Abstract
Background
Antitumor immune response of programmed cell death ligand (PD-L1) has shown clinical value not only in Hodgkin lymphoma and EBV-associated lymphomas but also in EBV-negative diffuse large B cell lymphoma (DLBCL) of non-germinal center B cell-like (non-GCB) subtype. Signal transducer and activator of transcription 3 (STAT3) is known to induce PD-L1 in immune cells and its activated form, phosphorylated STAT3 (pSTAT3), is also frequently expressed in non-GCB DLBCL. Herein, we investigated associations between PD-L1 expression/gene alteration, pSTAT3 expression and clinicopathologic variables in EBV-negative DLBCL.
Methods
In 107 cases of DLBCLs with non-GCB subtype (67%; 72/107), GCB subtype (25%; 27/107) and unclassifiable cases (8%; 8/107), we performed PD-L1 and pSTAT3 immunohistochemistry and fluorescence in situ hybridization for PD-L1 gene translocation and copy number gain/amplification.
Results
PD-L1 was expressed in tumor cells (PD-L1t) in 21% (23/107; 30% cutoff), immune cells (PD-L1i) in 36% (38/107; 20% cutoff), and pSTAT3 in tumor nuclei in 41% (44/107; 40% cutoff). PD-L1 gene alteration was observed in 10% (10/102) including translocation in 6% (6/102) and copy number gain/amplification in 4% (4/102). Non-GCB subtype was associated with PD-L1t and pSTAT3 (p = 0.006 and p = 0.042), and tended to have PD-L1 gene alteration (p = 0.058). Tumoral PD-L1 expression without gene alteration (PD-L1t+ GA−) correlated with pSTAT3-positive tumor cell proportions (%) (p = 0.033). In survival analysis, pSTAT3 expression independently predicted shorter PFS in total cohort (p = 0.017) and R-CHOP-treated group (p = 0.007), and in pSTAT3-negative R-CHOP-treated subset, PD-L1 expression in immune cells (PD-L1i) correlated with shorter PFS (p = 0.042).
Conclusions
Gene alteration and protein expression of PD-L1 and pSTAT3 expression were closely related in DLBCL and constituted features of non-GCB subtype. In addition to known clinical significance of pSTAT3, immune cell expression of PD-L1 (PD-L1i) had also clinical value in pSTAT3-dependent manner. These findings may provide an insight into immunotherapeutic strategy and risk stratification in DLBCL patients.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1689-y) contains supplementary material, which is available to authorized users.
Keywords: Diffuse large B cell lymphoma, PD-L1, pSTAT3, Microenvironment, Prognosis
Background
Immune escape and immune suppression has emerged as important mechanisms for the development of solid and hematologic malignancies. Research on immunotherapy has thus opened a new era in the treatment of many malignancies. Recently, impressive advancements in the treatment of hematologic malignancies with the immune checkpoint inhibitors targeting programmed cell death-1 (PD-1) receptor and its ligand (PD-L1) lead to the approval of nivolumab and pembrolizumab by the Food and Drug Administration for the treatment of classical Hodgkin lymphoma in relapsed or refractory cases.
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma in adults. The 5-year overall survival reaches only 60–70% when treated with standard chemotherapy of rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP) [1]. DLBCL is known to be heterogeneous in genetic and prognostic aspects. The subclassification of DLBCL based on cell-of-origin using the Hans algorithm proved useful in predicting prognosis as germinal center B-cell like (GCB) subtypes have better outcomes than non-GCB subtypes [2, 3].
PD-L1 research on DLBCL has found that not only tumor B-cells but also non-malignant immune cells in the microenvironment express PD-L1 [4–7]. In addition, many studies have reported that non-GCB DLBCL shows higher PD-L1 expression than GCB DLBCL [5, 8, 9], and that similar to Hodgkin lymphoma, Epstein–Barr virus (EBV) infection probably plays an important role in inducing PD-L1 expression in DLBCL [4, 10, 11].
The mechanisms of PD-L1 expression in DLBCL has not been elucidated clearly. In many subtypes of lymphoma, including classical Hodgkin lymphoma, primary mediastinal B-cell lymphoma and primary central nervous system lymphoma, genetic alterations such as copy-number alterations, amplification, and translocation were observed on the 24.1 locus of the short arm of chromosome 9. On this locus lie the genes encoding PD-L1, PD-L2 and JAK2, resulting in overexpression of PD-L1 when such genetic alterations occur [7, 11, 12]. In DLBCL, however, only a subgroup of less than 20% has these genetic changes, most of which are primary mediastinal large B cell lymphoma [8, 13]. Thus, other mechanisms are currently being investigated.
Infection by EBV upregulates PD-L1 expression in tumor cells either directly by binding to latent membrane protein (LMP-1) that triggers a signaling cascade involving JUN-B or indirectly by activating inflammatory cytokines [14]. Inflammatory cytokines, represented by IFN-γ and IL-10, in turn induce PD-L1 expression via the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway in Hodgkin and non-Hodgkin lymphoma. Other mechanisms also involve the JAK/STAT signaling pathway such as inhibition of Suppressor of cytokine signaling-1 (SOCS-1) or microRNA miR-135a [7].
Phosphorylated STAT3 (pSTAT3), a transcription factor, is a key player of the JAK/STAT pathway that regulates cell proliferation and survival [15]. Cytoplasmic protein STAT3 moves into the nucleus and binds to the promoter of PD-L1 gene when phosphorylated by JAK to enhance transcription [16, 17]. This process can be stimulated by LMP-1 of EBV, resulting in concomitant overexpression of pSTAT3 with increased PD-L1 expression in nasopharyngeal carcinoma [14]. Reed-Sternberg cells stain positive for both PD-L1 and pSTAT3, suggesting an association between these two markers in lymphoma [18]. A few reports discovered association of PD-L1 and pSTAT3 expression in natural killer/T cell lymphoma [17, 19], ALK-negative anaplastic large cell lymphoma [20] and cell lines of adult T cell leukemia/lymphoma [21]. In DLBCL, pSTAT3 is overexpressed, similar to PD-L1, in non-GCB subtype and shows higher expression than GCB subtypes [22–28]. The association between PD-L1 and pSTAT3 in DLBCL, however, has not yet been explored.
In this study, we evaluated the protein expression of PD-L1 in terms of its potential intrinsic and extrinsic mechanism, i.e., PD-L1 gene alteration and STAT3 activation, in EBV-negative DLBCLs. We also investigated the clinical significance of PD-L1/pSTAT3-related biomarkers in association with clinicopathologic parameters and prognosis in DLBCL patients.
Methods
Patients and samples
Our cohort consisted of 107 patients newly diagnosed with de novo DLBCL, not otherwise specified (NOS) between May 2005 and January 2013 at Seoul National University Bundang Hospital. Clinicopathological information was obtained from clinical records and pathology reports. Large B cell lymphomas with distinct entities including ‘EBV-positive DLBCL, NOS’, ‘DLBCL of the central nervous system’, ‘primary mediastinal large B cell lymphoma’ as well as transformed DLBCL from low grade lymphomas were excluded. The pathologic classification was based on the 2016 Revised 4th edition of World Health Organization classification [29] and further subtyped by Hans algorithm [2] by experienced hematopathologists. The study protocol was approved by the Institutional Review Board of Seoul National University Bundang Hospital.
Construction of tissue microarray (TMA)
Hematoxylin and eosin (H & E)-stained slides were reviewed in each case to confirm the original diagnosis and select the most representative sections. A tissue microarray (TMA) was constructed using 2 mm-diameter cores derived from the representative areas of the formalin-fixed paraffin-embedded tissue blocks from each case and from normal tonsils for controls by SuperBioChips Laboratories (Seoul, Korea), as previously described [30].
Immunohistochemical staining
TMAs were sectioned at a thickness of 4-μm and stained with Benchmark XT and Benchmark ULTRA (Roche Diagnostics) along with normal tonsil specimens as controls for the following antibodies (clones): anti-Bcl-2 (std32, 1:50, Dako), anti-Bcl-6 (std32, ready-to-use, Ventana), anti-CD10 (std32, ready-to-use, Ventana), anti-Mum-1 (mild32, 1:150, Dako), anti-PD-L1 (E1L3N, 1:100, Cell Signaling) and anti-pSTAT3 (#9131, 1:20, Cell Signaling). Using immunochemistry results, the Hans algorithm was applied to each case as described previously [31].
Evaluation of PD-L1 and pSTAT3 immunohistochemistry
The immunohistochemistry slides were scored by two pathologists (H.J.K and J.H.P). PD-L1 was scored both in tumor B-cells and in non-malignant immune cells. PD-L1 was considered positively expressed in tumor or non-malignant immune cells if membranous staining alone or membranous and cytoplasmic staining together was present. The percentage of stained tumor B-cells and non-malignant immune cells were estimated in each TMA core regardless of intensity. Tumor cells were distinguished from non-malignant immune cells by histologic clues such as nuclear enlargement and atypism, followed by comprehensive interpretation with other immunohistochemical markers including CD20, BCL2, BCL6, CD10 and MUM1. Since an optimal cut-off could not be determined by receiver operating characteristic (ROC) curve analysis, cases were classified by a 30% cutoff for tumor B-cells and 20% cutoff for non-malignant immune cells according to previous studies [6]. pSTAT3 expression was scored in tumor B-cells alone with cutoff value of 40%, which was set based on ROC curve analysis. In addition, the proportion (%) of pSTAT3-positive cells was digitally counted by using digital slide scanner and image analyzer (3DHISTECH, Budapest, Hungary) for correlation analysis.
