1. Introduction
Mood and anxiety disorders are strongly associated with somatic symptoms such as gastrointestinal distress (Bekhuis et al., 2014, Felice et al., 2015), and a high co-morbidity exists between stress-related neuropsychiatric symptoms and gastrointestinal disorders such as irritable bowel syndrome (Kanuri et al., 2016, Kennedy et al., 2012, Qin et al., 2014). One possibility is that stress impacts brain function and mental health via its effect on the gastrointestinal tract (for review see Dinan & Cryan, 2012, Dinan et al., 2015, Parashar & Udayabanu, 2016). Given that the available treatment strategies for a variety of stress-related neuropsychiatric disorders are inadequate for many, expanding our knowledge of a broader range of potential etiologic factors might lead to novel, more effective therapeutics (Culpepper et al., 2015, Haddad et al., 2015, Nestler et al., 2002).
The mammalian gastrointestinal tract houses over 100 trillion microorganisms (Eckburg et al., 2005), which are critical for vital functions such as processing and digestion of food, synthesis of vitamins, inhibition of pathogens, and immune system development and maturation (Ramakrishna, 2013). Thus, a stable and symbiotic relationship exists between these microorganisms, referred to as gut microbiota, and the host gastrointestinal system (Mayer et al., 2015). These microbiota are essential to homeostasis, and abrupt dysbiosis or absence of this vibrant community can compromise the physical and mental health of the host (Chang et al., 2008, Cryan, 2016, Luczynski et al., 2016, Turnbaugh et al., 2006). Recently, it has been demonstrated that there is bi-directional communication, referred to as the gut-brain axis, between the central nervous system and the gastrointestinal tract, and altering the gut microbiota during early development or adulthood changes both stress-related behavior and responsivity of the stress axis (Desbonnet et al., 2014, Diaz et al., 2011, Collins et al., 2013, Sudo et al., 2004).
Many neuropsychiatric disorders and symptoms that are associated with gastrointestinal dysfunction are also caused or exacerbated by exposure to stress (Agid et al., 2000, Kessler, 1997, Saveanu & Nemeroff, 2012), and stress has been associated with significant alterations in the gut microbial community in mammals, including humans (Lyte et al., 2011). These stress-induced alterations are associated with consequences ranging from inflammation to increased anxiety-like behavior (Bailey & Coe, 1999, Bailey et al., 2011). Exposure to social stress, in particular, can cause or exacerbate disabling neuropsychiatric disorders, including depression and PTSD (Bjorkqvist et al., 2001, Qiao et al., 2016). Relatively little is known, however, about the direct impact of social stress on the gut microbial community and how these microbes, in turn, may affect behavior. Bailey et al. (2011) demonstrated that group-housed mice exposed to 6, 2 hr bouts of social disruption stress exhibited alterations of the gut microbial community characterized by a reduction in microbial diversity and richness. A similar response was observed in mice that were exposed to a more severe, 10-day social defeat procedure (Bharwani et al., 2016). There is even some evidence that a single, 2 hr exposure to a social defeat stressor in mice impacts gut microbiota (Galley et al., 2014), suggesting that even acute social stress might have effects on the gut.
The current study utilizes a well-characterized resident-intruder model in Syrian hamsters (Jasnow et al., 2001, Potegal et al., 1993) to investigate whether exposure to social stress affects the commensal gut microbiota and, in particular, whether it does so differently in individuals that “win” a social conflict (i.e., become dominant) versus those that “lose” (i.e., become subordinate). Syrian hamsters are ideal candidates for the study of social stress because when weight- and age-matched conspecifics are paired, they readily produce aggressive and territorial behavior that rapidly results in the formation of a stable dominance relationship (Albers et al., 2002). This allows a direct comparison of commensal bacteria in dominants and subordinates. This comparison is not possible in mice because conspecifics generally do not fight; defeated mice are produced using a larger, more aggressive heterospecific (e.g., C57J/BL6 defeated by a CD-I mouse). An additional benefit of using hamsters is that their agonistic behavior during brief encounters is highly ritualized and rarely results in any tissue damage, allowing us to focus on the psychological, as opposed to physical, aspects of social stress (Huhman & Jasnow, 2005). Furthermore, no studies have examined whether the baseline composition of the gut microbiota alters behavioral responses to social stress. Thus, we also measured whether the baseline gut microbiota composition can predict whether an animal becomes dominant or subordinate after a subsequent agonistic encounter.
2. Materials and Methods
2.1. Animals
Adult male Syrian hamsters (Mesocricetus auratus), weighing between 120 and 130 g, were obtained from Charles River Laboratory (Kingston, NY) at approximately 3 months of age. Hamsters were individually housed in polycarbonate cages (24×33×20cm) with corncob bedding, cotton nesting material, and a wire mesh top in a temperature controlled colony room under a 14:10hr light/dark cycle, which is standard to maintain reproductive gonadal status in hamsters. Food and water were available ad libitum. All hamsters were handled daily for 7 days to acclimate them to handling stress before the beginning of the experiment. Individual housing is not stressful for Syrian hamsters (Ross et al., 2017), and with the exception of the agonistic pairings described in Section 2.2, hamsters remained separated throughout the experiment. All protocols and procedures were approved by the Georgia State University Institutional Animal Care and Use Committee prior to experimentation, and all methods align with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.2. Behavioral procedures and fecal collection
Two days before testing occurred, hamsters were weighed and randomly assigned to one of three weight-matched treatment groups: Resident (n=9), Intruder (n=9), or Home Cage Control (n=5). Control animals experienced the same treatment and fecal collection protocol throughout the experiment, with the exception that they were never paired with another hamster, to control for all environmental variables experienced by the animals besides social stress. For fecal collection, animals were transferred to a clean cage and fecal samples (approximately 5 fecal boli) were collected from the bedding of each hamster’s home cage just before the beginning of the active (dark) phase of the daily light: dark cycle. Collection from the home cage was done to avoid any additional stress to the animals. To help ensure that the samples were fresh, we collected fecal boli that were moist, on top of the bedding, and had no bedding stuck to them. These samples were collected in RNase-free microcentrifuge tubes and were immediately frozen and stored at −80° C until further processing. One day before testing began, all animals were transferred to a clean cage and fecal samples were collected 24hr later. Behavioral testing occurred over the next 5 days. All behavioral manipulations occurred during the first 3 hr of the dark phase of the daily light: dark cycle to control for circadian variation in microbiota and behavior and because this is when hamsters exhibit the majority of their agonistic behavior. Each day, all hamsters were moved into the behavioral suite 30 min prior to any manipulation to allow time to acclimate. Trials were run under dim red light and were recorded with a CCD camera for later scoring of behavior by observers blinded to the experimental condition. Home cage controls were not manipulated other than handling, transport to the testing suite, and cage changes. For agonistic encounters, an intruder was placed in the home cage of a resident for 15 min on Day 1 and twice a day for 5 min on Days 2-5, and a clear plastic lid was placed over the resident’s cage during each pairing to prevent escape. A dominance hierarchy was rapidly established during the first pairing, resulting in a winner (i.e., a dominant) who reliably attacked and defeated its losing partner (i.e., subordinate); the latter exhibited submissive and defensive behaviors such as upright defense, flee, and tail lift (Huhman et al., 1990). The scored behaviors were divided into four categories (i.e., social, aggression, submission, and nonsocial), as described in detail in Albers et al. (2002). Hamsters were transferred to a clean cage immediately after each agonistic interaction. Fecal samples were collected 24 hr after the first encounter from the animals’ cages immediately before the animals were paired again on Day 2. Two additional pairings per day were conducted on Days 2-5, one at the start of the dark phase, as on Day 1, and the second 4 hr later. The same resident/intruder pairings were used throughout the experiment. On Day 5, after the final pairing, all hamsters were immediately transferred into clean cages, and fecal samples were again collected 24 hr later to assess the effect of repeated agonistic interaction on gut microbiota. All hamsters were carefully observed during each agonistic encounter for coprophagia and for any injury. No coprophagia or tissue damage occurred during these encounters.
