Abstract
Heparanase, an endo-β-D-glucuronidase, cleaves cell surface and extracellular matrix heparan sulfate (HS) chains and plays important roles in cellular growth and metastasis. Heparanase assays reported to-date are labor intensive, complex and/or expensive. A simpler assay is critically needed to understand the myriad roles of heparanase. We reasoned that fluorescent heparin could serve as an effective probe of heparanase levels. Following synthesis and screening, a heparin preparation labeled with DABCYL and EDANS was identified, which exhibited a characteristic increase in signal following cleavage by human heparanase. This work describes the synthesis of this heparin substrate, its kinetic and spectrofluorometric properties, optimization of the heparanase assay, use of the assay in inhibitor screening, and elucidation of the state of heparanase in different cell lines. Our FRET-based assay is much simpler and more robust than all assays reported in the literature as well as a commercially available kit.
Keywords: Enzyme assay, Enzyme Inhibition, Fluorescent Heparin, FRET, Heparanase, Heparin
1. Introduction
The role of heparanase in tumor growth, angiogenesis, and metastasis is well established (Sanderson, Elkin, Rapraeger, Ilan, & Vlodavsky, 2017). Growing evidence also supports its role in inflammation (Goldberg et al., 2013). Although heparanase is generally recognized as an extracellular enzyme, it is also found in the cytoplasm and nucleus of cells (Chen & Sanderson, 2009; Santos et al., 2014) and appears to be a critical regulator in the production of exosomes (David & Zimmermann, 2016).
Human heparanase is an endo-β-D-glucuronidase that cleaves heparan sulfate (HS) chains present in proteoglycan form on cell surfaces and in the extracellular matrix (Dempsey, Brunn, & Platt, 2000; Fairbanks et al., 1999; Kussie et al., 1999; Sanderson et al., 2017; Toyoshima & Nakajima, 1999). Substrate specificity studies indicate that heparanase preferably cleaves the 1→4-inter-glycosidic bond between a glucuronic acid (GlcA) and glucosamine-N,6-disulfate (GlcNS6S) residues, although other sequences are also likely to be targeted (Peterson & Liu, 2013). Considering the domain structure of HS, heparanase cleavage of this polymer is expected to generate chains of variable sizes in the range of 5 to 15 residues, although much remains to be understood about the biological roles of such chains.
The importance of heparanase in biology has led to the development of several biochemical and biophysical assays over the past two decades. Starting with radioisotope-based assays developed in the late 1990s and early 2000s (Behzad & Brenchley, 2003; Freeman & Parish, 1997; Nardella & Steinkühler, 2004), newer assays have come to rely on either colorimetry or spectrofluorimetry (Enomoto, Okamoto, Numata, & Takemoto, 2006; Hammond, Li, & Ferro, 2010; Huang et al., 2004; Melo, Tersariol, Nader, Pinhal, & Lima, 2015; Pearson, Kiefel, Ferro, & von Itzstein, 2011; Tsuchida, Podyma-Inoue, & Yanagishita, 2004) (see Supplementary Table S1). A variety of substrates have been evaluated including native and modified forms of HS (Huang et al., 2004; Melo et al., 2015; Tsuchida et al., 2004), heparan sulfate proteoglycan (HSPG) (Enomoto et al., 2006), fondaparinux (Hammond et al., 2010; Melo et al., 2015) and synthetic disaccharides modified with chromogenic or fluorogenic groups (Pearson et al., 2011). Methods to detect heparanase activity have relied on either quantifying the levels of HS degradation products (Huang et al., 2004; Tsuchida et al., 2004), or uncleaved HS (Melo et al., 2015), or GlcA-terminated reducing ends (Hammond et al., 2010; Melo et al., 2015).
Despite the availability of numerous assays, no particular assay appears to have been broadly used to understand heparanase biology, pharmacology, and drug discovery. The assays typically suffer from the involvement of multiple steps. An easier, one-step assay would greatly help deduce inhibitors, understand substrate specificity, elucidate the mechanism of action, and clarify the enzymatic or non-enzymatic role in cellular systems. A more robust heparanase assay would be easier to implement (fewer steps, faster screening time, no immobilization, no post-assay signal development), be adaptable to microplate format, enable assaying active heparanase in cellular media, and perhaps help monitor heparanase in vivo.
Although it may not be possible to achieve all these attributes in one assay, we posited that a fluorescence resonance energy transfer (FRET)-based assay may offer a one-step solution that can address several of these attributes. This work documents our studies on developing a FRET-based assay for active heparanase. Our work shows that a FRET-enabled heparin chain can effectively serve as a substrate of heparanase, help detect an active enzyme in media, and help screen potential inhibitors.
2. Materials and Methods
2.1. Materials
All anhydrous organic solvents were purchased from Sigma-Aldrich (Milwaukee, WI) or Fisher (Pittsburgh, PA) and used as received. Heparin from porcine mucosa was purchased from Sigma-Aldrich (St. Louis, MO). EDANS and Dabcyl were from AnaSpec (Fremont, CA). Chemical reactions sensitive to air or moisture were carried out under nitrogen atmosphere in oven-dried amber glassware. All chemicals were of analytical grade unless otherwise specified. Heparinases I, II and III were from New England Bio-labs (Ipswich, MA).
2.2. Synthesis of Heparin-DE
Standard carbodiimide-based linking chemistry was used to synthesize DABCYL and EDANS labeled heparin, called heparin-DE. Unfractionated heparin (1 equivalent) was dissolved in dd H2O to give 0.25 % (w/v) solution. To this, a solution containing 2 equivalents each DABCYL C2 amine and EDANS in DMSO were added, followed by 20 and 4 equivalents of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) and N-hydroxy succinimide (NHS), respectively, and the final volume made up to 2 ml with dd H2O. The mixture was stirred for 20 h at 37° C, dialyzed against dd H2O (3500 MWCO), and concentrated by ultrafiltration using 3000 MWCO membrane. The double labeled heparin product so obtained, named as heparin-DE, was purified by G-15 Sephadex size exclusion chromatography (SEC) and characterized using NMR spectroscopy. Heparin-DE was stored at − 80 °C following lyophilization.
