Abstract
Nitrite-oxidizing bacteria (NOB) play a critical role in the mitigation of nitrogen pollution by metabolizing nitrite to nitrate, which is removed via assimilation, denitrification, or anammox. Recent studies showed that NOB are phylogenetically and metabolically diverse, yet most of our knowledge of NOB comes from only a few cultured representatives. Using cultivation and genomic sequencing, we identified four putative Candidatus Nitrotoga NOB species from freshwater sediments and water column samples in Colorado, USA. Genome analyses indicated highly conserved 16S rRNA gene sequences, but broad metabolic potential including genes for nitrogen, sulfur, hydrogen, and organic carbon metabolism. Genomic predictions suggested that Ca. Nitrotoga can metabolize in low oxygen or anoxic conditions, which may support an expanded environmental niche for Ca. Nitrotoga similar to other NOB. An array of antibiotic and metal resistance genes likely allows Ca. Nitrotoga to withstand environmental pressures in impacted systems. Phylogenetic analyses highlighted a deeply divergent nitrite oxidoreductase alpha subunit (NxrA), suggesting a novel evolutionary trajectory for Ca. Nitrotoga separate from any other NOB and further revealing the complex evolutionary history of nitrite oxidation in the bacterial domain. Ca. Nitrotoga-like 16S rRNA gene sequences were prevalent in globally distributed environments over a range of reported temperatures. This work considerably expands our knowledge of the Ca. Nitrotoga genus and suggests that their contribution to nitrogen cycling should be considered alongside other NOB in wide variety of habitats.
Introduction
Increasing anthropogenic sources of nitrogen have led to environmental risks including eutrophication, increased greenhouse gas emissions, and acidification. Nitrite-oxidizing bacteria (NOB) play a critical role in mitigating the harmful effects of nitrogen pollution by linking nitrogen sources to nitrogen removal. Specifically, nitrite pools from natural (e.g., ammonia oxidation) or anthropogenic (e.g., fertilizer) sources are converted into nitrate, which is assimilated or removed as inert nitrogen gas via denitrification or anammox pathways. Thus, understanding the diversity, metabolism, and environmental limits of NOB is important for controlling and managing elevated nitrogen in contaminated ecosystems.
Despite the environmental relevance of NOB, the group is understudied in part due to slow growth, taking as long as 12 years for isolation [1]. Assiduous cultivation efforts [2–7], as well as single-cell [8] and metagenomic [9] sequencing studies are beginning to illuminate the diversity of nitrite oxidizers. The known NOB belong to four phyla and seven different genera. Three genera were proposed within the last decade based on NOB enrichment (Candidatus Nitrotoga), isolation (Nitrolancea), or single-cell genomes recovered from the environment (Candidatus Nitromaritima) [2, 7, 8]. NOB are physiologically versatile, utilizing nitrite oxidation, complete ammonia oxidation (comammox within the Nitrospira) [5, 6], as well as organic carbon, hydrogen, and sulfur metabolisms to drive growth [10–12].
Ca. Nitrotoga is an understudied genus of NOB that may play a critical role in nitrogen removal in engineered and natural environments [13]. Recent 16S rRNA-based molecular surveys have identified Ca. Nitrotoga-like sequences in a wide range of habitats, including: glacial soils [14–17], an underground cave [18], a freshwater seep [19], drinking water [20, 21], a subglacial Antarctic lake [22], Yellow Sea seawater [23], salt marsh sediments [24], rivers [25, 26], and various wastewater treatment plants (WWTPs) or sequence batch reactors [13, 27–32]. The relative abundance of Ca. Nitrotoga-like sequences in several WWTPs and a subglacial lake were as high as 2–13% of the total bacterial community, and were occasionally the only observed NOB [13, 22, 32]. The distribution and relative abundance of Ca. Nitrotoga-like sequences suggest these organisms likely play a critical nitrogen cycling role in diverse environments; however, few studies have attempted to characterize their diversity or physiology.
Only four Ca. Nitrotoga cultures have been reported to date, with no confirmed isolates and no genome sequences are available within the genus. Ca. Nitrotoga arctica was enriched from permafrost [2]; Ca. Nitrotoga sp. HAM-1 from activated sludge [33]; Ca. Nitrotoga sp. HW29 from an aquaculture system [34]; and Ca. Nitrotoga sp. AM1 from coastal sand [35]. All Ca. Nitrotoga were enriched at low temperatures (4–17 °C), however temperature optima were slightly higher (13−22 °C) [2, 33–35]. Ca. Nitrotoga arctica has an intermediate nitrite affinity and is adapted to low nitrite concentrations (0.3 mM) compared to other NOB [36, 37].
Here, we describe near-complete draft genome sequences of four putative Ca. Nitrotoga species enriched from river water column and sediment samples. Each organism contained genomic capabilities for diverse metabolisms, which could support their growth in a wide range of habitats. This study represents the first reported cultivation of Ca. Nitrotoga from natural, freshwater systems and the first reported genome profiling from within the genus. These findings extend our understanding of freshwater nitrite oxidation and form the basis for further experimental work aimed at testing genomic predictions in culture and in the environment.
Methods
Culture inoculation and growth
Two surface sediment samples were collected from the urban-impacted Cherry Creek in downtown Denver, CO (samples MKT and LAW). Two water column samples were collected from two agriculturally-impacted rivers near Greeley, CO (about 100 km North of Denver, CO) (samples CP45 from the Cache la Poudre River and SPKER from the South Platte River). Sediment and water column samples were inoculated into freshwater nitrite oxidizer medium (FNOM) with 0.3 mM nitrite and incubated at room temperature in the dark (Supplemental Note). Enrichment cultures were transferred to new media approximately every two weeks. Enrichment was enhanced by serial dilution, rapid transfers at the beginning of nitrite consumption, and low volume transfers (as low as 0.1% inoculum vol./vol.). Nitrite consumption was regularly monitored in the cultures using a Griess nitrite color reagent [38]. Nitrite oxidation rates were determined in triplicate for each enrichment culture (Supplemental Note). Fluorescent in situ hybridization (FISH) was performed on enrichment cultures using Ca. Nitrotoga-specific probes [13] (Supplemental Note).
