Abstract
Background
Lung squamous cell carcinoma (LUSC) accounts for 20–30% of non-small cell lung cancers (NSCLCs). There are limited treatment strategies for LUSC in part due to our inadequate understanding of the molecular underpinnings of the disease. We performed whole-exome sequencing (WES) and comprehensive immune profiling of a unique set of clinically annotated early-stage LUSCs to increase our understanding of the pathobiology of this malignancy.
Methods
Matched pairs of surgically resected stage I-III LUSCs and normal lung tissues (n = 108) were analyzed by WES. Immunohistochemistry and image analysis-based profiling of 10 immune markers were done on a subset of LUSCs (n = 91). Associations among mutations, immune markers and clinicopathological variables were statistically examined using analysis of variance and Fisher’s exact test. Cox proportional hazards regression models were used for statistical analysis of clinical outcome.
Results
This early-stage LUSC cohort displayed an average of 209 exonic mutations per tumor. Fourteen genes exhibited significant enrichment for somatic mutation: TP53, MLL2, PIK3CA, NFE2L2, CDH8, KEAP1, PTEN, ADCY8, PTPRT, CALCR, GRM8, FBXW7, RB1 and CDKN2A. Among mutated genes associated with poor recurrence-free survival, MLL2 mutations predicted poor prognosis in both TP53 mutant and wild-type LUSCs. We also found that in treated patients, FBXW7 and KEAP1 mutations were associated with poor response to adjuvant therapy, particularly in TP53-mutant tumors. Analysis of mutations with immune markers revealed that ADCY8 and PIK3CA mutations were associated with markedly decreased tumoral PD-L1 expression, LUSCs with PIK3CA mutations exhibited elevated CD45ro levels and CDKN2A-mutant tumors displayed an up-regulated immune response.
Conclusion(s)
Our findings pinpoint mutated genes that may impact clinical outcome as well as personalized strategies for targeted immunotherapies in early-stage LUSC.
Keywords: LUSC, whole exome sequencing, immune profiles
Introduction
Lung squamous cell carcinoma (LUSC) is a major subtype of non-small cell lung cancer (NSCLC) that is mainly diagnosed in smokers and accounts for approximately 20–30% of new lung cancer cases and 40 000 annual deaths in the United States [1]. While targeted strategies have shown therapeutic benefit in lung adenocarcinoma, very limited personalized therapies have been developed for LUSC [1]. These contrasts are largely due to a more limited understanding of molecular targets (e.g. genomic alterations) in LUSC [1, 2]. Previous profiling efforts identified recurrent alterations in LUSC including mutations in TP53, CDKN2A, PIK3CA, DDR2, NFE2L2, KEAP1 and MLL2 [3–5], gain of the lineage-specific oncogene SOX2 [3, 6] and loss of the CDKN2A tumor suppressor [3, 7]. Mutations in TP53 represent the most frequent (> 60–70%) genomic alteration found in LUSC [3, 7]. While these alterations have increased our understanding of the molecular pathology of LUSC, their impact on clinical outcome and personalized treatment strategies in LUSC is still not clear.
Earlier work suggested that the immune system can suppress the progression of lung cancer [8, 9]. Evasion of immune surveillance by the tumor is crucial for oncogenesis and is largely due to decreased activation of effector T-cells and suppression of anti-cancer immunity by inhibitory checkpoints such as PD-1 receptor and its ligand PD-L1 [8, 9]. The interplay between the immune system and lung tumor has important clinical ramifications. For example, NSCLC patients with low levels of tumor-infiltrating effector T-cells display relatively poor prognosis [10]. Targeted therapies that re-engage the immune system (e.g. checkpoint-blockade immunotherapies) have demonstrated durable responses in NSCLC patients [11]. Of note, recent studies pointed to the association of pre-existing immunity with clinical response to immunotherapies [12]. Thus, understanding molecular phenotypes in the tumor that are linked to the host immune system may improve personalized treatment using immunotherapies.