2.3. Expression of Human Heparanase
A modified Sf9 insect cell expression protocol, based on the published protocol, was used to express human heparanase (HPSE). Briefly, a heparanase expression vector, pFastBac Dual, expressing 8 kDa and 50 kDa heparanase subunits was obtained from the Guido laboratory [University of York, UK]. The subunits were subcloned into BamHI/PstI sites of the pFastBac vector under the control of promoters PolH and p10, respectively. The expression of the ligated HPSE gene led to an extra DPG tripeptide sequence in the expressed polypeptide, as noted earlier (Wu, Viola, Brzozowski, & Davies, 2015a). The bacmid was prepared in DH10Bac and purified according to the standard protocols (Invitrogen). The baculovirus was produced by transfection of bacmid into Sf9 cells (Invitrogen) using cellfectin transfection reagent. After 72 hrs cells were harvested and centrifuged (4000 rpm, 15 min, 4 °C). The supernatant containing heparanase was collected, followed by addition of DTT (1 mM) and 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF, 0.1 mM) and then loaded onto a HiTrap Heparin HP column (5 mL) pre-equilibrated with 20 mM HEPES buffer, pH 7.4, containing 100 mM NaCl and 1 mM DTT. Heparanase was eluted from the affinity matrix using a linear salt gradient from 0.1 to 1.5 M NaCl at room temperature. Heparanase was detected using SDS-PAGE and appropriate fractions were pooled, exchanged with 20 mM sodium acetate buffer, pH 5.0, containing 100 mM NaCl, and stored at −80 °C.
2.4. Western Blot Analysis
Purified protein samples (20 μg) were separated on SDS-PAGE gels (Bio-Rad 8-14% gel) and transferred to nitrocellulose membrane using Criterion™ Blotter (Bio-Rad). Blocking was done with 5% BSA (large subunit) and 5% skim milk (small sub unit) for 1 hr followed by overnight incubation with primary antibody (dilution 8:1000). Antibodies used for analysis include anti-heparanase 1 (aa 301-331, Boster bio, PB9427) and anti-HPSE (aa34-115, LS Bio, LS-C294467). This was followed by incubation with appropriate secondary antibody and protein bands were visualized and captured using the Amersham Imager 600 (GE Healthcare).
2.5. Heparanase Activity
Heparanase activity measurements were performed using the assay buffer (20 mM sodium acetate buffer, pH 5.0, containing 100 mM NaCl) at 37 °C. Briefly, a 100 μL solution of recombinant heparanase and the labeled substrate was vigorously shaken for 4 h, following which the enzyme was inactivated by incubating on ice until the measurements were read. The mixture was transferred to a fluorescence cuvette in a QM4 spectrofluorometer (Photon Technology International, Birmingham, NJ) and emission read at 500 nm (λEX = 340 nm). The excitation and emission slits were set to 0.5 mm. The observed change in fluorescence (ΔF) was normalized to initial fluorescence (F0) to calculate the % change in signal at every addition of test sample.
2.6. Heparanase Inhibition Assay
Inhibition assays were performed using varying concentrations of heparin-DE. The hydrolysis of heparin-DE by HPSE in the presence of suramin, a known inhibitor, was monitored in 96-well plates (Corning® 3897) on a fluorescence microplate reader FlexStation III (Molecular Devices) in a final volume of 100 μL. Stock solutions of the inhibitor were prepared in the assay buffer. Heparanase (1 μM) and suramin were incubated for 30 min in the assay buffer at 37 °C. The hydrolytic reaction, initiated by introducing heparin-DE (66 μM final concentration), was constantly stirred for 4 h, after which the reaction was stopped by placing on ice and fluorescence measured at 500 nm (λEX = 340 nm) at 4 °C.
2.7. LC-MS Validation of Heparin-DE as a Substrate
Size exclusion chromatography (SEC) was used to validate heparin-DE as a substrate of heparanase. The SEC experiments were performed using a Shodex OH Pak SB-802.5 HQ column (8.0 mm × 300 mm; exclusion limit 10,000 Da) on a Shimadzu HPLC system equipped with an RF-10A fluorescence detector. The hydrolytic reaction mixture (5 μl) was injected into the column at RT, eluted with 0.5 ml/min of acetonitrile-water (0.3:1 v/v) and monitored at 490 nm (λEX = 330nm). RP-IP UPLC–ESI-MS analysis of reaction mixtures was performed at 40 °C. The digestion mixtures (1 mg/ml) were analyzed using Waters Acquity H-Class UPLC coupled to ESI-MS equipped with a triple quadrupole TQD detector. Reversed-phase ion pairing (RP-IP) LC using a BEH C18 1.7 μm (2.1 mm×150 mm) column and a linear gradient of 100% solvent A consisting of H2O:CH3CN (95:5, v/v) to 100% solvent B consisting of H2O:CH3CN (15:85, v/v) in 45 min at 40 °C (flow rate 0.1 ml/min) was employed to resolve oligosaccharides. Hexylamine (15 mM) was used as an ion-pairing agent and hexafluoroisopropanol (100 mM) was used as an organic modifier in both solvent systems. The ESI-MS peaks were acquired in the positive ionization mode with a capillary voltage of 3.2 kV, extractor voltage of 1 V, cone voltage between 20 and 180 V, the radio frequency of 0.1 V, source temperature of 150 °C, and desolvation temperature of 350 °C (Bhushan, Alabbas, Kuberan, Gupta, & Desai, 2017).