2.3. Fecal microbiota composition analysis by 16S rRNA gene sequencing
Fecal 16S rRNA gene amplification and sequencing were done using Illumina MiSeq technology following the protocol of the Earth Microbiome Project with their modifications to the MO BIO PowerSoil DNA Isolation Kit procedure for extracting DNA (www.earthmicrobiome.org/emp-standard-protocols), as described previously (Caporaso et al., 2012, Gilbert et al., 2010). In brief, bulk DNA was extracted from frozen feces using a PowerSoil-htp kit from MO BIO Laboratories (Carlsbad, California, USA) with mechanical disruption (bead-beating). The 16S rRNA genes, region V4, were PCR amplified from each sample using a composite forward primer and a reverse primer containing a unique 12-base barcode, designed using the Golay error-correcting scheme, which was used to tag PCR products from respective samples. We used the forward primer 515F 5’-AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTGTGCCAGCMGCCGCGGTAA-3‘: the italicized sequence is the 5’ Illumina adapter B, the bold sequence is the primer pad, the italicized and bold sequence is the primer linker and the underlined sequence is the conserved bacterial primer 515F. The reverse primer 806R used was 5’-CAAGCAGAAGACGGCATACGAGATXXXXXXXXXXXX AGTCAGTCAG CCGGACTACHVGGGTWTCTAAT-3’: the italicized sequence is the 3’ reverse complement sequence of Illumina adapter, the 12 X sequence is the golay barcode, the bold sequence is the primer pad, the italicized and bold sequence is the primer linker and the underlined sequence is the conserved bacterial primer 806R. PCR reactions consisted of Hot Master PCR mix (Five Prime), 0.2 μM of each primer, 10-100 ng template, and reaction conditions were 3 min at 95°C, followed by 30 cycles of 45 s at 95°C, 60 s at 50°C, and 90 s at 72°C on a Biorad thermocycler. Two independent PCRs were performed for each sample, then combined and purified with Ampure magnetic purification beads (Agencourt), and products were visualized by gel electrophoresis. Products were then quantified (BIOTEK Fluorescence Spectrophotometer). A master DNA pool was generated from the purified products in equimolar ratios. The pooled products were sequenced using an Illumina MiSeq sequencer (paired-end reads, 2 × 250 bp) at Cornell University, Ithaca.
2.4. 16S rRNA gene sequence analysis
The sequences were demultiplexed and quality filtered using the Quantitative Insights Into Microbial Ecology (QIIME, version 1.8.0) software package (Caporaso et al., 2010). Forward and reverse Illumina reads were joined using the fastq-join method (Aronesty, 2011, 2013). We used the QIIME default parameters for quality filtering as described in detail in Caporaso et al. (2010). Sequences were clustered using the UCLUST algorithm with a (Edgar, 2010) 97% homology threshold. Clusters were then classified taxonomically using the Green genes reference database, Version 13.5 (McDonald et al., 2012). Clusters that did not match any Green genes Operational taxonomic units (OTUs) were kept. A single representative sequence for each OTU was aligned and a phylogenetic tree was built using FastTree (Price et al., 2009).
The phylogenetic tree described above was used to assess beta and alpha diversity. Beta diversity measures the variation in microbiota composition between individual samples. Alpha diversity measures both the richness and evenness (or distribution) of unique microbial taxa within a sample (Mackos et al., 2017, Sekirov et al., 2010). Unweighted UniFrac distances between samples were computed, as done previously, to measure beta diversity (Lozupone et al., 2006, Lozupone & Knight, 2005) using rarefied OTU table count. Principal coordinates analysis (PCoA) plots were used to further assess and visualize beta diversity. Groups were compared for distinct clustering using PERMANOVA method using vegan R-package through QIIME. Kruskal-Wallis with Dunn’s multiple comparisons test was used to compare the unweighted UniFrac distances within or between groups. The phylogeny-based metric, phylogenetic diversity whole tree (PD whole tree) measurements were determined with QIIME using an OTU table rarefied at various depths and the non phylogeny-based metric, Shannon measurements were determined with QIIME using the alpha_diversity.py command line of the rarefied OTU table count. Area under the curve was calculated for each rarefaction curve and Kruskal-Wallis with Dunn’s multiple comparison test was used to determine differences among groups. Due to technical limitations, not all samples could be amplified; therefore, we were unable to run repeated measures for the comparisons. Lastly, LEfSE (Linear Discriminate Analysis Effect Size) was used to investigate bacterial taxa that drive differences between groups by comparing the abundance of specific taxa between each experimental group (Segata et al., 2011), and Mann-Whitney U tests were performed where appropriate. All statistics were done in Graph Pad Prism software, version 6.01 and IBM SPSS Statistics, version 22. Unprocessed sequencing data are deposited in the European Nucleotide Archive under accession numbers XXXXXX.
3. Results
3.1. Behavioral responses to social stress
Each resident/intruder pair rapidly formed a dominant/subordinate relationship on Day 1, whereon either the resident or the intruder was defeated during the initial encounter. Dominant hamsters produced 211.1 ± 49.2 s of aggression and 1.2 ± 1.1 s of submission while their opponents (subordinates) produced 6.3 ± 3.1 s of aggression and 264.4 ± 49.5 s of submission during the initial 15min pairing. The dominance relationship within each pairing, with the exception of one, remained stable throughout the experiment. The pair in which the dominance hierarchy reversed after the first pairing was removed from the study and was not included in the analyses. While residence often confers dominance in resident-intruder models, pairing weight-matched animals tends to even those odds and has done so in our previous work. Nonetheless, in this study, residents became the dominant animal more frequently (8 of 9 pairings) than expected for these weight-matched animals.