DNA extraction and sequencing
DNA was extracted from each culture at mid-phase to late-phase of exponential nitrite oxidation (Supplemental Note). Extracted DNA was sheared using a Covaris S220 (Covaris, Woburn, MA) and libraries were prepped with an insert size of 400 bp using an Ovation Ultralow System V2 (No. 0344) kit (Nugen, San Carlos, CA) by the University of Colorado Anschutz Medical Campus Genomics Core. DNA was sequenced on an Illumina HiSeq 2500 using V4 chemistry (Illumina, San Diego, CA) with 125 bp paired end reads.
Metagenome and Ca. Nitrotoga genome assembly and annotation
Metagenomes from each enrichment culture were assembled with quality filtered and trimmed reads using MEGAHIT [39] and contigs were binned using MetaBAT [40]. Ca. Nitrotoga genomes were assembled iteratively with SPAdes v3.9.0 [41] (Supplemental Note). Genome bin completeness and contamination estimates were calculated using the CheckM v1.0.11 lineage workflow [42] (Supplemental Note).
Final assemblies were filtered to remove contigs <2 kb, all of which had very low and uneven coverage estimates. The Ca. Nitrotoga genomes were aligned and contigs reordered with progressiveMauve [43], using the Ca. Nitrotoga species from the MKT culture as a reference due to its long contig length and simple assembly graph. Genomes were submitted to the DOE-JGI Microbial Genome Annotation Pipeline (MGAP) [44] for final contig trimming of ambiguous and low-complexity sequences. Gene annotations were evaluated based on results from MGAP including COG, KEGG, Pfam, and TIGRfam assignments, InterPro Scan, IMG term assignments, and final protein product assignments ([44] and references within), as well as a BLASTP search of all predicted CDS against the nr database and annotation with KASS [45]. RNAs were detected via MGAP with CRT, pilercr, tRNAscan, hmmsearch, BLASTN, and cmsearch ([44] and references within).
Ca. Nitrotoga comparative genomics
The Anvi’o pangenome pipeline [46] was used to cluster (mcl = 10) coding sequences from each Ca. Nitrotoga genome in order to establish a “core genome” of genes shared by all four putative species, and genes unique to each genome or shared by two or three putative species. Average nucleotide identity (ANI) and average amino acid identity (AAI) were calculated using the online enveomics tools [47].
Ca. Nitrotoga phylogenetic analyses
Near full-length Ca. Nitrotoga-like 16S rRNA gene sequences in the NCBI nt database were identified by a BLASTN search. Sequences were retained if they were ≥1300 bp in length and had ≥95% identity to any of the cultivated Ca. Nitrotoga 16S rRNA gene sequences from this study. 16S rRNA gene sequences from the BLASTN search were aligned with MAFFT [48] and manually trimmed. A maximum likelihood tree was generated using RAxML (version 8.2.9) with 100 rapid bootstraps and the GTRGAMMA model of nucleotide substitution. A single monophyletic clade was extracted from the resulting phylogenetic tree, which included all known Ca. Nitrotoga 16S rRNA gene sequences. Neighboring clades included members of different genera. Selected outgroup sequences were added to the extracted sequences and then were realigned and trimmed, and a maximum likelihood tree was generated with 1000 rapid bootstraps.
Protein sequences (≥853 amino acids in length) of Type II DMSO reductase family enzymes were compiled based on previous research [8, 49, 50] and a BLASTP search of the NCBI nr database. Sequences were aligned using MAFFT [48] and manually trimmed (alignment length was 1644 amino acid positions with gaps). A maximum likelihood tree was built using RAxML with 1000 rapid bootstraps and the LG likelihood model of amino acid substitution (based on PROTGAMMAAUTO selection). All trees were visualized and annotated in iTOL [51].
Distribution of Ca. Nitrotoga-like 16S rRNA gene sequences in the environment
Full-length 16S rRNA gene sequences from each Ca. Nitrotoga culture were submitted to the IMNGS online server [52] for searches against all 183,153 16S rRNA gene amplicon runs from the NCBI Sequence Read Archive (SRA) (March 2018 release). A minimum identity threshold of 97% (over an alignment of at least 200 bp) was chosen to represent Ca. Nitrotoga-like sequences at the genus level due to phylogenetic resolution of 16S rRNA gene sequences (see results). Samples were removed if they did not have at least 100 reads associated with Ca. Nitrotoga-like operational taxonomic units (OTUs). Data was summarized by SRA annotated environment categories. In some cases, SRA annotated categories were merged together: “aquatic”, “freshwater”, and “pond” were merged into “freshwater”; “freshwater_sediment” and “sediment” were merged into “sediment”; “biofilm” and “microbial_mat” were merged into “biofilm”; “soil”, “terrestrial”, and “peat” were combined into “soil”; and “plant”, “rhizosphere”, and “root” were merged into “plant-associated”.
Relative abundance of Ca. Nitrotoga in creek sediments and water column
Water column and sediment samples were collected from 18 sites along Bear Creek, 18 sites along Cherry Creek, and four sites along the South Platte River at the confluences of Bear Creek and Cherry Creek in Denver, Colorado, USA in Fall 2016 (Supplemental Note). DNA extracts were sent to the University of Illinois Roy J. Carver Biotechnology Center, Urbana, Illinois for total 16S rRNA amplicon sequencing using the 515F-Y and 926R primers [53, 54]. Libraries were prepared with the Fluidigm 48.48 Access Array IFC platform (Fluidigm Corporation, South San Francisco, CA) as previously described [55], and sequenced on an Illumina HiSeq with Rapid 250 bp paired-end reads (Illumina, San Diego, CA). Filtered reads were clustered into OTUs at 97% sequence identity and taxonomy was assigned with a BLASTN search against the SILVA 16S rRNA gene database (release 128) [56].
