Abstract
N6-methyladenosine (m6A) mRNA methylation is a gene regulatory mechanism affecting cell differentiation and proliferation in development and cancer. To study the roles of m6A mRNA methylation in cell proliferation and tumorigenicity, we investigated human endometrial cancer in which a hotspot R298P mutation is present in a key component of the methyltransferase complex (METTL14). We found ~70% of endometrial tumors exhibit reductions in m6A methylation that are likely due to either this METTL14 mutation or reduced expression of METTL3, another component of the methyltransferase complex. These changes lead to increased proliferation and tumorigenicity of endometrial cancer cells through activation of the AKT pathway. Reductions in m6A methylation lead to decreased expression of the negative AKT regulator PHLPP2 and increased expression of the positive AKT regulator mTORC2. Together, these results reveal reduced m6A mRNA methylation as an oncogenic mechanism in endometrial cancer and identify m6A methylation as a regulator of AKT signaling.
N6-methyladenosine (m6A) is the most prevalent mRNA modification in humans1,2. This modification is reversible3, and its biological effects are mostly mediated through “writer,” “eraser,” and “reader” proteins1,2. A writer complex, consisting of a core METTL3-METTL14 m6A methyltransferase along with regulatory subunits4–8, catalyzes the m6A methylation of mRNA. At least two eraser enzymes, FTO and ALKBH5, mediate the reversal of this methylation3,9. m6A methylated transcripts are recognized by reader proteins that regulate pre-mRNA processing10–14, translation15–19, and degradation10,19,20. m6A-dependent mRNA regulation is essential in mammals21, and defects in m6A methylation affect diverse biological processes1,2. In particular, m6A mRNA methylation regulates the self-renewal and differentiation of stem cells by affecting mRNA turnover during cell differentiation and plays critical roles in transcriptome switching during embryonic development8,21–23. Consistent with these roles, m6A mRNA methylation is emerging as a pathway affecting cancer initiation and progression in a variety of cancers24–35.
m6A mRNA methylation affects the growth and proliferation of stem cells and cancer cells8,21,22,26–35. However, how m6A methylation affects cell growth and which underlying pathways and mechanisms mediate these changes are still not fully elucidated. Herein, we study this question in endometrial cancer, where sequencing studies have identified frequent mutation of the m6A methyltransferase subunit METTL14(ref 36). We found that ~70% of endometrial tumors exhibit reduced m6A methylation compared to matched, normal endometrium. These reductions in m6A methylation were likely caused by either mutation of METTL14 or reduced expression of the METTL3 methyltransferase. Reducing m6A mRNA levels in endometrial cancer cells through either METTL14 mutation or METTL3 downregulation could enhance cell proliferation and tumorigenicity in vitro and in vivo. m6A-seq characterization of endometrial cancer patient tumors and cell lines revealed that reduced m6A mRNA methylation could promote cell proliferation by altering the expression of key enzymes that affect the AKT signaling pathway. Inhibition of AKT activation reversed the increased proliferation caused by reduced m6A methylation. Together, these results characterize and attribute a somatic mutation of the m6A methylation machinery as an important factor promoting cancer progression, reveal that reduced m6A mRNA methylation is most likely an oncogenic mechanism underlying a large portion of endometrial cancers, and identify m6A methylation as an important regulator of the AKT pathway and cell growth.
RESULTS
Loss of function METTL14 mutations in endometrial cancer.
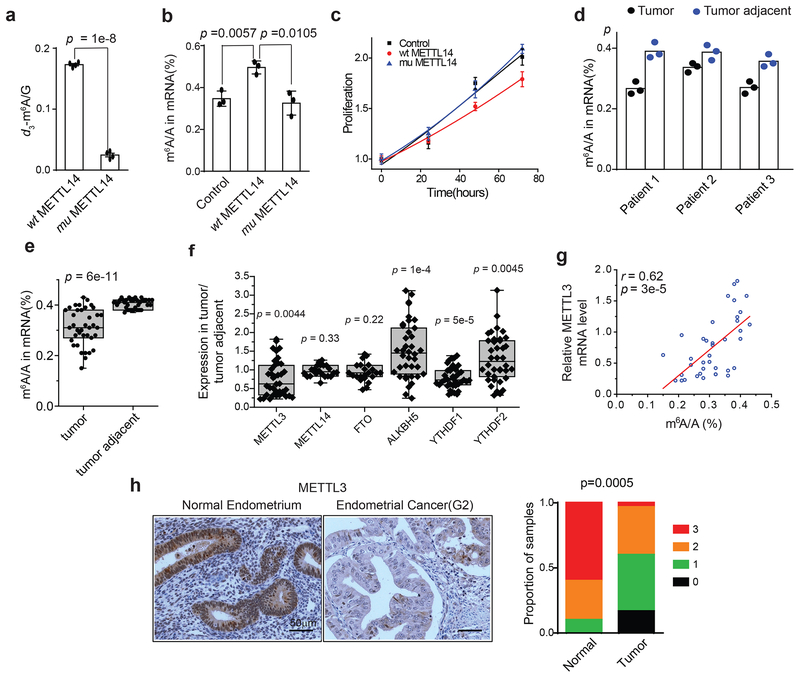
Sequencing studies have found that the METTL14 subunit of the core m6A methyltransferase complex is frequently mutated in endometrial tumors36, but the relevance of these mutations and of m6A mRNA methylation to the disease has not yet been established. The predominant mutation occurs at position 298 of METTL14, is more prevalent than other mutations in endometrial tumors and occurs in ~1.5% of endometrial cancer patients36. Crystal structures of the METTL3-METTL14 complex reveal that the R298 residue lies in the putative RNA-binding groove at the interface between the two subunits37–39. Consistent with previous observations38, we found that the R298P hotspot mutation significantly reduced the RNA methylation activity of the writer complex in vitro (Fig. 1a). Whereas overexpression of wild-type METTL14 promoted m6A methylation of cellular polyA RNAs in HEC-1-A endometrial cancer cells, the mutant METTL14 appeared inactive upon overexpression (Fig. 1b). While overexpression of wild-type METTL14 decreased cell proliferation, overexpression of the mutant had no noticeable effect on cell proliferation (Fig. 1c), suggesting that the METTL14 mutation is likely a loss of function allele that shows no evidence of further dominant negative effects on m6A methylation or cell proliferation.
Figure 1.
The METTL14(R298P) mutation and reduced METTL3 expression contribute to decreased m6A mRNA methylation in endometrial cancer patients. (a) The methyltransferase activity of the METTL3-METTL14 complex containing either the METTL14(R298P) mutant or wild-type METTL14 was determined by measuring the d3-m6A/G ratio by LC-MS/MS after incubation of the methyltransferase complex with RNA probe. We independently purified two batches of protein and performed two independent trials per protein preparation for a total of n = 4 independent trials. (b) LC-MS/MS quantification of the m6A/A ratio in polyA-RNA isolated from HEC-1-A cells overexpressing wild-type METTL14, mutant METTL14, or empty vector control. n = 3 biological replicates. (c) Cell proliferation of HEC-1-A cells was measured by MTS assay after transfection with the indicated reagents. n = 3 biological replicates. For panels a-c, error bars indicate mean ± s.e.m. (d) LC-MS/MS quantification of the m6A/A ratio in polyA-RNA isolated from three endometrial tumors with a METTL14(R298P) mutation and adjacent normal endometrium. The bar shows the mean from n = 3 technical replicates per patient. (e) Box plot of the relative m6A levels in polyA RNA isolated from endometrial tumor tissues versus tumor-adjacent tissues, n = 38 tumor-normal pairs. (f) Box plot of the expression levels of METTL3, METTL14, FTO, ALKBH5, YTHDF1 and YTHDF2 in tumor tissues relative to tumor-adjacent tissues, n = 22 tumor-normal pairs for METTL14 and FTO, and n = 38 tumor-normal pairs for the others. For panels a-c and e-f, the p-values were determined by two-tailed t-test. See Methods for box plot characteristics. (g) Scatter plot showing the correlation of m6A methylation level with the expression of METTL3. The linear best fit line shown in red. The Pearson correlation coefficient (r) and p-value (p) from a two-tailed t-test of r = 0 are shown, n = 38 tumor-normal pairs. (h) Left: Immunohistochemical staining of endometrial tissue microarray cores for METTL3. Right: Quantification of IHC staining in normal endometrium (n = 10 cores) and epithelial endometrial tumors (n = 30 cores). Staining was assessed using automated software55 and scored on a scale of 0 (no staining) to 3 (high staining). p-value determined by χ2-test.
To examine the consequence of the mutation in tumor tissue, we identified three endometrial tumor samples bearing the METTL14(R298P) mutation and purified mRNA from these tumors as well as from adjacent benign endometrial tissues (see Methods). Compared to mRNA from the wild-type adjacent normal tissues, mRNA from the three mutant tumors had reduced overall m6A methylation (p = 0.04, paired two-tailed t-test), suggesting that the METTL14(R298P) mutation inhibits m6A mRNA methylation in tumors (Fig. 1d).