3.2. Social stress alters gastrointestinal microbiota composition
Microbiota composition was analyzed by 16S rRNA Illumina sequencing of fecal DNA samples collected before any interaction (“baseline”), after the first (“acute”) pairing, and after nine (“repeated”) agonistic encounters. Microbiota composition was also analyzed from home cage control animals collected concurrently. 16S rRNA Illumina sequencing revealed that several samples had a low number of sequences and, therefore, were not included in the analyses. This included two samples from each group at baseline sampling, one home cage control at repeated sampling, and one subordinate at the acute sample. PERMANOVA analysis of the unweighted Unifrac distance (Lozupone & Knight, 2005) revealed that after acute and repeated pairing, microbiota composition of both dominant and subordinate hamsters was significantly altered when compared to their respective baseline values (Acute, dominant p = 0.002, subordinate p = 0.004; Repeated, dominant p = 0.001, subordinate p = 0.001) (Figure 1). It should be noted that before any social interaction there was no distinct clustering of the samples between home cage control and dominant animals (p = 0.844), between home cage control and subordinate animals (p = 0.781), nor between dominant and subordinate animals (p = 0.695) (Figure 2A). Further examination of the unweighted Unifrac distance revealed that, compared with home cage controls, both dominant and subordinate hamsters did not show a significant change in beta diversity after both an acute pairing (Figure 2B; dominant p = 0.911, subordinate p = 0.414) and repeated pairings (Figure 2C; dominant p = 0.321, subordinate p = 0.086) using the PERMANOVA test. This is likely due to the high variability between individuals. Further, Kruskal-Wallis showed no differences in the unweighted Unifrac distance at baseline (Figure 2D) or after acute pairing (Figure 2E); however, after repeated pairings, there was a significant increase in the unweighted Unifrac distance between subordinate animals when compared with home cage controls (Figure 2F; H(2,74) = 14.44, p < 0.001, Dunn’s multiple comparison test, p < .05) suggesting an increase in inter individual variation of the microbial composition in animals who lost. No differences in microbiota composition was observed between dominants versus subordinates after acute (Figure 2B; p = 0.413) or repeated stress (Figure 2C; p = 0.820).
Figure 1. Social stress alters intestinal microbiota composition within dominant and subordinate hamsters compared to their baseline values.
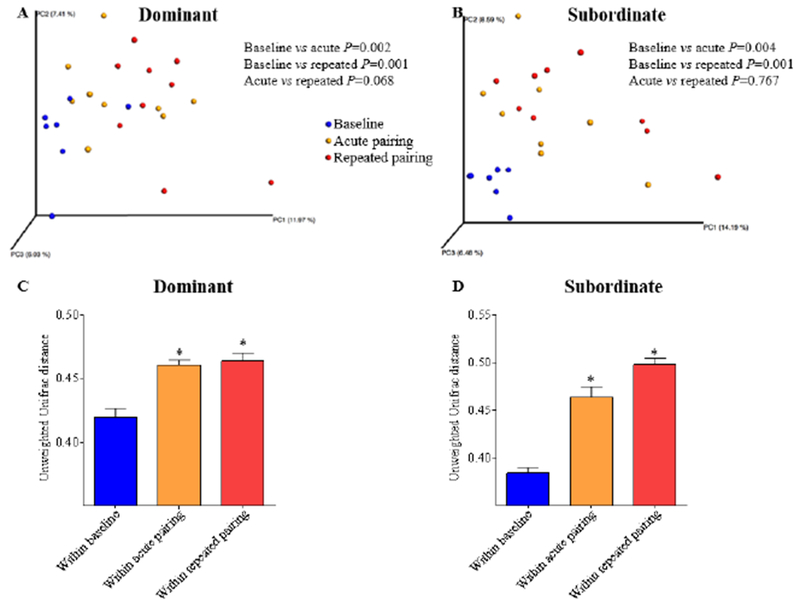
Principal coordinate analysis (PCoA) of the unweighted UniFrac distance, illustrating changes in beta diversity, for dominant (A) and subordinate (B) hamsters comparing before social stress (baseline, blue dots), after one (acute pairing, orange dots), and repeated (repeated pairing, red dots) exposure to social stress. P values were determined using PERMANOVA analysis (A-B). No significant change in beta diversity occurred in home cage controls over the three collection time points (data not shown). Rarefied count: 5468 for A, 9288 for B. Panels C and D show the average (means ± SEM) unweighted Unifrac distance for dominant (C) and subordinate (D) hamsters, illustrating that within each social status there was a significant increase in the variation among samples at each time point compared with baseline. P values for C-D were determined using Kruskal-Wallis with Dunn’s multiple comparison test. *denotes significantly greater than their respective baseline value (P<0.05).
Figure 2. Social stress alters intestinal microbiota composition similarly in dominants and subordinates compared to home cage controls.
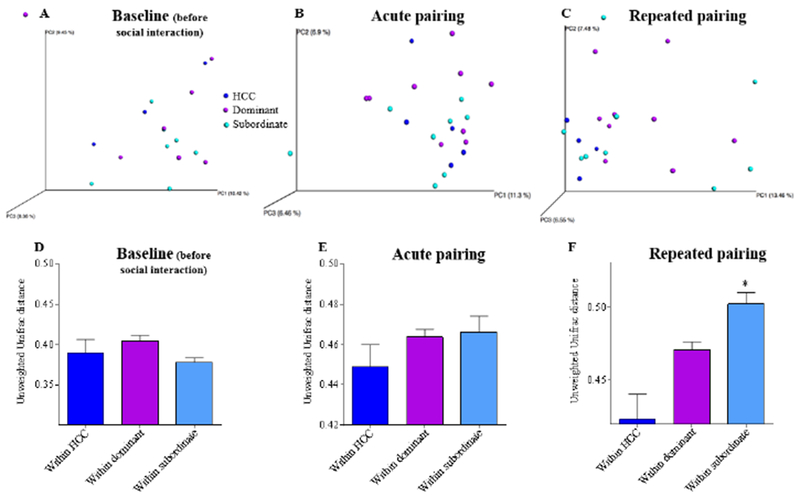
Principal coordinate analysis (PCoA) of the unweighted UniFrac distance at baseline (A), after acute pairing (B) and repeated pairing (C) for home cage control (blue dots), dominant (purple dots) and subordinate (light blue dots) hamsters. P values were determined using PERMANOVA analysis. Rarefied count: 5468 for A, 11242 for B, 9288 for C. Average (mean ± SEM) of the unweighted Unifrac distance within groups (control, dominant and subordinate) at baseline (D), after acute (E) or repeated pairing (F) illustrating that inter individual variation differed among groups after repeated pairing. P values in D-F were determined using Kruskal-Wallis with Dunn’s multiple comparison test. *denotes significantly greater than home cage controls (HCC; P<0.05).
The analysis of alpha diversity of the intestinal microbiota, reflecting the bacterial richness and evenness of the community, revealed a significant effect of stress using both phylogeny-based (PD whole tree) and non phylogeny-based (Shannon) measurements. Although home cage control animals demonstrated stable diversity over time (Figure 3A; H(2, 9) = 1.414, p = 0.542; Supplementary Figure 1; H(2,9) = 0.695, p = .707), post hoc analysis revealed that repeated stress reduced microbiota diversity in dominant animals compared with their baseline (Figure 3B; H(2, 22) = 7.559, p = 0.023, Dunn’s multiple comparisons test, p < .05); Supplementary Figure 1; H(2, 22) = 5.865, p = 0.053, Dunn’s multiple comparisons test, p < .05). The apparent reduction in microbiota diversity for subordinates following stress was not significant (Figure 3C; H(2, 21) = 3.517, p = 0.17; Supplementary Figure 1; H(2, 21) = 3.88, p = 0.14). Significance was not reached after an acute interaction (Figure 3D; H(2, 19) = 0.120, p = 0.948) when dominant, subordinate, and home cage control groups were compared; however, when these groups were compared following repeated interactions significance was reached (Figure 3E; H(2, 19) = 8.117, p = 0.017) with dominant hamsters harboring reduced microbiota diversity following repeated exposure to social stress compared with home cage controls (Figure 3E; Dunn’s multiple comparisons test, p < .05). Subordinate animals showed a similar pattern when compared to home cage controls, yet this observation was not significant (Figure 3E; Dunn’s multiple comparison test, p > .05).