Results and discussion
Enrichment and nitrite oxidation
The four nitrite-oxidizing cultures were enriched for 17 (CP45 and SPKER) or 20 (LAW and MKT) months via serial dilution, low volume transfers, and rapid transfers. 16S rRNA gene sequence analyses and PCR with NOB specific primers revealed that each culture contained only one NOB related to the Candidatus Nitrotoga genus (Betaproteobacteria class; Nitrosomonadales order; Gallionellaceae family). Ca. Nitrotoga was 16–24% enriched at the time of genomic sequencing (based on relative abundance of contigs encoding ribosomal protein S3), and approximately 65% enriched upon further purification (based on FISH estimates for CP45) (Supplementary Figure S1). The Ca. Nitrotoga cultures and genomes were named based on sampling location: Ca. Nitrotoga sp. CP45, Ca. Nitrotoga sp. LAW, Ca. Nitrotoga sp. MKT, and Ca. Nitrotoga sp. SPKER. Nitrite oxidation rates (calculated across three measurements during logarithmic nitrite oxidation) averaged 135.6 ± 21.5 µM NO2-/day (Supplementary Figure S1), which was similar to previously reported data for Ca. Nitrotoga arctica [37].
Metagenome assembly and binning
Metagenomic sequencing of the enrichment cultures showed that each culture contained 8–32 genome bins (CP45: 8; MKT: 14; LAW: 23; SPKER: 32), likely corresponding to a similar number of unique organisms in each enrichment culture. Attempted co-assemblies of all four metagenomes were not successful as each enrichment culture had different community compositions. Each metagenome assembly contained only one predicted NOB genome related to Ca. Nitrotoga spp. (all other NOB were absent, including Nitrospira, Nitrobacter, Nitrolancea, Nitrospina, Nitrococcus, and Ca. Nitromaritima).
One genome bin from the SPKER enrichment metagenome likely belonged to a protozoan in the Neobodonida order that may prey on bacteria in culture. Most of the other genomes in the enrichment metagenomes were related to Proteobacteria including several members of the families Pseudomonadaceae, Methylophilaceae, and Comamonadaceae (based on EMIRGE-assembled 16S rRNA genes [57] and the CheckM lineage workflow). Persistence of these organisms over 17–20 months suggests that they may form a mutualistic relationship with NOB (making it difficult to isolate NOB in pure culture). For instance, heterotrophic bacteria may feed on dead cells within the culture (since no external organic matter is added to the media) and NOB may utilize metabolites produced by heterotrophs to minimize energetic costs of biosynthesis.
Ca. Nitrotoga genome assembly
The Ca. Nitrotoga genome bins from each culture had very high average coverage (201–398×). Ca. Nitrotoga genome bins were predicted to be 99.8% complete with ≤0.3% contamination by CheckM (after manual assignment of some marker genes; Supplemental Note; Supplementary Table S1). These high-quality draft sequences are the first reported Ca. Nitrotoga genomes. The closest relatives with genome sequences were Sideroxydans lithotrophicus ES-1, Gallionella capsiferriformans ES-2, and Ca. Gallionella acididurans ShG14-8.
Ca. Nitrotoga genomes ranged in size from 2.707 to 2.982 Mbp, with 23–59 contigs and GC content between 47.5 and 48.8% (Supplementary Table S1). The number of coding sequences ranged from 2526–2799 with 36–39 tRNAs encoding all twenty amino acids. ANI values between the four Ca. Nitrotoga genomes ranged from 85.9–93.4% (Table 1), while AAI values had a similar range of 87.5–94.6%. These ANI values were indicative of each enrichment containing a distinct putative Ca. Nitrotoga species, as they fell below the assumed 95% ANI threshold that separates most bacterial species [58–61]. AAI values indicated these organisms were still highly conserved and likely shared many of the same traits. The SPKER Ca. Nitrotoga genome was considerably more divergent than the CP45, MKT, or LAW genomes.
Table 1.
Average nucleotide identity (ANI) between enriched Ca. Nitrotoga genomes and close relative genomes available on NCBI (unhighlighted cells; top right) and average amino acid identity (AAI) pairwise comparisons (highlighted cells; bottom left)

|
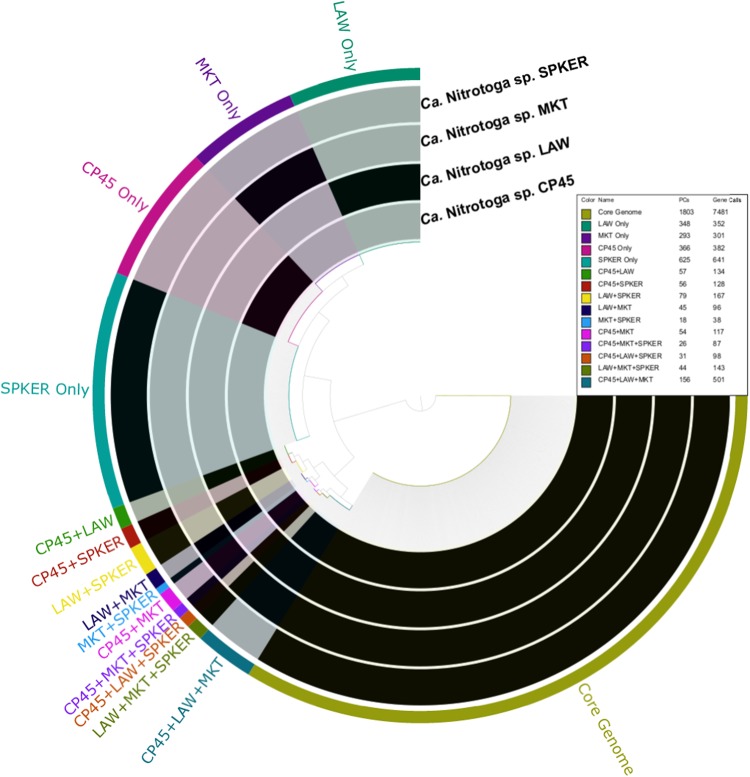
Anvi’o pangenome analysis showed that coding sequences from all four genomes (10,666 in total) grouped into 4001 protein clusters (PCs) (Fig. 1). The core genome was represented by 1803 PCs found in all four Ca. Nitrotoga genomes (45.1% of total). Each individual genome (e.g., MKT only) contained 293–625 unique PCs, while 566 PCs were shared among two or three genomes. Of the 2198 accessory genome PCs, 1041 were annotated as hypothetical proteins.
Fig. 1.