Fluorescence in situ hybridization for PD-L1 gene
For fluorescence in situ hybridization (FISH) staining, PD-L1 break-apart probe (9p24.1) (catalog # PDL1BA-20-ORGR) and PD-L1 (orange)/chromosome 9 (green) probe set (9p24.1/9p21.33) (catalog # PDL1-CHR09-20-ORGR) were purchased from empire genomics (Buffalo, NY). The FISH staining process was performed as previously described [32]. Briefly, after deparaffinization and dehydration, slides were immersed in 2 M HCl, boiled using a microwave oven in citrate buffer (pH 6.0), incubated in 1 M NaSCN for 40 min at 80 °C, immersed in a protease buffer and fixed in 10% neutral buffered formalin. After applying the DNA probe set to the slides, they were incubated in a humidified chamber at 83 °C for 3 min to denature the target DNA and probe, and subsequently incubated overnight at 37 °C to achieve hybridization. Following post-hybridization washing, 4,6-diamidino-2-phenylindole (DAPI) and an anti-fade compound (p-phenylenediamine) were applied to the slides as a counter-stain. An Olympus BX51TRF microscope (Olympus Corp., Tokyo, Japan) equipped with the appropriate filter sets was used to analyze the stained cells. For the interpretation of translocation and/or copy number gain/amplification, more than 200 tumor cells in non-overlapping intact nuclei were counted [17, 32]. Separation of orange and green signals in more than 15% of tumor cells were interpreted as translocation [31]. Copy number gain and amplification were defined as PD-L1 gene/chromosome 9 ratio > 2 and > 4, respectively.
Statistical analysis
Statistical analysis was carried out using SPSS Statistics ver. 19 (IBM, Chicago, IL). Chi square or Fisher’s exact test was used to compare clinicopathological characteristics. Pearson’s correlation test was performed to determine the association of immunohistochemical expressions. Progression-free survival (PFS) was defined as the time interval from the start date of treatment to the date of progression including radiologically confirmed progressive disease based on positron-emission tomography (PET)-computed tomography (CT) or CT, refractoriness to the first-line therapy in response evaluation after second cycle or completion of treatment, relapse or death. Overall survival (OS) was defined as the time interval from the date of diagnosis to the date of last follow-up or death. The Kaplan–Meier analysis was used to construct survival curves, with which the log-rank test compared the difference. A multivariate analysis by Cox proportional hazards regression modeling was performed. All p values reported are two-sided and statistical significance was accepted with those less than 0.05.
Results
Clinicopathologic characteristics
The characteristics of 107 patients with DLBCL are summarized on Table 1. Briefly, our cohort consisted predominantly of non-GCB subtype (67%; 72/107) compared to GCB subtype (25%; 27/107) or unclassifiable cases (8%; 8/107) by Hans algorithm. The total cohort mainly included cases with good Eastern Cooperative Oncology Group performance status (ECOG PS) (< 2; 91%; 97/107), absence of B symptoms (79%; 85/107), low international prognostic index (IPI; 66%; 71/107), less than 2 extranodal site involvements (76%; 81/107), absence of bone marrow involvement (79%; 84/107) and non-bulky masses (92%; 98/107). Most of the patients received R-CHOP chemotherapy (87%; 93/107). Compared to the patients with GCB subtype, the patients with non-GCB subtype frequently had elevated serum lactate dehydrogenase levels (p = 0.040). Non-GCB subtype also tended to be associated with a high IPI score (3–5; p = 0.056) and presence of B symptoms (p = 0.088), which did not reach statistical significance.
Table 1.
Clinicopathologic characteristics according to Hans classification in diffuse large B cell lymphoma patients
| Clinicopathologic characteristics | Total | GCB | Non-GCB | p-value |
|---|---|---|---|---|
| Age | ||||
| ≤ 60 | 53 (50%) | 17 (63%) | 31 (43%) | 0.078# |
| > 60 | 54 (50%) | 10 (37%) | 41 (57%) | |
| Sex | ||||
| Male | 61 (57%) | 7 (26%) | 34 (47%) | 0.055# |
| Female | 46 (43%) | 20 (74%) | 38 (53%) | |
| Primary site | ||||
| Nodal | 54 (50%) | 23 (85%) | 51 (71%) | 0.196† |
| Extranodal | 53 (49%) | 4 (15%) | 21 (29%) | |
| B symptoms | ||||
| Absent | 85 (79%) | 25 (93%) | 55 (76%) | 0.088† |
| Present | 22 (21%) | 2 (7%) | 17 (24%) | |
| ECOG PS | ||||
| < 2 | 97 (91%) | 26 (96%) | 64 (89%) | 0.437† |
| ≥ 2 | 10 (9%) | 1 (4%) | 8 (11%) | |
| Serum lactate dehydrogenase | ||||
| Normal | 54 (50%) | 18 (69%) | 32 (46%) | 0.040# |
| Elevated | 50 (47%) | 8 (31%) | 38 (54%) | |
| Unknown | 3 (3%) | |||
| No. of extranodal sites | ||||
| < 2 | 81 (76%) | 17 (63%) | 33 (46%) | 0.129# |
| ≥ 2 | 26 (24%) | 10 (37%) | 39 (54%) | |
| Ann Arbor stage | ||||
| I–II | 56 (52%) | 17 (63%) | 35 (49%) | 0.203† |
| III–IV | 51 (48%) | 10 (37%) | 37 (51%) | |
| International prognostic index | ||||
| 0–2 | 71 (66%) | 22 (81%) | 44 (61%) | 0.056# |
| 3–5 | 36 (34%) | 5 (19%) | 28 (39%) | |
| Bone marrow involvement | ||||
| Absent | 84 (79%) | 25 (96%) | 55 (85%) | 0.168† |
| Present | 14 (13%) | 1 (4%) | 10 (15%) | |
| Unknown | 9 (8%) | |||
| Bulky mass (cm) | ||||
| < 10 | 98 (92%) | 25 (93%) | 66 (92%) | > 0.999† |
| ≥10 | 9 (8%) | 2 (7%) | 6 (8%) | |
| Hans classification | ||||
| GCB | 27 (25%) | 27 (100%) | 0 (0%) | NA |
| Non-GCB | 72 (67%) | 0 (0%) | 72 (100%) | |
| Unclassifiable | 8 (8%) | |||
| BCL2 expression | ||||
| Negative | 42 (39%) | 13 (50%) | 24 (33%) | 0.133# |
| Positive | 65 (61%) | 13 (50%) | 48 (67%) | |
| BCL6 expression | ||||
| Negative | 60 (56%) | 8 (30%) | 44 (61%) | 0.005# |
| Positive | 47 (44%) | 19 (70%) | 28 (39%) | |
| CD10 expression | ||||
| Negative | 92 (86%) | 12 (44%) | 72 (100%) | < 0.001† |
| Positive | 15 (14%) | 15 (56%) | 0 (0%) | |
| MUM1 expression | ||||
| Negative | 41 (38%) | 25 (93%) | 8 (11%) | < 0.001† |
| Positive | 66 (62%) | 2 (7%) | 64 (89%) | |
| Treatment regimen | ||||
| R-CHOP | 93 (87%) | 23 (85%) | 64 (89%) | 0.731† |
| Others | 14 (13%) | 4 (15%) | 8 (11%) | |
| Total N (%) | 107 (100%) | |||
NA: not applicable; ECOG PS: the Eastern Cooperative Group Performance Status; GCB: germinal center B-cell like; R-CHOP: rituximab with cyclophosphamide–doxorubicin–vincristine–prednisone, GI: gastrointestinal
p values were calculated by using Fisher’s exact test (2-sided)† or Pearson’s Chi square test (2-sided)#
Association between clinicopathologic variables and protein expression and gene alteration of PD-L1 and pSTAT3 expression
We investigated the frequencies of PD-L1 expression, PD-L1 gene alteration, i.e., translocation, copy number gain and/or amplification, and pSTAT3 expression in tumor cell nuclei according to clinicopathologic parameters in DLBCL patients (Table 2). The PD-L1 expression was interpreted in two aspects as expression in tumor cells (PD-L1t) and expression in immune cells (PD-L1i). The prevalence of PD-L1 expression was 21% (23/107) in tumor cells and 36% (38/107) in non-malignant immune cells. All cases with PD-L1 gene alteration (PD-L1 GA+) expressed PD-L1 protein in tumor cells (PD-L1t+), and accounted for 10% (10/102) of DLBCLs which includes translocation in 6% (6/102) and gain/amplification in 4% (4/102). Nuclear expression of pSTAT3 (> 40% cutoff) was observed in 41% (44/107) of DLBCLs (Figs. 1 and 2).
Table 2.