Endometrial cancer is associated with low levels of m6A mRNA methylation.
Intriguingly, about 70% of all tumors we examined (including a majority of tumors with wild-type METTL14) exhibited reduced total m6A mRNA methylation compared to adjacent, normal endometrial tissues (Fig. 1e). Thus, we hypothesized that endometrial cancer could be more broadly associated with the altered expression of factors that regulate m6A mRNA methylation. To test this hypothesis, we evaluated the expression of m6A writer, erasers, and readers in tumor and adjacent normal endometrial tissues by RT-qPCR (Fig. 1f and Supplementary Fig. 1a). We found that a majority of endometrial cancers exhibited significantly reduced expression of the METTL3 m6A methyltransferase compared to adjacent normal tissues. Decreased METTL3 expression correlates with reduced m6A methylation in these tumor tissues (Fig. 1g). Immunohistochemistry of a tissue microarray with both normal endometrium and epithelial endometrial cancer specimens revealed a significant decrease in METTL3 expression in tumor tissue at the protein level (Fig. 1h). METTL14 mutation and decreased METTL3 expression appear to be mutually exclusive as all three of the tumors with the METTL14 mutation had normal expression of METTL3 relative to adjacent normal tissues. Analysis of the TCGA endometrial cancer dataset did not reveal any significant correlation between the mutation status of frequently mutated genes in endometrial cancer and low METTL3 expression (Supplementary Fig. 1b). Taken together, these results suggest that a large portion of human endometrial tumors are characterized by reduced m6A mRNA methylation, either through METTL14 loss of function mutation or decreased METTL3 expression.
Reduced m6A methylation promotes endometrial cancer cell proliferation.
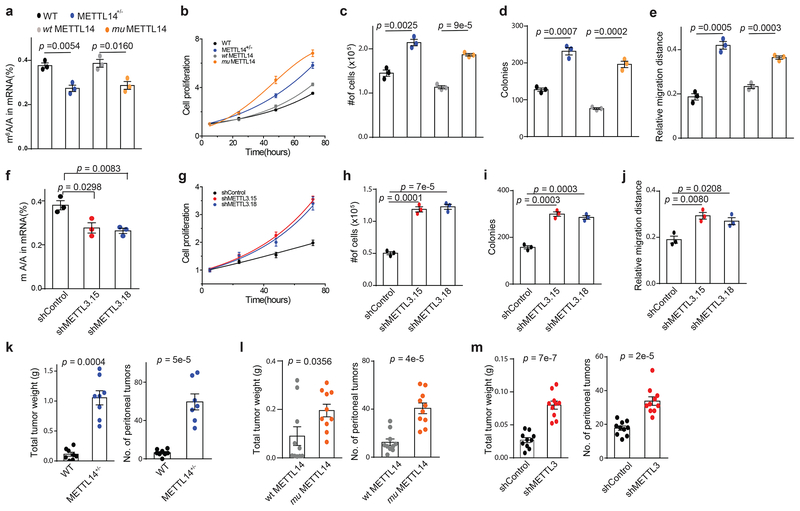
Considering that ~20–30% of all mRNAs are methylated inside most mammalian cells10,40, the observed global decrease in m6A mRNA methylation could have significant effects on cellular physiology, in particular if the methylation of key transcripts is dramatically affected25–31. Analysis of patients from the TCGA dataset showed that tumors with low METTL3 expression are associated with a slight increase in mortality, though this difference is not statistically significant (Supplementary Fig. 1c). However, analysis of other related cancer types (high grade serous ovarian cancer41 and pancreatic adenocarcinoma42) found statistically significant increases in mortality associated with decreased METTL3 expression (Supplementary Fig. 1c). Therefore, we next examined whether the reduced m6A methylation observed in the human endometrial tumor tissue samples affects functions associated with tumor progression in human tumor cells. To investigate the effects of METTL14 loss of function in endometrial cancer cell lines, we used CRISPR technology to delete METTL14 in HEC-1-A cells. We obtained clones exhibiting only heterozygous knockout of METTL14, reflecting the essential nature of the writer complex in mammals. Heterozygous knockout was confirmed by western blot (Supplementary Fig. 2a) and sequencing (see Methods). As expected, the METTL14+/– cells exhibited reduced m6A mRNA methylation, and the reductions in m6A methylation are similar to those observed in the tumor samples (Fig. 2a). Consistent with a role for METTL14 loss of function in endometrial cancer, heterozygous knockout of METTL14 increased cell proliferation, anchorage-independent growth, colony formation, cell migration, and invasion (Fig. 2b–e and Supplementary Fig. 2b). The reduced m6A methylation and changes to the cancer cell physiology could be partially rescued by stable expression of wild-type METTL14 but not mutant METTL14 (Fig. 2 and Supplementary Fig. 2b). We observed similar effects after shRNA knockdown of METTL14 versus control shRNA (Supplementary Fig. 2a, c–h).
Figure 2.
Reduced m6A methylation increases cell proliferation, anchorage-independent growth, and migration and in vivo tumor growth. (a) LC-MS/MS quantification of the m6A/A ratio in polyA-RNA from the indicated HEC-1-A cell lines. (b) Cell proliferation measured by MTS assay of wild-type HEC-1-A cells, METTL14+/− knockout cells, and knockout cells rescued by stable transfection of wild-type METTL14 or METTL14(R298P). Cell numbers were normalized to the MTS signal ~ 5 h after cell seeding. (c-e) Anchorage-independent cell growth (c), colony formation (d), cell migration in a wound healing experiment (e) were assessed for wild-type HEC-1A cells, METTL14+/− knockout cells, and knockout cells rescued with wild-type or mutant METL14. (f) LC-MS/MS quantification of the m6A/A ratio in polyA-RNA from the indicated HEC-1-A cell lines. (g) Cell proliferation measured by MTS assay of HEC-1-A cells stably expressing control shRNA versus shRNA targeting METTL3. Cell numbers were normalized to the MTS signal ~ 5 h after cell seeding. (h-j) Anchorage-independent cell growth (h), colony formation (i), cell migration in a wound healing assay (j) were assessed for HEC-1A cells stably expressing control shRNA or shRNA targeting METTL3. For panels a-j, n = 3 biological replicates. Error bars indicate mean ± s.e.m. p-values determined by two-tailed t-test. (k-m) Wild type HEC-1-A cells and METTL14+/− knockout cells (k), knockout cells rescued with wild-type or mutant METTL14 (l), and HEC-1-A cells with shRNA knockdown of METTL3 or control shRNA (m) were injected into mice. The total tumor weight (left) and the total number of tumors (right) were recorded after 2–3 weeks. For panel k, n – 8 and for panels l and m n = 10 mice per group. Error bars indicate mean ± s.e.m. p-values determined by two-tailed t-test.
To determine the effects of reduced METTL3 expression in endometrial cancer cells, METTL3 was stably knocked down by shRNA in HEC-1-A cells using two different shRNA sequences (Supplementary Fig. 2i). Similar to the expression of mutant METTL14, knockdown of METTL3 decreased the overall levels of m6A mRNA methylation and promoted cell proliferation, anchorage-independent growth, colony formation, migration and invasion relative to control cells (Fig. 2f–j and Supplementary Fig. 2j).
To corroborate the observations that reduced m6A methylation stimulates aggressive phenotypes of cancer cells in vitro, we investigated the roles of METTL14 and METTL3 in tumor growth in vivo. Wild-type HEC-1-A cells and METTL14+/− knockout cells were injected into the peritoneal cavity of nude mice, and tumor numbers and total mass were evaluated after 2–3 weeks. METTL14+/− knockout cells showed dramatically larger tumors and an increased number of metastases relative to wild-type HEC-1-A cells (Fig. 2k). Similar trends were observed when METTL14+/− cells rescued with wild-type and mutant METTL14 were compared (Fig. 2l) and when comparing METTL3 knockdown HEC-1-A cells to control (Fig. 2m). Taken together, these results reveal that the reduced m6A mRNA methylation observed in the patient endometrial tumor samples, whether induced by METTL14(R298P) mutation or reduced METTL3 expression, could promote the tumorigenicity of endometrial cancer cells and might play critical roles in the progression of endometrial cancer.
m6A-seq identifies transcripts with altered methylation in endometrial tumors.