Anvi’o pangenome analysis of four Ca. Nitrotoga genomes. Coding sequences for all four genomes (10,666) in total grouped into 4001 protein clusters (PCs), based on a pairwise BLAST of all coding sequences from all Ca. Nitrotoga genomes and clustering using the MCL algorithm (mcl = 10). The core genome was represented by 1803 PCs found in all four Ca. Nitrotoga genomes. Each individual genome (e.g., MKT only) contained 293–625 unique PCs not found in any other Ca. Nitrotoga genome
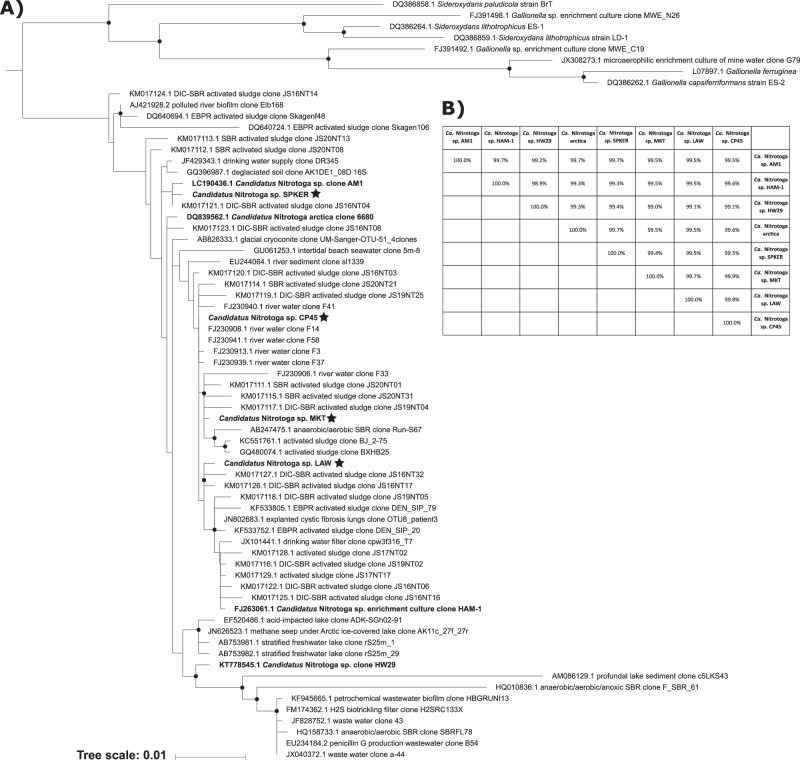
Despite genomic level differences, the Ca. Nitrotoga 16S rRNA gene sequences were highly conserved (Fig. 2; Supplementary Figure S2). 16S rRNA genes were present at double coverage in all Ca. Nitrotoga genomes, indicating a gene duplication. The 16S rRNA gene from the SPKER genome had three single nucleotide variants (SNVs) present in ~50% of mapped reads, likely indicating small differences between the duplicate copies. A consensus sequence was used here, as the variants could not be isolated with paired reads. Among the four Ca. Nitrotoga 16S rRNA gene sequences assembled in this study, the most divergent are 99.4% identical across a 1544 bp alignment (9 total mismatches). Pairwise comparisons of all 8 enriched Ca. Nitrotoga 16S rRNA gene sequences (four previous studies, four from this study) average 99.5% identical across near full-length genes (Fig. 2b) and a comparison of all 60 Ca. Nitrotoga-like sequences in Fig. 2, comprising the known Ca. Nitrotoga genus, averaged 98.7% (Supplemental Fig. 2). The highly conserved nature of Ca. Nitrotoga 16S rRNA gene sequences does not likely represent genome-level conservation across the genus, based on 16S rRNA:genome comparisons in the present study.
Fig. 2.
a Maximum likelihood phylogenetic tree of 16S rRNA gene sequences from representative Ca. Nitrotoga sequences and close relatives. Sequences were aligned across 1422 positions; phylogenetic trees were generated using RAxML (version 8.2.9) with 1000 rapid bootstraps and the GTRGAMMA model of nucleotide substitution. Nodes with bootstrap support values ≥50% are shown with a black circle. A midpoint root was used when selecting outgroup sequences. Bolded sequence names have been enriched in culture. Nodes with a star represent organisms presented in this study. b BLASTN comparisons of the near full-length 16S rRNA gene sequences from the eight known enriched Ca. Nitrotoga spp
Ca. Nitrotoga nitrogen metabolism
Nitrite oxidation
Nitrite oxidoreductase (NXR) is a heterotrimeric enzyme (NxrABC) that oxidizes nitrite to nitrate, liberating two electrons from a water molecule (Supplemental Note). NXR is a member of the Type II DMSO reductase family of molybdenum enzymes. NXR is bound to the cell membrane and can be classified into at least two distinct phylogenetic and functional groups based on orientation towards the cytoplasm or periplasm [50]. Oxidation kinetics studies have associated NXR orientation with ecological niche formation, as bacteria with cytoplasmic-facing NXR (Nitrobacter, Nitrococcus, and Nitrolancea) typically dominate in relatively high nitrite environments over bacteria with periplasmic-facing NXR (Nitrospira, Nitrospina, and Candidatus Nitromaritima) [7, 36, 37, 49, 50, 62–65]. Periplasmic-facing NXR have the energetic benefit of H+ release contributing to the proton motive force, while nitrite must be pumped into the cell for cytoplasmic-facing NXR.
Ca. Nitrotoga nxr genes were found on single contigs (6.7–9.5 kb) within each genome forming an nxrABC operon, with an additional nxrD chaperone (Supplemental Note). Based on the number of neighboring contigs in the assembly and read coverage estimates, the nxr genes in the CP45, LAW, and MKT genomes are thought to be duplicated. The SPKER nxr operon is likely present three times throughout the genome based on the same criteria.