In this study we analyzed the mutational landscape of 108 early-stage surgically resected LUSCs by whole-exome sequencing (WES) and assessed pre-existing immunity in 91 of these LUSCs using immunohistochemistry (IHC)- and imaging-based immunoprofiling. We utilized a well annotated specimen set that permits analysis of mutations, in single or in combination, with outcome. We describe genomic mutations that are significantly associated with clinical outcome of early-stage LUSCs as well as alterations that may impact personalized approaches for immune-based therapies.
Methods
Detailed methods and codes pertaining to statistical analyses are found in the supplementary Methods, available at Annals of Oncology online and supplementary sweave reports S1–4, available at Annals of Oncology online accompanying the manuscript.
Early-stage LUSC cohort
Early-stage surgically resected LUSCs and matched normal lung tissues (n = 108 pairs, supplementary Table S1, available at Annals of Oncology online) were analyzed in this study. All malignant and normal lung tissues were acquired from patients who were evaluated at MD Anderson Cancer Center (MD Anderson) following informed consent under protocols approved by the Institutional Review Board. Tumors were classified using the 2004 WHO classification system as described previously [13]. All samples were obtained snap-frozen. For each tissue sample, the percentage of malignant tissue was assessed by histological examination. The analyzed LUSCs comprised at least 30% malignant lung cells.
WES and identification of somatic point mutations and CNVs
Genomic DNA was captured on the NimbleGen 2.1M human exome array according to the manufacturer’s instructions. Sequencing was performed using 75-bp paired-end reads and the Illumina HiSeq2000 platform [14]. Reads were mapped to the reference genome (hg19) and mutations were identified as described previously [15, 16] (supplementary Methods, available at Annals of Oncology online). Somatic CNVs were determined by comparing and contrasting coverage depth of individual capture intervals (1 Mb intervals) between LUSCs and paired normal lung tissues.
IHC-based analysis of immune markers
Sequential histological tumor sections (4 µm in thickness) were prepared from 91 formalin-fixed paraffin-embedded LUSCs studied by WES. IHC was performed using an automated staining system (BOND-MAX; Leica Microsystems) using antibodies raised against 10 immune markers: PD-L1, PD-1, CD3, CD4, CD8, CD45RO, CD57, Granzyme B, FOXP3 and CD68 (supplementary Methods, available at Annals of Oncology online).
Results
Sequencing of early-stage LUSCs
We performed WES of 108 early-stage (stages I-III) LUSCs and paired normal lung tissues (supplementary Table S1, available at Annals of Oncology online). All LUSC patients were former or current smokers (n = 54 each). Median follow-up times to survival and recurrence were 42.8 and 28.9 months, respectively. Tumors were sequenced at a greater depth relative to paired normal lung tissues with the targeted sequences read an average of 189 X and 106 X, respectively (supplementary Table S2, available at Annals of Oncology online). We then computed differences in B-allele frequencies of common genomic variants in tumor-normal pairs in order to map loss-of-heterozygosity (LOH) segments. We found a median of 21.5 LOH events (supplementary Figure S1A, available at Annals of Oncology online) and an overall LOH rate of 42.6% across the tumor genome (supplementary Figure S1B, available at Annals of Oncology online).
The early-stage LUSCs harbored 22 530 somatic coding mutations (mean = 209) that consisted of 15 422 missense, 1167 nonsense, 5090 silent mutations, 374 small insertions and deletions (indels) and 477 alterations residing in exon-exon boundaries. The most common substitutions were C > A transversions (supplementary Figure S2, available at Annals of Oncology online), indicative of a smoking mutation signature [17]. Former and current smokers exhibited similar spectra of somatic base substitutions (supplementary Figure S2, available at Annals of Oncology online). There were no significant differences in somatic mutation burden and in total nonsynonymous mutations by stage (supplementary Figure S3A and B, available at Annals of Oncology online, respectively) as well as no significant associations between somatic mutation burden and overall (OS) or recurrence-free (RFS) survival (supplementary sweave report S1, available at Annals of Oncology online).