2.8. Assay for Heparanase Expression by Cells
The human mammary carcinoma cell line, MCF7 and HEK 293T were a generous gift from the Grossman lab (Massey Cancer Center, Richmond, VA). Both cell lines were maintained in monolayer conditions in Dulbecco's Modified Eagle Medium (DMEM) growth media (Invitrogen, USA), supplemented with 10% fetal bovine serum (FBS), and antibiotic-antimycotic liquid (AA) in 5% CO2 at 37°C, as recommended by ATCC. The cells were grown in 6-well tissue culture-treated plates in monolayer condition and passaged twice a week. MCF7 cells were initially grown as a monolayer in the growth media for 24 h to ensure efficient growth and adhesion. Subsequently, the media was replaced with FBS-free (0% serum), minimal FBS (2% serum) or 10% serum in growth media, and the cells were allowed to propagate for an additional 48 h in normoxia and hypoxia conditions. For hypoxic conditions, cells were grown in a humidified atmosphere at 37°C with a stabilized gas mixture input containing 94% N2/5% CO2/1% O2 in a HERAcell I50i incubator (ThermoScientific, USA). For normoxic conditions, cells were grown as mentioned above in a humidified atmosphere maintained at 37°C in 5% CO2/ 95% atmospheric air in Forma Series II, water jacketed CO2 incubator (Thermo Electron Corporation, USA). After 48 hours of incubation in the indicated conditions, the supernatants were collected and centrifuged at 3000 rpm for 10 mins at 4°C. For the time-based heparanase activity measurements, media was harvested at varying time points and frozen immediately at −20°C until the readings were to be recorded. The heparin-DE substrate was added to 100 μL media in 96-well plates (Corning® 3897) and HPSE activity of the media was measured directly.
3. Results and Discussion
3.1. Heparanase expression in Sf9 cells
The expression of human heparanase in insect cells has been reported earlier (McKenzie et al., 2003; Wu, Viola, Brzozowski, & Davies, 2015). We used essentially the same procedure to express and isolate active, mature heparanase in >95% purity, as demonstrated by SDS PAGE and Western blotting analysis (Supplementary Figure S1 and S2). The two subunits of human heparanase, called large and small subunits, were identified using two corresponding primary antibodies available from commercial sources. Human heparanase is not catalytically active without either of the subunits and we confirmed this for our enzyme (data not shown).
3.2. Rationale for developing a FRET-based assay
FRET-based assays are routinely used for assaying enzymatic activity. When the donor and acceptor fluorophores are incorporated into the substrate, even a small FRET signal is easier to detect over a darker background. Typically, this results in higher sensitivity, which is good for in vitro diagnostics and drug discovery applications. More importantly, FRET-based assays have been used for visualizing the sub-cellular action of proteins (Allen & Zhang, 2006; Lossi, Cocito, Alasia, & Merighi, 2016; Roderfeld et al., 2007). Although these assays have typically involved fluorescent proteins, in principle, the design of an appropriate, FRET-compatible heparanase substrate that can be transported to various sub-cellular locations may help in identifying spatiotemporal effects of enzymatically active heparanase.
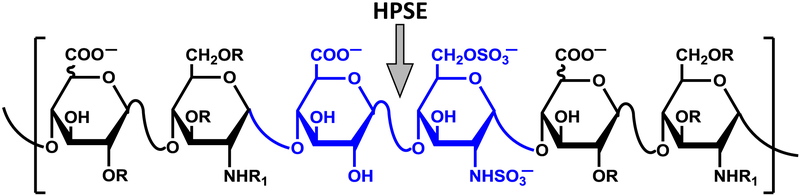
We reasoned that polymeric heparin, which is known to contain a few GlcA—GlcNS6S inter-glycosidic bonds (Figure 1), may serve as a heparanase substrate. Additionally, growing evidence indicates that polymeric heparin is internalized by cells (Eroglu, Oner, & Bostan, 2017; Maguire et al., 2014; Raman, Mencio, Desai, & Kuberan, 2013a; Wang, Wang, Xiang, & Yao, 2010; Yu et al., 2007). This implies that if heparanase is catalytically active at sub-cellular locations, a FRET-based heparin substrate that can be internalized may offer an effective probe of its spatiotemporal functions. Hence, we sought to develop a heparin analog modified with an appropriate donor-acceptor pair as a substrate of human heparanase.
Figure 1.
Generic structure of heparin (R = H or SO3−; R1 = H, COCH3 or SO3−) showing typical site (↓) cleaved by human heparanase (HPSE).
3.3. Selection of Fluorescence Labels and Labeled Heparin as Substrate
Several FRET donor-acceptor pairs are reported in the literature. We selected a few FRET pairs rhodamine 123 (λEX = 515 nm; λEM = 540 nm) and fluoresceinamine 1 (λEX = 495 nm; λEM = 515 nm) as the first FRET pair to test with regard to HPSE assay development. Unfortunately, following coupling and ascertaining its structure using 1H NMR spectroscopy, we discovered that the rhodamine–heparin–fluoresceinamine substrate failed to produce any FRET signal following HPSE treatment (data not shown). We reversed the positions of the two fluorophores, i.e., rhodamine 123 was coupled at the reducing terminus and fluoresceinamine 1 to a uronic acid so as to synthesize a fluoresceinamine–heparin–rhodamine substrate. Yet, the substrate did not yield any FRET signal (data not shown). Hence, we studied EDANS (λEX = 335 nm; λEM = 522 nm) and DABCYL C2 (λMAX = 472 nm) as a FRET pair (R0 = 33 Å). Here, DABCYL acts as a dark quencher, absorbing EDANS’ donor emission via vibrational energy, which could theoretically be more efficient when in close proximity.
This implies that it may be possible to monitor HPSE activity with even a small increase in EDANS signal. Simultaneous incubation of both fluorophores with heparin in the presence of EDC led to the formation of labeled heparin-–DE. Four heparin–DE samples, projected to carry either 1 or 2 molar ratios of the two probes per average chain of unfractionated heparin, were synthesized using varying levels of EDANS and DABCYL in the reaction mixture (Table 1). The Supplementary Figure S3 shows the schematic illustration of substrate synthesis and the FRET assay. Following SEC purification, 1H NMR spectra of the labeled products showed distinctive signals assignable to the aromatic scaffolds of the two labels (δ 8.37—7.04) and the anomeric protons (δ 5.43—5.22) of heparin (Szajek et al., 2016) (see Supplementary Figure S4). The ratio of signal integrals corresponding to these sets of protons was used to calculate the stoichiometry of each label per average chain of heparin (see Supplementary Material for additional information).
Table 1.