Figure 3. Social stress decreases intestinal microbiota diversity.
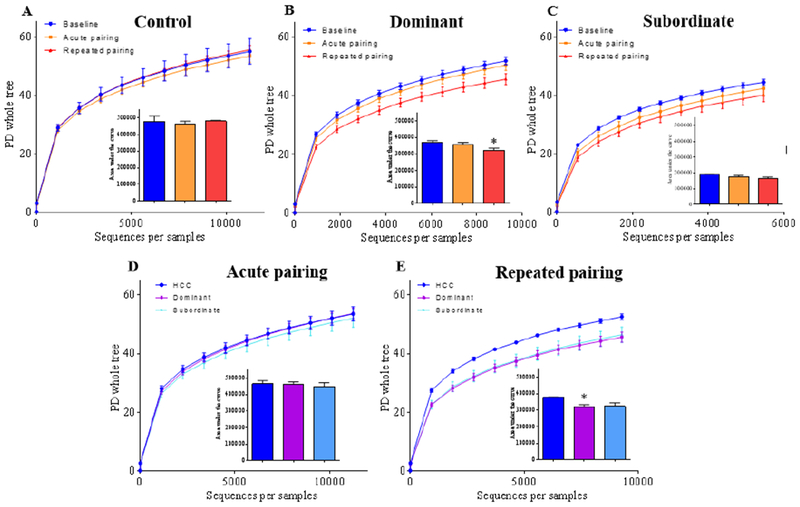
(A-C) Alpha diversity was determined by phylogenetic diversity whole tree measurement, as well as Shannon index (see Supplemental Figure 1) in control (A), dominant (B) and subordinate (C) hamsters before any social stress (baseline), after one (acute pairing) or repeated (repeated pairing) exposure to social stress. (D-E) PD whole tree and Shannon index measurements (see Supplemental Figure 1) were also used to compare alpha diversity across groups after one (acute pairing, D) or repeated (repeated pairing, E) exposure to social stress in home cage control (HCC), dominant and subordinate hamsters. Insets represent area under the curve for each group. Data are the means ± SEM; Comparisons done using Kruskal-Wallis with Dunn’s multiple comparison test (Panel 3B and 3E). * denotes significantly reduced (p<0.05) in repeated pairing compared to baseline or home cage controls.
LEfSE analysis (Linear Discriminant Analysis (LDA) Effect Size) was then used to find differentially abundant taxa between groups following social interactions, in order to identify which bacterial taxa drove changes in the microbiota community. A LDA threshold of 2 was used to infer significance (LDA > 2, p < 0.05). Bacterial taxa were largely unchanged in home cage controls between the baseline measurement and the subsequent sampling (Figure 4A and B), indicating that the microbiota remained stable in controls over the duration of the experiment. However, within both dominant and subordinate groups, numerous taxa of the intestinal bacterial community were significantly altered compared with their baselines following an acute pairing (Figure 4C and E, Supplementary Figure 2A), with more changes observed following repeated pairing (Figure 4D and F, Supplementary Figure 2B and C). Further, when comparing home cage controls to dominant and subordinate animals after repeated pairing, significant differences were also observed (Figure 5A-B, Supplementary Figure 3).
Figure 4. Identification of bacterial taxa altered following acute and repeated social stress compared to each group’s baseline value.
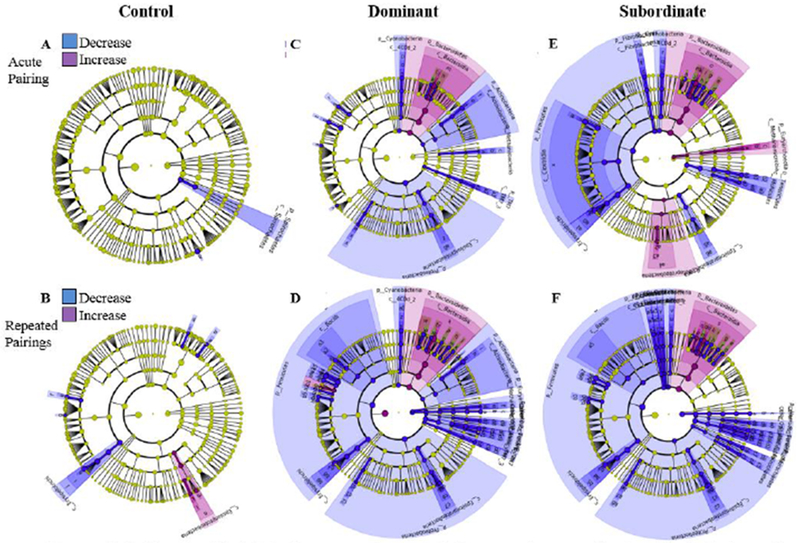
Acute Social Stress (top row): LEfSE (Linear discriminant analysis Effect Size) was used to investigate bacterial taxa that drive differences between samples at baseline and after acute pairing in control (A), dominant (C) and subordinate (E) hamsters. Repeated Social Stress (bottom row): bacterial taxa that drive differences between samples at baseline and after repeated pairing in control (B), dominant (D) and subordinate (F) hamsters. Red, taxa decreased after pairing compared with that group’s baseline value; green, taxa increased after pairing compared to baseline. Only taxa meeting an LDA significant threshold >2.0 are represented. Note: for a list of significantly altered taxa in panels where labels overlap (E, D, F) see Supplemental Figure 2.
Figure 5. Identification of bacterial taxa altered by repeated social stress compared to home cage controls.
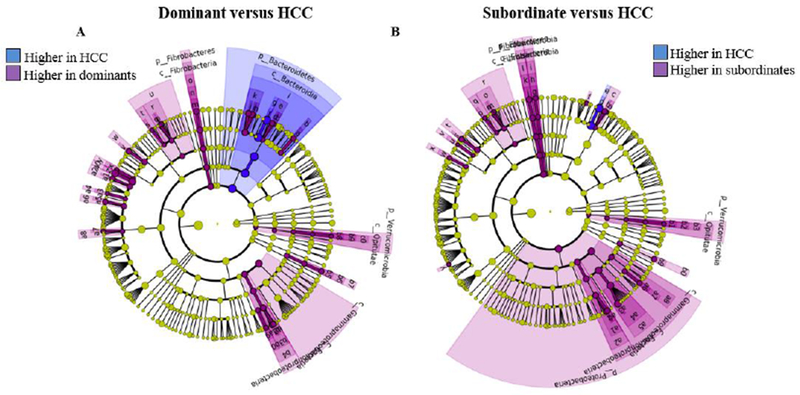
LEfSE (LDA Effect Size) was used to investigate bacterial taxa that drive differences between control and dominant hamsters (A) after repeated pairing and between control and subordinate hamsters after repeated pairing (B). Green, taxa decreased compared to control; red, taxa increased compared to control. Only taxa meeting an LDA significant threshold >2.0 are represented. Note: for a list of significantly altered taxa for both panels see Supplemental Figure 3.