Analysis of single-nucleotide variants and copy number alterations in early-stage LUSC
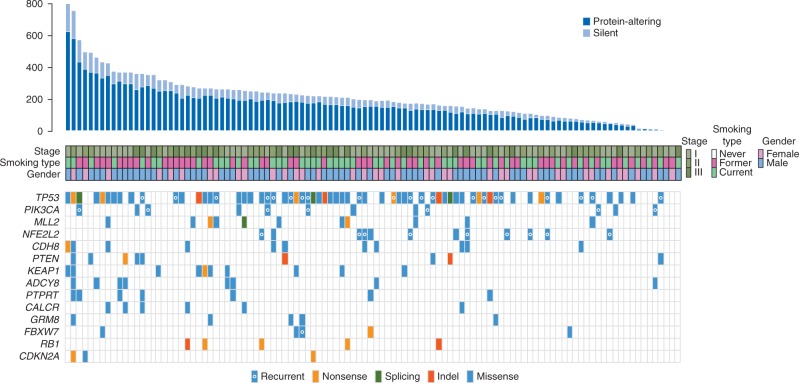
To increase our power of mutated gene detection, we first combined the MD Anderson cohort with 178 LUSCs profiled by the TCGA [3]. The probabilities of mutations in each gene were adjusted in the combined set to prioritize variants for analysis in the MD Anderson cohort (supplementary Methods, available at Annals of Oncology online). Fourteen genes exhibited significant overall mutation enrichment based on the genome-wide threshold of P < 2.4 × 10 − 6 (Figure 1; supplementary Table S3, available at Annals of Oncology online). These included genes that were previously reported [3, 4, 18] to be significantly mutated in LUSC: TP53, KEAP1, PIK3CA, NFE2L2, MLL2, PTEN, RB1, CDKN2A, FBXW7 and GRM8 (Figure 1). We also found additional significantly mutated genes: CDH8, ADCY8, PTPRT and CALCR (Figure 1). The distribution of mutated genes was not influenced by stage (Figure 1). TP53 was the most frequently mutated gene (60%) and mutations in NFE2L2 and in its inhibitor KEAP1 were mutually exclusive (Figure 1). With the exception of CDKN2A (MD Anderson, 2.8%; TCGA, 12.4%) and MLL2 (MD Anderson, 10.2%; TCGA, 17.4%), the genes exhibited similar mutation frequencies between both cohorts (supplementary Table S3, available at Annals of Oncology online).
Figure 1.
Genes with significant enrichment for mutations in the 108 early-stage LUSCs. Early-stage LUSCs are denoted by columns and are arranged from left to right by the number of exonic non-silent somatic mutations (top panel). Rows represent genes with statistical enrichment for mutations. The significantly mutated genes are ordered vertically in decreasing order of non-silent mutation frequency. Middle rows indicate smoking status (former and current), gender and pathological stage of the LUSC patients. Colored rectangles indicate mutation categories.
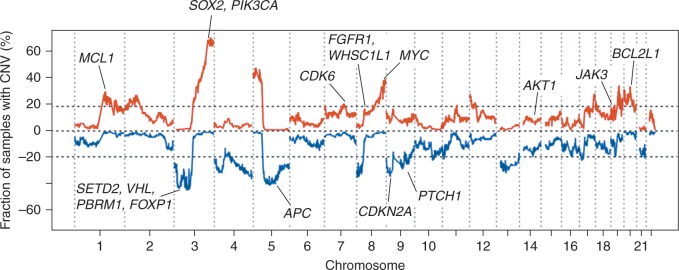
Consistent with previous reports [3, 6], gain of chromosome 3q, comprising SOX2, PIK3CA and TP63, was the most frequently observed CNV (70.4%) in this cohort (Figure 2; supplementary Table S4, available at Annals of Oncology online). We also noted broad gains in chromosomal regions harboring MYC, BCL2L1, MCL1, CDK6 and JAK3 (Figure 2). We observed focal gains of regions that include AKT1 as well as FGFR1 and WHSC1L1 (Figure 2). Copy number aberration of chromosome 3p, including the tumor suppressors SETD2 and VHL was the most frequently observed deletion (44.4%; Figure 2; supplementary Table S5, available at Annals of Oncology online). Additional deletions included the tumor suppressors CDKN2A, PTCH1 and APC. Global CNV patterns between MD Anderson and TCGA cohorts were significantly and positively correlated (R = 0.82, P < 2.2 × 10 − 16; supplementary Figure S4, available at Annals of Oncology online).
