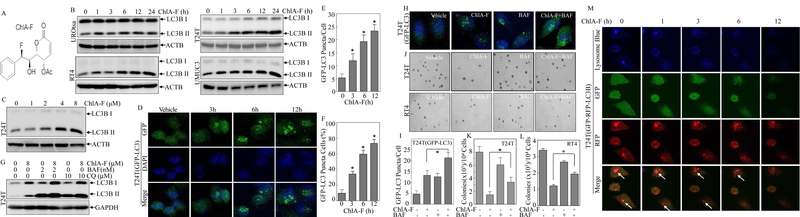
The chemical structure of ChlA-F shown in Fig. 1A is a natural product-based druggable compound as a new ChelA derivative [8]. Through structural modifications, ChlA-F has been found to have improved chemical stability and water solubility (Patents ZL201310034985.5 and PCT/CN2014/071751). To evaluate the potential involvement of autophagic responses in ChlA-F-treated human bladder cancer cells, we first evaluated the effects of ChlA-F on the conversion of LC3B I to LC3B II following ChlA-F treatment of normal urothelial cell line UROtsa in comparison to human BC cell lines, including RT4, T24T and UMUC3 [32]. The results indicated that ChlA-F treatment led to a substantial conversion of LC3B I to LC3B II in a time- and dose-dependent manners in human BC cells, whereas same treatment only show a slightly effect on normal UROtsa cells (Figs. 1B & 1C). Further, a well-established tandem GFP-LC3 fusion expression construct was stably transfected into T24T cells to monitor autophagosome formation using confocal laser scanning microscopy. As shown in Figs. 1D-1F, ChlA-F treatment induced formation of autophagosomes in a time-dependent manner. The increase of autophagic flux caused by ChlA-F was further confirmed by the co-treatment of cells with Bafilomycin A1 (BAF) or Chloroquine (CQ). BAF is an inhibitor that prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes, the late phase of autophagy [33]. CQ inhibits the fusion between autophagosomes and lysosomes, thus preventing the maturation of autophagosomes into autolysosomes, and blocking a late step of macroautophagy [33]. The results showed that co-treatment of cells with BAF or CQ and ChlA-F resulted in a significant increase in the conversion of LC3B I to LC3B II (Fig. 1G) and GFP-LC3 puncta formation (Figs. 1H & 1I), providing evidence that ChlA-F treatment increases the autophagy flux in human BC cells. Consistently, ChlA-F treatment resulted in a significant attenuation of anchorage-independent growth of T24T and RT4 cells, whereas this inhibition by ChlA-F was significantly reversed by co-treatment of cells with BAF (Figs. 1J -1L). Moreover, we performed live-cell imaging to monitor autophagic flux based on the use of a tandem monomeric GFP-RFP-tagged LC3B. GFP is a stably folded protein and relatively resistant to lysosomal proteases. However, the low pH inside the lysosome quenches the fluorescent signal of GFP. In contrast, RFP exhibits more stable fluorescence in acidic compartments, and RFP-LC3 can readily be detected in autolysosomes. By exploiting the lysosomal degradation with these two fluorescent proteins (lysosomal quenching of GFP fluorescence versus lysosomal stability of RFP fluorescence), live-cell imaging is used to morphologically trace with an GFP-RFP- LC3 tandem construct and lysosome blue staining. This method depends on the acidification and degradation capacity of the lysosome. The result showed that RFP-LC3B puncta was degraded in autophagosome over time following ChlA-F treatment (Fig. 1M). These results provide strong evidence indicating an autophagic induction by ChlA-F in human BC cells.
Fig. 1. ChlA-F induced autophagy and inhibited anchorage-independent growth in human invasive bladder cancer cells.
(A) The structure of ChlA-F. (B & C) UROtsa, RT4, T24T and UMUC3 cells were treated with 8µM of ChlA-F for the indicated periods (B) or ChlA-F at the indicated concentrations for 12h (C). The cell lysates were subjected to Western Blot assay using the indicated antibodies. (D) The GFP-LC3 construct was stably transfected into T24T cells, and the transfectants were treated with 8µM of ChlA-F for various times. LC3 puncta formation was observed and images were captured using fluorescence microscopy. (E & F) Percentage of cells with GFP-LC3B puncta (E) and number of puncta per positive cell (F) was calculated and presented as described in “Materials and Methods”. (G) T24T cells were treated with ChlA-F (8 µM), together with or without 2 nM BAF or 10 µM CQ for 12 h and the protein levels of LC3B were assessed by Western Blot. (H) T24T cells were transfected with the GFP-LC3 construct and the transfectants were treated with ChlA-F (8 µM), with or without 2 nM BAF for 12h. The representative images of GFP-LC3 puncta were captured using a confocal fluorescence microscope. (I) Number of puncta per GFP-LC3-positive cell was calculated and presented as described in “Materials and Methods”. (J) Representative images of colonies of T24T treated with either 8µM ChlA-F or 2nM BAF, or both, in soft-agar assay, were captured as described in “Materials and Methods”. (K & L) Colonies shown in Fig. 1J were counted under an inverted microscope with more than 32 cells of each colony. The results are presented as colonies per 104 cells. The bars show Mean ± SD from three in dependent experiments. The symbol (*) indicates a significant increase from the ChlA-F treatment group (p<0.05). (M) T24T(RFP-GFP-LC3B) cells were seeded into 35mM glass dish to perform Live-cell imaging. Image were presented as the indicated time following ChlA-F treatment.
3.2. ChlA-F upregulated SESN2 expression, which, in turn contributes to autophagy induction and anchorage-independent growth inhibition
Autophagy is tightly controlled by different regulators, that include: ATG5/ATG12, ATG7, BECN1, SQSTM1 and SESN2 [33]. To elucidate the molecular mechanisms that are responsible for ChlA-F-induced autophagy, the potential effect of ChlA-F on autophagy-related protein expression was evaluated in T24T and RT4 cells following ChlA-F treatment. The results show that ChlA-F treatment distinctly increased SESN2 protein expression, but no observable effect on the expression of other autophagy-related proteins, such as ATG5/ATG12, ATG7, BECN1 and SQSTM1 (Figs. 2A & 2B), suggesting that SESN2 may be involved in autophagic induction due to ChlA-F treatment. Our most recent study has also reveal that ISO treatment effectively mediates autophagic response in a SESN2-dependent manner in human BC cells [26]. We next evaluated whether SESN2 was responsible for ChlA-F-initiated autophagy induction. To this end, we transfected shRNA that specifically targets human SESN2 into T24T cells and the stable transfectant T24T(shSESN2) was established and identified, as shown in Fig. 2C. Knockdown of SESN2 resulted in the attenuation of autophagic responses following ChlA-F treatment in T24T cells (Fig. 2D). These data indicate that SESN2 does indeed play an essential role in the autophagy process as a result of ChlA-F treatment. Autophagy has been shown to function as a double-edged sword that either protects or suppresses human cancer, depending on the stimuli of autophagy, the stage of cancer, and the downstream mediators or effectors [12, 34–39]. In combination with various conventional chemotherapeutic drugs, induction of autophagy has emerged as an attractive and promising approach to sensitizing malignancies to chemotherapy in cancer therapy [40, 41]. Therefore, we evaluated the effects of SESN2 on the inhibition of anchorage-independent growth following ChlA-F treatment. The results indicated that SESN2 knockdown attenuated ChlA-F’s inhibition of anchorage-independent growth of T24T cells in comparison to that in the scramble vector transfectant, T24T(Nonsense) (Figs. 2E & 2F). These results provide consistent to demonstration of SESN2’s contributes to ChlA-F-induced autophagic responses and the inhibition of anchorage-independent growth in T24T cells.
Fig. 2. SESN2 induction was required for autophagic response and anchorage-independent growth inhibition by ChlA-F in human invasive bladder cancer cells.