Of note, microbiota from the order Lactobacillales and phyla Firmicutes were found to be significantly decreased, and microbiota from the phyla Bacteroidetes were found to be significantly increased following repeated social interactions in both dominant and subordinate animals (Figure 4D and F, Supplementary Figure 2B and C; LDA > 2, p < 0.05). The significant increase in phyla Bacteroidetes was also observed after the acute social interaction compared with baseline in both dominants and subordinates (Figure 4C and E, Supplementary Figure 2A; LDA > 2, p < 0.05). Differences between dominant and subordinate animals were also observed. Of particular interest, bacteria from the family Clostridiacea (Figure 4D, Supplementary Figure 2B; LDA > 2, p < 0.05) increased only in dominant animals, and bacteria from phyla Firmicutes (Figure 4E, Supplementary Figure 2A; LDA > 2, p < 0.05) significantly reduced after acute defeat only in subordinate animals.
3.3. Baseline microbiota composition can predict the outcome of a social conflict
We next investigated if the abundances of particular microbial taxa at baseline (i.e., before any social interaction) were different in animals that ultimately became dominant versus those that became subordinate. PERMANOVA analysis of the unweighted Unifrac distance revealed that there were no overall differences in microbiota community composition between future dominant or future subordinate hamsters before they were exposed to social conflict (Figure 2A; p = 0.695). However, LEfSE analysis revealed that the abundance of several individual bacterial taxa were significantly different at baseline in future dominant versus future subordinate hamsters (Figure 6A-F). Using a more stringent Mann-Whitney U test to compare individual OTUs, we observed significantly more Proteobacteria (Figure 6B; U(12) = 4, p = 0.007) and fewer Firmicutes (Figure 6F; U(12) = 9, p = 0.053) at baseline in future dominant animals compared with future subordinate animals. In addition, at the genus level, more Prevotella (Figure 6E; U(12) = 8, p = 0.038) was associated with future dominant status and more Allobaculum (Figure 6C; U(12) = 6, p = 0.018) with future subordinate status.
Figure 6. Identification of bacterial taxa that predict future dominant or subordinate status.
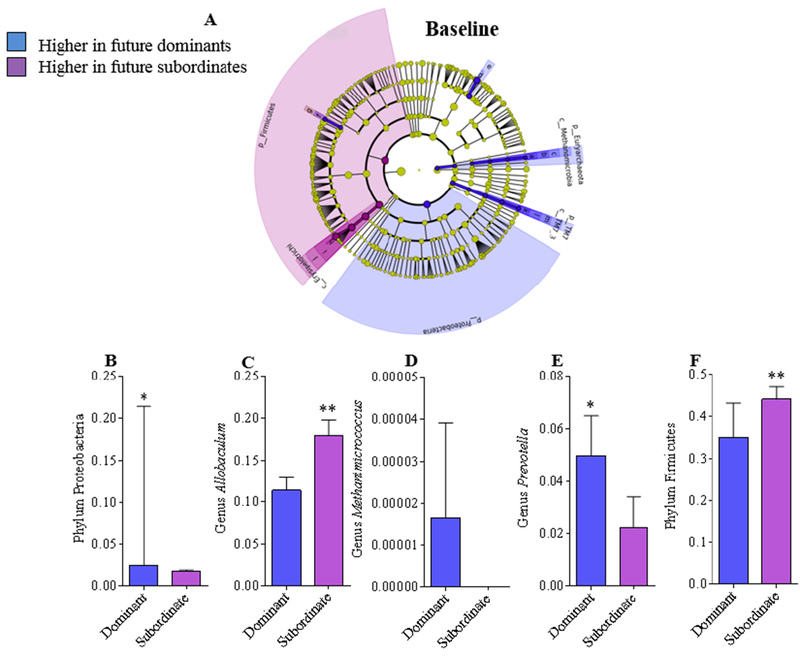
(A) LEfSE (LDA Effect Size) was used to investigate baseline bacterial taxa that predict dominant versus subordinate status upon pairing. Red, future dominant-enriched taxa; green, future subordinate-enriched taxa. Only taxa meeting an LDA significant threshold >2.0 are represented. Relative values of Phylum Proteobacteria (B), Genus Allobaculum (C), Genus Methanimicrococcus (D), Genus Prevotella (E), and Phylum Firmicutes (F) abundance at baseline in future dominant and subordinate hamsters. Data are the medians ± IQR. P values were determined using Mann-Whitney U test. * denotes significantly higher at baseline in future dominants; ** significantly higher at baseline in future subordinates (P<0.05).
4. Discussion
Using a well-characterized resident-intruder model in Syrian hamsters, we demonstrated that exposure to repeated and even to a single agonistic encounter causes alterations in gut microbiota in hamsters, whether they become dominant or subordinate. To our knowledge, this is the first time that the effects of social conflict on gut microbiota have been examined in hamsters, in animals that become dominant, and after both acute and repeated resident-intruder pairings in age- and weight-matched conspecifics. Intriguingly, we also found that certain microbes significantly differed in abundance between future dominants and subordinates, suggesting the possibility that baseline commensal gut bacteria in these animals could act as a predictor, or biomarker, of which animal would become dominant or subordinate during a subsequent social encounter. This exciting possibility would build on current evidence that gut microbiota can modulate social behavior and should be an area of future investigation.
PERMANOVA analysis revealed a significant shift from baseline values in the overall composition of the gut microbial community in hamsters after both one and repeated (e.g., nine) agonistic encounters. Interestingly, dominant and subordinate animals exhibited a similar increase in the inter individual variation of the microbial community after social conflict indicating that it is conflict, itself, that changes beta diversity independent of the outcome of the encounter; although it should be noted only subordinate animals reached significance when the unifrac distance was compared using Kruskal-Wallis. By contrast, home cage controls that experienced the same handling, transport, and cage changes, but no social interactions, exhibited little alteration in their gut microbes over the course of the study. Because we have previously observed a more pronounced hormonal response to social conflict in subordinates than in dominants (Huhman et al., 1990, 1991), we expected the microbial alterations in subordinate hamsters to be exacerbated compared to that observed in dominants. At this point it is unclear if the observed changes to gut microbiota following social conflict have functional relevance or if the similar patterns of change observed in dominants and subordinates could possibly have different functional consequences for each group.
Shifts in microbial richness and evenness of the intestinal microbiota (i.e., a decrease in alpha diversity) was found in hamsters exposed to both acute and repeated agonistic encounters compared to their respective baseline values and to home cage controls following repeated encounters. Disruption to the richness and evenness of the intestinal microbiota can compromise the gastrointestinal epithelium, and such a compromise can be associated with bacterial translocation and a pro-inflammatory immune response (Chassaing et al., 2015, Maes, 2008). This phenomenon has been reported in a mouse model of social disruption and has also been linked to pathophysiology underlying major depressive disorder in humans (Bailey et al., 2011, Maes et al., 2008). We did not examine gastrointestinal status or immune responses in the current study, but we suggest that hamsters are ideal for testing these endpoints particularly because social stress can be examined in dominants and subordinates, in both males and females (Rosenhauer et al., 2017), and in the absence of social stress-induced tissue damage. Removing the confound of injury would thus allow an assessment of changes to the gastrointestinal epithelium and inflammation following exposure to a largely psychological stressor.