Figure 2.
Genome-wide copy number gains and losses in the 108 early-stage LUSCs. Gains and losses within 1 Mb chromosomal intervals were identified as described in supplementary Methods, available at Annals of Oncology online. Frequency of chromosomal interval copy number gain (red) and loss (blue) were plotted along the genome. Bona fide driver genes within recurrent chromosomal interval gains and losses and in significant CNV peak regions are labeled.
Prognostic and predictive alterations in early-stage LUSCs
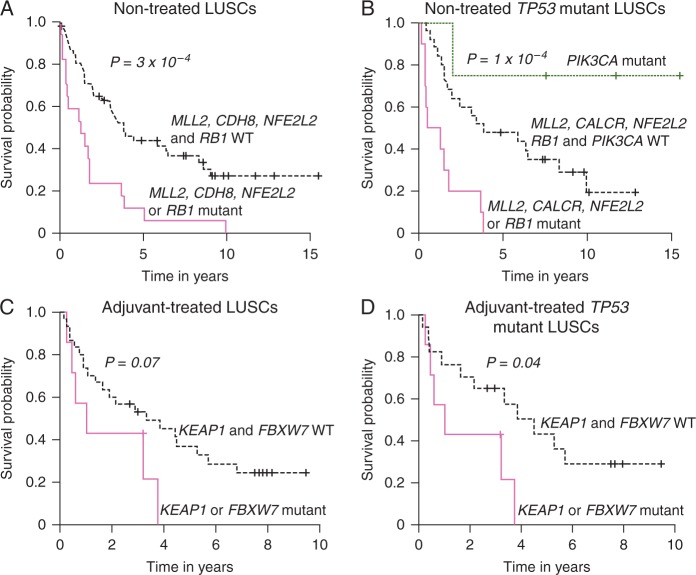
Next we sought to identify prognostic mutations and CNVs by analysis of LUSC patients who did not receive adjuvant therapy as well as predictive aberrations by probing patients who did receive therapy. We applied univariate models to prioritize altered genes and CNVs for inclusion in multivariate models (supplement Data, available at Annals of Oncology online). Akaike information criterion stepwise method was then employed to remove insignificant terms during building of multivariate models (supplementary Methods, available at Annals of Oncology online). Mutant MLL2 was the most statistically significant (P < 10 − 6) indicator of prognosis (worse RFS) in early-stage LUSC patients who did not receive adjuvant therapy (nontreated), irrespective of TP53 mutation status (supplementary Figure S5, available at Annals of Oncology online). Conversely, in patients treated with adjuvant therapy, MLL2 mutations were not predictive of RFS and response to therapy (supplementary Figure S6A, available at Annals of Oncology online). Mutations in MLL2, CDH8, NFE2L2 or RB1 were associated with significantly worse RFS (P < 0.001; Figure 3A). Among LUSCs with mutant TP53, mutations in MLL2, NFE2L2, CALCR or RB1 were associated with significantly worse RFS compared to patients who were WT for these genes or to patients with mutant PIK3CA (P < 0.001) (Figure 3B). Among LUSC patients who received adjuvant therapy, mutations in FBXW7 (P < 0.001) and KEAP1 (P = 0.07) were associated with poor response to therapy (supplementary Figure S6B and C, available at Annals of Oncology online, respectively). There was a trend for poor response in patients with mutations in FBXW7 or KEAP1 (Figure 3C). This association reached statistical significance (P = 0.04) in TP53 mutant LUSCs (Figure 3D). Of note, while being predictive of poor response to adjuvant therapy, FBXW7 and KEAP1 mutations were not significantly associated with prognosis (supplementary Figure S6D and E, available at Annals of Oncology online, respectively).