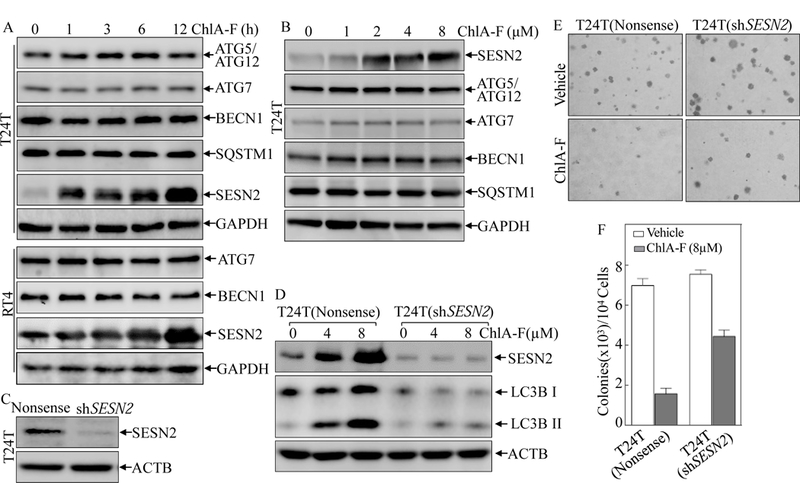
(A & B) treatment of T24T and RT4 cells with ChlA-F at 8µM for the time points indicated (A) or at the indicated concentrations for 12 h (B). The cell lysates were subjected to Western Blot to evaluate the expression of ATG5/ATG12, ATG7, BECN1, SQSTM1, and SESN2. (C) shRNA SESN2 were stably transfected into T24T cells and the stable transfectants were identified by Western Blot. (D) The indicated stable transfectants were subjected to Western Blot to determine SESN2 and LC3B levels in T24T cells following ChlA-F treatment for 12 h. (E & F) Representative images of colonies of T24T (shSESN2) and T24T (Nonsense) cells in soft-agar assay in the presence or absence of ChlA-F(8µM) were captured (E) or counted under an inverted microscope. The results are presented as colonies per 104 cells, and the bars show Mean ± SD from three independent experiments (F). The symbol (*) indicates a significant decrease from vehicle control (p<0.05).
3.3. SESN2 upregulation by ChlA-F was due to the promotion of both transcription and mRNA stability
SESN2 could be regulated at multiple levels, including transcription, mRNA stability, translation, and protein degradation. To test whether SESN2 is regulated at the level of either mRNA transcription or mRNA degradation, RT-PCR was employed to determine the effect of ChlA-F on SESN2 mRNA expression. Consistent with the results observed at the protein level, SESN2 mRNA expression was markedly upregulated following ChlA-F treatment (Fig. 3A). We next evaluated whether ChlA-F regulated SESN2 expression at the transcription level and/or mRNA stability level. We first transfected the wild-type SESN2 promoter-driven luciferase reporter into T24T cells [26]. The transfectants were then used to determinate ChlA-F on SESN2 transcription. As expected, ChlA-F treatment indeed led to a significant increase in SESN2 promoter transcription activity in a time-dependent manner (Fig. 3B). Next, we tried to exclude the possibility of ChlA-F regulation SESN2 mRNA stability. Unexpectedly, treatment of T24T cells with ChlA-F in the presence of actinomycin D (Act D) exhibited a reduction of the SESN2 mRNA degradation rate in comparison to that observed in cells treated with Act D alone (Fig. 3C). Taken together, our results suggest that ChlA-F upregulates SESN2 abundance via the promotion of both mRNA transcription and mRNA stability.
Fig. 3. ChlA-F treatment increased both SESN2 transcription and SESN2 mRNA stability.
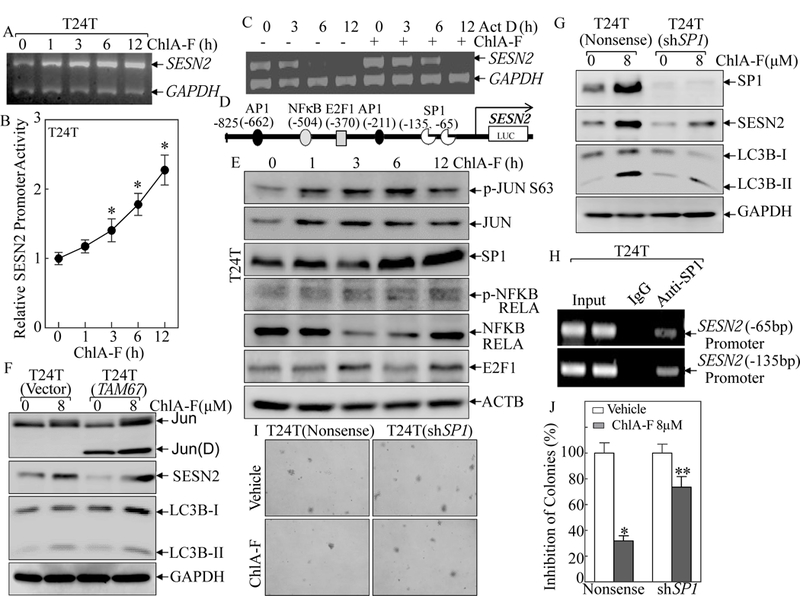
(A) Total RNAs were isolated from T24T cells treated with or without 8 µM ChlA-F for the indicated times. RT-PCR was performed to determine SESN2 mRNA levels. The gapdh mRNA levels were used as loading control. (B) T24T stably co-transfected with SESN2 promoter-driven luciferase reporter and pRL-TK was treated with 8µM ChlA-F for the indicated times to evaluate SESN2 promoter transcriptional activity. The induction fold was normalized to internal control pRL-TK. The results were presented as SESN2 promoter activity relative to vehicle control (relative SESN2 promoter activity). The bars indicate a Mean ± SD from three independent experiments. The symbol (*) indicates a significant increase from the vehicle control (p<0.05). (C) T24T cells were co-incubated with ChlA-F (8 µM) and Act D (20 µg/ml) for the indicated times. Total RNA was isolated and RT-PCR was then performed to determine SESN2 mRNA levels. The result is representative of three independent experiments. (D) Schematic representation of the putative transcription factor consensus binding sites in the SESN2 proximal promoter region was predicted using bioinformatics analysis. (E) The protein expressions of potential transcription factors were determined following ChlA-F (8 µM) treatment for the indicated periods using Western Blot. (F) TAM67 was stably transfected into T24T cells and the stable transfectants were then subjected to ChlA-F treatment for 12 h for identification of c-JUN(D) (TAM67) expression and determination of SESN2 and LC3B. (G) T24T Cells stably transfected with nonsense control or shRNA specifically targeting human Sp1 were treated with ChlA-F (8 µM) for 12 h, and the cell extracts were subjected to Western Blot as indicated. (H) ChIP assay was carried out using anti-Sp1 antibody to detect the interaction between Sp1 and the SESN2 promoter. (I & J) Anchorage-independent growth of T24T(Nonsense) and T24T(shSp1) cells was determined in soft agar in the absence or presence of ChlA-F. The number of colonies is scored and is presented as colonies per 104 cells. The symbol (*) indicates a significant increase in comparison to the nonsense transfectant (p < 0.05).
3.4. Sp1, but not c-Jun, was responsible for ChlA-F-induced SESN2 transcription
Our most recent study reveal that the regulatory effect of ISO on SESN2 expression occurs only at the transcriptional level in a c-Jun/AP-1-dependent manner [26]. Involvement of AP-1 in upregulation of SESN2 transcription is also found in LPS-treated cells [42]. To evaluate whether c-Jun is involved in ChlA-F-induced SESN2 transcription, the bioinformatics analysis of the SESN2 promoter was performed and the results showed several potential transcription factors binding sites, including Sp1, AP-1, E2F1, and NFκB in the SESN2 promoter region as shown in Fig. 3D. To identify the specific transcription factor(s) participating in the modulation of SESN2 transcription, the effect of ChlA-F on the related transcription factor protein abundance/activation was determined. The results showed that ChlA-F treatment specifically induced expression of Sp1 and c-Jun together with c-Jun phosphorylation at Ser63 and Ser73 (Fig. 3E), whereas it did not show any observable increases in phospho-NFκB (RELA), NFκB (RELA), or E2F1 (Fig. 3E), suggesting that Sp1 and c-Jun might be involved in the transcriptional activation of the SESN2 promoter that has been treated with ChlA-F. Thus, the stable dominant negative mutant c-Jun transfectant, T24T(TAM67) was used to address the role of c-Jun in ChlA-F-induced SESN2 expression. Unexpectedly, blockage of c-Jun activation by ectopic expression of TAM67 did not show any observable effect on SESN2 expression or the conversion of LC3B I to LC3B II followed ChlA-F treatment (Fig. 3F). In contrast, knockdown of Sp1 in T24T cells abolished the up-regulatory effects of ChlA-F on the expression of SESN2 and the conversion of LC3B I to LC3B II (Fig. 3G). These results demonstrate that the Sp1, but not c-Jun, is particularly crucial for SESN2 promoter transcription and autophagic induction followed by ChlA-F treatment. To further provide direct evidence showing of Sp1 specifically binding to the SESN2 promoter region, we performed a chromatin immunoprecipitation (ChIP) assay using an anti-Sp1 antibody. As shown in Fig. 3H, Sp1 did directly bind to the two different putative Sp1 binding sites in the SESN2 promoter (Fig. 4H). The soft agar assay results indicated that Sp1 knockdown also attenuated ChlA-F’s inhibition of anchorage-independent growth of T24T cells in comparison to that of its scramble nonsense transfectant (Figs. 4I & 2J). Taken together, our data demonstrate that ChlA-F induces SESN2 mRNA transcription via a Sp1-dependent and c-Jun-independent fashion, which is distinctly different from the molecular mechanism reported in our recent study, revealing that c-Jun is required for ISO-induced SESN2 transcription in human BC cells [26].