LEfSE analysis was used to identify specific microbial taxa driving changes to gut microbiota in dominants and subordinates. One of the taxa driving the altered composition in both dominants and subordinates following repeated fighting was the order Lactobacillales, which was significantly reduced following stress compared to baseline. One genus of this order, Lactobacillus, is often examined in the literature and a reduction in this genus is observed following an acute and a prolonged social disruption stressor (Bailey et al., 2011, Galley et al., 2014). Although increases in certain members of lactobacilli have been associated with pathology, the majority of these bacteria are thought to be non-pathogenic or beneficial (Marin et al., 2017). Bailey et al., (2011), and Jones & Versalovic (2009) point out that many members of the genus Lactobacillus prevent the bacterial translocation that can trigger the production of pro-inflammatory cytokines. Certain members of lactobacilli are also thought to impact the behavior of their hosts (Dinan & Cryan, 2012). Supplementation with probiotic strains of lactobacilli in rodents reduces anxiety-like and depressive-like behavior and suppresses corticosterone release (Ait-Belgnaoui et al., 2014, Bravo et al., 2011). In humans, probiotics containing strains of lactobacilli reduce symptoms of anxiety in patients diagnosed with chronic fatigue syndrome and alleviate psychological distress in healthy human volunteers (Messaoudi et al., 2011, Rao et al., 2009). Given these findings, it is possible that the reduction within the order Lactobacillales observed after social stress is anxiogenic and impacts behavioral responses to subsequent stressors, however; this possibility requires further investigations into the particular strains driving the change in this order.
In addition to the potential effects discussed above, a reduction in certain strains of lactobacilli can also increase permeability of the gut epithelium to other bacteria, such as genus Clostridium (Bailey et al., 2011). Clostridium has been shown to increase in abundance following stress and is linked to gastrointestinal disease and inflammation (Aguilera et al., 2013, Brook, 2008, Libby & Bearman, 2009). Clostridium may act by producing propionic acid, which is thought to stimulate anxiety-like behavior, to further compromise an adaptive response to future stressors (Hanstock et al., 2004). Interestingly, both dominant hamsters in the current study and mice subjected to a prolonged social disruption stressor (Bailey et al., 2011) exhibit increases in genus Clostridium.
Phyla Firmicutes and Bacteroidetes make up 90% of the bacteria in the gut of humans (Eckburg et al., 2005). Patients diagnosed with major depressive disorder and systemic lupus erythematosus consistently exhibit relatively lower Firmicutes and higher Bacteroidetes than do healthy controls (He et al., 2016, Jiang et al., 2015). Interestingly, LEfSe analysis revealed that Firmicutes were significantly lower and Bacteroidetes were significantly higher in both dominants and subordinates after repeated pairings compared to baseline. This finding extends the results of Bailey et al. (2011), who reported modest reductions in Firmicutes and increases in Bacteroidetes in mice following 6, 2hr bouts of social disruption stress. Further, the current study indicates that these shifts in microbial abundance are not specific only to hamsters that are being attacked, but are also observed in individuals on the winning end of a social conflict experience. Of course, we cannot rule out the possibility that the observed changes following social encounters may be due to social contact, itself, and not due to the agonistic behavior. The fact that we observed some differences in taxa changed between dominant and subordinate animals, however, suggests that the outcome of the agonistic encounters has at least some effect on gut microbiota.
The baseline abundance of certain microbial members has been previously associated with physiological and behavioral responses to subsequent stressors (Mika et al., 2016, Thompson et al., 2016). In this study, baseline abundance of some microbial species was differentially enriched in hamsters that would subsequently become dominant or subordinate in a social conflict situation. For example, bacteria of the phylum Proteobacteria and genus Prevotella were significantly higher in hamsters that went on to become dominant compared with future subordinates. Interestingly, Thompson et al. (2016) found higher levels of Proteobacteria predicted greater REM sleep recovery following inescapable tail shock. Further, taxa from the genus Prevotella are thought to drive overall composition of the microbial community (Yatsunenko 2012) and reductions in these taxa has been associated with physiological consequences such as irritable bowel syndrome (Seksik 2003), eczema (Mah 2006), and rheumatoid arthritis (Vaahtovuo 2008). Therefore, increased levels of these taxa may be beneficial when presented with different physiological and psychological stressors. In contrast, phylum Firmicutes was significantly higher in hamsters that went on to lose their agonistic encounters compared with those that won. While this post-facto association would require replication to assess the notion that microbiota composition can predict an animal’s likelihood of becoming dominant or subordinate following a resident-intruder interaction, it is in accord with recent data suggesting that gut microbiota can influence social behavior (Dinan et al., 2015, Parashar & Udayabanu, 2015). One possible explanation could be that microbiota composition influences, or is influenced by, hormones that control aggressive/subordinate behavior. These possibilities should be pursued in future studies.
In conclusion, this study used a social conflict model in Syrian hamsters to examine the effect of social conflict stress on commensal gut microbiota. We used this model in part because brief exposure to social conflict in this species causes pronounced and well-characterized responses in brain and behavior and because it is also possible to directly compare the responses of weight and age-matched hamsters that become both dominant and subordinate. Here, we demonstrated that 1) one agonistic encounter is sufficient to induce significant changes to gut microbiota, 2) the opportunity to engage in social interaction induces alterations to gut microbiota, although the particular microbial taxa that are altered are not completely overlapping in dominants and subordinates, and 3) certain microbial taxa may predict the outcome of an agonistic encounter. Our study only examined the effect of agonistic encounters after one or nine pairings over the course of 5 days. Further studies should extend this timeline to assess chronic effects of social stress on the microbiome. Although the mechanistic link between gut bacteria and future social rank were not assessed in this study, our findings do raise a number of intriguing questions about how the gut might influence brain and behavior to shape responses to social stress. Future work should investigate the functional consequence of fecal transplant or manipulation of relevant bacteria in animals before exposure to social stress to further elucidate the role of gut microbiota in social conflict-related behavior.