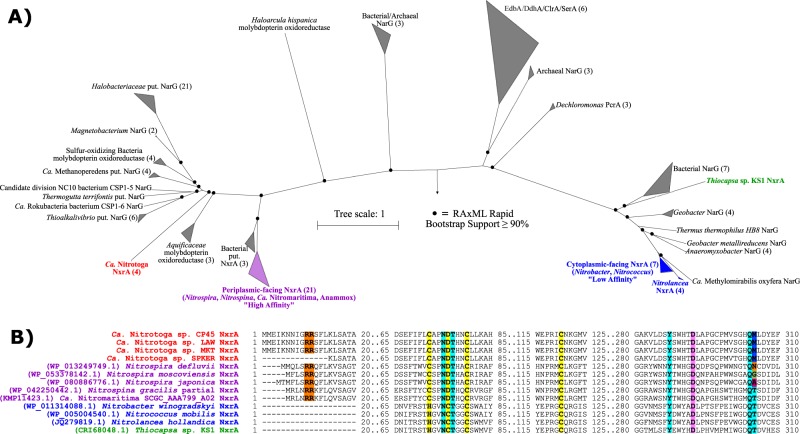
Ca. Nitrotoga NxrA protein sequences were divergent from all other members of the Type II DMSO reductase enzyme family, and the closest relatives were putative bacterial and archaeal nitrate reductase (NarG) proteins (Fig. 3a). The deeply branching Ca. Nitrotoga NxrA protein sequences may represent a fourth evolutionary development of nitrite oxidation, separate from the cytoplasmic-facing, periplasmic-facing, and phototrophic Thiocapsa nitrite oxidizers [50, 66]. Interestingly, some contigs surrounding the Ca. Nitrotoga nxr operons contained putative transposase and integrase genes, possibly suggesting that the genes were horizontally transferred similar to the theory of periplasmic-facing nxr gene development [50]. The Ca. Nitrotoga genus also represents the only known Betaproteobacterial lineage of nitrite-oxidation.
Fig. 3.
Phylogenetic and structural analysis of the alpha subunit of nitrite oxidoreductase (NxrA). a Phylogeny of 118 members of the Type II DMSO reductase protein family aligned and manually trimmed to 1644 amino acid positions. References were selected to include both cytoplasmic-facing “Low Affinity” and periplasmic-facing “High Affinity” NxrA [8], as well as other family members: PcrA, perchlorate reductase; EbdA, ethylbenzene dehydrogenase; DdhA, dimethylsulfide dehydrogenase; ClrA, chlorate reductase; SerA, selenite reductase; and NarG, nitrate reductase. Putative enzymes are marked as “put.”. The number of sequences in collapsed nodes are shown in parentheses. RAxML rapid bootstrap support values ≥90% are marked with a black circle. b Partial alignment of selected important residues in Ca. Nitrotoga, periplasmic-facing, and cytoplasmic-facing NxrA (including Thiocapsa). Highlights represent Tat signal peptides (Orange), Fe-S cluster binding residues (Yellow), molybdenum coordinating residues (Pink), and nitrite/nitrate binding residues (Light Blue)
Ca. Nitrotoga NxrA from the CP45, LAW, and MKT genomes had a predicted twin-arginine translocation (Tat) signal peptide on the N-terminus (Fig. 3b), which supports the translocation of this enzyme into the periplasm [67]. The SPKER NxrA was lacking a signal peptide, although an alignment showed that the first 14 amino acids may be missing, as the entire protein was otherwise aligned without gaps. The gene was located at the end of a contig and was missing a start codon.
Conserved residues were previously predicted to play a role in binding a molybdopterin cofactor, a [4Fe–4S] cluster, and the nitrite/nitrate substrate in NxrA and nitrate reductase NarG [50, 68]. All Ca. Nitrotoga NxrA shared the conserved residues for the molybdopterin, iron-sulfur cluster, and four of the five nitrite/nitrate binding residues. However, an analysis of the fifth residue revealed a lack of conservation among all NOB with at least five different residues predicted in known NxrA sequences, including variations within nxrA copies from a single genome (e.g., Nitrospira nitrosa has two nxrA copies, one with asparagine and one with glycine in this same position) (Fig. 3b). This residue may not play a critical role in nitrite/nitrate binding, or may be involved in the variable nitrite oxidation kinetics observed previously [36, 37].
Ca. Nitrotoga NxrB were lacking a signal peptide, but may be translocated into the periplasm via a “hitch-hiker” method as described previously [50, 68]. All NxrB had coordinating residues for three [4Fe–4S] and one [3Fe–4S] cluster for electron conductance (Supplemental Fig. 3).
One Ca. Nitrotoga NxrC predicted protein sequence was found in each genome with an N-terminal signal peptide for translocation. Signal peptides were previously observed in Nitrospina gracilis and Candidatus Nitromaritima NxrC [8, 49], however the predicted signal peptide cleavage site was located in the middle of a predicted transmembrane region. Biochemical studies confirmed the membrane-association of the NXR holoenzyme in Nitrospina and Nitrospira [69], indicating that NxrC were not fully translocated into the periplasm [50]. Ca. Nitrotoga NxrC did not have predicted transmembrane regions, and were more similarly related to the soluble periplasmic gamma subunit of ethylbenzene dehydrogenase, which likely interacts with cytochrome c proteins to shuttle electrons to the membrane [70]; similar observations were made in the analysis of Nitrospina gracilis and Candidatus Nitromaritima genomes [8, 49].
Overall, sequence and phylogenetic analyses indicated that Ca. Nitrotoga encode a form of NXR that is divergent from known NOB. Phylogenetically, the Ca. Nitrotoga NXR alpha subunit (NxrA) was deeply branched near putative bacterial and archaeal NarG, but still maintained all necessary residues for nitrite oxidation. The NxrA and NxrC subunits had signal peptides for translocation to the periplasm, but all subunits were lacking transmembrane domains for anchoring in the cytoplasmic membrane. This may suggest that Ca. Nitrotoga have a soluble NXR periplasmic holoenzyme, which has not been reported in any other NOB.