Clinicopathologic characteristics according to protein expression and genetic alteration of PD-L1 and STAT3 activation in EBV-negative diffuse large B cell lymphoma patients
| Clinicopathologic characteristics | PD-L1 expression in tumor cells (PD-L1t) | PD-L1 expression in immune cells (PD-L1i) | PD-L1 genetic alteration in tumor cells (PD-L1 GA) | pSTAT3a expression in tumor cells | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PD-L1t− | PD-L1t+ | p value | PD-L1i− | PD-L1i+ | p value | PD-L1 GA− | PD-L1 GA+ | p value | pSTAT3− (< 40%) |
pSTAT3+ (> 40%) |
p value | |
| Age | ||||||||||||
| ≤ 60 | 40 (48%) | 13 (57%) | 0.449# | 38 (55%) | 15 (39%) | 0.122# | 43 (47%) | 7 (70%) | 0.196† | 34 (54%) | 19 (43%) | 0.272# |
| > 60 | 44 (52%) | 10 (43%) | 31 (45%) | 23 (61%) | 49 (53%) | 3 (30%) | 29 (46%) | 25 (57%) | ||||
| Sex | ||||||||||||
| Male | 49 (58%) | 12 (52%) | 0.597# | 40 (58%) | 21 (55%) | 0.787# | 52 (57%) | 6 (60%) | 1.000† | 33 (52%) | 28 (64%) | 0.247# |
| Female | 35 (42%) | 11 (48%) | 29 (42%) | 17 (45%) | 40 (43%) | 4 (40%) | 30 (48%) | 16 (36%) | ||||
| Primary site | ||||||||||||
| Nodal | 44 (52%) | 10 (43%) | 0.449# | 33 (48%) | 21 (55%) | 0.462# | 50 (54%) | 2 (20%) | 0.049† | 27 (43%) | 27 (61%) | 0.060# |
| Extranodal | 40 (48%) | 13 (57%) | 36 (52%) | 17 (45%) | 42 (46%) | 8 (80%) | 36 (57%) | 17 (39%) | ||||
| B symptoms | ||||||||||||
| Absent | 68 (81%) | 17 (74%) | 0.561† | 55 (80%) | 30 (79%) | 0.926# | 77 (84%) | 6 (60%) | 0.087† | 48 (76%) | 37 (84%) | 0.320# |
| Present | 16 (19%) | 6 (26%) | 14 (20%) | 8 (21%) | 15 (16%) | 4 (40%) | 15 (24%) | 7 (16%) | ||||
| ECOG PS | ||||||||||||
| < 2 | 77 (92%) | 20 (87%) | 0.445† | 61 (88%) | 36 (95%) | 0.489† | 85 (92%) | 8 (80%) | 0.214† | 56 (89%) | 41 (93%) | 0.520† |
| ≥ 2 | 7 (8%) | 3 (13%) | 8 (12%) | 2 (5%) | 7 (8%) | 2 (20%) | 7 (11%) | 3 (7%) | ||||
| Serum lactate dehydrogenasea | ||||||||||||
| Normal | 45 (56%) | 9 (39%) | 0.164# | 34 (52%) | 20 (53%) | 0.913# | 47 (51%) | 5 (50%) | 1.000† | 32 (52%) | 22 (49%) | 0.896# |
| Elevated | 36 (44%) | 14 (61%) | 32 (48%) | 18 (47%) | 42 (49%) | 5 (50%) | 29 (48%) | 21 (51%) | ||||
| No. of extranodal sites | ||||||||||||
| < 2 | 65 (77%) | 16 (70%) | 0.439# | 54 (78%) | 27 (71%) | 0.405# | 71 (78%) | 5 (50%) | 0.118† | 51 (81%) | 30 (68%) | 0.130# |
| ≥ 2 | 19 (23%) | 7 (30%) | 15 (22%) | 11 (29%) | 21 (22%) | 5 (50%) | 12 (19%) | 14 (32%) | ||||
| Ann Arbor stage | ||||||||||||
| I–II | 45 (54%) | 11 (48%) | 0.625# | 38 (55%) | 18 (47%) | 0.445# | 50 (%) | 5 (50%) | 1.000† | 37 (59%) | 19 (43%) | 0.113# |
| III–IV | 39 (46%) | 12 (52%) | 31 (45%) | 20 (53%) | 42 (%) | 5 (50%) | 26 (41%) | 25 (57%) | ||||
| International prognostic index | ||||||||||||
| 0–2 | 57 (68%) | 14 (61%) | 0.530# | 47 (68%) | 24 (63%) | 0.603# | 64 (70%) | 5 (50%) | 0.286† | 44 (70%) | 27 (61%) | 0.361# |
| 3–5 | 27 (32%) | 9 (39%) | 22 (32%) | 14 (37%) | 28 (30%) | 5 (50%) | 19 (30%) | 17 (39%) | ||||
| Bone marrow involvementa | ||||||||||||
| Absent | 66 (87%) | 18 (82%) | 0.553† | 54 (86%) | 30 (86%) | 1.000 | 76 (89%) | 7 (70%) | 0.283† | 51 (88%) | 33 (83%) | 0.450# |
| Present | 10 (13%) | 4 (18%) | 9 (14%) | 5 (14%) | 9 (11%) | 2 (20%) | 7 (12%) | 7 (17%) | ||||
| Bulky mass (cm) | ||||||||||||
| < 10 | 78 (93%) | 20 (87%) | 0.401† | 64 (93%) | 34 (89%) | 0.718† | 84 (91%) | 9 (90%) | 1.000† | 56 (89%) | 42 (95%) | 0.302† |
| ≥ 10 | 6 (7%) | 3 (13%) | 5 (7%) | 4 (11%) | 8 (9%) | 1 (10%) | 7 (11%) | 2 (5%) | ||||
| Hans classificationa | ||||||||||||
| GCB | 26 (34%) | 1 (5%) | 0.006† | 20 (32%) | 7 (19%) | 0.186 | 26 (30%) | 0 (0%) | 0.058† | 20 (35%) | 7 (17%) | 0.042# |
| Non-GCB | 51 (66%) | 21 (95%) | 43 (68%) | 29 (81%) | 61 (70%) | 10 (100%) | 37 (65%) | 35 (83%) | ||||
| BCL2 expressiona | ||||||||||||
| Negative | 33 (40%) | 8 (35%) | 0.665# | 31 (46%) | 10 (26%) | 0.051# | 35 (38%) | 2 (20%) | 0.321† | 31 (50%) | 10 (23%) | 0.004# |
| Positive | 50 (60%) | 15 (65%) | 37 (54%) | 28 (74%) | 57 (62%) | 8 (80%) | 31 (50%) | 34 (77%) | ||||
| BCL6 expression | ||||||||||||
| Negative | 43 (51%) | 17 (74%) | 0.052# | 36 (52%) | 24 (63%) | 0.273# | 46 (50%) | 9 (90%) | 0.019† | 39 (62%) | 21 (48%) | 0.146# |
| Positive | 41 (49%) | 6 (26%) | 33 (48%) | 14 (37%) | 46 (50%) | 1 (10%) | 24 (38%) | 23 (52%) | ||||
| CD10 expression | ||||||||||||
| Negative | 69 (82%) | 23 (100%) | 0.037† | 57 (83%) | 35 (92%) | 0.248† | 77 (84%) | 10 (100%) | 0.351† | 52 (83%) | 40 (91%) | 0.268† |
| Positive | 15 (18%) | 0 (0%) | 12 (17%) | 3 (8%) | 15 (16%) | 0 (0%) | 11 (17%) | 4 (9%) | ||||
| MUM1 expression | ||||||||||||
| Negative | 36 (43%) | 5 (22%) | 0.065# | 28 (41%) | 13 (34%) | 0.517# | 35 (38%) | 1 (10%) | 0.094† | 31 (49%) | 10 (23%) | 0.006# |
| Positive | 48 (57%) | 18 (78%) | 41 (59%) | 25 (66%) | 57 (62%) | 9 (90%) | 32 (51%) | 34 (77%) | ||||
| Total N (%) | 84 (100%) | 23 (100%) | 69 (100%) | 38 (100%) | 92 (100%) | 10 (100%) | 63 (100%) | 44 (100%) | ||||
NA: not applicable; ECOG PS: the Eastern Cooperative Group Performance Status; GCB: germinal center B-cell like; R-CHOP: rituximab with cyclophosphamide–doxorubicin–vincristine–prednisone; GI: gastrointestinal; GA: genetic alteration; IHC: immunohistochemistry; SD: standard deviation
aThese variables excluded unclassifiable (or unknown) cases
p values were calculated by using Fisher’s exact test (2-sided)† or Pearson’s Chi square test (2-sided)#
Fig. 1.