Supplementary Material
6. Acknowledgements
The authors acknowledge the helpful contribution of Hao Tran, Anna Rosenhauer, Taylor Kahl, and Alejandro Guzman Bambaren. This work was supported by the National Institutes of Health R01MH062044 awarded to KLH, a Brains and Behavior Seed Grant to KLH and KEM, a Brains and Behavior Fellowship awarded to KAP, and a Next Generation New Scholars Fellowship awarded to LQB by the Center for Behavioral Neuroscience at GSU. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Brains and Behavior Program, the Center for Behavioral Neuroscience, or Georgia State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
5. Conflicts of interest
None
References
- Agid O, Kohn Y, & Lerer B (2000). Environmental stress and psychiatric illness. Biomed Pharmacother , 54(3), 135–141. doi:10.1016/s0753-3322(00)89046-0 [DOI] [PubMed] [Google Scholar]
- Aguilera M, Vergara P, & Martinez V (2013). Stress and antibiotics alter luminal and wall-adhered microbiota and enhance the local expression of visceral sensory-related systems in mice. Neurogastroenterol Motil, 25(8), e515–529. doi:10.1111/nmo.12154 [DOI] [PubMed] [Google Scholar]
- Ait-Belgnaoui A, Durand H, Cartier C, Chaumaz G, Eutamene H, Ferrier L, … Theodorou V (2012). Prevention of gut leakiness by a probiotic treatment leads to attenuated HPA response to an acute psychological stress in rats. Psychoneuroendocrinology, 37(11), 1885–1895. doi:10.1016/j.psyneuen.2012.03.024 [DOI] [PubMed] [Google Scholar]
- Albers HE, Huhman KL, & Meisel RL (2002). 6 - Hormonal Basis of Social Conflict and Communication A2 - Pfaff, Donald W In Arnold AP, Fahrbach SE, Etgen AM, & Rubin RT (Eds.), Hormones, Brain and Behavior (pp. 393–433). San Diego: Academic Press. [Google Scholar]
- Aronesty E (2011). ea-utils: Command-line tools for processing biological sequencing data. Expression Analysis, Durham, NC. [Google Scholar]
- Aronesty E (2013). Comparison of sequencing utility programs. The Open Bioinformatics Journal, 7(1). [Google Scholar]
- Bailey MT, & Coe CL (1999). Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol, 35(2), 146–155. [PubMed] [Google Scholar]
- Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, & Lyte M (2011). Exposure to a Social Stressor Alters the Structure of the Intestinal Microbiota: Implications for Stressor-Induced Immunomodulation. Brain Behav Immun, 25(3), 397–407. doi:10.1016/j.bbi.2010.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekhuis E, Boschloo L, Rosmalen JG, & Schoevers RA (2015). Differential associations of specific depressive and anxiety disorders with somatic symptoms. J Psychosom Res, 78(2), 116–122. doi:10.1016/j.jpsychores.2014.11.007 [DOI] [PubMed] [Google Scholar]
- Bharwani A, Mian MF, Foster JA, Surette MG, Bienenstock J, & Forsythe P (2016). Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Psychoneuroendocrinology, 63, 217–227. doi:10.1016/j.psyneuen.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Bjorkqvist K (2001). Social defeat as a stressor in humans. Physiol Behav, 73(3), 435–442. [DOI] [PubMed] [Google Scholar]
- Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, … Cryan JF (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A, 108(38), 16050–16055. doi:10.1073/pnas.1102999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I (2008). Infective endocarditis caused by anaerobic bacteria. Arch Cardiovasc Dis, 101(10), 665–676. doi:10.1016/j.acvd.2008.08.008 [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Knight R (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods, 7(5), 335–336. doi:10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, … Knight R (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme j, 6(8), 1621–1624. doi:10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, & Young VB (2008). Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis, 197(3), 435–438. doi:10.1086/525047 [DOI] [PubMed] [Google Scholar]
- Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, & Gewirtz AT (2015). Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature, 519, 92. doi:10.1038/nature14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SM, Kassam Z, & Bercik P (2013). The adoptive transfer of behavioral phenotype via the intestinal microbiota: experimental evidence and clinical implications. Curr Opin Microbiol, 16(3), 240–245. doi:10.1016/j.mib.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Cryan JF (2016). Stress and the Microbiota-Gut-Brain Axis: An Evolving Concept in Psychiatry. Can J Psychiatry, 61(4), 201–203. doi:10.1177/0706743716635538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culpepper L, Muskin PR, & Stahl SM (2015). Major Depressive Disorder: Understanding the Significance of Residual Symptoms and Balancing Efficacy with Tolerability. Am J Med, 128(9 Suppl), S1–s15. doi:10.1016/j.amjmed.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Desbonnet L, Clarke G, Shanahan F, Dinan TG, & Cryan JF (2014). Microbiota is essential for social development in the mouse. Mol Psychiatry, 19(2), 146–148. doi:10.1038/mp.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, … Pettersson S (2011). Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA, 108(7), 3047–3052. doi:10.1073/pnas.1010529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinan TG, & Cryan JF (2012). Regulation of the stress response by the gut microbiota: implications for psychoneuroendocrinology. Psychoneuroendocrinology, 37(9), 1369–1378. doi:10.1016/j.psyneuen.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Dinan TG, Stilling RM, Stanton C, & Cryan JF (2015). Collective unconscious: how gut microbes shape human behavior. Journal Of Psychiatric Research, 63, 1–9. doi:10.1016/j.jpsychires.2015.02.021 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, … Relman DA (2005). Diversity of the human intestinal microbial flora. Science, 308(5728), 1635–1638. doi:10.1126/science.1110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. doi:10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Felice VD, Moloney RD, Cryan JF, Dinan TG, & O’Mahony SM (2015). Visceral Pain and Psychiatric Disorders. Mod Trends Pharmacopsychiatri, 30, 103–119. doi:10.1159/000435936 [DOI] [PubMed] [Google Scholar]
- Galley JD, Nelson MC, Yu Z, Dowd SE, Walter J, Kumar PS, … Bailey MT (2014). Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol, 14, 189. doi:10.1186/1471-2180-14-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Jansson JK, & Knight R (2014). The Earth Microbiome project: successes and aspirations. BMC Biol, 12, 69. doi:10.1186/s12915-014-0069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad PM, Talbot PS, Anderson IM, & McAllister-Williams RH (2015). Managing inadequate antidepressant response in depressive illness. British Medical Bulletin, 115(1), 183–201. doi:10.1093/bmb/ldv034 [DOI] [PubMed] [Google Scholar]
- Hanstock TL, Clayton EH, Li KM, & Mallet PE (2004). Anxiety and aggression associated with the fermentation of carbohydrates in the hindgut of rats. Physiol Behav, 82(2-3), 357–368. doi:10.1016/j.physbeh.2004.04.002 [DOI] [PubMed] [Google Scholar]
- He Z, Shao T, Li H, Xie Z, & Wen C (2016). Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog, 8, 64. doi:10.1186/s13099-016-0146-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhman KL, Bunnell BN, Mougey EH, & Meyerhoff JL (1990). Effects of social conflict on POMC-derived peptides and glucocorticoids in male golden hamsters. Physiol Behav, 47(5), 949–956. [DOI] [PubMed] [Google Scholar]
- Huhman KL, & Jasnow AM (2005). Conditioned defeat. Biology of aggression, 295–326. [Google Scholar]
- Huhman KL, Moore TO, Ferris CF, Mougey EH, & Meyerhoff JL (1991). Acute and repeated exposure to social conflict in male golden hamsters: increases in plasma POMC-peptides and cortisol and decreases in plasma testosterone. Horm Behav, 25(2), 206–216. [DOI] [PubMed] [Google Scholar]