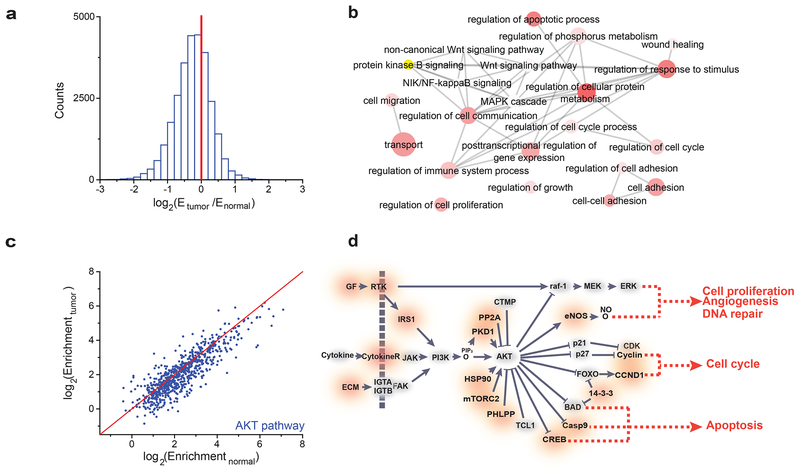
Next, we performed m6A-seq analysis of human endometrial tumor tissues versus normal, tumor-adjacent tissues from five patients. All five tumors exhibited low total m6A levels; one carried the METTL14 mutation and the four others exhibited low METTL3 expression (Supplementary Fig. 3a,b). Consistent with previous m6A-seq results10,40, the m6A peaks we identified were enriched near the start and stop codons and were characterized by the canonical GGACU motif (Supplementary Fig. 3c,d). In the normal tissue samples, we identified on average ~20,000 significant m6A peaks (FDR < 0.05) in ~8,000 transcripts, and the identified transcripts show good agreement between samples (Supplementary Fig. 3e). Among the m6A peaks detected in over half of the patient samples, we found that their m6A mRNA methylation was reduced globally in the tumor compartment compared to adjacent, normal control tissues (Fig. 3a). The transcripts exhibiting decreased m6A methylation were fairly consistent between tissue samples (Supplementary Fig. 3f), and the transcripts showing decreased m6A methylation in at least two samples were enriched for GO terms related to cell migration, proliferation, growth, adhesion, and cell death (Fig. 3b). m6A-seq experiments revealed similar global decreases in m6A methylation and GO term enrichment in HEC-1-A METTL3 knockdown and mutant METTL14 cells relative to controls (Supplementary Fig. 3e–g and Supplementary Fig. 4a).
Figure 3.
m6A-seq of tumors with reduced m6A methylation. (a) Histogram showing the changes in m6A enrichment between normal and tumor samples of all peaks showing enrichment in the normal tissue. The change in enrichment is the median of n = 5 tumor-normal pairs. (b) GO term analysis of transcripts with reduced m6A in tumor tissues versus adjacent normal tissues. (c) Scatter plot of the m6A enrichment in normal, tumor-adjacent and tumor tissue for m6A peaks in genes involved in the PI3K/AKT pathway. The red line is the y = x line. 650/765 of the m6A peaks examined show greater enrichment in the normal sample than the tumor sample. The enrichment values are the median of n = 5 patient samples [AU: please indicate which statistical analysis was performed]. (d) Diagram of the PI3K/AKT pathway with genes affected by m6A marked in red. Diagram is based on KEGG annotations63.
m6A methylation regulates activation of AKT.
The GO term analysis identified the AKT/Protein Kinase B signaling pathway as being significantly altered by reduced m6A methylation in both the patient samples (p-value = 1.51e–8) and the endometrial cancer cell lines (p-value = 1.02e–8) (Fig. 3b and Supplementary Fig. 4a). Because the AKT signaling pathway promotes cell survival and growth and is frequently activated through oncogenic mutations in endometrial cancer and other cancers43–45, we hypothesized that reduced m6A methylation might promote tumor growth through activation of the AKT pathway. Indeed, many of the genes involved in the AKT pathway showed reduced m6A methylation in tumors compared to tumor-adjacent tissues (Fig. 3c,d). We evaluated a subset of these transcripts in the mutant METTL14 and METTL3 knockdown cells by m6A-immunoprecipitation (m6A-IP) followed by RT-qPCR, which confirmed their reduced m6A methylation (Supplementary Fig. 4b–e).
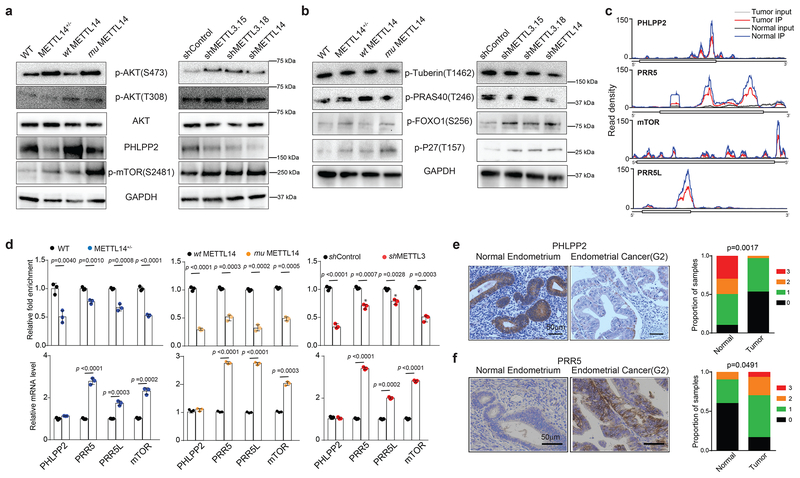
We next determined if reduced m6A methylation in endometrial cancer cells affects AKT signaling by investigating the phosphorylation status of AKT. Our METTL14 loss of function HEC-1-A cell lines (METTL14+/−, mutant METTL14 rescue, and shMETTL14) showed increased phosphorylation of AKT at Ser-473 compared to the relevant control cell lines (wild-type HEC-1-A cells, wild-type METTL14 rescue, and shControl, respectively) (Fig 4a). Similar increases in AKT(S473) phosphorylation were seen in the METTL3 knockdown HEC-1-A cell lines relative to control knockdown cells (Fig. 4a). In contrast, phosphorylation at Thr-308 and the total AKT protein expression remained unchanged (Fig. 4a). To assess whether these changes in AKT phosphorylation stimulate AKT signaling, we assessed the phosphorylation status of downstream effectors of AKT (Fig. 4b). Both FOXO1 and p27 showed increased phosphorylation in the METTL14 loss of function and METTL3 knockdown cells relative to control. Two other AKT substrates, Tuberin and PRAS40, showed no consistent changes in phosphorylation, congruent with previous reports that only a subset of AKT targets are affected by changes to Ser-473 phosphorylation without difference in Thr-303 phosphorylation46. These results suggest that reducing m6A methylation activates the AKT pathway.
Figure 4.
Reduced m6A methylation activates AKT. (a) Immunoblot analyzing levels of AKT phosphorylation and expression of proteins that regulate AKT phosphorylation in HEC-1-A cells with the indicated perturbations to m6A methylation. (b) Immunoblot examining the phosphorylation of AKT target proteins in HEC-1-A cells with the indicated perturbations to m6A methylation. Quantification of the immunoblots in panels a and b are presented in Supplementary Fig. 4f. Not all panels shown are from the same immunoblot, and raw gel images with the appropriate loading controls are provided in Supplementary Figure 8. (c) The average read density from m6A-seq experiments on n = 5 tumor-normal pairs showing the m6A peaks identified in the PHLPP2, PRR5, mTOR, and PRR5L transcripts. (d) m6A IP combined with RT-qPCR was used to quantify the relative m6A level (top) and mRNA levels (bottom) of PHLPP2, PRR5, PRR5L and mTOR transcripts in the wild type, METTL14+/−, wild-type METTL14, mutant METTL14, shControl, and shMETTL3 HEC-1-A cells. Error bars indicate mean ± s.e.m from n = 3 biological replicates. p-values determined by two-tailed t-test. (e,f) Left: Immunohistochemical staining of tissue microarray cores for PHLPP2 (e) and PRR5 (f). Right: Quantification of IHC staining in normal endometrium (n = 10) and endometrial tumors (n = 30). Staining was assessed using automated software55 and scored on a scale of 0 (no staining) to 3 (high staining). The p-value was determined by a χ2-test.
m6A methylation controls the expression of regulators of AKT activation.