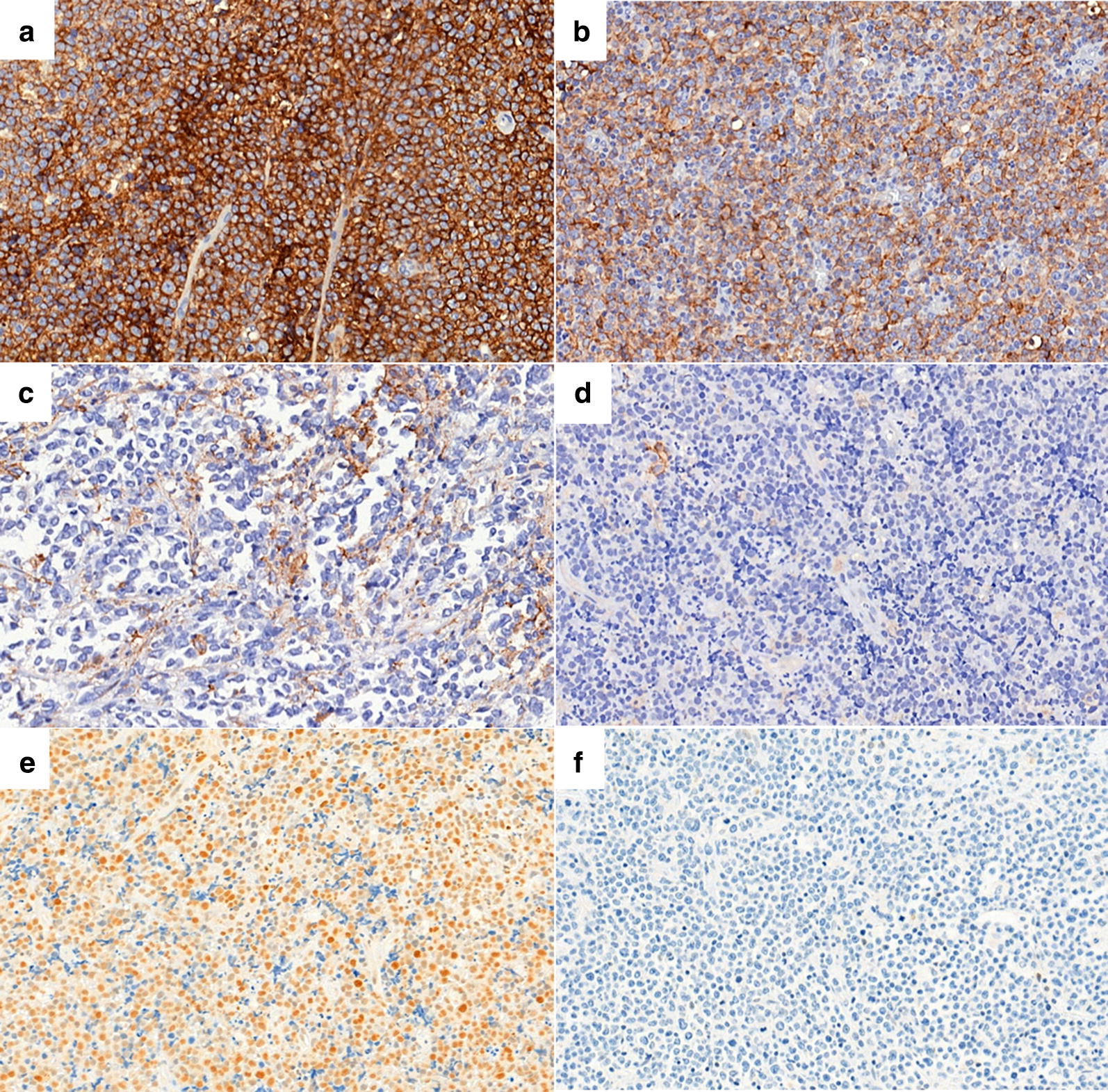
Immunohistochemical expression patterns of PD-L1 and pSTAT3 in diffuse large B cell lymphoma. PD-L1 is expressed in both tumor and immune cells (a), only in tumor (b) or only in immune cells (c). PD-L1 is not expressed in either tumor cells or immune cells (d). Tumor cell nuclei are positive for pSTAT3 (e), compared to a pSTAT3-negative case (f) (×400 magnification)
Fig. 2.
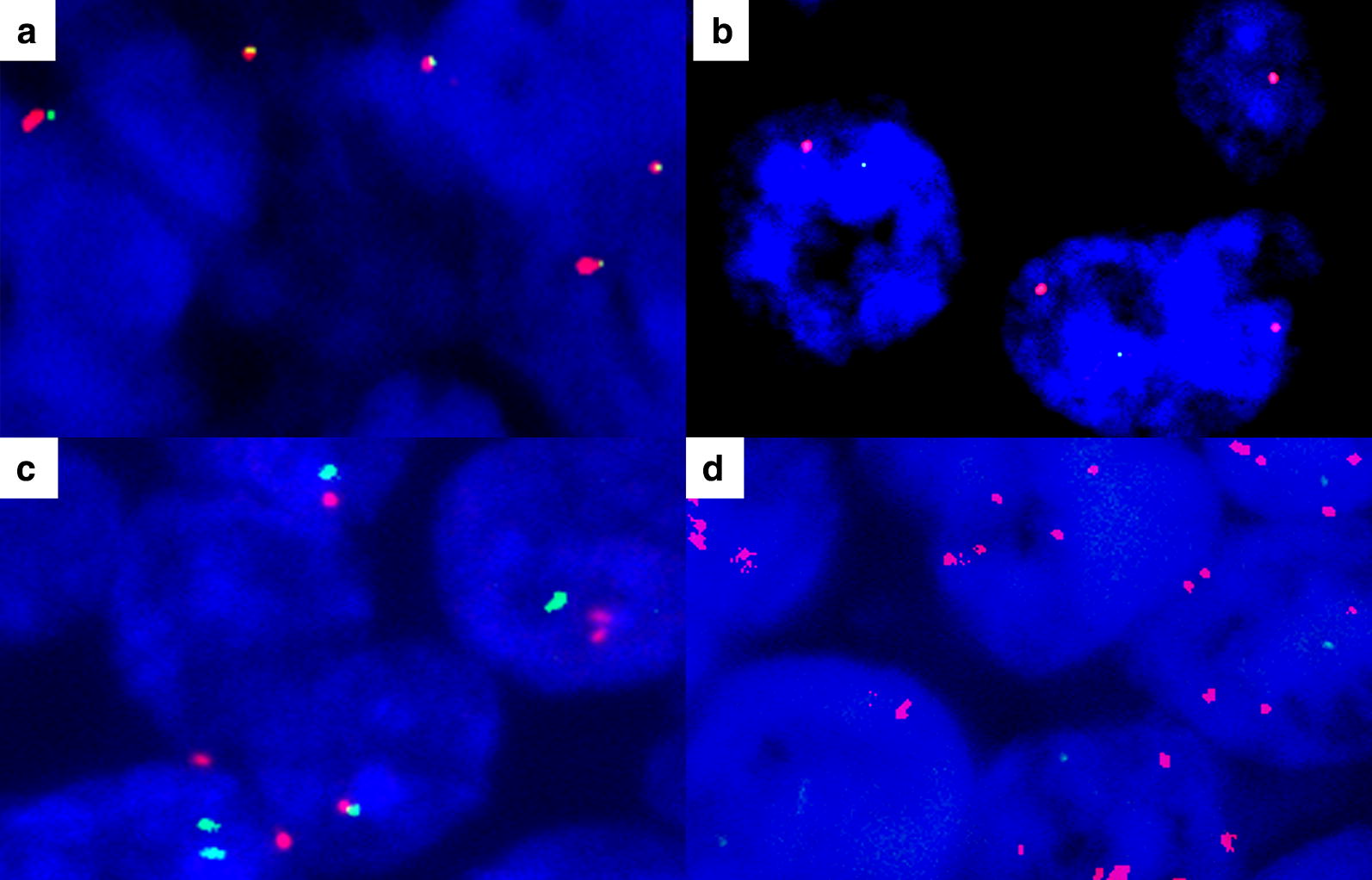
Fluorescence in situ hybridization analysis of PD-L1 gene in diffuse large B cell lymphoma. By using dual-color orange/green break-apart probe, in contrast to non-split fused yellow signals showing no translocation (a), separate orange and green signals indicate translocation of PD-L1 gene (b). Copy number analysis probes containing orange (PD-L1 gene) and green (chromosome 9) signals show nearly 1:1 ratio of orange to green signals, indicating no increase of copy number of PD-L1 gene (c). In contrast, orange signals are amplified, compared to green signals (d)
In the analysis with clinicopathologic variables (Table 2), PD-L1 GA+ was more frequent in primary extranodal DLBCLs than nodal cases (p = 0.049), and all PD-L1 GA+ cases (n = 10) belonged to non-GCB subtype according to Hans algorithm, while not reaching statistical significance (p = 0.058). As for protein expression, non-GCB subtype also showed more frequent PD-L1 expression (PD-L1t+), as well as pSTAT3 expression, in tumor cells than GCB subtype (p = 0.006 and p = 0.042, respectively). In the analysis with Bcl-2 and each component of Hans algorithm, PD-L1 GA and PD-L1t was significantly or marginally associated with lack of expression of Bcl-6 and/or CD10 (p = 0.019 for PD-L1 GA vs. Bcl-6; p = 0.052 for PD-L1t vs Bcl-6; p = 0.037 for PD-L1t vs CD10), while pSTAT3 expression was mainly related with MUM1 expression (p = 0.006). Of note, PD-L1i was marginally associated with Bcl-2 expression in tumor cells (p = 0.051). Taken together, PD-L1 and/or pSTAT3 signaling pathways are frequently activated in non-GCB subtype or extranodal DLBCLs.
Association within PD-L1/pSTAT3-related markers
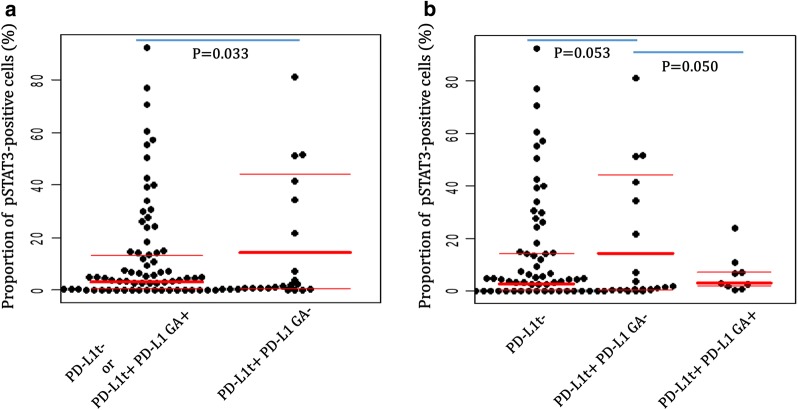
To clarify the associations between PD-L1 gene/protein status and pSTAT3 expression in DLBCLs, we next performed correlation analysis among PD-L1/pSTAT3-related markers including PD-L1t, PD-L1i, PD-L1 GA and pSTAT3 expression with dichotomized variable by 40% cutoff and continuous variable (%) (Table 3). To effectively recognize associations between PD-L1-related markers and pSTAT3, pSTAT3 was analyzed as two different modes of variables: (1) a digitally counted proportional variable (%) of pSTAT3-positive tumor cells and (2) a conventionally interpreted dichotomized variable with 40% cutoff value. In this way, PD-L1t was positively correlated with PD-L1i, PD-L1 translocation and PD-L1 gain/amplification, while PD-L1i was associated only with PD-L1 translocation. Notably, tumor cells of DLBCLs with PD-L1 expression but no PD-L1 gene alteration (PD-L1t+ PD-L1 GA−) had a higher proportion (%) of pSTAT3-positive tumor cells than the rest of the subset (PD-L1t− or PD-L1t+ PD-L1 GA+) (p = 0.033; Fig. 3a), and in comparison to both PD-L1t− subset (p = 0.053) and PD-L1t+ PD-L1 GA+ subset (p = 0.050) with marginal significance as well (Fig. 3b). These results suggest that PD-L1 protein expression in DLBCL tumor cells lacking intrinsic PD-L1 gene activation mechanism, i.e., translocation and gain/amplification, may be induced by STAT3-mediated signaling pathway.