- Jasnow AM, Drazen DL, Huhman KL, Nelson RJ, & Demas GE (2001). Acute and chronic social defeat suppresses humoral immunity of male Syrian hamsters (Mesocricetus auratus). Horm Behav, 40(3), 428–433. doi:10.1006/hbeh.2001.1708 [DOI] [PubMed] [Google Scholar]
- Jiang H, Ling Z, Zhang Y, Mao H, Ma Z, Yin Y, … Ruan B (2015). Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun, 48, 186–194. doi:10.1016/j.bbi.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Jones SE, & Versalovic J (2009). Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol, 9, 35. doi:10.1186/1471-2180-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanuri N, Cassell B, Bruce SE, White KS, Gott BM, Gyawali CP, & Sayuk GS (2016). The impact of abuse and mood on bowel symptoms and health-related quality of life in irritable bowel syndrome (IBS). Neurogastroenterol Motil. doi:10.1111/nmo.12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy PJ, Murphy AB, Cryan JF, Ross PR, Dinan TG, & Stanton C (2016). Microbiome in brain function and mental health. Trends in Food Science & Technology. [Google Scholar]
- Kessler RC (1997). The effects of stressful life events on depression. Annu Rev Psychol, 48, 191–214. doi:10.1146/annurev.psych.48.1.191 [DOI] [PubMed] [Google Scholar]
- Libby DB, & Bearman G (2009). Bacteremia due to Clostridium difficile--review of the literature. Int J Infect Dis, 13(5), e305–309. doi: 10.1016/j.ijid.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, & Knight R (2006). UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics, 7, 371. doi:10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, & Knight R (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol, 71(12), 8228–8235. doi:10.1128/aem.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczynski P, Whelan SO, O’Sullivan C, Clarke G, Shanahan F, Dinan TG, & Cryan JF (2016). Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur J Neurosci. doi:10.1111/ejn.13291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyte M, Vulchanova L, & Brown DR (2011). Stress at the intestinal surface: catecholamines and mucosa-bacteria interactions. Cell Tissue Res, 343(1), 23–32. doi:10.1007/s00441-010-1050-0 [DOI] [PubMed] [Google Scholar]
- Mackos AR, Maltz R, & Bailey MT (2017). The role of the commensal microbiota in adaptive and maladaptive stressor-induced immunomodulation. Horm Behav, 88, 70–78. doi:10.1016/j.yhbeh.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes M (2008). The cytokine hypothesis of depression: inflammation, oxidative & nitrosative stress (IO&NS) and leaky gut as new targets for adjunctive treatments in depression. Neuro Endocrinol Lett, 29(3), 287–291. [PubMed] [Google Scholar]
- Maes M, Kubera M, & Leunis JC (2008). The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuro Endocrinol Lett, 29(1), 117–124. [PubMed] [Google Scholar]
- Mah KW, Bjorksten B, Lee BW, van Bever HP, Shek LP, Tan TN, … Chua KY (2006). Distinct pattern of commensal gut microbiota in toddlers with eczema. Int Arch Allergy Immunol, 140(2), 157–163. doi:10.1159/000092555 [DOI] [PubMed] [Google Scholar]
- Marin IA, Goertz JE, Ren T, Rich SS, Onengut-Gumuscu S, Farber E, … Gaultier A (2017). Microbiota alteration is associated with the development of stress-induced despair behavior. Scientific Reports, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EA, Tillisch K, & Gupta A (2015). Gut/brain axis and the microbiota. J Clin Invest, 125(3), 926–938. doi:10.1172/jci76304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, … Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme j, 6(3), 610–618. doi:10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, … Cazaubiel JM (2011). Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr, 105(5), 755–764. doi:10.1017/s0007114510004319 [DOI] [PubMed] [Google Scholar]
- Mika A, Rumian N, Loughridge AB, & Fleshner M (2016). Exercise and Prebiotics Produce Stress Resistance: Converging Impacts on Stress-Protective and Butyrate-Producing Gut Bacteria. Int Rev Neurobiol, 131, 165–191. doi:10.1016/bs.irn.2016.08.004 [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Barrot M, DiLeone RJ, Eisch AJ, Gold SJ, & Monteggia LM (2002). Neurobiology of Depression. Neuron, 34(1), 13–25. http://dx.doi.org/10.1016/S0896-6273(02)00653-0 [DOI] [PubMed] [Google Scholar]
- Parashar A, & Udayabanu M (2016). Gut microbiota regulates key modulators of social behavior. Eur Neuropsychopharmacol, 26(1), 78–91. doi:10.1016/j.euroneuro.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Potegal M, Huhman K, Moore T, & Meyerhoff J (1993). Conditioned defeat in the Syrian golden hamster (Mesocricetus auratus). Behavioral and Neural Biology, 60(2), 93–102. doi:http://dx.doi.org/10.1016/0163-1047(93)90159-F [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, & Arkin AP (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol, 26(7), 1641–1650. doi:10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Li MX, Xu C, Chen HB, An SC, & Ma XM (2016). Dendritic Spines in Depression: What We Learned from Animal Models. Neural Plast, 2016, 8056370. doi:10.1155/2016/8056370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin HY, Cheng CW, Tang XD, & Bian ZX (2014). Impact of psychological stress on irritable bowel syndrome. World J Gastroenterol, 20(39), 14126–14131. doi:10.3748/wjg.v20.i39.14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna BS (2013). Role of the gut microbiota in human nutrition and metabolism. J Gastroenterol Hepatol, 28 Suppl 4, 9–17. doi:10.1111/jgh.12294 [DOI] [PubMed] [Google Scholar]
- Rao AV, Bested AC, Beaulne TM, Katzman MA, Iorio C, Berardi JM, & Logan AC (2009). A randomized, double-blind, placebo-controlled pilot study of a probiotic in emotional symptoms of chronic fatigue syndrome. Gut Pathog, 7(1), 6. doi:10.1186/1757-4749-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenhauer AM, McCann KE, Norvelle A, & Huhman KL (2017). An acute social defeat stressor in early puberty increases susceptibility to social defeat in adulthood. Horm Behav. doi:10.1016/j.yhbeh.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Ross AP, Norvelle A, Choi DC, Walton JC, Albers HE, & Huhman KL (2017). Social housing and social isolation: Impact on stress indices and energy balance in male and female Syrian hamsters (Mesocricetus auratus). Physiol Behav, 777(Supplement C), 264–269. doi: https://doi.org/10.1016/i.physbeh.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu RV, & Nemeroff CB (2012). Etiology of depression: genetic and environmental factors. Psychiatr Clin North Am, 35(1), 51–71. doi:10.1016/j.psc.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, & Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome Biol, 72(6), R60. doi:10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, & Finlay BB (2010). Gut microbiota in health and disease. Physiol Rev, 90(3), 859–904. doi:10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, … Dore J (2003). Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut, 52(2), 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, … Koga Y (2004). Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol, 558(Pt 1), 263–275. doi:10.1113/jphysiol.2004.063388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RS, Roller R, Mika A, Greenwood BN, Knight R, Chichlowski M, … Fleshner M (2016). Dietary Prebiotics and Bioactive Milk Fractions Improve NREM Sleep, Enhance REM Sleep Rebound and Attenuate the Stress-Induced Decrease in Diurnal Temperature and Gut Microbial Alpha Diversity. Front Behav Neurosci, 70, 240. doi:10.3389/fnbeh.2016.00240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, & Gordon JI (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444(7122), 1027–1031. doi:10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, & Toivanen P (2008). Fecal microbiota in early rheumatoid arthritis. J Rheumatol, 35(8), 1500–1505. [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, … Gordon JI (2012). Human gut microbiome viewed across age and geography. Nature, 486(7402), 222–227. doi:10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