Figure 3.
Prognostic and predictive significantly mutated genes and CNVs in early-stage LUSC. (A) Mutations in MLL2, CDH8, NFE2L2 or RB1 were associated with significantly poorer RFS. (B) Among TP53 mutant LUSCs, mutations in MLL2, CALCR, NFE2L2 or RB1 were associated with poor RFS whereas mutations in PIK3CA predicted better RFS. Among those who received adjuvant chemotherapy, mutations in KEAP1 or FBXW7 exhibited a trend for predicting poor response to therapy (C) which reached statistical significance when assessed in TP53 mutant LUSCs (D). P-values were obtained using the log-rank test and the Kaplan–Meier method for estimation of survival probability.
Associations between mutational and immune phenotypes in early-stage LUSC
We analyzed associations between mutational phenotypes of LUSCs and markers of immune response. Using imaging-based IHC, we examined the protein expression (intratumoral or peritumoral) of PD-1, CD3, CD4, CD8, CD45ro, CD57, CD68, FOXP3 and Granzyme B as well as tumoral levels of PD-L1 in 91 out of the 108 early-stage LUSCs. Among the markers, only peritumoral expression of CD57 was significantly correlated (positively; P = 0.006) with total number of exonic somatic mutations (supplementary Table S6, available at Annals of Oncology online).
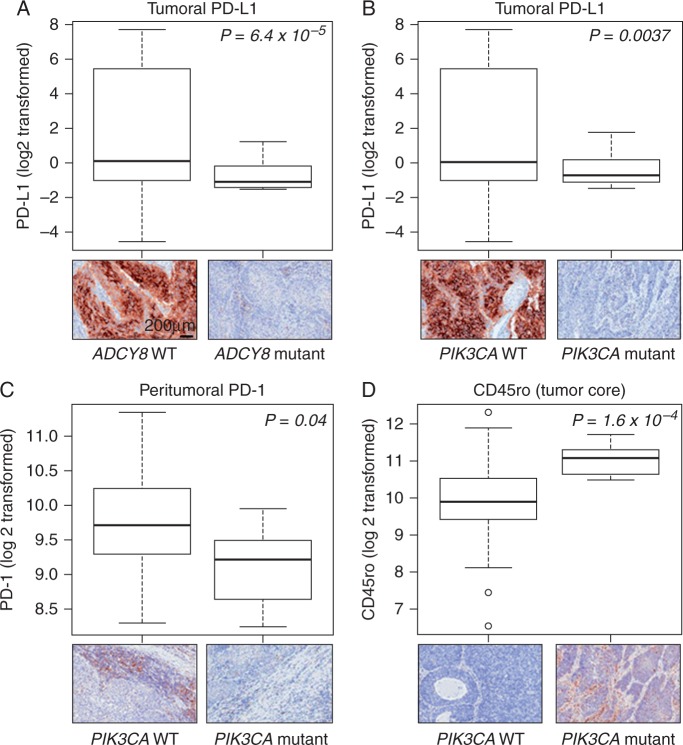
In the context of the significantly mutated genes, mutations in ADCY8 were significantly associated with suppressed tumoral PD-L1 expression (Figure 4A;P < 0.001). Moreover, tumoral PD-L1 (P = 0.004), and peritumoral levels of its receptor PD-1 (P = 0.04), were significantly decreased in PIK3CA mutant relative to WT LUSCs (Figure 4B and C, respectively). PIK3CA mutant LUSCs also exhibited significantly elevated intratumoral expression of the memory T cell marker CD45ro (P < 0.001; Figure 4D). CDKN2A mutated LUSCs exhibited an overall up-regulated immune response whereas NFE2L2 mutant tumors displayed reduced expression of various markers (supplementary Table S7, available at Annals of Oncology online). Among TP53 mutant LUSCs, co-occurring mutations in PIK3CA were significantly associated with reduced PD-L1 expression (supplementary Figure S7, available at Annals of Oncology online; P < 10 − 7). Among TP53 WT tumors, mutations in PIK3CA and MLL2 were significantly associated with decreased peritumoral and intratumoral PD-1, respectively (both P < 0.05; supplementary Figure S8, available at Annals of Oncology online).