To determine the mechanisms underlying increased AKT activation upon reduced m6A methylation, we examined PHLPP2, a phosphatase regulating AKT(S473) phosphorylation47, and mTORC2, a kinase that phosphorylates AKT(S473)48. Transcripts encoding PHLPP2 and three components of the mTORC2 complex (PRR5, PRR5L, and mTOR) showed decreased m6A methylation in patient samples (Fig. 4c). These transcripts also showed decreased m6A methylation in the METTL14 loss-of-function and METTL3 knockdown HEC-1-A cell lines (Fig. 4d). In these cell lines, we observed decreased expression of PHLPP2 protein, while its mRNA levels were not noticeably altered; in contrast, we observed increased mRNA expression of PRR5 PRR5L and mTOR in addition to increased protein levels of mTOR and p-mTOR(S2481), a marker for mTORC2(ref 49) (Fig. 4a, d). These changes are consistent with increased AKT phosphorylation and activity. To investigate whether these changes in protein expression occur in tumor cells, we performed immunohistochemical staining in normal endometrium and endometrial tumors in a tissue microarray (Fig. 4e,f and Supplementary Fig 4g,h). PHLPP2 was indeed downregulated in human endometrial tumors compared to benign endometrial glands. We observed increased staining for PRR5, PRR5L, and phospho-mTOR(S2481) in a subset of tumors, though the increases were not always statistically significant, suggesting that additional factors may be influencing mTORC2 expression in tumors compared to our cell lines.
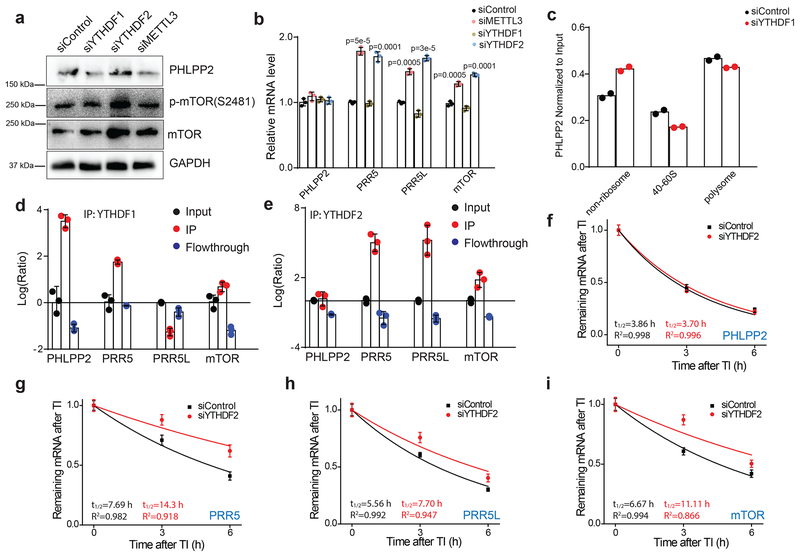
We next explored the mechanism for how m6A methylation regulates the expression of PHLPP2 and mTORC2. Because m6A methylation appeared to promote the expression of PHLPP2, we hypothesized that PHLPP2 transcripts are targets of YTHDF1, the m6A reader protein that promotes translation of m6A methylated transcripts16. Consistent with this hypothesis, siRNA knockdown of YTHDF1 in HEC-1-A cells decreased expression of PHLPP2 to a similar extent as knockdown of METTL3 (Fig. 5a). These changes in PHLPP2 expression are not due to changes in the abundance of the PHLPP2 transcript but the association of the PHLPP2 transcript with actively transcribing ribosomes (Fig. 5b,c and Supplementary Fig. 5b,c). mTORC2, on the other hand, appeared to be downregulated by m6A methylation, suggesting that it is a target of YTHDF2, the m6A reader protein that promotes the decay of m6A methylated transcripts20. Consistent with this hypothesis, siRNA knockdown of YTHDF2 increased the abundance of the PRR5, PRR5L and mTOR transcripts and these transcripts showed decreased RNA decay rates upon knockdown of YTHDF2 (Fig. 5b,g–i). Knockdown of YTHDF2 also resulted in higher protein levels of mTOR and p-mTOR(S2481) (Fig. 5a). The effects of the reader proteins appeared to be specific to these sets of transcripts as knockdown of YTHDF2 did not appreciably affect the expression or decay of PHLPP2 (Fig. 5a,b,f), while knockdown of YTHDF1 does not appreciably affect the expression of mTOR or p-mTOR(S2481) nor the association of PRR5, PRR5L or mTOR transcripts with ribosomes (Fig. 5a and Supplementary Fig 5d–f). The specificity for reader function likely occurs at the level of RNA binding as PHLPP2 transcripts interacted more strongly with YTHDF1 than YTHDF2 while PRR5, PRR5L and mTOR transcripts interacted more strongly with YTHDF2 than YTHDF1 (Fig. 5d,e).
Figure 5.
Regulation of AKT pathway genes by m6A reader proteins. (a) Immunoblot analyzing the levels of PHLPP2, mTOR, and p-mTOR(S2481) in HEC-1-A cells upon transient siRNA knockdown of YTHDF1, YTHDF2 or METTL3. Quantification of this immunoblot is shown in Supplementary Fig. 5a. Raw gel images are provided in Supplementary Fig. 8. (b) RT-qPCR was used to quantify the relative levels of PHLPP2, PRR5, PRR5L and mTOR upon transient siRNA knockdown of YTHDF1, YTHDF2, or METTL3 in HEC-1-A cells. Error bars indicate mean ± s.e.m from n = 3 biological replicates. p-values determined by two-tailed t-test. (c) Polysome profiling was used to examine the distribution of PHLLP2 transcripts among non-ribosomal, ribosome-associated and polysome-associated fractions. n = 2 biological replicates. (d-e) YTHDF1 (d) and YTHDF2 (e) were immunoprecipitated and RIP-qPCR was used to assess the association of the indicated transcripts with each protein. n = 3 biological replicates. Error bars indicate mean ± s.e.m. (f-i) RNA lifetime for PHLPP2 (f), PRR5 (g), PRR5L (h), and mTOR (i) in HEC-1-A cells transfected with control siRNA or siRNA targeting YTHDF2. n = 3 biological replicates, and error bars indicate mean ± s.e.m. For details on the determination of the decay half-lives, see the Methods. [AU: for d-i, please indicate the statistical assays].
To better understand the effects of m6A-mediated changes to PHLPP2 and mTORC2 expression on AKT activation, we examined the time course of AKT activation upon stimulation of HEC-1-A cells. Consistent with previous studies47, we observed transient phosphorylation of AKT in our knockdown control cells after EGF stimulation, which decreased after 0.5–1 h; however, in the METTL3 knockdown cell line, AKT activation persisted for much longer periods of time (Supplementary Fig 6a), consistent with decreased PHLPP2 expression. Similar results were observed for the wild-type METTL14 versus mutant METTL14 HEC-1-A cells (Supplementary Fig. 6b), though the kinetics of dephosphorylation were slower in the wild-type METTL14 cell line than in the knockdown control cell line, perhaps reflecting incomplete rescue of the HEC-1-A METTL14+/− cells.
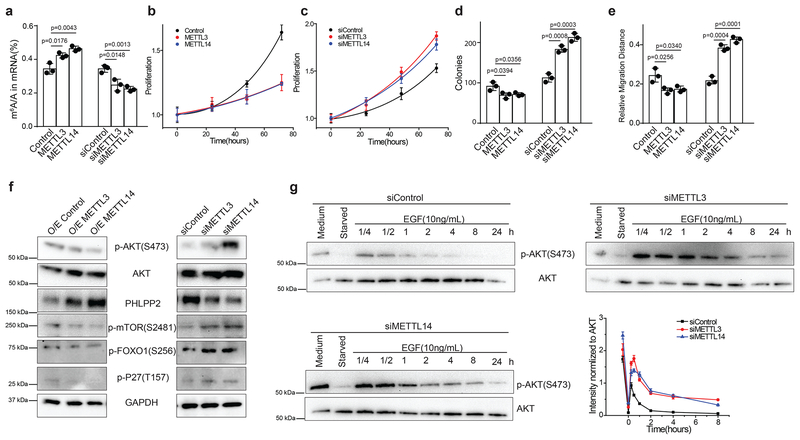
To confirm that our results extend beyond the HEC-1-A endometrial cancer cell line, we tested the effects of knockdown and overexpression of METTL3 and METTL14 in a second endometrial cancer cell line, RL95–2, as well as hTert-immortalized human endometrial stromal cells (T-HESCs), a normal non-transformed cell line, and found similar m6A-mediated changes to cell proliferation and AKT signaling (Fig. 6 and Supplementary Fig. 6c–f). Thus, decreased m6A methylation leads to diminished translation of PHLPP2, a negative regulator of AKT activation, and precludes the YTHDF2-promoted decay of mRNAs encoding the mTORC2 complex, a positive regulator of AKT. Altogether, these experiments reveal that reduced m6A mRNA methylation affects multiple AKT pathway components to stimulate AKT activation.
Figure 6.