Table 3.
Associations between PD-L1, pSTAT3 and other clinicopathologic factors in diffuse large B cell lymphoma (n = 107)
| PD-L1i | PD-L1 translocation (A) | PD-L1 gain/amplification (B) | PD-L1 gene alteration (A + B) | PD-L1 non-genetic expressionb | pSTAT3a (continuous) | pSTAT3 (> 40%) | |
|---|---|---|---|---|---|---|---|
| PD-L1t | R = 0.325 p = 0.001 |
R = 0.477 p < 0.001 |
R = 0.386 p < 0.001 |
R = 0.629 p < 0.001 |
R = 0.698 p < 0.001 |
R = 0.079 p = 0.429 |
R = 0.071 p = 0.466 |
| PD-L1i | R = 0.239 p = 0.015 |
R = 0.159 p = 0.109 |
R = 0.291 p = 0.003 |
R = 0.166 p = 0.092 |
R = 0.081 p = 0.414 |
R = 0.015 p = 0.879 |
|
| PD-L1 translocation (A) | R = − 0.051 p = 0.614 |
R = 0.758 p < 0.001 |
R = − 0.090 p = 0.364 |
R = − 0.049 p = 0.628 |
R = − 0.037 p = 0.714 |
||
| PD-L1 gain/amplification (B) | R = 0.613 p < 0.001 |
R = − 0.073 p = 0.464 |
R = − 0.117 p = 0.242 |
R = − 0.072 p = 0.470 |
|||
| PD-L1 gene alteration (A + B) | R = − 0.120 p = 0.228 |
R = − 0.117 p = 0.245 |
R = − 0.021 p = 0.835 |
||||
| PD-L1 non-genetic expressionb | R = 0.205 p = 0.033 |
R = 0.120 p = 0.224 |
Correlation coefficients (R) and p values by Pearson’s correlation
a Indicates the proportion (%) of tumor cells expressing pSTAT3 by using digital image analyzer
b Indicates PD-L1t+ cases with no alteration (translocation or gain/amplification) of PD-L1 gene
Fig. 3.
Dot plots for proportions of pSTAT3-positive tumor cells according to PD-L1 protein expression in tumor cells and gene alteration status in diffuse large B cell lymphoma. PD-L1t+ PD-L1 GA− cases have higher proportions (%) of pSTAT3-positive tumor cells than the rest cases (PD-L1t− or PD-L1t+ PD-L1 GA+) (a), and this tendency was preserved in comparison of each subset (PD-L1t+ PD-L1 GA− vs. PD-L1t−; PD-L1t+ PD-L1 GA− vs. PD-L1t+ PD-L1 GA+) (b)
Associations between PD-L1 expression and gene alteration and primary sites
Frequent gene alteration and expression of PD-L1 were known in DLBCLs arising in certain specific sites such as mediastinum and brain [33, 34], both of which were excluded in our cohort. In our cohort, we further analyzed the PD-L1 according to the primary sites, focusing on gastrointestinal tract, testis and adrenal gland. As shown in Table 4, primary testis cases (n = 14) had a higher frequency of PD-L1 gene alteration (29%; 4/14), mainly translocation (21%; 3/14), than non-testis cases (p = 0.029 for PD-L1 GA; p = 0.031 for PD-L1 translocation). Notably, one of the three adrenal gland cases (33%) had a gain/amplification of PD-L1, though with limited significance due to low incidence (p = 0.112). These data suggest that DLBCLs of specific anatomic sites might have preferential alteration patterns of PD-L1 gene.
Table 4.
Associations between genetic alteration and protein expression status of PD-L1 and primary sites
| Primary site | GI | Testis | Adrenal gland | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-GI | GI | p value | Non-testis | Testis | p value | Non-adrenal | Adrenal | p value | |
| PD-L1t | |||||||||
| Absent | 67 | 17 | 0.354 | 74 | 10 | 0.494 | 83 | 1 | 0.116 |
| Present | 21 | 2 | 19 | 4 | 21 | 2 | |||
| PD-L1i | |||||||||
| Absent | 55 | 14 | 0.435 | 59 | 10 | 0.766 | 68 | 1 | 0.287 |
| Present | 33 | 5 | 34 | 4 | 36 | 2 | |||
| PD-L1 translocation (A) | |||||||||
| Absent | 80 | 18 | 0.587 | 87 | 11 | 0.031 | 95 | 3 | 1.000 |
| Present | 6 | 0 | 3 | 3 | 6 | 0 | |||
| PD-L1 gain/amplification (B) | |||||||||
| Absent | 81 | 19 | 1.000 | 87 | 13 | 0.444 | 98 | 2 | 0.112 |
| Present | 4 | 0 | 3 | 1 | 3 | 1 | |||
| PD-L1 genetic alteration (A + B) | |||||||||
| Absent | 75 | 18 | 0.203 | 83 | 10 | 0.029 | 91 | 2 | 0.266 |
| Present | 10 | 0 | 6 | 4 | 9 | 1 | |||
p values were calculated by using Fisher’s exact test (2-sided)
GI: gastrointestinal; GA: genetic alteration
Survival analysis
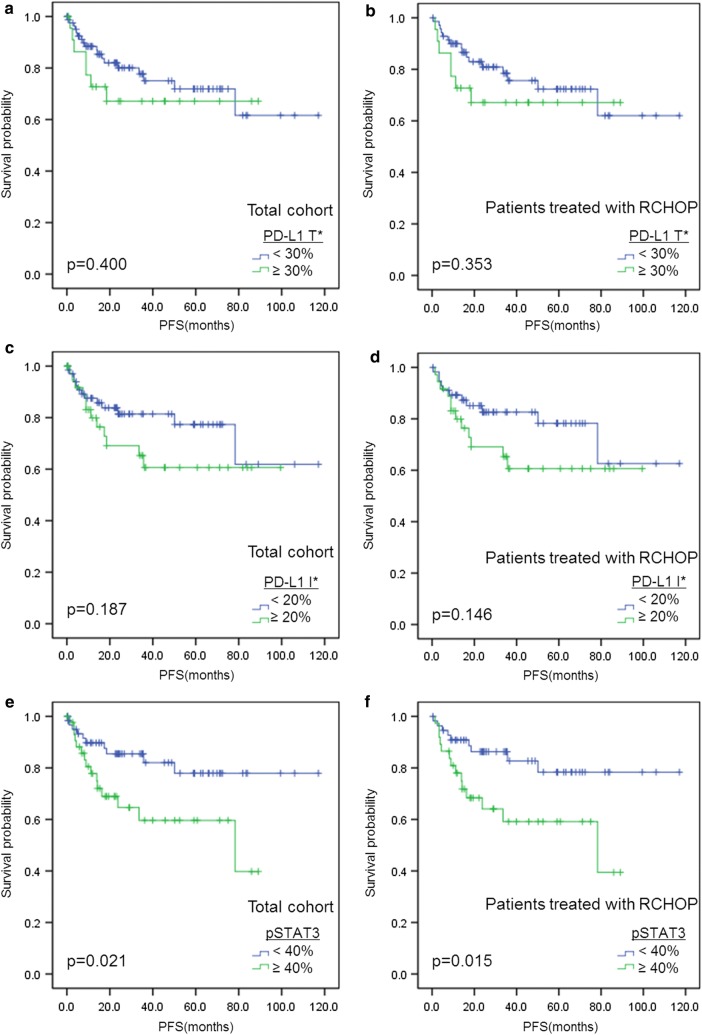
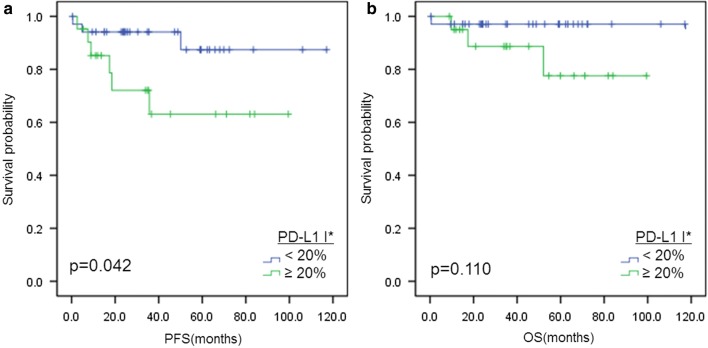
Survival analysis was conducted in total cohort and separately in the subgroup of patients treated with R-CHOP (n = 93; Table 5). No significance was found in clinical outcome with PD-L1 expression alone in either tumor (PD-L1t) or non-malignant immune cells (PD-L1i) (Fig. 4a–d). However, pSTAT3 expression (> 40%) was significantly associated with inferior progression-free survival in the total cohort (p = 0.021; Fig. 4e) and in the R-CHOP-treated group (p = 0.015; Fig. 4f). Multivariate cox regression that followed revealed pSTAT3 to be an independent prognostic factor in both the total cohort (p = 0.017, HR = 2.724) and R-CHOP-treated patients (p = 0.007, HR = 3.510). Other significant prognostic factors in this multivariate analysis were IPI (p = 0.001, HR = 4.910 in total cohort; p = 0.002, HR = 4.823 in the R-CHOP treated group), Eastern Cooperative Group Performance Status (p = 0.010, HR = 3.717 in the total cohort; p = 0.031, HR = 3.286 in R-CHOP treated group) and Bulky mass (p = 0.010, HR = 4.664 in the R-CHOP treated group). When the R-CHOP cohort (n = 93) was divided into pSTAT3-negative (n = 56) and pSTAT3-positive subgroups (n = 37), PD-L1 expression of non-malignant immune cells (PD-L1i) correlated with poor progression-free survival in pSTAT3-negative patients who received R-CHOP regimen by log-rank test in Kaplan–Meier analysis (p = 0.042; Fig. 5). It did not, however, turn out to be an independent prognostic factor (p = 0.089; Table 6), while ECOG PS was the only significant prognostic factor (p < 0.001, HR = 21.553). Analysis with overall survival revealed no significant results (Additional file 1: Figure S1).