Figure 4.
Differential expression of immune markers in early-stage LUSCs with significantly mutated genes. Significantly decreased expression of tumoral PD-L1 expression in ADCY8 (A) and PIK3CA (B) mutant LUSCs compared with wild-type tumors. (C) Significantly attenuated peritumoral PD-L1 expression in PIK3CA mutant relative to wild type LUSCs. (D) Significantly elevated CD45ro expression in immune cells infiltrating TP53 mutant relative to wild type LUSCs. Upper panels represent graphical summaries (box plots) of the log (base 2) transformed expression of the indicated immune markers. Lower panels comprise representative photomicrographs (200 × magnification) of the immunohistochemical expression of the immune markers. P-values were obtained by t-tests; values < 0.05 were considered statistically significant.
Discussion
In the present study, we probed molecular and immunophenotypes in 108 surgically resected LUSCs generating a unique dataset of early-stage LUSC comprising both WES and immune marker profiling data. Our WES analysis identified significantly mutated genes and recurrent CNVs in early-stage LUSC some of which were predictive of prognosis (e.g. mutated MLL2) or response to therapy (e.g. mutated FBXW7). Our analysis also pointed to putative immune markers that are significantly altered based on somatic mutation status. Of note, PIK3CA mutant LUSCs, relative to WT tumors, exhibited substantially reduced expression of the immune inhibitory checkpoint PD-L1 in tumor cells as well as reduced expression of the receptor PD-1 in surrounding (peritumoral) immune cells. Our findings point to mutations and CNVs that could play a key role in the clinical outcome and in engaging the host immune system of early-stage LUSC patients.
Sequence analysis of this cohort (Figures 1 and 2) identified significant mutated genes and CNVs that corroborated previous reports [3–7, 18, 19]. Also similar to previous studies [3, 4], TP53 was the most frequently mutated gene. Our WES analysis pointed to additional genes with significant enrichment for mutation: CDH8, ADCY8, PTPRT, CALCR and GRM8. Importantly, we found that these five genes exhibited similar mutation frequencies between the MD Anderson and TCGA cohorts (supplementary Table S3, available at Annals of Oncology online). Of note, mutations in the glutamate receptor GRM8 were reported in LUSCs (∼ 8%) [18], metastatic melanomas [20] and endometrial cancers [21]. Also, the protein tyrosine phosphatase PTPRT was found to be significantly mutated in head and neck squamous cell carcinomas [22] and in metastatic melanoma [20]. Our sequencing analyses in the context of previously reported findings, underscore the plausible roles of the identified significantly mutated genes in the pathogenesis of LUSC.
Univariate analysis revealed that mutant MLL2, a histone lysine N-methyltransferase also known as KMT2D, was the alteration most significantly associated with poor prognosis. These findings are in close agreement with the previous report by Kim and colleagues on genomic analysis of East Asian LUSCs [4]. They also suggest that aberrant epigenetic regulators such as MLL2 may represent viable targets in LUSC. Among adjuvant-treated LUSCs, mutant FBXW7 was the most significant predictor of poor response to therapy. Moreover, our multivariate analysis revealed that among TP53 mutant early-stage LUSC patients, co-occurring mutations in either KEAP1 or FBXW7 significantly predicted poor response to adjuvant therapy. It is noteworthy that the tumor suppressive function of FBXW7 was shown to be crucial for chemosensitivity of lung carcinoma cell lines [23]. Also, our data on KEAP1 mutations are in accordance with the previously reported association of mutations in KEAP1 with poor response to adjuvant chemotherapy in the study by Solis and colleagues [24]. Additionally, in a companion manuscript by Kadara and colleagues [25], KEAP1 mutations in early-stage lung adenocarcinomas (LUADs) were also found to be significantly associated with poor response to chemotherapy. Our coupled WES and clinicopathological analysis revealed mutations and CNVs with key roles in the clinical outcome of early-stage LUSC patients. It is noteworthy that our analysis is not only limited by the cohort size but also by the number of patients exhibiting particular mutated genes. Our findings on the association of genomic alterations with the clinical outcome and chemosensitivity of early-stage LUSC are hypotheses generating and warrant further examination in future studies.