Effects of m6A methylation on non-transformed T-HESC endometrial cell line. (a-g) Effects of alterations to m6A methylation on non-transformed T-HESC endometrial cells were examined after transient transfection with control siRNA, siRNA targeting METTL3, siRNA targeting METTL14, empty vector, plasmid encoding METTL3 or plasmid encoding METTL14. (a) LC-MS/MS quantification of the m6A/A ratio in polyA-RNA after transient transfection of T-HESC cells after the indicated treatments. (b,c) Cell proliferation measured by MTS assay of T-HESCs transfected with the indicated reagents. Cell numbers were normalized to the MTS signal ~ 5 h after cell seeding. (d) Colony formation of T-HESCs transformed with the indicated reagents. (e) Migration in a wound-healing assay. For panels a-e, n = 3 biological replicates and error bars indicate mean ± s.e.m. p-values determined by two-tailed t-test. (f) Immunoblot showing the effects of the indicated perturbations to m6A methylation on the expression and phosphorylation of proteins involved in the AKT pathway in T-HESCs. Three independent experiments have been repeated with similar results. (g) Immunoblots showing the time course of AKT(S473) phosphorylation after EGF stimulation in T-HESCs treated with control siRNA or siRNAs targeting METT3 or METTL14 for 48 h. Plots quantifying the time-course of EGF activation show mean ± s.e.m. from n = 3 biological replicates. Raw gel images for panels f,g are provided in Supplementary Fig. 8.
Increased AKT signaling mediates the effects of reduced m6A methylation on cell proliferation.
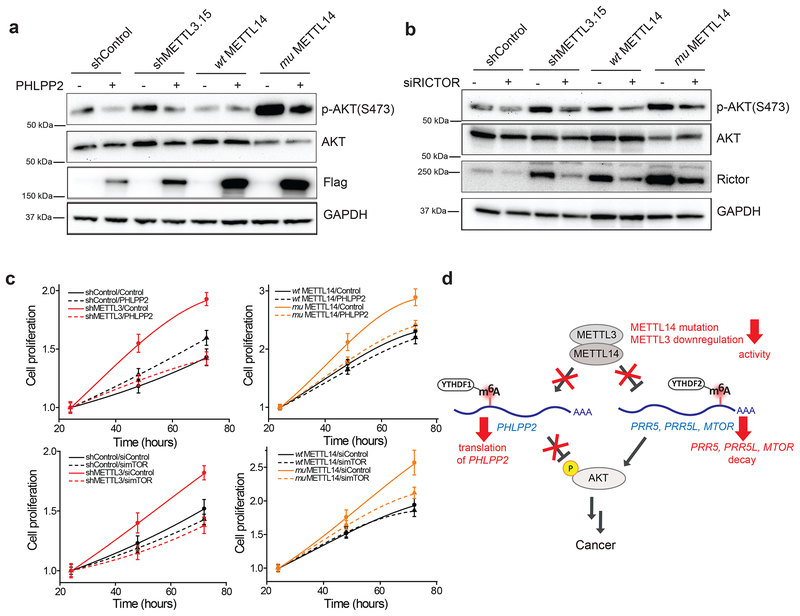
To determine if enhanced AKT activation underlies the increased proliferation observed upon reducing m6A methylation in endometrial cancer cells, we attempted to rescue this phenotype by either overexpressing PHLPP2 or by inhibiting mTORC2 through knockdown of the mTORC2-specific subunit RICTOR. Consistent with previous results47,48, overexpression of PHLPP2 (Fig. 7a and Supplementary Fig. 7a) and knockdown of RICTOR (Fig. 7b and Supplementary Fig. 7b) both decreased the levels of p-AKT(S473) in METTL3 knockdown, METTL14 mutant, and METTL14+/− HEC-1-A cells. PHLPP2 overexpression or RICTOR knockdown in METTL3 knockdown cells reduced cell proliferation rates, whereas these treatments had much smaller effects on the control cells (Fig. 7c). Importantly, the cell proliferation rates of METTL3 knockdown cells after PHLPP2 overexpression or RICTOR knockdown were comparable to those of the control cells with normal METTL3 expression. Similar results were observed in the METTL14 mutant cells (Fig. 7c) as well as the METTL14+/− cells (Supplementary Fig. 7c,d). Similar results were also seen when using a small molecule inhibitor of the AKT enzyme (Supplementary Fig. 7e,f). Thus, genetic or pharmacologic suppression of AKT reverses the increased proliferation observed in METTL3 knockdown and METTL14 loss of function cells.
Figure 7.
The AKT pathway mediates the changes in cell proliferation from reduced m6A methylation. (a,b) Immunoblots analyzing the effect of FLAG-PHLPP2 overexpression (a) or RICTOR knockdown (b) on AKT phosphorylation in HEC-1-A cells. Three independent experiments have been replicated with similar results. Raw gel images are provided in Supplementary Fig. 8. (c) Proliferation measured by MTS assay of METTL3 knockdown versus control knockdown cells (left) or wild-type METTL14 versus mutant METTL14 HEC-1-A cells (right). Cells were transiently transfected with a PHLPP2 overexpression plasmid versus empty vector (top) or siRNAs targeting RICTOR versus negative control siRNAs (bottom), n = 3 biological replicates; error bars indicate mean ± s.e.m. [AU: please indicate the statistical assays] (d) Model showing how reduced m6A methylation alters AKT signaling to contribute to tumor progression.
DISCUSSION
The PI3K/AKT pathway plays important roles in a variety of biological processes, and dysfunctional AKT signaling contributes to diseases such as cancer, diabetes, and autoimmune disease44,45. In this study, we discovered that m6A mRNA methylation regulates the AKT pathway to control cell proliferation in endometrial cancer (Fig. 7d). m6A methylation normally attenuates AKT activity in the endometrium by promoting the m6A-dependent translation of PHLPP2 and m6A-dependent degradation of transcripts encoding subunits of mTORC2. Upregulation of the PHLPP2 phosphatase and downregulation of the mTORC2 kinase both contribute to the inhibition of AKT activity by maintaining dephosphorylation of AKT(S473). Reduced m6A methylation disrupts the regulation of these transcripts, leading to decreased PHLPP2 expression, increased mTORC2 expression, and increased AKT activity (Fig. 7d).
This mechanism likely contributes to a large fraction of endometrial tumors as ~70% of tumor samples from endometrial cancer patients exhibited decreased m6A levels due to either decreased expression of METTL3 or loss of function mutation in METTL14. Using cultured endometrial cancer cells, we revealed that either mutation of METTL14 or downregulation of METTL3 reduced m6A mRNA methylation and enhanced proliferation and tumorigenicity. Our m6A-seq results from endometrial tumors and matched normal tissue along with our mechanistic studies in endometrial cancer cells reveal regulation of AKT activation as an important mediator of these changes to cell proliferation. Increased AKT activation is likely one of the main mediators of increased proliferation in cells with reduced m6A methylation, as inhibition of AKT is sufficient to rescue the changes in cell proliferation. However, we cannot rule out the involvement of other signaling pathways that could be altered directly or indirectly by changes to m6A methylation.
Because AKT is known to be an important regulator of cell proliferation, growth and survival in many cancers, these findings may be applicable beyond endometrial cancer to other cancers driven by increased AKT signaling. Other types of tumors could exploit aberrant RNA methylation to gain survival and growth advantages via AKT activation in addition to other proposed mechanisms24–35. Indeed, others have observed increased proliferation of stem cells and cancer cells with reduced m6A methylation21,22,26–31, and while this paper was under review, m6A methylation was reported to affect AKT activity in AML33, renal cell carcinoma30, and T-cell differentiation50. Although our results suggest that decreased m6A methylation promotes tumorigenesis in the endometrium, other cancers are associated with high METTL3 expression and increased m6A methylation and could involve different mechanisms24,32–34. Nevertheless, our results suggest that regulation of AKT activity through m6A methylation could be a general growth control mechanism that affects a range of other biological processes, which will be a new direction to explore in the future.
METHODS
Cell lines, antibodies, siRNA knockdown and plasmid transfection.
The HEC-1-A cells used in this study were purchased from ATCC (HTB-112) and grown in McCoy’s 5A medium (Gibco, 16600) supplemented with 10% FBS (Gibco), and 1% Penicillin-Streptomycin (Gibco, 15140). Cells were free of mycoplasma (IDEXX STAT-Myco). The RL95–2 cells used in this study were purchased from ATCC (CRL-1671) and grown in DMEM:F-12 medium (Gibco, 11320) supplemented with 10% FBS, 0.005 mg/mL insulin (Sigma I0516), and 1% Penicillin-Streptomycin. The T-HESCs used in this study were purchased from ATCC (CRL-4003) and grown in DMEM:F-12 medium supplemented with 10% FBS, 1% ITS-premix (Corning 354352), 1 mM pyruvate (Gibco, 11360), 0.5 μg/mL puromycin (Gibco, A11138), and 1% Penicillin-Streptomycin. The HEC-1-A cell line was authenticated with STR profiling (IDEXX Cell Check 9 Plus). The other cell lines were not authenticated.