Table 5.
Survival analysis with progression-free survival in total cohort and in patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone (n = 93)
| Clinicopathologic variables | Total cohort | Patients treated with R-CHOP | ||||
|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||
| p value | p value | HR (95% CI) | p value | p value | HR (95% CI) | |
| Age | ||||||
| > 60 vs. ≤ 60 | 0.035 | 0.181 | 0.074 | |||
| Sex | ||||||
| Male vs. female | 0.544 | 0.661 | ||||
| ECOG PS | ||||||
| ≥ 2 vs. < 2 | <0.001 | 0.010 | 3.717 (1.371–10.080) | < 0.001 | 0.031 | 3.286 (1.116–9.677) |
| B symptoms | ||||||
| Present vs. absent | 0.049 | 0.807 | 0.112 | |||
| Serum LDH | ||||||
| Elevated vs. normal | 0.003 | 0.328 | 0.033 | 0.319 | ||
| IPI | ||||||
| 3–5 vs. 0–2 | < 0.001 | 0.001 | 4.910 (1.888–12.767) | < 0.001 | 0.002 | 4.823 (1.763–13.191) |
| Primary site | ||||||
| Extranodal vs. nodal | 0.488 | 0.523 | ||||
| Extranodal site | ||||||
| ≥ 2 vs. < 2 | < 0.001 | 0.649 | 0.001 | 0.978 | ||
| Bone marrow involvement | ||||||
| Present vs. absent | 0.308 | 0.359 | ||||
| Bulky mass | ||||||
| ≥ 10 cm vs. < 10 cm | 0.136 | 0.048 | 0.010 | 4.664 (1.449–15.017) | ||
| Ann Arbor stage | ||||||
| III–IV vs. I–II | < 0.001 | 0.682 | < 0.001 | 0.812 | ||
| Hans classification | ||||||
| Non-GCB vs. GCB | 0.075 | 0.135 | ||||
| Treatment regimen | ||||||
| RCHOP vs. other | 0.676 | – | ||||
| BCL2 expression | ||||||
| Positive vs. negative | 0.391 | 0.699 | ||||
| BCL6 expression | ||||||
| Positive vs. negative | 0.247 | 0.545 | ||||
| CD10 expression | ||||||
| Positive vs. negative | 0.543 | 0.778 | ||||
| MUM1 expression | ||||||
| Positive vs. negative | 0.138 | 0.211 | ||||
| PD-L1 tumor cell expression | ||||||
| Positive vs. negative | 0.400 | 0.353 | ||||
| PD-L1 immune cell expression | ||||||
| Positive vs. negative | 0.187 | 0.146 | ||||
| PD-L1 gene alteration | ||||||
| Present vs. absent | 0.900 | 0.871 | ||||
| pSTAT3 expression (> 40%) | ||||||
| Positive vs. negative | 0.021 | 0.017 | 2.724 (1.196–6.204) | 0.015 | 0.007 | 3.510 (1.418–8.692) |
PD-L1: programmed cell death-ligand 1; pSTAT3: phosphorylated signal transducer and activator of transcription 3; ECOG PS: the Eastern Cooperative Group Performance Status; GCB: germinal center B-cell like; R-CHOP: rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone
Fig. 4.
Kaplan–Meier curves of progression-free survival in total and R-CHOP-treated diffuse large B cell lymphoma patients. Survival analysis for progression-free survival (PFS) according to PD-L1t in total cohort (n = 107) (a) and R-CHOP cohort (n = 93) (b), and according to PD-L1i in total cohort (c) and R-CHOP cohort (d). Survival analysis for PFS according to pSTAT3 (40% cutoff) in total cohort (e) and R-CHOP cohort (f). *T: tumor cell; I: immune cell
Fig. 5.
Kaplan–Meier curves of progression-free and overall survival in pSTAT3-negative diffuse large B cell lymphoma patients treated with R-CHOP. Survival analysis for progression-free survival (PFS) according to PD-L1i in pSTAT3-negative R-CHOP treated subset (n = 57) (a) and survival analysis for overall survival in pSTAT3-negative R-CHOP treated subset (n = 57) (b). * I: non-malignant immune cell
Table 6.
Survival analysis with progression-free survival in patients treated with rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone (n = 56)
| Clinicopathologic variables | Univariate | Multivariate | |
|---|---|---|---|
| p value | p value | HR (95% CI) | |
| Age | |||
| > 60 vs. ≤ 60 | 0.158 | ||
| Sex | |||
| Male vs. female | 0.702 | ||
| ECOG PS | |||
| ≥ 2 vs. < 2 | < 0.001 | < 0.001 | 21.553 (4.682–99.208) |
| B symptoms | |||
| Present vs. absent | 0.099 | ||
| Serum LDH | |||
| Elevated vs. normal | 0.189 | ||
| IPI | |||
| 3–5 vs. 0–2 | 0.002 | 0.238 | |
| Primary site | |||
| Extranodal vs. nodal | 0.204 | ||
| Extranodal site | |||
| ≥ 2 vs. < 2 | < 0.001 | 0.167 | |
| Bone marrow involvement | |||
| Present vs. absent | 0.476 | ||
| Bulky mass | |||
| ≥ 10 cm vs. < 10 cm | 0.279 | ||
| Ann Arbor stage | |||
| III–IV vs. I–II | 0.037 | 0.610 | |
| Hans classification | |||
| Non-GCB vs. GCB | 0.503 | ||
| BCL2 expression | |||
| Positive vs. negative | 0.983 | ||
| BCL6 expression | |||
| Positive vs. negative | 0.600 | ||
| CD10 expression | |||
| Positive vs. negative | 0.600 | ||
| MUM1 expression | |||
| Positive vs. negative | 0.409 | ||
| PD-L1 tumor cell expression | |||
| Positive vs. negative | 0.209 | ||
| PD-L1 immune cell expression | |||
| Positive vs. negative | 0.042 | 0.089 | |
| PD0L1 gene alteration | |||
| Present vs. absent | 0.285 | ||
PD-L1: programmed cell death-ligand 1; pSTAT3: phosphorylated signal transducer and activator of transcription 3; ECOG PS: the Eastern Cooperative Group Performance Status; GCB: germinal center B-cell like; R-CHOP: rituximab with cyclophosphamide, doxorubicin, vincristine and prednisone
Discussion
The roles of PD-L1 expression and gene alteration are relatively unclear in EBV-negative DLBCLs, compared to EBV-positive DLBCLs. In this study, we observed that (1) immunopositivity for PD-L1 was 21% in tumor cells which included all the cases of PD-L1 gene alteration that accounts for 10%, and 36% in immune cells, while pSTAT3 positivity was 41% in tumor cells, (2) the non-GCB subtype showed higher PD-L1 and pSTAT3 tumor cell expression and a tendency for PD-L1 gene alteration, (3) PD-L1 expression without gene alteration correlated with pSTAT3 expression, (4) pSTAT3 expression independently predicted shorter PFS in total cohort and R-CHOP-treated group and (5) PD-L1 expression in immune cells correlated with shorter PFS in pSTAT3-negative R-CHOP-treated subset, while influenced by ECOG PS.
Research on PD-L1 in DLBCL has started rather recently and the evaluation methods or standards of PD-L1 immunohistochemistry in DLBCL has not yet met consensus. Studies vary in the PD-L1 antibodies used, the method of evaluation (manual or digital) and the cutoff values to determine positivity. The prevalence of PD-L1 expression ranges greatly from 10.5 to 61.1% in tumor cells and 15.3 to 30% in non-malignant immune cells [4–6, 35, 36], which are similar to our observation.
In the present study, tumor cell expression of PD-L1, but not the non-malignant immune cell expression, significantly associated with non-GCB DLBCL. This is in concordance with previous studies [6–8]. Siddiqi and colleagues provided more concrete evidence of the association between PD-L1 expression and Hans algorithm classification by conducting next generation sequencing on primary DLBCL specimens [9]. In their study, the positive correlation of PD-L1 expression and mutations associated with non-GCB DLBCL such as FAT2 contrasted with the negative correlation of mutations associated with GCB DLBCL such as BCL2, FOXO1 and KMT2D. Considering that PD-L1 gene tended to be frequently altered in non-GCB or extranodal DLBCLs, PD-L1 gene alteration may be one of the important genetic features of non-GCB DLBCLs, and these findings may provide an insight into the understanding of the pathogenesis of non-GCB DLBCLs.