Understanding the interplay between molecular and immune phenotypes may help improve strategies for personalized immunotherapy [11, 12]. Towards this, we first probed associations between immune marker expression and the somatic mutation burden and found that CD57 was the only examined immune marker that was significantly correlated with the somatic mutation burden. Of note, this finding is in contrast to the companion study by Kadara and colleagues demonstrating elevated expression of various immune markers, including PD-L1, with increased mutational burden in smoker LUADs [25]. Moreover, a higher non-silent (NS) mutation burden in NSCLCs was associated with improved objective response to treatment with the anti-PD-1 antibody pembrolizumab [26] suggesting that expression of immunotherapy targets such as PD-L1 are elevated in tumors with increased levels of somatic mutations. However, it is noteworthy the late-stage (stage IV) NSCLC cohort in the study by Rizvi and colleagues [26] was mainly comprised of LUADs (29 out of 34) and the few LUSCs mainly exhibited negative or weak PD-L1 immunohistochemical expression. Thus, based on findings from our present study and on published data from others, it is plausible that the impact of the exonic somatic mutation burden on the immune system and response to anti-PD-1 treatment may be restricted to non-squamous NSCLCs.
We also found that PIK3CA mutations were associated with significantly reduced expression of tumoral PD-L1 and peritumoral PD-1. These findings suggest that PD-L1/PD-1 immune checkpoint signaling is attenuated in LUSCs with mutations in the PIK3CA oncogene. Notably, in the companion study by Kadara and colleagues, tumoral PD-L1 expression was also markedly reduced in PIK3CA mutant LUADs [25]. Since objective responses to the PD-L1-targeting antibody atezolizumab were reported to be markedly associated with high levels of tumoral PD-L1 [12], it is tempting to speculate that mutations in PIK3CA might serve as viable biomarkers for relatively poor response to immunotherapies such as PD-L1-targeting antibodies. This supposition warrants further efforts to ascertain the role of these somatic mutations in personalized anti-PD-L1 immunotherapy.
Our coupled WES and clinicopathological analysis identified prognostic and predictive somatic genomic alterations in early-stage LUSC. Combined immunoprofiling and WES analyses revealed mutational profiles in early-stage LUSC that may alter engagement of the host immune system, and thus, the response of the tumors to immunotherapy. Additionally, our findings indicate mutated genes that could serve as candidate biomarkers and play key roles in personalized strategies for targeted immunotherapies, such as therapies currently being investigated in the Lung Master Protocol [27].
Funding
This work was supported by Yale Gilead Program [R.S.H., R.P.L. and J.S.]; Department of Defense (DoD) PROSPECT grant W81XWH-07-1-0306 [I.I.W.]; The Cancer Prevention and Research Institute of Texas (CPRIT) multi-investigator research award RP120713 [I.I.W.]; The University of Texas Specialized Programs of Research Excellence (SPORE) in lung cancer grant P50CA70907 [I.I.W]; Yale University SPORE in lung cancer grant P50CA196530 [R.S.H. and E.K.] and R01CA155196 [R.S.H].