The primary antibodies were purchased from commercial sources, and information about the antibodies are given in Supplementary Table 1. Actinomycin D (A9415) was purchased from Sigma, recombinant human EGF (PHG0311) was purchased from Thermo Fisher Scientific, MK-2206 (S1078) was purchased from Selleckchem, and cyclohexamide (C7698) was purchased from Sigma.
The pcDNA3-HA-PHLPP2 plasmid was a gift from Alexandra Newton (Addgene plasmid #22403)47. Construction of the pcDNA3 plasmids for the expression of METTL3 and METTL14 in mammalian cells was described previously5. All the siRNAs were ordered from QIAGEN. Allstars negative control siRNA (1027281) was used as siRNA control. Sequences for the other siRNA are: METTL3, 5’-CGTCAGTATCTTGGGCAAGTT-3’; YTHDF1, 5’- CCGCGTCTAGTTGTTCATGAA-3’; YTHDF2, 5’-AAGGACGTTCCCAATAGCCAA-3’; RICTOR, 5’-TAGGTGCATTGACATACAACA-3’. Transfection was achieved by using Lipofectamine RNAiMAX (Invitrogen) for siRNA, or Lipofectamine 2000 (Invitrogen) for the plasmids following manufacturer’s protocols.
Patient sample collection, genomic DNA and RNA extraction, and genotyping.
All samples were obtained with informed consent under a protocol approved by the University of Chicago Institutional Review Board or the ethics committee of Zhongnan Hospital of Wuhan University, China. The study is compliant with all relevant ethical regulations regarding research involving human participants. Information about the patient sex, age, and tumor characteristics are given in Supplementary Table 2. For fresh tissues, endometrial tumor and adjacent normal endometrium were separately dissected at the time of surgery and immediately transferred to RNAlater (Thermo Fisher, AM7021). Tissues were homogenized in Trizol reagent (Thermo Fisher, 15596026) with a Tissure-Tearor (Biospec Products, Inc, 985–370). RNA and DNA were extracted following the manufacturer’s instructions. For archival formalin-fixed, paraffin-embedded (FFPE) specimens, 10 × 10 μm scrolls were collected from FFPE blocks at the University of Chicago Human Tissue Resource Center. Samples were processed with a High Pure FFPE RNA Micro Kit (Roche, 04823125001) following the manufacturer’s instructions. We used the TOPO-TA cloning kit (Invitrogen) to assess METTL14 mutation status in patient samples following the manufacturer’s instructions. Primers used for subcloning shown here: Forward, 5’- ATCCCAAAGATTCCGAGAAATGAGG-3’; Reverse, 5’- TGAGGTCCTACCTGGTCGAATTGT-3’.
In vitro assay for m6A methyltransferase activity.
The recombinant, FLAG-tagged proteins METTL3, METTL14 and METTL14(R298P) were expressed in insect cells using the Bac-to-Bac baculovirus expression system and purified through FLAG-tag immunoprecipitation according to a previously published procedure5. Protein purity was assessed by SDS-PAGE, and protein concentration was determined by UV absorbance at 280 nm.
We performed an in vitro methyltransferase activity assay in a 50 μL reaction mixture containing the following components: 0.15 nmol RNA probe, 0.15 nmol each fresh recombinant protein (METTL3 combination with an equimolar ratio of METTL14 or mutant METTL14), 0.8 mM d3-SAM, 80 mM KCl, 1.5 mM MgCl2, 0.2 U μL−1 RNasin, 10 mM DTT, 4% glycerol and 15 mM HEPES (pH 7.9). The reaction was incubated for 12 h at 16°C, RNA was recovered by phenol/chloroform (low pH) extraction followed by ethanol precipitation and was digested by nuclease P1 and alkaline phosphatase for LC-MS/MS detection. The nucleosides were quantified by using the nucleoside-to-base ion mass transitions of 285 to 153 (d3-m6A) and 284 to 152 (G).
Construction of the stable cell lines.
To construct the METTL14 mutant cell line, we first used the CRISPR-Cas9 genome editing system from IDT to generate a METTL14+/− cell line in HEC-1-A cells following the manufacturer’s protocols. The guide RNA sequencing targeting METTL14 was 5’-GCTCCCGGATCTCCTGCAAGCGG-3’. Heterozygous METTL14 knockout cells were identified by western blotting for METTL14 and targeted Sanger sequencing. Sanger sequencing identified a 16nt deletion removing sequences encoding amino acids M1 to Q6, eliminating the start codon. Next, we transfected the METTL14+/− cell lines with a PiggyBac Transposon System (SBI) encoding FLAG-tagged wild-type METTL14 or METTL14(R298P). Stable transformants were selected with 1 μg/mL puromycin and confirmed by western blotting for the FLAG tag and METTL14.
To construct the METTL3 knockdown and control cell lines, we used the TRC Lentiviral Human shRNA system encoding a control shRNA or a shRNAs targeting METTL3 (RHS4533-EG56339, Dharmacon). Lentivirus was generated by transfection of 293T cells with shRNA constructs, VSV-G (Addgene, 8454) and gag/pol (pCMVΔR8.2; Addgene, 8455) vectors51. Viral supernatants were collected at 48 and 72 h post-transfection, filtered, and added to target cells for 4 h. Stable transformants were selected with 1 μg/mL puromycin and confirmed by immunoblotting for METTL3.
RNA isolation.
Total RNA was isolated with TRIZOL reagent (Invitrogen). mRNA was extracted from the total RNA using the Dynabeads® mRNA Purification Kit (Invitrogen), followed by removal of contaminating rRNA with the RiboMinus transcriptome isolation kit (Invitrogen). mRNA concentration was measured by UV absorbance at 260 nm.
Total RNA samples used for RT-qPCR were isolated by using the RNeasy kit (Qiagen) with an additional on-column DNase-I digestion step.
RT-qPCR.
Quantitative reverse transcription PCR (RT-qPCR) was used to assess the relative abundance of mRNA. Total RNA or purified mRNA was reverse transcribed with SuperScript II Reverse Transcriptase (Invitrogen) using poly(dA) primers to obtain cDNA. qPCR was performed by using SYBR Premix Ex Taq II (Takara). GAPDH (for mRNA expression level) and HPRT1 (for mRNA stability and m6A IP) were used as an internal control. Primers used for RT-qPCR are:
METTL3_For, CTATCTCCTGGCACTCGCAAGA;
METTL3_Rev, GCTTGAACCGTGCAACCACATC;
YTHDF1_For, CAAGCACACAACCTCCATCTTCG;
YTHDF1_Rev, GTAAGAAACTGGTTCGCCCTCAT;
YTHDF2_For, TAGCCAGCTACAAGCACACCAC;
YTHDF2_Rev, CAACCGTTGCTGCAGTCTGTGT;
FTO_For, CCAGAACCTGAGGAGAGAATGG;
FTO_Rev, CGATGTCTGTGAGGTCAAACGG;
ALKBH5_For, CCAGCTATGCTTCAGATCGCCT;
ALKBH5_Rev, GGTTCTCTTCCTTGTCCATCTCC;
CCND1_For, TCTACACCGACAACTCCATCCG;
CCND1_Rev, TCTGGCATTTTGGAGAGGAAGTG;
IRS1_For, AGTCTGTCGTCCAGTAGCACCA;
IRS1_Rev, ACTGGAGCCATACTCATCCGAG;
HSP90AA1_For, TCTGCCTCTGGTGATGAGATGG;
HSP90AA1_Rev, CGTTCCACAAAGGCTGAGTTAGC;
HPRT1_For, CATTATGCTGAGGATTTGGAAAGG;
HPRT1_Rev, CTTGAGCACACAGAGGGCTACA;
GAPDH_For, AGAAGGCTGGGGCTCATTTG;
GAPDH_Rev, AGGGGCCATCCACAGTCTTC;
IGF1R_For, CCTGCACAACTCCATCTTCGTG;
IGF1R_Rev, CGGTGATGTTGTAGGTGTCTGC;
SGK1_For, GCTGAAATAGCCAGTGCCTTGG;
SGK1_Rev, GTTCTCCTTGCAGAGTCCGAAG;
IGFR1_For, ATACAGGTGCCAGAGAGGTCTC;
IGFR1_Rev, CCAGCTTATCCTTCCACGCATG;
PDGFB_For, GAGATGCTGAGTGACCACTCGA;
PDGFB_Rev, GTCATGTTCAGGTCCAACTCGG;
PHLPP2_For, CCTTCCAACACTGGTAGAGCAC;
PHLPP2_Rev, CGGATGGTAAAGACTCCAGACTA;
PRR5_For, GTGCTGAGGTTCACAGTGACGT;
PRR5_Rev, GGTTGTAGAGCCTCTGGATCTC;
PRR5L_For, CGCATTGAGGTTCTGGCTGAAG;
PRR5L_Rev, CCTTCAGCAAGACTAGGTCTCG;
mTOR_For, AGCATCGGATGCTTAGGAGTGG;
mTOR_Rev, CAGCCAGTCATCTTTGGAGACC;
RICTOR_For, GCCAAACAGCTCACGGTTGTAG;
RICTOR_Rev, CCAGATGAAGCATTGAGCCACTG.