Fig. 4. ChlA-F treatment promoted SESN2 mRNA 3’UTR activity accompanied by attenuation of miR-27a expression in human invasive BC cells.
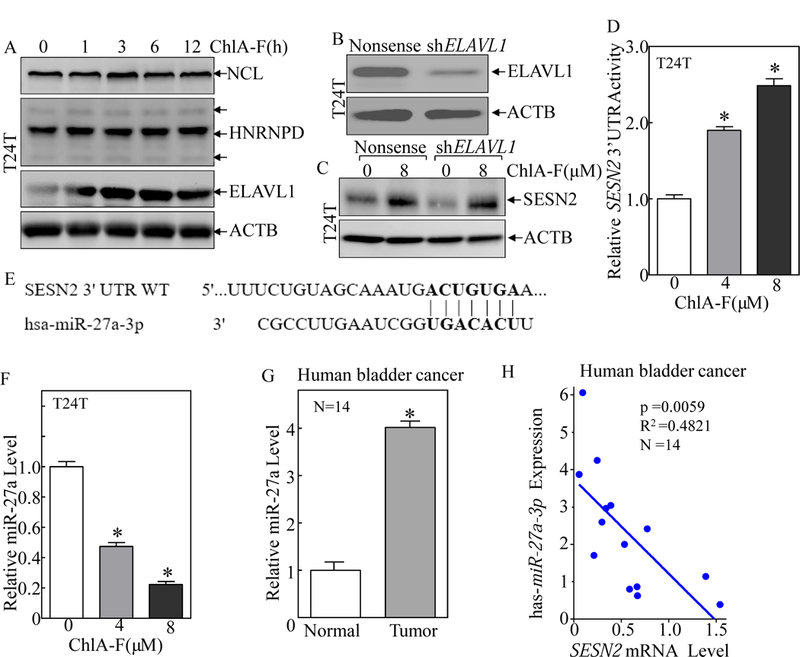
(A) T24T cells were treated with ChlA-F (8 µM) for the indicated periods. The total cell extracts were subjected to Western Blot analysis of expression of HNRNPD, ELAVL1, NCL. (B & C) T24T cells were transfected with an shRNA specifically targeting human ELAVL1 or nonsense control constructs. The cell extracts from stable transfectants were subjected to Western Blot analysis for determination of ELAVL1 expression (B).The stable transfectants were treated with ChlA-F for 12 h, and the cell extracts were subjected to Western blot analysis for SESN2 expression (C). (D) SESN2 3’UTR luciferase reporter was transiently transfected into T24T cells, and the transfectants were subjected to Western Blot to determine the effect of ChlA-F on SESN2 mRNA 3’UTR activity in T24T cells following ChlA-F treatment for 12h. The symbol (*) indicated a significant increase of SESN2 3’UTR activity in comparison to the vehicle control (P<0.05). The results are shown as Mean ± SD from three independent experiments. (E) The binding site of miR-27a in 3’UTR of human SESN2 mRNA was analyzed and presented as indicated. (F & G) The relative expression level of miR-27a was evaluated by quantitative real-time PCR in T24T cells following ChlA-F treatment at different concentrations for 12 h (F) and in human bladder cancer tissues as compared to the corresponding adjacent normal bladder tissues (G). The symbol (*) indicates a significant increase in comparison to normal bladder tissues. (H) The correlation analysis of pathophysiological association between miR-27a and SESN2 mRNA in human BC tissues.
3.5. Downregulation of miR-27a contributed to SESN2 mRNA stabilization, autophagic response and anchorage-independent growth inhibition following treatment with ChlA-F
Several RNA-binding proteins, including NCL, HNRNPD and ELAVL1, have been reported to bind to their targeted mRNA and regulate their mRNA stability [43–47]. To test the potential contribution of these RNA-binding proteins to ChlA-F upregulation of SESN2 mRNA stability, the effects of ChlA-F on the expression of NCL, HNRNPD and ELAVL1 were evaluated. As shown in Fig. 4A, ChlA-F treatment led to upregulation of ELAVL1 protein,whereas it did not show an observable effect on the expression of either NCL or HNRNPD. To determine the role of ELAVL1 in the regulation of SESN2 mRNA stability in T24T cells, shRNA specifically targeting human ELAVL1 was employed to knockdown ELAVL1 expression in T24T cells (Fig. 4B). Much to our surprise, knockdown of ELAVL1 had no observable effect on ChlA-F-induced SESN2 expression (Fig. 4C). Given that microRNAs (miRNAs) have been reported to regulate their target gene expression by either suppressing protein translation and/or regulating mRNA degradation usually through imperfect complementary base pairing to the target gene mRNA 3’UTR [48, 49], we tested the potential effect of ChlA-F on SESN2 mRNA 3’UTR activity. The results indicated that ChlA-F treatment promoted SESN2 mRNA 3’UTR activity (Fig. 4D). In light of this finding, we further used the TargetScan database to analyze potential miRNA binding sites in the 3’UTR region of SESN2 mRNA. miR-27a is one of the most matched miRNAs that is likely to target the SESN2 mRNA 3’UTR region (Fig. 4E). We next used real-time PCR to evaluate the possibility that ChlA-F treatment alters miR-27a expression in T24T cells. As expected, ChlA-F treatment resulted in a dramatic reduction of miR-27a in T24T cells (Fig. 4F), suggesting that miR-27a downregulation might be involved in ChlA-F stabilizing SESN2 mRNA. Furthermore, we compared the expression of miR-27a in 14 pairs of human bladder cancer tissues and matched adjacent normal bladder tissues. As shown in Fig. 4G, the real-time PCR result of miR-27a expression was greatly upregulated in human bladder cancer tissues as compared to the corresponding adjacent normal bladder tissues. Moreover, we determined miR-27a expression levels and its target gene SESN2 mRNA levels in the same 14 pairs of human BC tissues and matched adjacent normal bladder tissues (Fig. S2). The correlation of miR-27a and SESN2 mRNA levels in these human tissues were analyzed and the results indicated that there was a negative pathophysiological association between these two molecules as shown in Fig. 4H.
To determine the role of miR-27a in the regulation of ChlA-F-induced SESN2 expression, miR-27a expressing construct was stably transfected into T24T cells and the stable transfectant was identified as shown in Fig. 5A. Ectopic expression of miR-27a resulted in a remarkable inhibition of SESN2 expression, autophagy induction, as well as reversing the inhibition of anchorage-independent growth following ChlA-F treatment in T24T cells (Figs. 5B-5D). To determine whether miR-27a inhibition was due to specific targeting of miR-27a to 3’UTR of SESN2 mRNA, we constructed wild-type (WT) SESN2 mRNA 3’UTR luciferase reporter and we point mutated the miR-27a binding site in WT SESN2 mRNA 3’UTR luciferase reporter as displayed in Fig. 5E. Both WT and mutant of SESN2 mRNA 3’UTR luciferase reporter were stably transfected into T24T cells, respectively, and the stable transfectants were used to test whether the miR-27a binding site in SESN2 mRNA 3’UTR luciferase reporter is critical for ChlA-F inhibition of SESN2 mRNA 3’UTR activity. As shown in Fig. 5F, ChlA-F treatment profoundly promoted WT SESN2 mRNA 3’UTR activity; whereas mutation of miR-27a binding site impaired SESN2 mRNA 3’UTR activity followed ChlA-F treatment, indicating that miR-27a binding site is required for ChlA-F inhibition of SESN2 mRNA 3’UTR activity. Given these results showing that ChlA-F is able to stabilize SESN2 mRNA, we evaluated the effect of ectopic expression of miR-27a on the stability of SESN2 mRNA. Consistent with the role of miR-27a in SESN2 protein expression, miR-27a overexpression dramatically reduced SESN2 mRNA stability in T24T cells in comparison to scramble nonsense transfectant following ChlA-F treatment (Fig. 5G). Collectively, our results clearly indicate that ChlA-F treatment inhibits miR-27a expression, which results in the reduction of miR-27a binding to SESN2 mRNA 3’UTR and the subsequent increased stabilization of SESN2 mRNA, thereby promoting autophagy and further mediating the inhibition of anchorage-independent growth of human BC cells.