Maximal fluorescence enhancement following cleavage of heparin-DE with human heparanase and bacterial heparinases I, II and III.a
| Proportions used in synthesis of heparin–DEb |
Stoichiometry per heparin–DE chainc |
Maximal Fluorescence Enhancement (%)d | |||
|---|---|---|---|---|---|
| Heparanase | Hep I | Hep I, II, III | |||
| DABCYL (D) | EDANS (E) | D:Ee | |||
| 1 | 1 | 0.49:0.54 | 6.1 ± 1.2f | 18.4 ± 2.0 | 7.0 ± 1.0 |
| 2 | 2 | 0.97:1.03 | 14.5 ± 2.2 | 28 ± 2.5 | 22 ± 3.0 |
| 1 | 2 | 0.33:0.65 | 4.2 ± 1.0 | 12.4 ± 2.2 | 6.4 ± 0.5 |
| 2 | 1 | 0.60:0.37 | 12.2 ± 2.0 | 6.9 ± 1.2 | 4.9 ± 1.0 |
Labeled heparin (1 mg/ml) was incubated with either HPSE (1 μM), Hep 1 (2 IU) or Hep I, II, III (5 mU each) at 37 °C for 4 hr followed by measurement of emission at 500 nm.
Represents proportion per average chain of unfractionated heparin.
Calculated from 1H NMR spectra (see Supplementary Figure 4), as described in the main text.
Measured from emission at 500 nm.
D = DABCYL, E = EDANS
Represents standard deviation from at least 5 measurements.
Structurally, both fluorophores are attached to carboxyl groups present on uronic acid residues of heparin. The reaction conditions employed in the synthesis of heparin-DE ensure that no other group, e.g., the reducing end aldehyde, the N-acetyl, the N- & O- sulfates, the free hydroxyl groups, present on heparin can be modified except for the carboxylic acid groups. Yet, the nature of parent heparin polymer ensure that the heparin-DE produce is also heterogeneous and either probe (DABCYL or EDANS) could be placed anywhere along the chain. The key idea behind preparing this heterogeneous polymer was to assess whether heparin-DE chains can serve as substrate and whether a reasonable FRET signal could arise from its cleavage with heparanase.
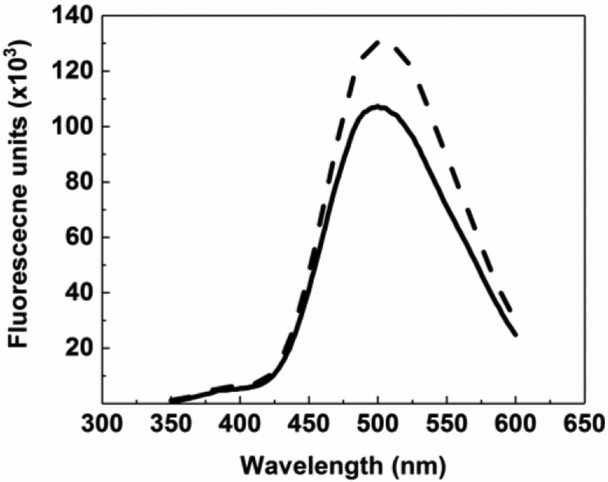
The four potential substrates were studied for their FRET signal enhancement following cleavage with heparanase in pH 5.0 buffer at 37 °C. Heparin–DE, carrying one molecule of EDANS and DABCYL, was found to display the most increase in FRET signal at 500 nm (~15%) as compared to other substrates (4 – 10%) (Figure 2). We also evaluated whether heparin–DE could serve as a substrate for bacterial heparin degrading enzymes, heparinases. Treatment of heparin– DE with heparinase I resulted in FRET enhancement of ~28%, which was similar to that observed when using a mixture of, heparinases I, II and III (~22%). This implied that heparinases degraded heparin–DE more than that observed with heparanase. This is not too surprising considering that heparanase tends to generate longer chains (Rabenstein, 2002; Vreys & David, 2007), whereas the three heparinases together tend to generate disaccharides (Silva & Dietrich, 1975).
Figure 2.
FRET-based assay for human heparanase. Heparin-DE (1 mg/ml) was incubated with HPSE (1 μM) at 37°C in 20 mM sodium acetate buffer, pH 5.0. Fluorescence emission spectra (λEX = 340 nm) at (bold line) 0 h, (dotted line) 4 h.
To assess whether the FRET signal arises from cleavage of larger chains, we utilized SEC–HPLC, which showed an increase of ~24% in the proportion of smaller oligosaccharides and a decrease of ~50% for the longer oligosaccharides (see Supplementary Figure S5). This implied that heparin–DE was cleaved by heparanase to a large extent. The results also show that many polymeric chains remain because the proportion of smaller oligosaccharides did not rise to 100%.
3.4. Optimization of Assay
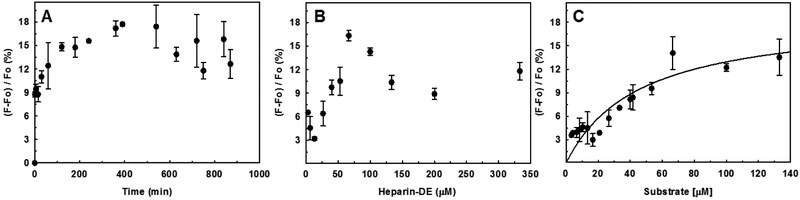
To identify optimal conditions for assaying heparanase, we measured kinetic properties of the substrate. We studied a range of heparanase levels and incubation time periods. Figures 3A and 3B show time- and heparanase- dependence of FRET signal generation at pH 5.0 and 37 °C, respectively. The results indicated that a heparanase concentration of 1 μM was necessary to produce a good FRET signal within a shorter time period (e.g., 4 hr), which is important for screening a library of potential inhibitors. It was possible to reduce the heparanase levels by 100-fold or so if the incubation time was correspondingly increased (e.g., overnight) (not shown).
Figure 3.