Among various extranodal DLBCLs, several site-specific variants or distinct entities have been described [29]. Primary testicular DLBCLs are well-known to have unique clinicopathologic features with frequent PD-L1 gene alteration [34]. In accordance with these reports, we observed frequent PD-L1 gene alteration of testicular DLBCLs in our cohort as well, and translocation was the predominant form of alteration. Primary adrenal DLBCL is another very rare extranodal subset with unique clinical features [37]. However, the underlying genetic alteration is not well characterized. In our observation, one of the three adrenal DLBCL harbored PD-L1 gain/amplification, although the clinical significance could not yet be determined. This finding might provide a clue to adrenal lymphomagenesis and lead to further investigation of PD-L1 gene alteration in adrenal DLBCLs.
The 41% positivity of pSTAT3 immunohistochemistry of this study is in agreement with most reports [27, 38–40]. Increased expression of pSTAT3 also showed significant association with non-GCB DLBCL. Constitutive activation of STAT3 in non-GCB DLBCL has been investigated more in depth than PD-L1. Higher levels of STAT3 mRNA was detected in the non-GCB subtype than the GCB subtype using DLBCL cell lines [22, 28] and pSTAT3 appeared to bind to promoters of different genes in each subtype, wherein genes regulating cell proliferation and survival are mostly upregulated in non-GCB DLBCL [28]. The preferential activation of STAT3 in non-GCB DLBCL may in part be explained by Bcl-6-induced down-regulation of STAT3 in non-GCB DLBCL cells [23]. It is of note that Bcl-6 is a typical GCB marker and its expression level is generally low in non-GCB DLBCL, possibly leading to STAT3 activation.
Activated PD-1/PD-L1 axis plays the role of tumor evasion from host tumor-specific T-cell immunity. Conceptually, the activation mechanism of PD-L1 gene may be divided into the intrinsic alteration of PD-L1 gene and activation of its upstream signaling pathway, which may contain STAT3 signaling [17]. This might be supported by our observation that the frequency of PD-L1 protein expression (21%) is higher than PD-L1 gene alteration (10%). In the present study, cases with no alteration of PD-L1 gene (PD-L1t+ PD-L1 GA−) tended to have higher proportions (%) of pSTAT3-positive cells than PD-L1t− subset and PD-L1t+ PD-L1 GA+ subset. This finding suggests that STAT3-mediated PD-L1 expression and genetically activated PD-L1 expression might partly be mutually exclusive, and STAT3-mediated signaling might be an alternative mechanism for PD-L1 expression, which remains to be clarified further.
Our finding in survival analysis for pSTAT3 expression as an independent prognostic factor for shorter progression-free survival is consistent with the previous reports in DLBCLs [19, 25, 39, 40]. Huang and colleagues [40] took a step further in taking DLBCL cell lines to gene expression profiling analysis and found an 11-gene STAT3 signature including CD48, IRF1 and IL10, which correlated well with inferior clinical outcomes. In an experimental animal model, microenvironmental immature dendritic cells coproducing IL-10 and PD-L1 enhanced anti-tumor immune reaction [41]. This finding suggests the cooperative immunosuppressive role of IL-10 and PD-L1, which may prevail in the STAT3-skewed microenvironment of non-GCB DLBCLs. Considering that IL-10 is also produced by B cells via Toll-like receptor/MyD88/STAT3 pathway in immune reaction [42], the mechanism of interplay between neoplastic B cells and non-malignant immune cells with activated STAT3- and PD-L1-related signaling in the milieu of IL-10 may be more complex than solid tumor models. In this context, the effects of PD-L1 on clinical outcome need to be carefully analyzed with distinctive interpretation of its expression on tumor cells and immune cells with consideration of activation status of the STAT3-related signaling pathway.
Few have investigated the prognostic value of PD-L1 in DLBCL and the results are controversial. Kiyasu and colleagues [6] reported that PD-L1 expression of DLBCL tumor cells was associated with poor clinical outcome whereas that of non-malignant stromal cells showed no significant difference in prognosis. Siddiqi’s group [9] also found PD-L1 tumor cell expression to be associated with inferior survival while Kwon and colleagues [35] reported no significant association to clinical outcome in DLBCL. In the present study, though PD-L1 tumor cell expression had no prognostic significance, immune cell expression of PD-L1 was associated with poor outcome in the pSTAT3-negative R-CHOP-treated subset in univariate analysis. It is not clear why this prognostic effect of PD-L1 expressing immune cell was observed in this subset. One explanation might be that paucity of STAT3-related signature could make the immune microenvironment more dependent on PD-L1 signaling. Furthermore, the pSTAT3-positive subset may have robust STAT3-driven survival signaling of tumor cells that can override the effect of PD-L1-mediated immune evasion [23, 30, 43]. In another point of view, the prognostic role of immune cell PD-L1 may be related with tumoral Bcl-2 expression in our study, where both markers of different cell types had marginal association. The intrinsic mechanism of cancer cell may influence anti-tumor immunity [44, 45], where Bcl-2 protein may act as a tumor-associated antigen [46], although further studies are required for support. In this context, proper isolation of clinicopathologic subsets may provide chances for efficient therapeutic application in targeting PD-L1 signaling in DLBCL patients.
Conclusion
Our study revealed the association between gene alteration and protein expression of PD-L1 and pSTAT3 expression, both of which constituted features of non-GCB DLBCLs. We also observed that DLBCL patients with pSTAT3-positive tumors had an independently inferior clinical outcome, while in those with pSTAT3-negative tumors, PD-L1 immune cell expression was predictive of poor prognosis. These findings may open another potential immunotherapeutic strategy for the treatment of DLBCL.
Additional file
Additional file 1: Figure S1. Kaplan–Meier curves of overall survival in total and R-CHOP-treated diffuse large B cell lymphoma patients. *T, tumor cell; I, immune cell.
Authors’ contributions
HJK and JHP conceived and designed the experiments. HJK, YJM and JHP performed the experiments and analyzed pathologic data. JSL and JOL collected and analyzed clinical data. HJK and JHP performed clinicopathologic analysis and wrote the manuscript. All authors edited the final manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The Institutional Review Board of Seoul National University Bundang Hospital (IRB of SNUBH) approved this study, and informed consent was waived by IRB of SNUBH because this study used archived materials with retrospective design.
Funding
This work was supported by Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (NRF-2016R1D1A1B03935507).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DLBCL
diffuse large B cell lymphoma
- PD-L1
programmed cell death ligand 1
- pSTAT3
phosphorylated signal transducer and activator of transcription 3
- GCB
germinal center B cell like
- R-CHOP
rituximab combined with cyclophosphamide–doxorubicin–vincristine–prednisone
- EBV
Epstein–Barr virus
- PD-L1t
PD-L1 expression in tumor cells
- PD-L1i
PD-L1 expression in immune cells
- PD-L1 GA
PD-L1 gene alteration
Contributor Information
Hyun Jung Kwon, Email: treeish@naver.com.
Jeong Mi Yang, Email: didwjdal1210@nate.com.
Jeong-Ok Lee, Email: deafkeller@snubh.org.
Jong Seok Lee, Email: jslee@snubh.org.
Jin Ho Paik, Phone: +82-31-787-7713, Email: paikjh@snu.ac.kr.