Dissimilatory and assimilatory nitrogen metabolism
All Ca. Nitrotoga genomes had genes for a NirK dissimilatory nitrite reductase for the reduction of nitrite to nitric oxide [71–73], which has been found in all other NOB genomes except Nitrolancea hollandica [7, 49]. The ultimate role of NirK in Ca. Nitrotoga is still unclear, but nitric oxide production might help manage the redox state of the cell as suggested in Nitrobacter [74]. NirK has been proposed to play a role in denitrification in Nitrospira, but function has not been confirmed [49, 50, 62]. In ammonia-oxidizing bacteria (AOB), NirK may play a role in aiding the efficient oxidation of ammonia to nitrite [75, 76]. In ammonia-oxidizing archaea (AOA), NirK seems to play an important role in ammonia oxidation, but the location and specific mechanism in the pathway is still debated [77–79]. The Ca. Nitrotoga CP45, LAW, and MKT genomes also encoded genes for a nitric oxide dioxygenase (Hmp), which is predicted to catalyze the conversion of nitric oxide to nitrate (possibly contributing to redox balance with NirK), and is evolutionarily related to an O2-binding protein similar to hemoglobin [80]. The Ca. Nitrotoga genomes encoded genes for transport of nitrite/nitrate (narK), formate/nitrite, and ammonium (amtB). The predicted periplasmic orientation of Ca. Nitrotoga NXR excludes the need for nitrite import into the cytosol, however genes for an assimilatory NirBD nitrite reductase and cytochrome c551/c552 to catalyze the reduction of nitrite to ammonia were found. Ca. Nitrotoga may also assimilate ammonia released during cyanide detoxification (Supplemental Note). A Ca. Nitrotoga enrichment culture from coastal sands grew faster when ammonium was added to the culture medium [35], likely due to the reduced need for assimilatory nitrite reduction.
Ca. Nitrotoga energy metabolism and reverse electron flow
Nitrogen energetics
A complete electron transport chain was present in the Ca. Nitrotoga genomes (Supplemental Fig. 4). After electrons are passed from NXR to cytochrome c, they are transferred to oxygen via a terminal oxidase (Complex IV). All Ca. Nitrotoga genomes contained genes for a cbb3-type cytochrome c oxidase, a member of the C-class heme-copper oxidases with an exceptionally high affinity for oxygen [81]. These genes did not form a distinct operon, however there were no other candidate terminal oxidase genes in most Ca. Nitrotoga genomes (see below). Organisms possessing cbb3-type oxidases, including the NOB Nitrospina gracilis and the phototrophic nitrite-oxidizer Thiocapsa KS1, are likely capable of growth in microoxic environments [49, 66, 82]. For instance, Nitrospina-like bacteria have been found to play a crucial role in carbon fixation in marine oxygen minimum zones, due in part to their cbb3-type terminal oxidases [11, 83, 84]. The capacity to encode a cbb3-type terminal oxidase indicates that Ca. Nitrotoga spp. may continue aerobic metabolisms at nanomolar O2 concentrations, allowing an incredibly wide habitat range (e.g., sediments, biofilms, marshes). Ca. Nitrotoga may also use alternative metabolisms (e.g., sulfur oxidation; see below) in low oxygen environments similar to Nitrococcus mobilis [11]. The Ca. Nitrotoga genomes encoded several O2 binding proteins including protoglobin, hemerythrin, and potentially nitric oxide dioxygenase, which may facilitate survival under low oxygen conditions.
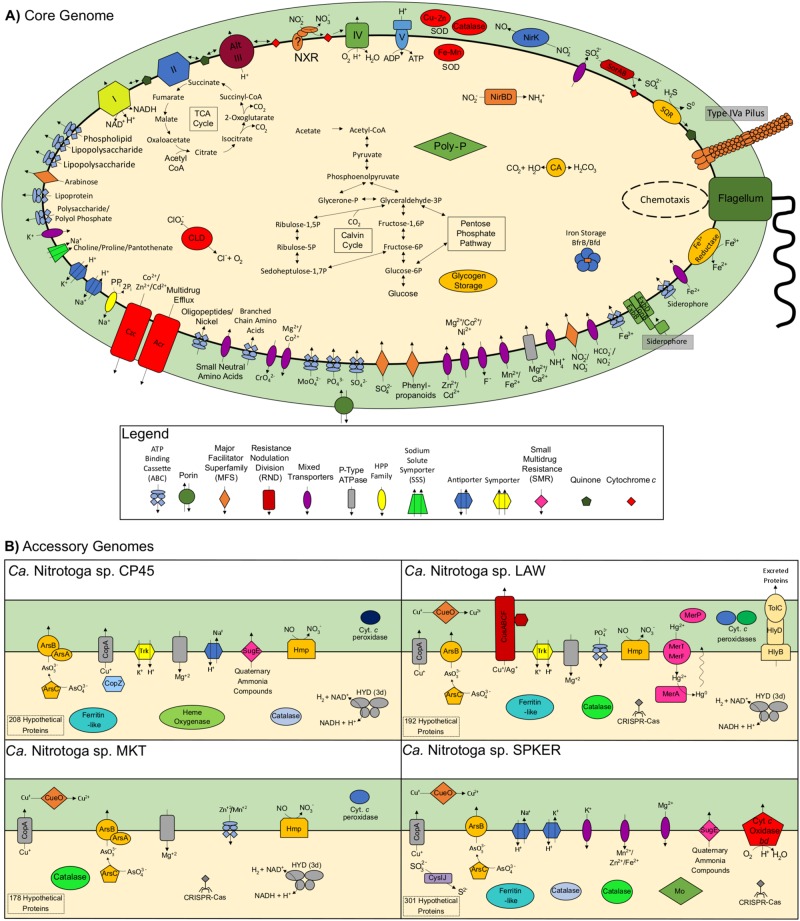
Fig. 4.
Schematic representation of Ca. Nitrotoga genome features. a The core Ca. Nitrotoga genome including predicted functions shared by all four genomes. The question mark located in the NxrC subunit symbolizes the uncertainty in whether or not the holoenzyme is anchored to the cell membrane. b Predicted function of individual genomic features found within one or more genomes, but not shared among all four. Each genome had 178–301 unique hypothetical proteins, and an additional 171 hypothetical proteins were shared among two or three genomes
Intriguingly, the SPKER genome contained genes for an additional bd-type terminal cytochrome c oxidase, which was also observed in the genomes of two close relatives: Sideroxydans lithotrophicus ES-1 and Gallionella capsiferriformans ES-2 [85]. This terminal oxidase may serve an additional role in energy conservation from organic carbon sources (see below), as bd-type terminal oxidases only receive electrons from the quinone pool, while cbb3-type terminal oxidases can receive electrons from cytochromes or the quinone pool [81, 86].
Genes for an F-type ATPase (Complex V) were present in all four Ca. Nitrotoga genomes for ATP generation via the proton motive force. The periplasmic oxidation of nitrite contributes to the proton gradient, as two protons are released in the reaction. Inorganic phosphate for ATP generation can likely be stored as polyphosphate in Ca. Nitrotoga and released by polyphosphate kinase and inorganic pyrophosphatase enzymes predicted within the genomes.