Optimization of heparanase assay using heparin-DE as substrate. (A) Labeled heparin (1 mg/ml) was incubated 1 μM HPSE at 37 °C for indicated times followed by measurement of emission at 500 nm. (B) Labeled heparin (1 mg/ml) was incubated with varying concentrations HPSE at 37 °C for 4 h followed by fluorescence measurement. (C) Michaelis–Menten kinetics of HPSE cleavage of heparin–DE using the optimized FRET quenching assay. The cleavage reactions were performed in microplate format (100 μL) in 20 mM sodium acetate buffer, pH 5.0, containing 1 mg/mL heparin–DE and 1 μM HPSE at 37 °C for 4 h. Solid lines represent curve fitting to the standard Michaelis equation. Error bars show variation from at least 3 measurements. F and F0 are fluorescence signals corresponding to the test sample and blank, respectively.
A quick word on the kinetics of heparin-DE cleavage by heparanase is in order. Both plots (Figures 3A and 3B) show that heparanase cleavage of heparin-DE follows standard Michaelis kinetics. As heparanase cleaves the heparin-DE, product forms in linear manner until the substrate starts to get exhausted and product formation plateaus. Likewise, the formation of product at a fixed time point (4 h) as a function of heparin-DE concentration increases in a linear manner, until the point where the enzyme is saturated with the substrate. Interestingly, heparanase–heparin-DE reaction shows a decrease in FRET signal, which could arise from photobleaching, inner filter effect and/or substrate inhibition of heparin.
The optimized assay conditions were used to measure the kinetic profile of heparanase cleavage of heparin–DE (Figure 3C). The KM calculated from this assay was 46±12 μM, which implies a rather weak overall affinity of heparin–DE for the enzyme. Although most studies reported in the literature do not disclose KM of substrates, fondaparinux, which is a synthetic, homogeneous pentasaccharide, displays a comparable affinity (46 ± 14 μM) (Hammond et al., 2010). A final part of this study was to assess repeatability. We performed these studies multiple times with different batches of heparin-DE and found identical FRET signal and Michaelis parameters (Vmax and KM).
1.5. Application of the Assay for Inhibition Studies
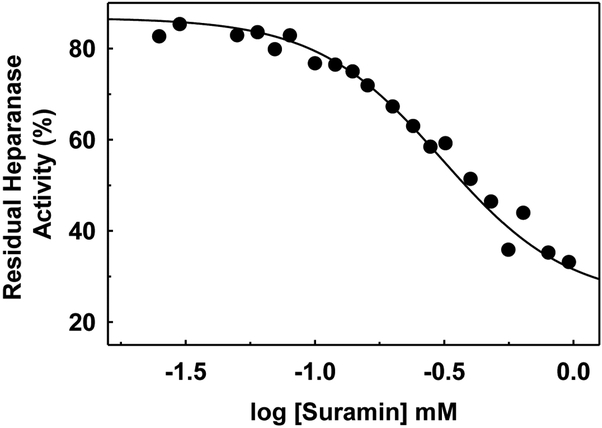
The FRET assay was utilized to measure the potency of suramin inhibition of heparanase. Suramin is a known inhibitor of human heparanase. The assay was implemented in a 96-well microplate format at 37 °C with vigorous stirring for 4 h and direct measurement of FRET signal at 500 nm using a fluorescence microplate reader. Suramin concentration was varied over nearly 3-orders of magnitude. Over several independent measurements, the FRET signal was found to offer sufficiently sensitive measurement of the activity of heparanase (Figure 4). The IC50 calculated for suramin was 330±2.5 μM, which was similar to IC50s of 224 and 332 μM measured using ovarian cancer cells (HO-8910PM) and cervical cancer cells (HeLa), respectively, in cell-based assays (Li et al., 2015).
Figure 4.
Suramin inhibition of HPSE using the one-step FRET quenching assay. The experiments were performed in microplate format (100 μL) in 20 mM sodium acetate buffer, pH 5.0, containing 1 mg/mL heparin–DE and 1 μM HPSE at 37 °C for 4 h in the presence of varying concentrations of suramin. Solid lines represent curve fitting to standard dose-response equation.
We also measured the IC50 of suramin inhibition using Cisbio heparanase kit, a commercially available product. It is a two-step assay and utilizes HS-labeled substrate containing biotin and Eu3+ cryptate. Active heparanase enzyme cleaves the substrate resulting in a loss of energy transfer, which is detected by addition of streptavidin-XL665. An IC50 of 170±9.1 μM was calculated for suramin inhibition of human heparanase (Supplementary Figure S6).
3.6. Application of the Assay to Detect Heparanase Activity in Biological Samples
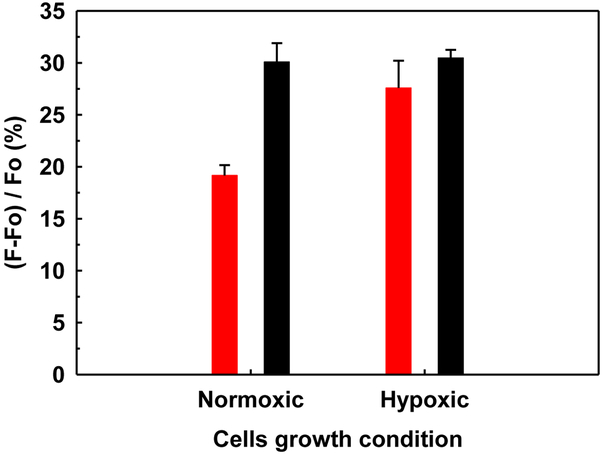
A majority of cancer cells including breast cancer cells secrete active heparanase, which plays a critical role in tumor development, metastasis and neo-angiogenesis (Parish, Freeman, & Hulett, 2001; Zcharia et al., 2001). Hence, we conducted additional studies to determine the usefulness of our new FRET assay in monitoring the activity of the heparanase secreted by the cancer cells. Growth and/or aggressive behavior, e.g., invasion and metastasis of the cancer cells, is dictated by access to nutrients and oxygen. In fact, cancer cells are exposed to different oxygen pressure and nutrients depending on the location within the tumor. To mimic conditions experienced by cancer cells in vivo, human breast cancer cell line MCF7 was grown in both normoxic (20% O2) and hypoxic (5% O2) conditions in the presence of varying amount of fetal bovine serum (0%, 2%, and 10%). For cells grown in fetal bovine serum (FBS)-free media, heparanase activity was found to be nearly 10% higher under hypoxic conditions as compared to normoxic conditions (Figure 5). In contrast, cells grown in 2% FBS did not show much difference in activity between hypoxic and normoxic conditions. Interestingly, high serum levels (10% FBS) suppressed heparanase activity for normoxic and hypoxic conditions (data not shown). These results are on par with an earlier report demonstrating increased levels of heparanase in MCF7 cells grown under hypoxia and FBS-free conditions (Poupard et al., 2017).