Relative changes in expression were calculated using the ΔΔCt method.
LC-MS/MS quantification of m6A in poly(A)-mRNA.
100–200 ng of mRNA was digested by nuclease P1 (2 U) in 25 μL of buffer containing 25 mM of NaCl, and 2.5 mM of ZnCl2 at 42 °C for 2 h, followed by the addition of NH4HCO3 (1 M, 3 μL) and alkaline phosphatase (0.5 U) and incubation at 37 °C for 2 h. The sample was then filtered (0.22 μm pore size, 4 mm diameter, Millipore), and 5 μL of the solution was injected into the LC-MS/MS. The nucleosides were separated by reverse phase ultra-performance liquid chromatography on a C18 column with online mass spectrometry detection using Agilent 6410 QQQ triple-quadrupole LC mass spectrometer in positive electrospray ionization mode. The nucleosides were quantified by using the nucleoside-to-base ion mass transitions of 282 to 150 (m6A), and 268 to 136 (A). Quantification was performed by comparison with a standard curve obtained from pure nucleoside standards run with the same batch of samples. The ratio of m6A to A was calculated based on the calibrated concentrations.
Cell proliferation assay.
5000 cells were seeded per well in a 96-well plate. The cell proliferation was assessed by assaying the cells at various time points using the CellTiter 96® Aqueous One Solution Cell Proliferation Assay (Promega) following the manufacturer’s protocols. For each cell line tested, the signal from the MTS assay was normalized to the value observed ~5 or 24 h after seeding.
Wound healing assay.
Cells were seeded on collagen I-coated six-well culture dishes at a density of 1×106 cells/well in media without serum to prevent cell proliferation and induce migration. A wound was made in the center of the culture 24 h after seeding, and cells were imaged directly after making the wound and 48 h later. Migration distance was calculated as follows:
where w0 is the width of the wound at time zero and w48 is the width of the wound after 48 h.
Soft Agar colony formation assay.
Anchorage-independent cell growth was assessed by using the CytoSelect™ 96-Well Cell Transformation Assay kit (Cell Biolabs) following the manufacturer’s protocols. 5000 cells were seeded per well and allowed to grow for 7 days. Cell numbers were determined by CyQUANT staining and quantified by comparison to a standard curve.
Colony formation assay.
500 cells were seeded per well in 6-well culture dishes. After 7 to 10 days, the culture medium was removed and the cells were washed twice with PBS, fixed with 4% paraformaldehyde for 20 minutes, stained with 0.1% crystal violet (in 25% methanol) for 20 minutes, washed with water, and dried. Colonies were counted manually.
Transwell migration and invasion assay.
For invasion assays, cell culture inserts (0.8 um, Falcon #353097) were coated with collagen type I (10 ug/insert, BD Biosciences #354236) in molecular grade water and dried overnight. For migration assays, inserts were not coated. Inserts were rehydrated with Opti-MEM (Invitrogen #31985–070) and fibronectin (4 μg/insert) for 2 h and 40,000 cells per insert were seeded in Opti-MEM Reduced Serum Media. Complete media (20% FBEssence in OptiMEM with fibronectin 3.75 μg/well) was used in the lower chamber. Following 24–48 h of migration or invasion, respectively, cells were fixed in 4% paraformaldehyde for 30 mins, treated with RNase A (Invitrogen #12091021), and cells visualized with SYBR Safe (1:5000, Invitrogen #233102) in PBS. Images were collected with a Nikon Eclipse Ti2 with NIS Elements Imaging Software (Version 5.02) and images analyzed with ImageJ (Version 1.51i).
Animal experiments.
Mice were housed at five mice per cage under pathogen-free conditions per the NIH Guide for the Care and Use of Laboratory Animals. All animal care and experiments were approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC), and the study is compliant with all relevant ethical regulations regarding animal research. 4×106 HEC-1-A endometrial cancer cells (shCtrl, shMETTL3, wild-type, METTL14+/−, or METTL14+/− rescued with wild-type or mutant METTL14) were injected intraperitoneally into 5 week old female athymic nude mice (Foxn1nu, Harlan; n=10 per group). Mice were sacrificed 2–3 weeks after injection and total tumor burden (weight) and number of tumor implants quantified. Organs involved included omentum, liver, intestines, spleen, ovaries, and uterine horns. Sample size was determined based on previous experience with intraperitoneal models of ovarian cancer52,53 and literature reports54. No animals were excluded from the study. Investigators were blinded to group allocation during intraperitoneal injections and when assessing outcome. Mice were not randomized.
Immunohistochemistry and tissue microarray analysis.
Tissue microarrays (TMAs) encompassing 11 benign endometrial tissues and 32 endometrial cancer specimens were purchased from US Biomax (EMC961). For METTL3 (1:200 dilution; Proteintech 15073–1-AP), PRR5 (1:200; Proteintech 17948–1-AP), PRR5L (1:200; LSBio LS-C144364–50), and phospho-mTOR-Ser2448 (1:100 dilution; Cell Signaling 49F9), heat-mediated antigen retrieval was performed with 10 mM sodium citrate, 0.05% Tween 20, pH 6; for PHLPP2 (1:100; Abcam ab71973), antigen retrieval was performed with 10 mM Tris base, 1 mM EDTA, 0.05% Tween 20, pH9. Slides were processed with the VECTASTAIN Elite ABC HRP kit and DAB Substrate Kit (Vector Laboratories). Slides were counterstained with hematoxylin and dehydrated through graded alcohols and xylene. A total of 10 normal and 30 tumor samples had sufficient tissue for unambiguous analyses; all analyses were limited to the epithelial component of both normal and tumor samples. Images were processed with the ImageJ plugin IHC Profiler using the author’s recommendations55. All tissues were assigned a score based on staining intensity in the epithelial compartment (0=no staining; 1=low positive; 2=positive; 3=high positive). The percentage of cells that were stained positive or negative was consistently uniform throughout the cores.
m6A-seq.
Total RNA was isolated from patient samples or HEC-1-A stable cell lines. Polyadenylated RNA was further enriched from total RNA by using Dynabeads® mRNA Purification Kit (Invitrogen). RNA fragmentation, m6A-IP, and library preparation were performed according to previously published protocols10. Sequencing was performed at the University of Chicago Genomics Facility on an Illumina HiSeq2500 machine in single-read mode with 50 bp per read.
m6A-seq data analyses.
m6A-seq data were analyzed according to the protocol described by Meng et al56. Briefly, Tophat2 (version 2.2.1) with Bowtie1 support57,58 was run to align the sequence reads to reference genome and transcriptome (hg19). Then exomePeak R/Bioconductor package (version 3.7)56 was used to find m6A peaks. Significant peaks with false discovery rate (fdr) less than 0.05 were annotated to RefSeq database (hg19). Sequence motifs were identified by using Homer (version 4.9)59, and DAVID (version 6.8) was used to perform GO term enrichment analysis60. Gene expression was calculated by Cufflinks (version 2.2.1) using the sequencing reads from input samples61. Cuffdiff was used to find the differentially expressed genes62. To assess global changes to m6A methylation, we identified all m6A peaks showing significant enrichment in at least half of the normal tissue samples, and enrichment values were averaged over all tumor or normal samples. Annotations for the PI3K/Akt pathway were taken from the KEGG Database63.
Measurement of RNA lifetime.
HEC-1-A cells were seeded in 10-cm plates at 50% confluency. After 24 h, each 10-cm plate was re-seeded into three 6-cm plates. After 48 h, actinomycin D was added to 5mg/ml at 6 h, 3 h, and 0 h before trypsinization and collection. The total RNA was purified by RNeasy kit with an additional DNase-I digestion step on column. RNA quantities were determined by RT-qPCR. The degradation rate of RNA (k) was estimated by plotting Nt/N0 against time and fitting to the following equation:
where t is the transcription inhibition time, and Nt and N0 are the RNA quantities at time t and time 0. The RNA lifetime (t1/2) can be calculated from the degradation rate as follows:
Quantification of mRNA methylation with m6A-IP and RT-qPCR.
We performed m6A-IP enrichment followed by RT-qPCR to quantify the changes to m6A methylation of a certain target gene. 3 μg purified cytosol mRNA extracted from the HEC-1-A stable cells was incubated with 5 μg m6A antibody for 4 h at 4 ℃, then pulled-down by Protein A Dynabeads (Invitrogen) for 2 h at 4℃. RNA was extracted from the bead and flow-through fractions by acid phenol/chloroform extraction, then subjected to RT-qPCR. The HPRT1 gene was used as a reference gene when performing qPCR. For each gene, the ΔCt of input (Δi), IP (Δe) and flow-thorugh (Δd) were calculated relative the reference. Given the following set of equations:
where Ea and Ra are the expression and methylation ratio of the gene of interest, and Er and Rr are the expression and methylation ratio of the reference gene, we can solve for Ra to obtain the following expression:
Polysome profiling.