Disclosure
IIW is an advisor to Genentech/Roche, Genecentric, GlaxoSmithKline, GE Healthcare, Lilly, Clovis Oncology, Bristol-Myers Squibb and Celgene and has received funding from Genetech, Merck, OncoPlex Diagnostics, Myriad Genetics, Bayer and Jounce Therapeutics outside the submitted work. RSH is an advisor to Koltan pharmaceuticals, DaiTech Oncology, and Biothera, received funding from Genentech and Gilead Sciences and has received Honoraria from Lily, Merck, Boehringer Ingelheim, Pfizer, AstraZeneca/MedImmune, Genetech and Bristol-Myers Squibb outside the submitted work. DR reports stock in Metamark Genetics, received honoraria from Amgen, ACD Inc, Astrazeneca and Bristol-Myers Squibb, is an advisor to Biocept, Cernostics, Genoptix and Perkin Elmer and received funding from Gilead Sciences, Cepheid, Koltan Pharmaceuticals, OncoPlex Diagnostics and Perkin Elmer outside the submitted work. SS is an advisor to GlaxoSmithKline outside the submitted work. JPT reports stock or ownership in Penrose Care, is an advisor to Merck, Sanofi Pasteur and Vitality outside the submitted work and has received funding from Gilead Sciences. ZM and SGG received funding from Gilead Sciences. JF received funding from Astellas Pharma outside the submitted work. TJL reports stock or ownership in and received honoraria from Bristol-Myers Squibb outside the submitted work. All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Derman BA, Mileham KF, Bonomi PD. et al. Treatment of advanced squamous cell carcinoma of the lung: a review. Transl Lung Cancer Res 2015; 4: 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cardarella S, Johnson BE. The impact of genomic changes on treatment of lung cancer. Am J Respir Crit Care Med 2013; 188: 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim Y, Hammerman PS, Kim J. et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol 2014; 32: 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hammerman PS, Sos ML, Ramos AH. et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov 2011; 1: 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bass AJ, Watanabe H, Mermel CH. et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009; 41: 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rooney M, Devarakonda S, Govindan R. Genomics of squamous cell lung cancer. Oncologist 2013; 18: 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dranoff G. Immunotherapy at large: balancing tumor immunity and inflammatory pathology. Nat Med 2013; 19: 1100–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quezada SA, Peggs KS, Simpson TR, Allison JP. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol Rev 2011; 241: 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schalper KA, Brown J, Carvajal-Hausdorf D. et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 2015; 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brahmer JR. Immune checkpoint blockade: the hope for immunotherapy as a treatment of lung cancer? Semin Oncol 2014; 41: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Herbst RS, Soria JC, Kowanetz M. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515: 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang X, Kadara H, Behrens C. et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res 2011; 17: 2434–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choi M, Scholl UI, Ji W. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 2009; 106: 19096–19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi M, Scholl UI, Yue P. et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 2011; 331: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao S, Choi M, Overton JD. et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci USA 2013; 110: 2916–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lawrence MS, Stojanov P, Polak P. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013; 499: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kan Z, Jaiswal BS, Stinson J. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 2010; 466: 869–873. [DOI] [PubMed] [Google Scholar]
- 19. Sakashita S, Sakashita M, Sound Tsao M. Genes and pathology of non-small cell lung carcinoma. Semin Oncol 2014; 41: 28–39. [DOI] [PubMed] [Google Scholar]
- 20. Ding L, Kim M, Kanchi KL. et al. Clonal architectures and driver mutations in metastatic melanomas. PLoS One 2014; 9: e111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang H, Cheung LW, Li J. et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res 2012; 22: 2120–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lui VW, Peyser ND, Ng PK. et al. Frequent mutation of receptor protein tyrosine phosphatases provides a mechanism for STAT3 hyperactivation in head and neck cancer. Proc Natl Acad Sci USA 2014; 111: 1114–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yokobori T, Yokoyama Y, Mogi A. et al. FBXW7 mediates chemotherapeutic sensitivity and prognosis in NSCLCs. Mol Cancer Res 2014; 12: 32–37. [DOI] [PubMed] [Google Scholar]
- 24. Solis LM, Behrens C, Dong W. et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 2010; 16: 3743–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kadara H, Choi M, Zhang J. et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Annals of Oncology 2017; 28: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rizvi NA, Hellmann MD, Snyder A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herbst RS, Gandara DR, Hirsch FR. et al. Lung Master Protocol (Lung-MAP)- A Biomarker-Driven Protocol for Accelerating Development of Therapies for Squamous Cell Lung Cancer: SWOG S1400. Clin Cancer Res 2015; 21: 1514–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.