Figure 5.
Detection of heparanase activity in the monolayer MCF7 cells expression medium of under normoxic and hypoxic conditions. The cells were cultured for 48 h in FBS-free (red bars) and 2% FBS (black bars). Results are presented as the mean±SD from at least three independent measurements.
3.7. Application to Identify Optimal Conditions of Heparanase Expression
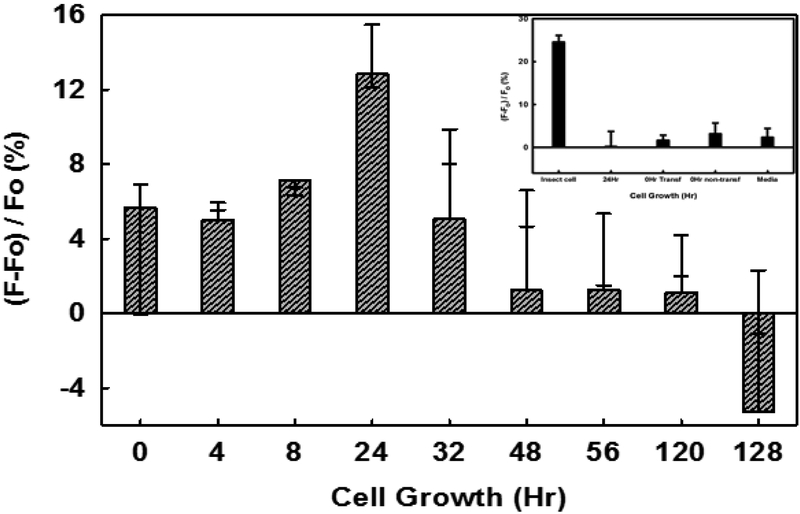
For assessing expression of native, wild-type heparanase in different cell lines, we utilized MCF7 and HEK 293T cell lines, which are representative of cancerous and non-cancerous cell lines, respectively. It is widely known that heparanase expression is affected in cancerous cells as compared to non-cancerous cells. We followed HPSE expression as a function of time using FRET assay (Figure 6). MCF7 cells showed a gradual increase in the heparanase activity from 0 to 24 h, which was followed by a rapid decrease in activity. In contrast, HPSE activity measured in HEK 293T cell culture media was minimal. The results with our FRET-based assay were also duplicated using the Cisbio kit (data not shown).
Figure 6.
Expression of active heparanase by MCF7 cells under normoxic conditions. Inset shows the HEK cells expressed HPSE activity measurement. Results are presented as the mean±SD (n>3).
4. Conclusions and Significance
The major conclusion of this work is that full-length heparin, when appropriately modified with donor and acceptor probes, can serve as a heparanase substrate and help quantify heparanase activity in one step. This is especially useful because heparanase assays reported to date, including the commercially available kits, are multi-step processes that involve expensive reagents, long incubation times, and are limited in terms of applicability. Heparin is a readily available chemical that can be modified with two common probes in a single step to generate heparin-DE, the substrate studied in this work, in good yields and reproducibility. Assay conditions developed in this work (37 °C, 4 h, pH 5.0, 1 μM HPSE, 1 mg/mL heparin-DE, microplate format) were directed to enable screening of a library of potential inhibitors but could be also adapted for rapid and/or continuous assay of cells for heparanase production purposes. A key potential advantage of this assay will be to assess whether intracellular heparanase is active because heparin chains are known to be internalized (Raman, Mencio, Desai, & Kuberan, 2013b; Zheng et al., 2017). Although maturation of heparanase occurs extracellularly, recent studies indicate that intracellular heparanase may exhibit catalytic activity (Leiser, Shilo, El Naaj, & Rachmiel, 2014; Yang et al., 2015). Thus, a cell penetrating heparanase substrate would be particularly useful for continuous monitoring of intracellular heparanase enzymatic activity.
Despite the advantages of this assay, it is important to note some limitations. Heparin-DE is a heterogeneous substrate. This means that if a commercially relevant product is to be produced, every batch of heparin-DE will have to be monitored with sophisticated array of tests. In our work, we prepared several batches of heparin-DE and found equivalent results in terms of FRET signal and label proportions in heparin-DE. This suggests excellent repeatability and offers confidence in further development of such as heterogeneous agent as a probe of heparanase. However, it would also be advisable to develop a heparin–DE chain that is homogeneous. In addition to ease of monitoring batch-to-batch purity, a homogeneous heparin-DE chain could perhaps also offer higher FRET signal. The design of such a heparin substrate is in progress.
Supplementary Material
Highlights.
Development of polymeric heparin as a FRET substrate of heparanase
The FRET substrate enables rapid, one-step screening of heparanase inhibitors
The FRET substrate enables monitoring of heparanase activity in biological media
Acknowledgments
We thank Prof. Davies (York University, UK) for the gift of heparanase cDNA and Dr. Akul Mehta, (Harvard Medical School, Boston) for help with initial heparin labeling experiments. This work was supported by grants from NIH HL107152 and HL128639 to URD.