Fig. 5. miR-27a downregulation was critical for ChlA-F-induced stabilization of SESN2 mRNA, autophagy induction, and anchorage-independent growth inhibition in human invasive BC cells.
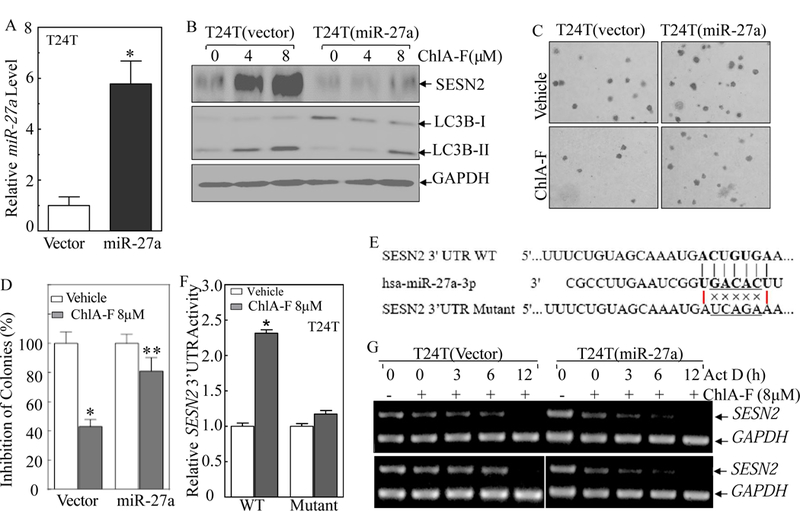
(A) Overexpression of miR-27a in T24T cells was identified by real-time PCR assay. (B) Ectopic expression of miR-27a reversed ChlA-F-induced SESN2 abundance and autophagy induction in T24T cells. (C & D) Representative images of colony formation of T24T(miR-27a) and T24T(Nonsense) cells in soft-agar assay in the presence or absence of ChlA-F (8 µM) were captured (C) and scored (D) under an inverted microscope. The results are presented as colonies per 104cells.The bars show Mean ± SD from three independent experiments. The symbol (*) indicates a significant decrease from vehicle control (p<0.05) (D). (E) Schematic of the miR-27a binding sites and its mutant of the pMIR-report-SESN2 3’UTR luciferase reporter. (F) Attenuation of ChlA-F induction of SESN2 mRNA 3’UTR activity in the miR-27a binding site mutant of pMIR-report-SESN2 3’UTR transfectants. (G) T24T cells were incubated with Act D (20 µg/ml) for the indicated time periods with or without ChlA-F (8 µM). Total RNA was isolated and RT-PCR was then performed to determine SESN2 mRNA levels. The result are representative of three independent experiments.
3.6. ChlA-F treatment attenuated Dicer and in turn reduced miR-27a maturation and expression
In order to characterize the molecular mechanism underlying the modulation of miR-27a expression by ChlA-F treatment, the miR-27a promoter-driven luciferase reporter was transfected into T24T cells and the miR-27a promoter activity in the transfectant was determined in the cells treated with ChlA-F. As seen in Fig. 6A, ChlA-F treatment had no significant effect on miR-27a promoter activity, excluding the possibility of ChlA-F inhibiting miR-27a transcription. It is known that miRNAs possess differential stability in human cells [50], the potential effect of ChlA-F on miR-27a stability was therefore determined. T24T cells were treated with ChlA-F with or without Act D (20 µg/ml) for the indicated time periods and Real-time PCR was employed to evaluate miR-27a abundance. The result show that in comparison to miR-27a levels observed in cells treated with Act D alone, the cells treated with the combination of Act D and ChlA-F did not show observable changes in the miR-27a degradation rate (Fig. 6B). These results led us to explore the possibility of ChlA-F modulation of pre-mir-27a maturation. We compared levels of pre-mir-27a and miR-27a expression in T24T cells treated with ChlA-F. As expected, pre-mir-27a was remarkably induced in ChlA-F-treated T24T cells, whereas matured miR-27a was significantly downregulated in the same ChlA-F-treated cells (Fig. 6C), demonstrating that ChlA-F exhibited an inhibitory effect on pre-mir-27a maturation. Pre-miRNA maturation needs successive enzymatic digestions of precursor miRNAs to produce pri-miRNAs by RNase-III-type enzyme Drosha, then pre-miRNAs by Dicer [51, 52]. We next tested whether Dicer was involved in ChlA-F attenuation of pre-mir-27a maturation. The results showed that Dicer protein was almost completely abolished without affecting its mRNA levels in the same T24T cells treated with ChlA-F (Figs. 6D & 6E). These results demonstrate that attenuation of pre-mir-27a maturation by ChlA-F might be through inhibition of Dicer protein abundance in human BC cells.
Fig. 6. ChlA-F treatment impaired miR-27a maturation accompanied by specific attenuation of Dicer protein, but not mRNA expression in human invasive BC cells.
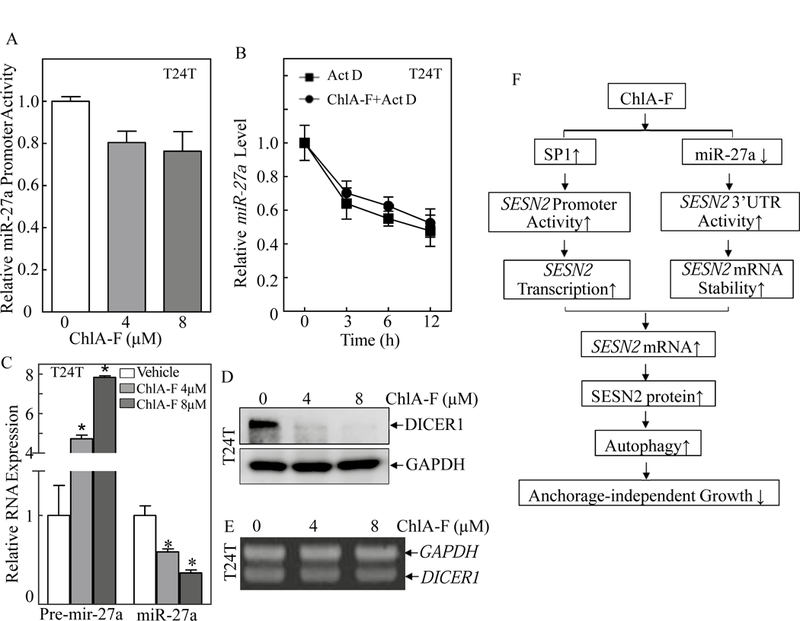
(A) T24T stably transfected with miR-27a promoter-driven luciferase reporter was treated with ChlA-F at different concentrations for 12 h to determine the potential effect of ChlA-F on miR-27a promoter transcriptional activity. (B) T24T cells were co-incubated with ChlA-F (8 µM) and Act D (20 µg/ml) for the indicated time periods. Total RNA was isolated and quantitative real-time PCR was then performed to determine miR-27a levels. The fold change was normalized to internal control gapdh. The result is representative of three independent experiments. (C) The relative expression levels of pre-mir27a and miR-27a were evaluated by quantitative real-time PCR in T24T followed by ChlA-F treatment at different concentrations for 12 h. The symbol (*) indicates a significant change from vehicle control (p<0.05). (D) T24T cells were treated with various concentrations of ChlA-F for 12h. The cell lysates were subjected to Western Blot to detect Dicer protein expression. (E) Total RNA was isolated from the T24T cells treated with the indicated concentrations of ChlA-F for 12 hours. RT-PCR was performed to determine dicer mRNA levels. The gapdh mRNA levels were used as loading control. (F) The schematic summary of molecular mechanisms underlying anticancer activity of ChlA-F on human bladder cancers.