HEC-1-A cells were subjected to siRNA knockdown for 48 h and treated with cycloheximide (CHX) at 100 μg ml−1 for 7 min before collection. Cells were pelleted, lysed on ice in lysis buffer (20 mM HEPES pH 7.6, 100 mM KCl, 5 mM MgCl2, 1% Triton X-100, 100μg/mL CHX, supplemented with protease inhibitor, and RNase inhibitor) then centrifuged. The supernatant (~1.2 ml) was collected and loaded onto a 10/50% (w/v) sucrose gradient prepared in a lysis buffer without Triton X-100. The gradients were centrifuged at 4°C for 3 h at 27,500 rpm (Beckman, rotor SW28). The sample was then fractioned and analyzed by Gradient Station (BioCamp) equipped with an ECONO UV monitor (BioRad) and fraction collector (FC203B, Gilson). The fractions were mixed with TRIzol reagent for purification of total RNA, which was analyzed by RT–PCR.
RIP-qPCR.
60 million HEC-1-A cells were collected from three 15-cm plates by gentle scraping, pelleted by centrifuge for 5 min at 1,000g and washed once with cold PBS (10 mL). The cell pellet was re-suspended with 2 volumes of lysis buffer (150 mM KCl, 10 mM HEPES pH 7.6, 2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, 1:100 protease inhibitor cocktail, 400 U ml–1 RNase inhibitor), pipetted up and down several times, and then the mRNP lysate was incubated on ice for 5 min and flash-frozen at with liquid nitrogen. The mRNP lysate was thawed on ice and centrifuged at 15,000g for 15 min to clear the lysate. The lysate was further cleared by filtering through a 0.22 μm membrane syringe. 50 μl cell lysate was saved as input, mixed with 1 ml TRIzol. Cell lysate was mixed with YTHDF1 (ProteinTech 17479–1-AP) or YTHDF2 (Aviva ARP67917_P50) antibody and then rotated continuously at 4 °C overnight. Protein G magnetic beads (Invitrogen, 20 μl per 1 μg antibody) was washed with a 600 μl NT2 buffer (200 mM NaCl, 50 mM HEPES pH 7.6, 2 mM EDTA, 0.05% NP-40, 0.5 mM DTT, 200 U ml–1 RNase inhibitor) four times and then re-suspended in 50 μl ice-cold NT2 buffer. The mixture (cell lysate and antibody) were mixed with prepared protein G magnetic beads and then rotated continuously at 4 °C for 2 h. The supernatant was saved as the flowthorugh fraction and mixed with 2ml TRIzol. The beads were collected, washed eight times with 1 ml ice-cold NT2 buffer. The beads were mixed with 1 ml TRIzol and saved as the IP sample. Total RNA isolated by TRIzol reagent was analyzed by RT–PCR.
Statistics and reproducibility.
At least three biological replicates were used in each experiment unless otherwise stated. Data are presented as the mean ± standard error of the mean (s.e.m.) or standard deviation (s.d.). Two-tailed Student’s t-tests were performed to assess the statistical significance of differences between groups. Pearson correlation coefficients (r) were calculated to assess correlation and statistical significance was assessed by a two-tailed t-test of r = 0. The statistical significance of differences in IHC scores of tumor and normal tissue were assessed by χ2-test. Immunoblots are the representative images of at least three independent experiments. For box plots, the center line represents the median, the box limits show the upper and lower quartiles, whiskers represent 1.5× the interquartile range, and outliers are represented as individual data points.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Martin Aryee (Massachusetts General Hospital) for initial discussions and Dr. Angela Andersen for editing the manuscript. This work was supported by a Marsha Rivkin Foundation award (M.A.E.); University of Chicago Institute for Biophysical Dynamics Yen Fellowship (B.T.H.); National Natural Science Foundation of China grants 81472023 and 81271919 (S.L.); The National Basic Research Program grants 2012CB720600 and 2012CB720605 (S.L.); National Key Research and Development Program of China 2017YFA0506800 (Jz. L.); Thousands Young Talents Plan of China and Hundreds Talents Program of Zhejiang University (Jz. L.); National Cancer Institute grants CA111882 (E.L.) and F32 CA221007 (B.T.H); National Institutes of Health grants R01 HG008688 and RM1 HG008935 (C.H); and University of Chicago Cancer Center Support Grant P30CA014599. M.A.E. thanks the Harris Family Foundation for their generous support. C. H. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Data availability. The RNA-seq and m6A-seq data generated by this study have been deposited in the GEO database under the accession number GSE93911. A summary of the m6A peaks identified by the m6A-seq experiments in the patient samples and cell lines can be found in Supplementary Tables 3 and 4. The human data for high grade serous ovarian cancer and pancreatic adenocarcinoma, as well as some data for endometrial cancer, were derived from the TCGA Research Network: http://cancergenome.nih.gov/. Source data for Figs 1–7 and Supplementary Figs 1–7 are provided in Supplementary Table 5. Unprocessed immunoblot scans are presented in Supplementary Fig. 8. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
COMPETING FINANCIAL INTERESTS
C.H. is a scientific founder of Accent Therapeutics and a member of its scientific advisory board. All other authors declare no competing financial interests.
MATERIALS & CORRESPONDENCE
Correspondence and requests for materials should be addressed to C.H. (chuanhe@uchicago.edu) and E.L. (elengyel@uchicago.edu).
REFERENCES
- 1.Liu N & Pan T N6-methyladenosine–encoded epitranscriptomics. Nat Struct Mol Biol 23, 98–102 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zhao BS, Roundtree IA & He C Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18, 31–42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia G et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7, 885–887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P & Rottman F Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem 269, 17697–17704 (1994). [PubMed] [Google Scholar]
- 5.Liu J et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10, 93–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ping X-L et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24, 177–189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz S et al. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep 8, 284–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng G et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49, 18–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominissini D et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Zhao X et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24, 1403–1419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao W et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 61, 507–519 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Roundtree IA et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer KD et al. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 163, 999–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li A et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 27, 444–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 27, 315–328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geula S et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Batista PJ et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao BS et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542, 475–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S, Choe J, Du P, Triboulet R & Gregory RI The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 62, 335–345 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang C et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A 113, E2047–2056 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma JZ et al. METTL14 suppresses the metastatic potential of HCC by modulating m6 A-dependent primary miRNA processing. Hepatology 65, 529–543 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Li Z et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Q et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep 18, 2622–2634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X et al. The M6A methyltransferase METTL3: acting as a tumor suppressor in renal cell carcinoma. Oncotarget 8, 96103–96116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X et al. Reduced m(6)A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget 8, 98918–98930 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen M et al. RNA N6-methyladenosine methyltransferase METTL3 promotes liver cancer progression through YTHDF2 dependent post-transcriptional silencing of SOCS2. Hepatology (2017). [DOI] [PubMed] [Google Scholar]
- 33.Vu LP et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23, 1369–1376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng H et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 22, 191–205 e199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su R et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172, 90–105 e123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandoth C et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sledz P & Jinek M Structural insights into the molecular mechanism of the m(6)A writer complex. Elife 5, e18434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P, Doxtader KA & Nam Y Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell 63, 306–317 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X et al. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Meyer KD et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 32, 185–203 e113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Le Gallo M & Bell DW The emerging genomic landscape of endometrial cancer. Clin Chem 60, 98–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vivanco I & Sawyers CL The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2, 489–501 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Jacinto E et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 127, 125–137 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Brognard J, Sierecki E, Gao T & Newton AC PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell 25, 917–931 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Sarbassov DD, Guertin DA, Ali SM & Sabatini DM Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Copp J, Manning G & Hunter T TORC-specific phosphorylation of mammalian target of rapamycin (mTOR): phospho-Ser2481 is a marker for intact mTOR signaling complex 2. Cancer Res 69, 1821–1827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li HB et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart SA et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9, 493–501 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y et al. Reversal of Chemoresistance in Ovarian Cancer by Co-Delivery of a P-Glycoprotein Inhibitor and Paclitaxel in a Liposomal Platform. Mol Cancer Ther 15, 2282–2293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kenny HA et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat Commun 6, 6220 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi K et al. Cetuximab inhibits growth, peritoneal dissemination, and lymph node and lung metastasis of endometrial cancer, and prolongs host survival. Int J Oncol 35, 725–729 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Varghese F, Bukhari AB, Malhotra R & De A IHC Profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS One 9, e96801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng J et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods 69, 274–281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanehisa M, Sato Y, Kawashima M, Furumichi M & Tanabe M KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44, D457–462 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.