Abbreviations
- DABCYL
4'-dimethylaminoazobenzene-4-carboxylate C2 amine
- EDANS
5-[(2-aminoethyl)amino]naphthalene-1-sulfonic acid
- EDC
1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide
- FRET
Fluorescence resonance energy transfer
- GAG
Glycosaminoglycan
- HS
heparan sulfate
- DMF
Dimethylformamide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen MD, & Zhang J (2006). Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochemical and Biophysical Research Communications, 348(2), 716–721. https://doi.org/10.1016/j.bbrc.2006.07.136 [DOI] [PubMed] [Google Scholar]
- Behzad F, & Brenchley PEC (2003). A multiwell format assay for heparanase. Analytical Biochemistry, 320(2), 207–213. https://doi.org/10.1016/S0003-2697(03)00358-0 [DOI] [PubMed] [Google Scholar]
- Bhushan I, Alabbas A, Kuberan B, Gupta RB, & Desai UR (2017). Immobilization alters heparin cleaving properties of heparinase I. Glycobiology, (October), 1–5. https://doi.org/10.1093/glycob/cwx074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, & Sanderson RD (2009). Heparanase regulates levels of syndecan-1 in the nucleus. PLoS ONE, 4(3), 1–6. https://doi.org/10.1371/journal.pone.0004947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G, & Zimmermann P (2016). Heparanase tailors syndecan for exosome production. Molecular & Cellular Oncology, 3(3), el047556 https://doi.org/10.1080/23723556.2015.1047556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey LA, Brunn GJ, & Platt JL (2000). Heparanase, a potential regulator of cell–matrix interactions. Trends in Biochemical Sciences, 25(8), 349–351. https://doi.org/10.1016/S0968-0004(00)01619-4 [DOI] [PubMed] [Google Scholar]
- Enomoto K, Okamoto H, Numata Y, & Takemoto H (2006). A simple and rapid assay for heparanase activity using homogeneous time-resolved fluorescence. Journal of Pharmaceutical and Biomedical Analysis, 41(3), 912–917. https://doi.org/10.1016/j.jpba.2006.01.032 [DOI] [PubMed] [Google Scholar]
- Eroglu MS, Oner ET, & Bostan ECM and M. S. (2017). Sugar Based Biopolymers in Nanomedicine; New Emerging Era for Cancer Imaging and Therapy. Current Topics in Medicinal Chemistry. https://doi.org/http://dx.doi.org/10.2174/1568026616666161222101703 [DOI] [PubMed] [Google Scholar]
- Fairbanks MB, Mildner AM, Leone JW, Cavey GS, Mathews WR, Drong RF, et al. (1999). Processing of the human heparanase precursor and evidence that the active enzyme is a heterodimer. Journal of Biological Chemistry, 274(42), 29587–29590. https://doi.org/10.1074/jbc.274.42.29587 [DOI] [PubMed] [Google Scholar]
- Freeman C, & Parish CR (1997). A rapid quantitative assay for the detection of mammalian heparanase activity. The Biochemical Journal, 325 (Pt 1, 229–37. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1218550&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R, Meirovitz A, Hirshoren N, Bulvik R, Binder A, Rubinstein AM, & Elkin M (2013). Versatile role of heparanase in inflammation. Matrix Biology, 32(5), 234–240. https://doi.org/10.1016/j.matbio.2013.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond E, Li CP, & Ferro V (2010). Development of a colorimetric assay for heparanase activity suitable for kinetic analysis and inhibitor screening. Analytical Biochemistry, 396(1), 112–116. https://doi.org/10.1016/j.ab.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Huang K. Sen, Holmgren J, Reik L, Lucas-McGady D, Roberts J, Liu CM, & Levin W (2004). High-throughput methods for measuring heparanase activity and screening potential antimetastatic and anti-inflammatory agents. Analytical Biochemistry, 333(2), 389–398. https://doi.org/10.1016/j.ab.2004.06.023 [DOI] [PubMed] [Google Scholar]
- Kussie PH, Hulmes JD, Ludwig DL, Patel S, Navarro EC, Seddon AP, et al. (1999). Cloning and functional expression of a human heparanase gene. Biochemical and Biophysical Research Communications 261(1), 183–187. [DOI] [PubMed] [Google Scholar]
- Leiser Y, Shilo D, El Naaj IA, & Rachmiel A (2014). Heparanase, a Potential Marker for Premalignant Oral Cavity Cancer. In Vivo, 25(5), 769–777. Retrieved from http://iv.iiarjournals.org/content/28/5/769.abstract [PubMed] [Google Scholar]
- Li H, Li H, Qu H, Zhao M, Yuan B, Cao M, & Cui J (2015). Suramin inhibits cell proliferation in ovarian and cervical cancer by downregulating heparanase expression. Cancer Cell International, 75(1), 52 https://doi.org/10.1186/sl2935-015-0196-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossi L, Cocito C, Alasia S, & Merighi A (2016). Ex vivo imaging of active caspase 3 by a FRET-based molecular probe demonstrates the cellular dynamics and localization of the protease in cerebellar granule cells and its regulation by the apoptosis-inhibiting protein survivin. Molecular Neurodegeneration, 11(1), 34 https://doi.org/10.1186/s13024-016-0101-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire CM, Mahfoud OK, Rakovich T, Gerard VA, Prina-Mello A, Gun’ko Y, & Volkov Y (2014). Heparin conjugated quantum dots for in vitro imaging applications. Nanomedicine: Nanotechnology, Biology, and Medicine, 10(8), 1853–1861. https://doi.org/10.1016/j.nano.2014.04.009 [DOI] [PubMed] [Google Scholar]
- McKenzie E, Young K, Hircock M, Bennett J, Bhaman M, Felix R, et al. (2003). Biochemical characterization of the active heterodimer form of human heparanase (Hpa1) protein expressed in insect cells. The Biochemical Journal, 373, 423–435. https://doi.org/10.1042/BJ20030318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo CM, Tersariol ILS, Nader HB, Pinhal MAS, & Lima MA (2015). Development of new methods for determining the heparanase enzymatic activity. Carbohydrate Research, 412, 66–70. https://doi.org/10.1016/j.carres.2015.04.020 [DOI] [PubMed] [Google Scholar]