4. Discussion
ChlA-F is a new modified conformation-derivative of ChelA that has never been reported before in its biological effect. In our current study, we tested the effect of ChlA-F treatment on autophagy induction in human normal bladder cell line UROtsa vs. human BC cell lines (RT4, T24T and UMUC3), and in human BC cell lines with WT-p53 vs. mutated-p53. We found that ChlA-F treatment significantly induced autophagy and subsequently inhibited anchorage-independent growth in human BC cells, while it only showed slightly effect on autophagic induction in UROtsa cells. These results suggest that ChlA-F have anti-cancer activity with relative cancer cell selectivity. We also found that ChlA-F treatment inhibited human BC cells with both WT-p53 and mutated-p53 cells, revealing that ChlA-F mainly targets p53-independent pathways, which is distinct from its parental compound ChelA (Patent PCT/CN2014/071751). This notion is also supported by the results showing that ChelA treatment induces autophagy in human BC cells [53] with no observable effect on SESN2 protein expression in T24T cells (Fig. S1B). It is also very interesting to note that SESN2 expression were similarly up-regulated by ChlA-F in both RT4 (WT-p53) and T24T (mutated-p53) cells, although high basal level of SESN2 is observed in RT4 (Fig. 2A). Our further studies showed that ChlA-F treatment specifically induced SESN2 expression via increasing its transcription and mRNA stability. On one hand, ChlA-F attenuates Dicer protein abundance, in turn abolishing miR-27a maturation and further relieving miR-27a binding directly to SESN2 mRNA 3’UTR, thereby promoting SESN2 mRNA stabilization. On the other hand, ChlA-F treatment promoted Sp1 abundance and consequently mediated SESN2 transcription. These findings are also distinct from previous report that p53 is a transcription factor for regulating SESN2 transcription [54]. The increase of SESN2 expression in turn leads to cell autophagy and anchorage-independent growth inhibition by ChlA-F. Our results not only clarify the role of autophagy in the anti-cancer activity of ChlA-F, but also further provide significant molecular insights into the understanding of autophagy induction upon ChlA-F treatment.
Autophagy is a multi-step process and numerous signaling pathways have been reported to be involved in the regulation of autophagy either at the early stage or at advanced stages of tumor development [55]. At the early stage of oncogenesis, autophagy is often inhibited; leading to the speculation that autophagy functions as a tumor suppressor by preventing the accumulation of damaged organelles and aggregated proteins [55, 56]. Given that autophagy is a tumor-suppression mechanism, and that autophagy defects cause a predisposition to cancer and other diseases, the prospect of stimulating autophagy as a disease prevention measure is promising [57]. It has been reported that multiple autophagic pathways are involved in an anti-cancer effect in various cancers. Tamoxifen (TAM), which targets the estrogen receptor, induces autophagic cell death in breast cancer cells [58]. The natural product resveratrol has anti-neoplastic activities and induces autophagy, thus inhibiting cell growth in ovarian cancer [59]. The anti-cancer compound fangchinoline is reported to induce autophagy through the activation of the p53/SESN2/AMPK signaling pathway in human hepatocellular carcinoma cells [54]. Our recent study demonstrates that the natural compound ISO induces autophagic growth inhibition via increasing SESN2 transcription in a c-Jun-dependent manner[26]. The current study illuminates the anti-cancer activity and the molecular mechanisms of ChlA-F in its inhibition of BC cells and defines its remarkable inhibition of anchorage-independent growth through inducing autophagy in a SESN2-dependent fashion. We first found that ChlA-F treatment activated the autophagy process in human BC cells. Using loss-of-function approach, we found that SESN2 is specifically critical for ChlA-F-triggered autophagy. Further studies reveal that ChlA-F promotes both Sp1-mediated SESN2 mRNA transcription and miR-27a directly binding-mediated stabilization of SESN2 mRNA. This novel finding indicates that the function of ChlA-F on BC inhibition is distinctly different from the mechanism that our lab identified for ISO’s anti-cancer activity, which indicates that ISO treatment only increases SESN2 transcription via c-Jun binding to the putative AP-1 binding site in the SESN2 promoter region [26]. In the current studies, blockage of c-Jun phosphorylation at Ser63 in T24T cells by ectopic expression of c-Jun dominant negative mutant, TAM67, did not show any observable inhibition of SESN2 expression following ChlA-F although ChlA-F treatment initiates c-Jun phosphorylation at Ser63, revealing that c-Jun activation is not involved in SESN2 transcription by ChlA-F. In contrast to c-Jun, knockdown of Sp1 not only attenuated the up-regulatory effect of ChlA-F on SESN2 protein expression, but also impaired ChlA-F inhibition of anchorage-independent growth of BC cells. Moreover, ectopic expression of miR-27a showed a suppression of SESN2 mRNA 3’UTR activity and protein expression in T24T cells, whereas mutation of the miR-27a binding site in SESN2 mRNA 3’UTR luciferase reporter blocked miR-27a inhibition of SESN2 mRNA 3’UTR activity, indicating that mi-27a specifically targets SESN2 mRNA 3’UTR for its inhibition of SESN2 mRNA stability. Sp1 is a ubiquitously expressed transcription factor in various cancer cells and tumor tissues [60]. It may activate and suppress a number of essential oncogenes and tumor suppressors [60–63]. The studies we present here demonstrate that ChlA-F treatment activates Sp1, which is able to bind to two Sp1 binding sites of the SESN2 promoter region, consequently resulting in transactivation and expression of SESN2, as well as autophagy in human BC cells. Therefore, the current studies define distinct mechanistic cascades that mediate the autophagic induction and anti-cancer activities of ChlA-F.
miR-27a is up-regulated and functions as an oncogenic miRNA in several types of human cancers [64–67]. For examples,miR-27a is reported to play a role in proliferation, migration and invasion of Hepatocellular carcinoma (HCC) cells [64].It can also act as an androgen-regulated oncogenic miRNA in prostate cancer via the increase of expression of AR-regulated genes, thereby promoting prostate cancer cell growth [66]. In the current study, we found that miR-27a is significantly upregulated in human bladder cancer tissues as compared to the corresponding adjacent normal bladder tissues, whereas SESN2 mRNA levels show a opposite expression in same human BC tissues. Statistically, miR-27a showed a negative correlation with SESN2 mRNA expression in same set of human bladder cancer tissues. Moreover, we also found that miR-27a overexpression is able to inhibit SESN2 expression, autophagy induction and reverse the inhibition of anchorage-independent growth followed ChlA-F treatment in T24T cells via its binding to 3’UTR region of SESN2 mRNA. These results reveal a novel mechanism underlying the oncogenic function of miR-27a in human BC cell biology. This function has never been reported in previous studies. Furthermore, our current studies show that ChlA-F treatment almost completely abolished Dicer protein expression accompanied by attenuating miR-27a maturation and expression without affecting dicer mRNA level in T24T cells, indicating that it’s most likely that ChlA-F exhibits its anti-cancer activity on human BC cells most likely through its retardation of pre-mir-27a maturation processing into miR-27a by abolished targeting of either Dicer protein translation and/or degradation. Further elucidating the molecular mechanisms underlying ChlA-F’s inhibition of Dicer protein expression will provide significant insight into understanding the action of this anti-cancer compound in human BCs.
In summary, our studies demonstrate that ChlA-F exhibits its anti-cancer activity on human bladder cancer via promotion of both Sp1-mediated SESN2 mRNA transcription and miR-27a-mediated stabilization of SESN2 mRNA, both of which lead to SESN2 protein expression, in turn triggering autophagic responses and subsequently resulting in the inhibition of anchorage-independent growth of human BC cells. The study provides important insights into the understanding of the molecular mechanism(s) that are responsible for the anti-cancer effect of ChlA-F, as well as revealing that elevating SESN-2-mediated autophagy activity might be a promising approach to treat human invasive/metastatic BC patients.
Supplementary Material
The anti-cancer activity and its potential mechanisms underlying ChlA-F action is first explored.
ChlA-F treatment significantly inhibited anchorage-independent growth of human BC cells by inducing autophagy in a SESN2-dependent fashion.
ChlA-F treatment specifically induced SESN2 expression via increasing its transcription and mRNA stability.
The study of the anti-cancer effect of ChlA-F on BC provides therapeutic alternatives against human BC.
5. Acknowledgments
We thank Dr. Gang Chen (University of Kentucky, Lexington, KY, USA) for providing the GFP-LC3 construct. This work was partially supported by grants from NIH/NCI CA217923, CA177665 and CA165980, and NIH/NIEHS ES000260; the Natural Science Foundation of China (NSFC91773391) and Key Project of Science and Technology Innovation Team of Zhejiang Province (2013TD10).