When nitrite is the only energy source, Ca. Nitrotoga must generate NADH via reverse electron flow to support normal cellular processes. Genes for a canonical cytochrome bc1 (Complex III), typically found in proteobacteria, were missing from Ca. Nitrotoga genomes. However, a suite of genes encoding an alternative complex III (actAB1B2CDEF) [87] was found in all Ca. Nitrotoga genomes and in the related iron-oxidizing bacteria of the Gallionellaceae family [85]. Electrons are distributed from the alternative complex III via quinones to succinate dehydrogenase (Complex II) for biochemical intermediate production, or to NADH dehydrogenase (Complex I) for the reduction of NAD+ (Supplemental Fig. 4).
In addition to nitrite oxidation, NXR mediated nitrate reduction with electrons derived from alternative donors has been observed in members of the Nitrobacter, Nitrospira, Nitrolancea, and Nitrococcus when oxygen is excluded [7, 11, 62, 88, 89]. Given the genomic potential for survival in low-oxygen environments, we predict that Ca. Nitrotoga are also capable of nitrate reduction via NXR when oxygen is not available. Alternative electron donors including organic carbon, reduced sulfur compounds, or hydrogen gas (see below) could contribute to the electron transport with NXR functionally replacing the terminal oxidase (Supplemental Fig. 4).
Sulfur energetics
Genomic pathways indicated that Ca. Nitrotoga may be capable of sulfur oxidation. The recently described Nitrococcus mobilis Nb-231 genome encoded genes for aerobic sulfur oxidation, and their activity was confirmed in pure culture [11]. Ca. Nitrotoga is predicted (based on genomic content) to harness the same sulfur sources using a periplasmic sulfite dehydrogenase (SOR) to oxidize sulfite (SO3-) to sulfate (SO42−) with electron donation to cytochrome c, and a sulfide:quinone oxidoreductase (SQR) that couples hydrogen sulfide (H2S) oxidation to elemental sulfur (S0) and reduction of quinone (Supplemental Fig. 4).
Hydrogen energetics
The catalytic subunit of a predicted Group 3d [NiFe]-hydrogenase was identified in the CP45, LAW, and MKT genomes with the HydDB online tool [90]. All other requisite genes were found nearby in the respective genomes. Group 3d [NiFe]-hydrogenases typically act as an NADH oxidoreductase, reducing NAD+ to NADH with concomitant oxidation of H2 to water [91]. Many sequenced NOB have a Group 3b [NiFe]-hydrogenase [5, 11, 49] that is also capable of elemental sulfur or polysulfide reduction to H2S, an advantageous enzyme that is apparently lacking in Ca. Nitrotoga genomes. Nitrospira moscoviensis utilizes a Group 2a [NiFe]-hydrogenase, and growth on H2 was confirmed in culture [12], while Nitrolancea hollandica encodes a group 1 h/5 hydrogenase [5, 7]. Hydrogen oxidation could serve as yet another energy metabolism for Ca. Nitrotoga cells.
Ca. Nitrotoga carbon metabolism
Carbon fixation
Ca. Nitrotoga genomes had genes encoding for the complete Calvin cycle (Supplemental Note), supporting CO2 fixation for autotrophic growth. A sedoheptulose-bisphosphatase gene, an important intermediate enzyme needed to regenerate ribulose-1,5-bisphosphate, was missing but is also missing in the NOB Nitrobacter winogradskyi, and the ammonia-oxidizing betaproteobacteria Nitrosomonas europaea, and Nitrosospira multiformis [65, 92, 93]. Fructose 1,6-bisphosphatase, which typically plays a role in gluconeogenesis, is thought to fill the same role in organisms lacking sedoheptulose-bisphosphatase [94, 95] and the gene was present in Ca. Nitrotoga genomes. Nitrolancea, Nitrococcus, and Nitrobacter also utilize the Calvin cycle [7, 11, 74].
Organic carbon utilization
Ca. Nitrotoga genomes encoded all genes for glycolysis, gluconeogenesis, the pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle (Fig. 4a). All necessary biochemical intermediates could be generated via these pathways, whether carbon is fixed via the Calvin cycle or imported into the cell. Ca. Nitrotoga genomes had genes for polysaccharide storage and exopolysaccharide synthesis, which may facilitate growth in biofilms (Supplemental Note).
Various sugar transporter genes are present in many NOB, including Nitrococcus mobilis, Nitrospira inopinata, Nitrospina gracilis, and Nitrolancea hollandica [5, 7, 11, 49], but none were identified in Ca. Nitrotoga genomes. However, several genes involved in the phosphotransferase system (PTS) to import and phosphorylate sugars were found. PTS genes were also found in Nitrobacter winogradskyi and Thiocapsa sp. KS1 [65, 66]. Genes encoding the EI and HPr subunits of the PTS were both found in Ca. Nitrotoga, but a complete set of substrate-specific EIIA, EIIB, and EIIC components were not identified. Only ascorbate-specific EIIA and nitrogen regulatory EIIA subunit genes were found. Ca. Nitrotoga genomes did encode genes related to a recently purified ABC transporter (AfuABC) that was found to import phosphorylated sugars instead of iron as originally reported [96].
Ca. Nitrotoga genomes had genes for an acetyl-CoA synthetase, which produces acetyl-CoA from acetate, suggesting the use of small organic carbon molecules if they can enter the cytosol. The incomplete Ca. Nitrotoga genomes, as well as otherwise complete organic carbon oxidation pathways, leaves potential for future identification of organic carbon transport. Preliminary physiology tests showed a dramatically increased rate of nitrite oxidation when acetate and dextrose were provided to the cultures (data not shown).