- Nardella C, & Steinkühler C (2004). Radiolabeled heparan sulfate immobilized on microplate as substrate for the detection of heparanase activity. Analytical Biochemistry, 332(2), 368–375. https://doi.org/10.1016/j.ab.2004.05.050 [DOI] [PubMed] [Google Scholar]
- Parish CR, Freeman C, & Hulett MD (2001). Heparanase: a key enzyme involved in cell invasion. Biochimica et Biophysica Acta, 1471(3), M99–108. https://doi.org/10.1016/S0304-419X(01)00017-8 [DOI] [PubMed] [Google Scholar]
- Pearson AG, Kiefel MJ, Ferro V, & von Itzstein M (2011). Synthesis of simple heparanase substrates. Organic & Biomolecular Chemistry, 9(12), 4614 https://doi.org/10.1039/clob05165b [DOI] [PubMed] [Google Scholar]
- Peterson SB, & Liu J (2013). Multi-faceted substrate specificity of heparanase. Matrix Biology, 32(5), 223–227. https://doi.Org/10.1016/j.matbio.2013.02.006 [DOI] [PubMed] [Google Scholar]
- Poupard N, Badarou P, Fasani F, Groult FL, Bridiau N, Sannier F, et al. , (2017). Assessment of heparanase-mediated angiogenesis using microvascular endothelial cells: Identification of λ-Carrageenan derivative as a potent anti angiogenic agent. Marine Drugs, 15(5), 1–18. https://doi.org/10.3390/mdl5050134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabenstein DL (2002). Heparin and heparan sulfate: structure and function. Natural Product Reports, 19(3), 312–331. https://doi.org/10.1039/bl00916h [DOI] [PubMed] [Google Scholar]
- Raman K, Mencio C, Desai UR, & Kuberan B (2013a). Sulfation patterns determine cellular internalization of heparin-like polysaccharides. Molecular Pharmaceutics, 10(4), 1442–1449. https://doi.org/10.1021/mp300679a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K, Mencio C, Desai UR, & Kuberan B (2013b, April). Sulfation patterns determine cellular internalization of heparin-like polysaccharides. Molecular Pharmaceutics. https://doi.org/10.1021/mp300679a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderfeld M, Graf J, Giese B, Salguero-Palacios R, Tschuschner A, Müller-Newen G, & Roeb E (2007). Latent MMP-9 is bound to TIMP-1 before secretion. Biological Chemistry, 388(11), 1227–1234. https://doi.org/10.1515/BC.2007.123 [DOI] [PubMed] [Google Scholar]
- Sanderson RD, Elkin M, Rapraeger AC, Ilan N, & Vlodavsky I (2017). Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS Journal, 284(1), 42–55. https://doi.org/10.llll/febs.13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T. C. F. dos, Gomes AM, Paschoal MEM, Stelling MP, Rumjanek VMBD, Junior A. do R. et al. , (2014). Heparanase expression and localization in different types of human lung cancer. Biochimica et Biophysica Acta (BBA) - General Subjects, 1840(8), 2599–2608. https://doi.org/10.1016/j.bbagen.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Silva ME, & Dietrich CP (1975). Structure of heparin. The Journal of Biological Chemistry, 250(17), 6841–6846. [PubMed] [Google Scholar]
- Szajek AY, Chess E, Johansen K, Gratzl G, Gray E, Keire D et al. , (2016). The US regulatory and pharmacopeia response to the global heparin contamination crisis. Nature Biotechnology, 34(6), 625–630. https://doi.org/10.1038/nbt.3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, & Nakajima M (1999). Human heparanase. Purification, characterization, cloning, and expression. The Journal of Biological Chemistry, 274(34), 24153–60. https://doi.org/10.1074/jbc.274.34.24153 [DOI] [PubMed] [Google Scholar]
- Tsuchida S, Podyma-Inoue KA, & Yanagishita M (2004). Ultrafiltration-based assay for heparanase activity. Analytical Biochemistry, 331(1), 147–152. https://doi.org/10.1016/j.ab.2004.04.033 [DOI] [PubMed] [Google Scholar]
- Vreys V, & David G (2007). Mammalian heparanase: What is the message? Journal of Cellular and Molecular Medicine, 11(3), 427–452. https://doi.org/10.1111/j.1582-4934.2007.00039.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Xiang J, & Yao K (2010). Target-Specific Cellular Uptake of Taxol-Loaded Heparin-PEG-Folate Nanoparticles, Biomacromolecules, 11(12), 3531–3538. doi: 10.1021/bml01013s [DOI] [PubMed] [Google Scholar]
- Wu L, Viola CM, Brzozowski AM, & Davies GJ (2015a). Structural characterization of human heparanase reveals insights into substrate recognition. Nature Structural & Molecular Biology, 22(12), 1016–1022. https://doi.org/10.1038/nsmb.3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Viola CM, Brzozowski AM, & Davies GJ (2015b). Structural characterization of human heparanase reveals insights into substrate recognition. Nature Structural & Molecular Biology, 22(12), 1016–1022. https://doi.org/10.1038/nsmb.3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gorzelanny C, Bauer AT, Halter N, Komljenovic D, Bäuerle T et al. , (2015). Nuclear heparanase-1 activity suppresses melanoma progression via its DNA-binding affinity. Oncogene, 34(47), 5832–5842. https://doi.org/10.1038/onc.2015.40 [DOI] [PubMed] [Google Scholar]
- Yu MK, Lee DY, Kim YS, Park K, Park SA, Son DH, et al. , (2007). Antiangiogenic and apoptotic properties of a novel amphiphilic folate-heparin-lithocholate derivative having cellular internality for cancer therapy. Pharmaceutical Research, 24(4), 705–714. https://doi.org/10.1007/sll095-006-9190-3 [DOI] [PubMed] [Google Scholar]
- Zcharia E, Metzger S, Chajek-Shaul T, Friedmann Y, Pappo O, Aviv A, et al. ,. (2001). Molecular properties and involvement of heparanase in cancer progression and mammary gland morphogenesis. Journal of Mammary Gland Biology and Neoplasia, 6(3), 311–322. https://doi.org/10.1023/A:1011375624902 [DOI] [PubMed] [Google Scholar]
- Zheng S, Kummarapurugu AB, Afosah DK, Sankaranarayanan NV, Boothello RS, Desai UR, et al. , (2017). 2-O, 3-O Desulfated Heparin Blocks High Mobility Group Box 1 Release by Inhibition of p300 Acetyltransferase Activity. American Journal of Respiratory Cell and Molecular Biology, 56(1), 90–98. https://doi.org/10.1165/rcmb.2016-00690C [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.