Abbreviations:
- Act D
actinomycin D
- ATG
autophagy-related
- BAF
bafilomycin A1
- BC
bladder cancer
- ChlA-F
Cheliensisine A-fluoride
- CQ
chloroquine
- DAPI
40,6-diamidino-2-phenylindole
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- IgG
immunoglobulin G
- AP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 beta
- miRNA
microRNA
- RFP
Red fluorescent protein
- RT-PCR
reverse transcription-polymerase chain reaction
- SESN2
sestrin2
- shRNA
short hairpin RNA
- 3’-UTR
3’-Untranslated Regions
- WT
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. Conflicts of Interest Statement
Neither this paper nor any similar paper has been or will be submitted to or published in any other scientific journal. All authors are aware and agree to the content of the paper and to their being listed as an author on the manuscript. There is no conflict of interest or competing financial interests for all authors.
7. References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2015, CA Cancer J Clin, 65 (2015) 5–29. [DOI] [PubMed] [Google Scholar]
- [2].Baudino TA, Targeted Cancer Therapy: The Next Generation of Cancer Treatment, Curr Drug Discov Technol, 12 (2015) 3–20. [DOI] [PubMed] [Google Scholar]
- [3].Ortiz R, Melguizo C, Prados J, Alvarez PJ, Caba O, Rodriguez-Serrano F, Hita F, Aranega A, New gene therapy strategies for cancer treatment: a review of recent patents, Recent Pat Anticancer Drug Discov, 7 (2012) 297–312. [DOI] [PubMed] [Google Scholar]
- [4].Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CM, Pryer N, Daniel D, Hwang ES, Rugo HS, Coussens LM, Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells, Cancer Cell, 26 (2014) 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, Yang H, Amigorena S, Ryffel B, Barrat FJ, Saftig P, Levi F, Lidereau R, Nogues C, Mira JP, Chompret A, Joulin V, Clavel-Chapelon F, Bourhis J, Andre F, Delaloge S, Tursz T, Kroemer G, Zitvogel L, Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy, Nat Med, 13 (2007) 1050–1059. [DOI] [PubMed] [Google Scholar]
- [6].Li C-M, Mu Q, Sun H-D, Xu B, Tang W-D, Zheng H-L, Tao G-D, A new anti-cancer constituent of Goniothalamus cheliensis, Acta Bot. Yunnan, 20 (1998) 102–104. [Google Scholar]
- [7].Zhao D, Gong T, Fu Y, Nie Y, He L-L, Liu J, Zhang Z-R, Lyophilized Cheliensisin A submicron emulsion for intravenous injection: Characterization, in vitro and in vivo antitumor effect, Int. J. Pharm, 357 (2008) 139–147. [DOI] [PubMed] [Google Scholar]
- [8].Deng X, Su J, Zhao Y, Peng L-Y, Li Y, Yao Z-J, Zhao Q-S, Development of novel conformation-constrained cytotoxic derivatives of cheliensisin A by embedment of small heterocycles, Eur. J. Med. Chem, 46 (2011) 4238–4244. [DOI] [PubMed] [Google Scholar]
- [9].Ro SH, Semple IA, Park H, Park H, Park HW, Kim M, Kim JS, Lee JH, Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1, FEBS J, 281 (2014) 3816–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mowers EE, Sharifi MN, Macleod KF, Autophagy in cancer metastasis, Oncogene, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou H, Yuan M, Yu Q, Zhou X, Min W, Gao D, Autophagy regulation and its role in gastric cancer and colorectal cancer, Cancer Biomark, 17 (2016) 1–10. [DOI] [PubMed] [Google Scholar]
- [12].Mathew R, Karantza-Wadsworth V, White E, Role of autophagy in cancer, Nat Rev Cancer, 7 (2007) 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karantza-Wadsworth V, White E, Role of autophagy in breast cancer, Autophagy, 3 (2007) 610–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guo JY, Xia B, White E, Autophagy-Mediated Tumor Promotion, Cell, 155 (2013) 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kimura T, Takabatake Y, Takahashi A, Isaka Y, Chloroquine in Cancer Therapy: A Double-Edged Sword of Autophagy, Cancer Research, 73 (2013) 3–7. [DOI] [PubMed] [Google Scholar]
- [16].Wirawan E, Vanden Berghe T, Lippens S, Agostinis P, Vandenabeele P, Autophagy: for better or for worse, Cell Res, 22 (2012) 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hu YL, Jahangiri A, DeLay M, Aghi MK, Tumor Cell Autophagy as an Adaptive Response Mediating Resistance to Treatments Such as Antiangiogenic Therapy, Cancer Research, 72 (2012) 4294–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].White E, DiPaola RS, The double-edged sword of autophagy modulation in cancer, Clin Cancer Res, 15 (2009) 5308–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Levine B, Kroemer G, Autophagy in the pathogenesis of disease, Cell, 132 (2008) 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brech A, Ahlquist T, Lothe RA, Stenmark H, Autophagy in tumour suppression and promotion, Mol Oncol, 3 (2009) 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lei YY, Zhang D, Yu J, Dong H, Zhang JW, Yang SM, Targeting autophagy in cancer stem cells as an anticancer therapy, Cancer Letters, 393 (2017) 33–39. [DOI] [PubMed] [Google Scholar]
- [22].Hu HJ, Shi ZY, Lin XL, Chen SM, Wang QY, Tang SY, Upregulation of Sestrin2 Expression Protects Against Macrophage Apoptosis Induced by Oxidized Low-Density Lipoprotein, DNA and Cell Biology, 34 (2015) 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee JH, Bodmer R, Bier E, Karin M, Sestrins at the crossroad between stress and aging, Aging-Us, 2 (2010) 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sahni S, Merlot AM, Krishan S, Jansson PJ, Richardson DR, Gene of the month: BECN1, J Clin Pathol, 67 (2014) 656–660. [DOI] [PubMed] [Google Scholar]
- [25].Maiuri MC, Malik SA, Morselli E, Kepp O, Criollo A, Mouchel PL, Carnuccio R, Kroemer G, Stimulation of autophagy by the p53 target gene Sestrin2, Cell Cycle, 8 (2009) 1571–1576. [DOI] [PubMed] [Google Scholar]
- [26].Liang Y, Zhu J, Huang H, Xiang D, Li Y, Zhang D, Li J, Wang Y, Jin H, Jiang G, Liu Z, Huang C, SESN2/sestrin 2 induction-mediated autophagy and inhibitory effect of isorhapontigenin (ISO) on human bladder cancers, Autophagy, (2016) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang Y, Xu J, Gao G, Li J, Huang H, Jin H, Zhu J, Che X, Huang C, Tumor-suppressor NFkappaB2 p100 interacts with ERK2 and stabilizes PTEN mRNA via inhibition of miR-494, Oncogene, 35 (2016) 4080–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhong L, Li CM, Hao XJ, Lou LG, Induction of leukemia cell apoptosis by cheliensisin A involves down-regulation of Bcl-2 expression, Acta Pharmacologica Sinica, 26 (2005) 623–628. [DOI] [PubMed] [Google Scholar]
- [29].Fang Y, Cao Z, Hou Q, Ma C, Yao C, Li J, Wu XR, Huang C, Cyclin d1 downregulation contributes to anticancer effect of isorhapontigenin on human bladder cancer cells, Mol Cancer Ther, 12 (2013) 1492–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zeng X, Xu Z, Gu J, Huang H, Gao G, Zhang X, Li J, Jin H, Jiang G, Sun H, Huang C, Induction of miR-137 by Isorhapontigenin (ISO) Directly Targets Sp1 Protein Translation and Mediates Its Anticancer Activity Both In Vitro and In Vivo, Mol Cancer Ther, 15 (2016) 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Song L, Gao M, Dong W, Hu M, Li J, Shi X, Hao Y, Li Y, Huang C, p85alpha mediates p53 K370 acetylation by p300 and regulates its promoter-specific transactivity in the cellular UVB response, Oncogene, 30 (2011) 1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E, Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy, Autophagy, 1 (2005) 84–91. [DOI] [PubMed] [Google Scholar]
- [33].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Arozena AA, Adachi H, Adams CM, Adams PD,Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J,Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An ZY, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK,Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J,Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Banhegyi G, Bao HJ, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolome A,Bassham DC, Bassi MT, Bast RC, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR, Becker C, Beckham JD, Bedard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GMN, Behrns KE, Bejarano E, Belaid A, Belleudi F, Benard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S,Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B,Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Bockler S, Boes M,Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouche M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N,Braga VMM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultman SJ, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Butikofer P, Caberlotto L, Cadwell K, Cahova M, Cai DS, Cai JJ, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao LZ, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LAM, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Cena V, Cenci S,Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EYW,Chan MTV, Chandra D, Chandra P, Chang CP, Chang RCC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che YS, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen GQ, Chen HZ, Chen JW, Chen JK, Chen M,Chen MZ, Chen PW, Chen Q, Chen Q, Chen SD, Chen S, Chen SSL, Chen W, Chen WJ, Chen WQ, Chen WL, Chen XM, Chen YH, Chen YG, Chen Y, Chen YY, Chen YS, Chen YJ, Chen YQ, Chen YJ, Chen Z, Chen Z, Cheng A, Cheng CHK, Cheng H, Cheong HS, Cherry S, Chesney