References
- 1.Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50:74–87. doi: 10.1016/j.pathol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 3.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 4.Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MG, Xu ML, Yu H, Fletcher CD, Freeman GJ, Shipp MA, et al. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res. 2013;19:3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurent C, Charmpi K, Gravelle P, Tosolini M, Franchet C, Ysebaert L, Brousset P, Bidaut A, Ycart B, Fournie JJ. Several immune escape patterns in non-Hodgkin’s lymphomas. Oncoimmunology. 2015;4:e1026530. doi: 10.1080/2162402X.2015.1026530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiyasu J, Miyoshi H, Hirata A, Arakawa F, Ichikawa A, Niino D, Sugita Y, Yufu Y, Choi I, Abe Y, et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood. 2015;126:2193–2201. doi: 10.1182/blood-2015-02-629600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gravelle P, Burroni B, Pericart S, Rossi C, Bezombes C, Tosolini M, Damotte D, Brousset P, Fournie JJ, Laurent C. Mechanisms of PD-1/PD-L1 expression and prognostic relevance in non-Hodgkin lymphoma: a summary of immunohistochemical studies. Oncotarget. 2017;8:44960–44975. doi: 10.18632/oncotarget.16680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgiou K, Chen L, Berglund M, Ren W, de Miranda NF, Lisboa S, Fangazio M, Zhu S, Hou Y, Wu K, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016;127:3026–3034. doi: 10.1182/blood-2015-12-686550. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqi IN, Thodima V, Friedman J, Violeta A, Tulpule A, Shaknovich R, Houldsworth J. PD-L1 expression identifies high risk diffuse large B-cell lymphoma and is associated with several genomic markers. Blood. 2016;128:153-. doi: 10.1182/blood-2016-05-716043. [DOI] [PubMed] [Google Scholar]
- 10.Nicolae A, Pittaluga S, Abdullah S, Steinberg SM, Pham TA, Davies-Hill T, Xi L, Raffeld M, Jaffe ES. EBV-positive large B-cell lymphomas in young patients: a nodal lymphoma with evidence for a tolerogenic immune environment. Blood. 2015;126:863–872. doi: 10.1182/blood-2015-02-630632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodman A, Patel SP, Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat Rev Clin Oncol. 2017;14:203–220. doi: 10.1038/nrclinonc.2016.168. [DOI] [PubMed] [Google Scholar]
- 12.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tirado CA, Chen W, Garcia R, Kohlman KA, Rao N. Genomic profiling using array comparative genomic hybridization define distinct subtypes of diffuse large B-cell lymphoma: a review of the literature. J Hematol Oncol. 2012;5:54. doi: 10.1186/1756-8722-5-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang W, Zhang J, Hong S, Zhan J, Chen N, Qin T, Tang Y, Zhang Y, Kang S, Zhou T, et al. EBV-driven LMP1 and IFN-gamma up-regulate PD-L1 in nasopharyngeal carcinoma: implications for oncotargeted therapy. Oncotarget. 2014;5:12189–12202. doi: 10.18632/oncotarget.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 16.Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51:221–228. doi: 10.1016/j.oraloncology.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Marzec M, Zhang Q, Goradia A, Raghunath PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1) Proc Natl Acad Sci USA. 2008;105:20852–20857. doi: 10.1073/pnas.0810958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song TNM, Lim J, Nagarajan S, Pang JW, Laurensia Y, Wijaya GC, Jing T, Ong C. Oncogenic activation of STAT3 pathway drives PD-L1 expression in natural killer/T cell lymphoma. Hematol Oncol. 2017;35:163–164. doi: 10.1002/hon.2438_20. [DOI] [Google Scholar]
- 20.Atsaves V, Tsesmetzis N, Chioureas D, Kis L, Leventaki V, Drakos E, Panaretakis T, Grander D, Medeiros LJ, Young KH, et al. PD-L1 is commonly expressed and transcriptionally regulated by STAT3 and MYC in ALK-negative anaplastic large-cell lymphoma. Leukemia. 2017;31:1633–1637. doi: 10.1038/leu.2017.103. [DOI] [PubMed] [Google Scholar]
- 21.Ma C, Horlad H, Pan C, Yano H, Ohnishi K, Fujiwara Y, Matsuoka M, Lee A, Niidome T, Yamanaka R, et al. Stat3 inhibitor abrogates the expression of PD-1 ligands on lymphoma cell lines. J Clin Exp Hematopathol. 2017;57:21–25. doi: 10.3960/jslrt.17006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, Chan JW, Rosenwald A, Gascoyne RD, Staudt LM. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111:3701–3713. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y, Cattoretti G, Ye BH. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood. 2008;111:1515–1523. doi: 10.1182/blood-2007-04-087734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scuto A, Kujawski M, Kowolik C, Krymskaya L, Wang L, Weiss LM, Digiusto D, Yu H, Forman S, Jove R. STAT3 inhibition is a therapeutic strategy for ABC-like diffuse large B-cell lymphoma. Cancer Res. 2011;71:3182–3188. doi: 10.1158/0008-5472.CAN-10-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ok CY, Chen J, Xu-Monette ZY, Tzankov A, Manyam GC, Li L, Visco C, Montes-Moreno S, Dybkaer K, Chiu A, et al. Clinical implications of phosphorylated STAT3 expression in de novo diffuse large B-cell lymphoma. Clin Cancer Res. 2014;20:5113–5123. doi: 10.1158/1078-0432.CCR-14-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohgami RS, Ma L, Monabati A, Zehnder JL, Arber DA. STAT3 mutations are present in aggressive B-cell lymphomas including a subset of diffuse large B-cell lymphomas with CD30 expression. Haematologica. 2014;99:e105–e107. doi: 10.3324/haematol.2013.101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia. 2012;26:1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardee J, Ouyang Z, Zhang Y, Kundaje A, Lacroute P, Snyder M. STAT3 targets suggest mechanisms of aggressive tumorigenesis in diffuse large B-cell lymphoma. G3 Bethesda. 2013;3:2173–2185. doi: 10.1534/g3.113.007674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swerdlow SH, Campo E, Harris NL. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press; 2017. [Google Scholar]
- 30.Paik JH, Nam SJ, Kim TM, Heo DS, Kim CW, Jeon YK. Overexpression of sphingosine-1-phosphate receptor 1 and phospho-signal transducer and activator of transcription 3 is associated with poor prognosis in rituximab-treated diffuse large B-cell lymphomas. BMC Cancer. 2014;14:911. doi: 10.1186/1471-2407-14-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paik JH, Go H, Nam SJ, Kim TM, Heo DS, Kim CW, Jeon YK. Clinicopathologic implication of A20/TNFAIP3 deletion in diffuse large B-cell lymphoma: an analysis according to immunohistochemical subgroups and rituximab treatment. Leuk Lymphoma. 2013;54:1934–1941. doi: 10.3109/10428194.2012.762511. [DOI] [PubMed] [Google Scholar]
- 32.Go H, Cho HJ, Paik JH, Park CM, Oh YH, Jung KC, Kim CW, Jeon YK. Thymic extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue: a clinicopathological and genetic analysis of six cases. Leuk Lymphoma. 2011;52:2276–2283. doi: 10.3109/10428194.2011.596968. [DOI] [PubMed] [Google Scholar]
- 33.Twa DD, Chan FC, Ben-Neriah S, Woolcock BW, Mottok A, Tan KL, Slack GW, Gunawardana J, Lim RS, McPherson AW, et al. Genomic rearrangements involving programmed death ligands are recurrent in primary mediastinal large B-cell lymphoma. Blood. 2014;123:2062–2065. doi: 10.1182/blood-2013-10-535443. [DOI] [PubMed] [Google Scholar]
- 34.Chapuy B, Roemer MG, Stewart C, Tan Y, Abo RP, Zhang L, Dunford AJ, Meredith DM, Thorner AR, Jordanova ES, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127:869–881. doi: 10.1182/blood-2015-10-673236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon D, Kim S, Kim PJ, Go H, Nam SJ, Paik JH, Kim YA, Kim TM, Heo DS, Kim CW, et al. Clinicopathological analysis of programmed cell death 1 and programmed cell death ligand 1 expression in the tumour microenvironments of diffuse large B cell lymphomas. Histopathology. 2016;68:1079–1089. doi: 10.1111/his.12882. [DOI] [PubMed] [Google Scholar]
- 36.Menter T, Bodmer-Haecki A, Dirnhofer S, Tzankov A. Evaluation of the diagnostic and prognostic value of PDL1 expression in Hodgkin and B-cell lymphomas. Hum Pathol. 2016;54:17–24. doi: 10.1016/j.humpath.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Rashidi A, Fisher SI. Primary adrenal lymphoma: a systematic review. Ann Hematol. 2013;92:1583–1593. doi: 10.1007/s00277-013-1812-3. [DOI] [PubMed] [Google Scholar]
- 38.Sung J-Y, Lim S-J, Kim YW, Lee J. Prognostic significance of pSTAT3 and Survivin expression in diffuse large B-cell lymphoma. Basic Appl Pathol. 2010;3:7–13. doi: 10.1111/j.1755-9294.2009.01063.x. [DOI] [Google Scholar]
- 39.Wu ZL, Song YQ, Shi YF, Zhu J. High nuclear expression of STAT3 is associated with unfavorable prognosis in diffuse large B-cell lymphoma. J Hematol Oncol. 2011;4:31. doi: 10.1186/1756-8722-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Meng B, Iqbal J, Ding BB, Perry AM, Cao W, Smith LM, Bi C, Jiang C, Greiner TC, et al. Activation of the STAT3 signaling pathway is associated with poor survival in diffuse large B-cell lymphoma treated with R-CHOP. J Clin Oncol. 2013;31:4520–4528. doi: 10.1200/JCO.2012.45.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Llopiz D, Ruiz M, Infante S, Villanueva L, Silva L, Hervas-Stubbs S, Alignani D, Guruceaga E, Lasarte JJ, Sarobe P. IL-10 expression defines an immunosuppressive dendritic cell population induced by antitumor therapeutic vaccination. Oncotarget. 2017;8:2659–2671. doi: 10.18632/oncotarget.13736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu BS, Cao Y, Huizinga TW, Hafler DA, Toes RE. TLR-mediated STAT3 and ERK activation controls IL-10 secretion by human B cells. Eur J Immunol. 2014;44:2121–2129. doi: 10.1002/eji.201344341. [DOI] [PubMed] [Google Scholar]
- 43.Lu L, Zhu F, Zhang M, Li Y, Drennan AC, Kimpara S, Rumball I, Selzer C, Cameron H, Kellicut A, et al. Gene regulation and suppression of type I interferon signaling by STAT3 in diffuse large B cell lymphoma. Proc Natl Acad Sci USA. 2018;115:E498–E505. doi: 10.1073/pnas.1715118115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spranger S, Gajewski TF. Mechanisms of tumor cell—intrinsic immune evasion. Annu Rev Cancer Biol. 2018;2:213–228. doi: 10.1146/annurev-cancerbio-030617-050606. [DOI] [Google Scholar]
- 45.Wellenstein MD, de Visser KE. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48:399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 46.Straten PT, Andersen MH. The anti-apoptotic members of the Bcl-2 family are attractive tumor-associated antigens. Oncotarget. 2010;1:239–245. doi: 10.18632/oncotarget.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Kaplan–Meier curves of overall survival in total and R-CHOP-treated diffuse large B cell lymphoma patients. *T, tumor cell; I, immune cell.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.