J, Cheung CHA, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GNC, Cho DH, Cho SG, Cho WC, Cho YY,Cho YS, Choi AMK, Choi EJ, Choi EK, Choi JY, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung HW, Chung TJ, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Claria J, Clarke PGH, Clarke R, Clementi E,Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EEW, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT,Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Chagin AS, Cossarizza A, Costelli P, Costes S, Costes S, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui TX, Cutillas PR,Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai CS, Dai WJ, Dai Y, Dalby KN, Valle LD, Dalmasso G, D’Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Davila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P,de Figueiredo RCBQ, de la Fuente J, De Martino L, De Matteis A, De Meyer GRY, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LMD, Deldicque L, Delorme-Axford E, Deng YZ, Dengjel J, Denizot M, Dent P, Der CJ,Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding ZF, Dini L, Distler JHW, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RCJ, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong XN, Dong Z, Donohue TM, Doran KS, D’Orazi G, Dorn GW, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du LH, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA, Dupont N, Dupuis L, Duran RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L,Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJH, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R,Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martinez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Faergeman NJ, Faggioni A, Fairlie WD, Fan CH, Fan DP, Fan J, Fang SY, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng YB, Feng YC, Ferguson TA,Fernandez AF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernandez-Lopez A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, Francois A, Frankel LB, Fraser IDC, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu DX, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannage M, Gao FB, Gao F, Gao JX, Nannig LG, Vescovi EG, Garcia-Macia M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge PF, Ge SF, Gean PW, Gelmetti V, Genazzani AA, Geng JF, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F,Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E,Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gomez-Sanchez R, Goncalves DAP, Goncu E, Gong QQ, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, Gonzalez-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo CY, Guo L, Guo M, Guo WJ, Guo XG, Gust AA, Gustafsson AB,Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hamid Q, Han F, Han WD, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U,Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He CC, He CY, He FT, He G, He RR, He XH, He YW, He YY, Heath JK, Hebert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her CT, Herman PK, Hernandez A, Hernandez C, Hernandez-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Hoglinger GU, Hohfeld J, Holz MK, Hong YG, Hood DA, Hoozemans JJM, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu GC, Hu HM, Hu HB, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang CH, Huang HL, Huang KH, Huang KY, Huang SL, Huang SQ, Huang WP, Huang YR, Huang Y, Huang YF, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SNA, Hussain S, Hwang JJ, Hwang SM, Hwang TIS, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe KI, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AKV, Izquierdo JM, Izumi Y, Izzo V, Jaattela M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia KL, Jia LJ, Jiang H, Jiang HC, Jiang LW, Jiang T, Jiang XY, Jiang XJ, Jiang XJ, Jiang Y, Jiang YJ, Jimenez A, Jin C, Jin HC, Jin L, Jin MY, Jin SK, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GVW, Johnson JD, Jonasch E, Jones C, Joosten LAB, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju DW, Ju JF, Juan HF, Juenemann K, Juhasz G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kagedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut ID, Khambu B, Khan MM, Khandelwal VKM, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knaevelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Kohler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong DX, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou YJ, Koukourakis MI, Koumenis C, Kovacs AL, Kovacs T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJR, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BYK, Law HKW, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei JZ, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng SL, Lenz G, Lenzi P, Lerman LO, Barbato DL, Leu JIJ, Leung HY, Levine B, Lewis PA, Lezoualc’h F, Li C, Li FQ, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li XT, Li YM, Lian JQ, Liang CY, Liang QR, Liao YL, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin FM, Lin FC, Lin K, Lin KH, Lin PH, Lin TW, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu JX, Liu JJ, Liu JL, Liu K, Liu LY, Liu L, Liu QT, Liu RY, Liu SM, Liu SW, Liu W, Liu XD, Liu XG, Liu XH, Liu XF, Liu X, Liu XQ, Liu Y, Liu YL, Liu ZX, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, Lopez-Otin C, Lopez-Vicario C, Lorente M, Lorenzi PL, Lorincz P, Los M, Lotze MT, Lovat PE, Lu BF, Lu B, Lu J, Lu Q, Lu SM, Lu SY, Lu YY, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo HL, Luo J, Luo SQ, Luparello C, Lyons T, Ma JJ, Ma Y, Ma Y, Ma ZY, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magarinos M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manie SN, Manzoni C, Mao K, Mao ZX, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Marino G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martin-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martinez-Velazquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei YD, Meier UC, Meijer AJ, Melendez A, Melino G, Melino S, de Melo EJT, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng LS, Meng LH, Meng SS, Menghini R, Menko AS, Menna-Barreto RFS, Menon MB, Meraz-Rios MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Moller AB, Mollereau B, Mollinedo F, Monick MM, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CEH, Mpakou VE, Mukhtar H, Levy JMM, Muller S, Munoz-Moreno R, Munoz-Pinedo C, Munz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HTT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VCO, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nurnberger T, O’Donnell VB, O’Donovan T, O’Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O’Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JHJ, Outeiro TF, Ouyang DY, Ouyang HJ, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palkova Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan HM, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KBS, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstadt H, Pavone F, Pedrozo Z, Pena FJ, Penalva MA, Pende M, Peng JX, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJS, Pereira PC, la Cruz VP, Perez-Perez ME, Perez-Rodriguez D, Perez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muinos FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Poggeler S, Poirot M, Polcic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu ZW, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren XC, Renna M, Reusch JEB, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CMP, Rodriguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MIG, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roue G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AMJ, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schonenberger MJ, Schonthal AH, Schorderet DF, Schroder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Segui-Simarro JM, Segura-Aguilar J, Seiliez I, Seki E, Sell C, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CKJ, Shen CC, Shen HM, Shen SB, Shen WL, Sheng R, Sheng XY, Sheng ZH, Shepherd TG, Shi JY, Shi Q, Shi QH, Shi YG, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJN, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninova I, Slavov N, Smaili SS, Smalley KSM, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song CJ, Song FY, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, St Clair D, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Strom AL, Stromhaug P, Stulik J, Su YX, Su ZL, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui XB, Sukseree S, Sulzer D, Sun FL, Sun JR, Sun J, Sun SY, Sun Y, Sun Y, Sun YJ, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Sward K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SWG, Takacs-Vellai K, Takahashi Y, Takats S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang DL, Tang DZ, Tang GM, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thome MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TLM, Tian L, Till A, Ting JPY, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu SP, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RFB, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcategui NL, Vaccari T, Vaccaro MI, Vachova L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJW, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, Campagne MV, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PST, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HLA, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan XB, Wang B, Wang CH, Wang CY, Wang CS, Wang CR, Wang CH, Wang D, Wang F, Wang FX, Wang GH, Wang HJ, Wang HC, Wang HG, Wang HM, Wang HD, Wang J, Wang JJ, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang XJ, Wang Y, Wang Y, Wang Y, Wang YJ, Wang YP, Wang Y, Wang YT, Wang YQ, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei JW, Weide T, Weihl CC, Weindl G, Weis SN, Wen LP, Wen X, Wen YF, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VKW, Woodcock EA, Wright KL, Wu CL, Wu DF, Wu GS, Wu J, Wu JF, Wu M, Wu M, Wu SZ, Wu WKK, Wu YH, Wu ZL, Xavier CPR, Xavier RJ, Xia GX, Xia T, Xia WL, Xia Y, Xiao HY, Xiao J, Xiao S, Xiao WH, Xie CM, Xie ZP, Xie ZL, Xilouri M, Xiong YY, Xu CS, Xu CF, Xu F, Xu HX, Xu HW, Xu J, Xu JZ, Xu JX, Xu L, Xu XL, Xu YQ, Xu Y, Xu ZX, Xu ZH, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang MH, Yang PM, Yang P, Yang Q, Yang WN, Yang WY, Yang XS, Yang Y, Yang Y, Yang ZF, Yang ZH, Yao MC, Yao PJ, Yao XF, Yao ZY, Yao ZY, Yasui LS, Ye MX, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida KI, Yoshimori T, Young KH, Yu HM, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu ZP, Yuan JY, Yuan ZM, Yue BYJT, Yue JB, Yue ZY, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng JS, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang HB, Zhang J, Zhang J, Zhang JW, Zhang JH, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang XN, Zhang XD, Zhang Y, Zhang Y, Zhang YJ, Zhang YM, Zhang YJ, Zhao M, Zhao WL, Zhao XN, Zhao YG, Zhao Y, Zhao YC, Zhao YX, Zhao ZD, Zhao ZZJ, Zheng DX, Zheng XL, Zheng XX, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou GF, Zhou HP, Zhou SF, Zhou XJ, Zhu HX, Zhu H, Zhu WG, Zhu WH, Zhu XF, Zhu YH, Zhuang SM, Zhuang XH, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM, Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition), Autophagy, 12 (2016) 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dickstein RJ, Nitti G, Dinney CP, Davies BR, Kamat AM, McConkey DJ, Autophagy limits the cytotoxic effects of the AKT inhibitor AZ7328 in human bladder cancer cells, Cancer Biol Ther, 13 (2012) 1325–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lin C, Tsai SC, Tseng MT, Peng SF, Kuo SC, Lin MW, Hsu YM, Lee MR, Amagaya S, Huang WW, Wu TS, Yang JS, AKT serine/threonine protein kinase modulates baicalin-triggered autophagy in human bladder cancer T24 cells, Int J Oncol, 42 (2013) 993–1000. [DOI] [PubMed] [Google Scholar]
- [36].Lin YC, Lin JF, Wen SI, Yang SC, Tsai TF, Chen HE, Chou KY, Hwang TI, Inhibition of High Basal Level of Autophagy Induces Apoptosis in Human Bladder Cancer Cells, J Urol, (2015). [DOI] [PubMed] [Google Scholar]
- [37].Mani J, Vallo S, Rakel S, Antonietti P, Gessler F, Blaheta R, Bartsch G, Michaelis M, Cinatl J, Haferkamp A, Kogel D, Chemoresistance is associated with increased cytoprotective autophagy and diminished apoptosis in bladder cancer cells treated with the BH3 mimetic (−)-Gossypol (AT-101), BMC Cancer, 15 (2015) 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F, Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer, Mol Biosyst, 9 (2013) 407–411. [DOI] [PubMed] [Google Scholar]
- [39].Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E, Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis, Genes Dev, 21 (2007) 1621–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Thorburn A, Thamm DH, Gustafson DL, Autophagy and cancer therapy, Mol Pharmacol, 85 (2014) 830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang ZJ, Chee CE, Huang S, Sinicrope FA, The Role of Autophagy in Cancer: Therapeutic Implications, Molecular Cancer Therapeutics, 10 (2011) 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kim MG, Yang JH, Kim KM, Jang CH, Jung JY, Cho IJ, Shin SM, Ki SH, Regulation of Toll-like receptor-mediated Sestrin2 induction by AP-1, Nrf2, and the ubiquitin-proteasome system in macrophages, Toxicol Sci, 144 (2015) 425–435. [DOI] [PubMed] [Google Scholar]
- [43].Abdelmohsen K, Gorospe M, Posttranscriptional regulation of cancer traits by HuR, Wiley Interdiscip Rev RNA, 1 (2010) 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gratacos FM, Brewer G, The role of AUF1 in regulated mRNA decay, Wiley Interdiscip Rev RNA, 1 (2010) 457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Abdelmohsen K, Gorospe M, RNA-binding protein nucleolin in disease, RNA Biol, 9 (2012) 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang J, Ouyang W, Li J, Zhang D, Yu Y, Wang Y, Li X, Huang C, Suberoylanilide hydroxamic acid (SAHA) inhibits EGF-induced cell transformation via reduction of cyclin D1 mRNA stability, Toxicol Appl Pharmacol, 263 (2012) 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jin H, Yu Y, Hu Y, Lu C, Li J, Gu J, Zhang L, Huang H, Zhang D, Wu XR, Gao J, Huang C, Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines, Oncotarget, 6 (2015) 522–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wu X, Brewer G, The regulation of mRNA stability in mammalian cells: 2.0, Gene, 500 (2012) 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Svoronos AA, Engelman DM, Slack FJ, OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer, Cancer Res, 76 (2016) 3666–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M, Differential regulation of microRNA stability, RNA, 16 (2010) 1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yamasaki T, Onishi M, Kim EJ, Cerutti H, Ohama T, RNA-binding protein DUS16 plays an essential role in primary miRNA processing in the unicellular alga Chlamydomonas reinhardtii, Proc Natl Acad Sci U S A, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kim YK, Kim B, Kim VN, Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis, Proceedings of the National Academy of Sciences of the United States of America, 113 (2016) E1881–E1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhang R, Che X, Zhang J, Li Y, Li J, Deng X, Zhu J, Jin H, Zhao Q, Huang C, Cheliensisin A (Chel A) induces apoptosis in human bladder cancer cells by promoting PHLPP2 protein degradation, Oncotarget, 7 (2016) 66689–66699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang N, Pan W, Zhu M, Zhang M, Hao X, Liang G, Feng Y, Fangchinoline induces autophagic cell death via p53/sestrin2/AMPK signalling in human hepatocellular carcinoma cells, Br J Pharmacol, 164 (2011) 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kondo Y, Kanzawa T, Sawaya R, Kondo S, The role of autophagy in cancer development and response to therapy, Nat Rev Cancer, 5 (2005) 726–734. [DOI] [PubMed] [Google Scholar]
- [56].Rao S, Yang H, Penninger JM, Kroemer G, Autophagy in non-small cell lung carcinogenesis: A positive regulator of antitumor immunosurveillance, Autophagy, 10 (2014) 529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E, Principles and current strategies for targeting autophagy for cancer treatment, Clin Cancer Res, 17 (2011) 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bursch W, Ellinger A, Kienzl H, Torok L, Pandey S, Sikorska M, Walker R, Hermann RS, Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: the role of autophagy, Carcinogenesis, 17 (1996) 1595–1607. [DOI] [PubMed] [Google Scholar]
- [59].Opipari AW Jr., Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR, Resveratrol-induced autophagocytosis in ovarian cancer cells, Cancer Res, 64 (2004) 696–703. [DOI] [PubMed] [Google Scholar]
- [60].Beishline K, Azizkhan-Clifford J, Sp1 and the ‘hallmarks of cancer’, FEBS J, 282 (2015) 224–258. [DOI] [PubMed] [Google Scholar]
- [61].Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC, Hung JJ, Sp1 expression regulates lung tumor progression, Oncogene, 31 (2012) 3973–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Biggs JR, Kudlow JE, Kraft AS, The role of the transcription factor Sp1 in regulating the expression of the WAF1/CIP1 gene in U937 leukemic cells, J Biol Chem, 271 (1996) 901–906. [DOI] [PubMed] [Google Scholar]
- [63].Wei M, Liu B, Gu Q, Su L, Yu Y, Zhu Z, Stat6 cooperates with Sp1 in controlling breast cancer cell proliferation by modulating the expression of p21(Cip1/WAF1) and p27 (Kip1), Cell Oncol (Dordr), 36 (2013) 79–93. [DOI] [PubMed] [Google Scholar]
- [64].Wu XJ, Li Y, Liu D, Zhao LD, Bai B, Xue MH, miR-27a as an Oncogenic microRNA of Hepatitis B Virus-related Hepatocellular Carcinoma, Asian Pacific Journal of Cancer Prevention, 14 (2013) 885–889. [DOI] [PubMed] [Google Scholar]
- [65].Liu T, Tang H, Lang YY, Liu M, Li X, MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin, Cancer Letters, 273 (2009) 233–242. [DOI] [PubMed] [Google Scholar]
- [66].Fletcher CE, Dart DA, Sita-Lumsden A, Cheng H, Rennie PS, Bevan CL, Androgen-regulated processing of the oncomir MiR-27a, which targets Prohibitin in prostate cancer, Human Molecular Genetics, 21 (2012) 3112–3127. [DOI] [PubMed] [Google Scholar]
- [67].Guttilla IK, White BA, Coordinate Regulation of FOXO1 by miR-27a, miR-96, and miR-182 in Breast Cancer Cells, Journal of Biological Chemistry, 284 (2009) 23204–23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.