Ca. Nitrotoga iron acquisition
Iron is often a limiting nutrient in oligotrophic freshwater environments. Ca. Nitrotoga genomes encoded an impressive array of genes useful for iron scavenging, including a complete TonB-dependent transport system with up to four copies of the outer membrane transport energization protein complex (ExbB/ExbD/TonB) and up to 16 TonB-dependent outer membrane siderophore receptors (CP45: 10; LAW: 7; MKT: 6; SPKER: 16). All of the TonB-dependent transporters (TBDTs) fell within the CirA and/or Fiu superfamilies (based on BLASTP and conserved domain results), which have been shown to transport a wide variety of siderophores including citrate, aerobactin, enterobactin, and salmochelin [97]. Eight TBDTs (MKT: 0; CP45: 1; LAW: 1; SPKER: 5) were preceded by FecI-FecR transcription factors known to monitor ferric dicitrate concentrations within the cell and control transcription of TBDTs in E. coli [98]. Siderophore synthesis seems unlikely within Ca. Nitrotoga (based on gene predictions; Supplemental Note), however Ca. Nitrotoga may scavenge siderophores from other bacteria within the community. Additionally, genes encoding several iron pumps and iron storage proteins were present (Supplemental Note). Iron is a critical cofactor in the NXR enzyme and the varied iron acquisition methods of Ca. Nitrotoga may offer a competitive advantage for these organisms in iron-limited environments.
Ca. Nitrotoga heavy metal transport and defense
Molybdate (MoO42−) transport genes (modABC) were found in the CP45, LAW, and MKT genomes; while the SPKER genome had two modA copies but no modB or modC. However, the SPKER genome encoded a pair of molybdenum storage genes (mosAB) for intracellular storage of molybdenum [99], not found in other Ca. Nitrotoga genomes. Molybdate is the naturally occurring form of molybdenum, which is a necessary cofactor for NXR, so Ca. Nitrotoga cells must have at least one method of acquiring molybdenum.
Genes encoding heavy metal efflux systems were prevalent in all Ca. Nitrotoga genomes. A predicted apaG cobalt and magnesium efflux protein, as well as complete cobalt-zinc-cadmium resistance transporters (czcABC) were found in all genomes. A chromate transporter and divalent cation tolerance protein may help protect against heavy metal accumulation in Ca. Nitrotoga.
Arsenate (AsO43−) can enter cells through normal phosphate transport systems, and Ca. Nitrotoga had genes encoding proteins for two different mechanisms for arsenate removal (Fig. 4b). The LAW and SPKER genomes had genes to encode ArsC and ArsB responsible for reducing arsenate to arsenite (AsO33−), and pumping arsenite from the cytoplasm, respectively. The CP45 and MKT genomes had genes to encode an additional subunit, ArsA, which acts as an ATPase to provide energy for export, while the ArsB pump can work alone with energy from the proton motive force.
All Ca. Nitrotoga genomes encoded the alpha (cusA) and beta (cusB) portions of the Cus Cu(I)/Ag(I) efflux system, composing the inner membrane transporter and periplasmic membrane fusion protein, respectively. The outer membrane factor channel protein, cusC, was only found in a complete operon in the LAW genome along with the cusF metallochaperone and cusR-cusS two-component signaling system. The MKT and SPKER genomes had cusR and cusS genes preceding a TolC-like outer membrane protein that could be CusC, but these were found separate from cusAB. A second copper exporter belonging to the Cue system was found in the LAW, MKT, and SPKER genomes consisting of a Cu2+ exporter (copA) and multicopper oxidase (cueO). The CP45 genome alone had a copZ gene that serves as a chaperone for Cu+ export via CopA.
Chromate (CrO42−) can enter cells via normal sulfate uptake systems. Genes encoding a putative soluble chromate reductase were found in all Ca. Nitrotoga genomes that can reduce the Cr(VI) found in chromate to the less toxic Cr(III), producing harmful reactive oxygen species (ROS) in the process which must be dealt with by other defenses [100]. In addition, a chromate transporter gene (chrA) was found in all genomes for the export of chromate ions from the cytoplasm.
A variety of other genes for heavy metal defense were found in individual Ca. Nitrotoga genomes (Fig. 4b). For instance, the CP45 genome encoded a gold/copper resistance efflux pump belonging to the resistance nodulation division (RND) superfamily. The LAW genome encoded a mercury resistance system (merRTPFA) for Hg(II) transport and reduction to volatile Hg(0), and the LAW and SPKER genomes encoded terC-like proteins associated with tellurite resistance but appear to be lacking other required genes for tellurite efflux and/or reduction. Heavy metal transport is critical for cofactor acquisition (e.g., iron, molybdenum) and for defense against metals that are toxic at even low concentrations (e.g., mercury, arsenic). Ca. Nitrotoga genomes encoded varied metal transporters, which could help support cells in the contaminated rivers sampled in this study.
Other defense mechanisms in Ca. Nitrotoga
In support of the aerobic metabolisms of Ca. Nitrotoga, all four genomes contained necessary catalase, superoxide dismutase (Cu–Zn and Fe–Mn families), and peroxiredoxin genes to combat ROS (Fig. 4). Cytochrome c peroxidase genes were found in the CP45, LAW, and MKT genomes to intercept peroxides in the periplasm with electrons donated from cytochrome c. Additionally, genes for a rubrerythrin protein used to combat ROS specifically in anaerobic bacteria were found, a reminder of the likely microaerophilic or anaerobic ancestry of Ca. Nitrotoga within the Gallionellaceae.
A few antibiotic resistance genes were present among the Ca. Nitrotoga genomes, including a generic antibiotic biosynthesis monooxygenase, VanZ-like family proteins that may confer low-level antibiotic resistance, and a broad-spectrum multidrug efflux system (AcrAB-TolC). An erythromycin esterase homolog was found in the LAW genome, which may confer resistance to erythromycin. Preliminary physiology tests indicated that Ca. Nitrotoga cultures continued nitrite oxidation in the presence of various antibiotics (data not shown).
The SPKER genome encoded a suite of genes responsible for DNA phosphorothionation (iscS, dndBCDE) (swapping a non-bridging oxygen atom for a sulfur atom) that could offer resistance to a related restriction-modification system (dptHGF) [101] found nearby on the genome. DNA phosphorothionation was recently found to expand microbial growth range under multiple stresses due to its intrinsic antioxidant function [102]. This ability may greatly expand the environments in which Ca. Nitrotoga sp. SPKER can proliferate, adding to its already wide potential range.
Motility and chemotaxis
All Ca. Nitrotoga genomes carried genes necessary for flagellar assembly and operation, as well as signal transducing pathways to stimulate gliding or twitching motility via a type IV pilus assembly. All genomes had general chemotaxis genes, but the SPKER genome was notably missing an aerotaxis receptor (aer) capable of detecting oxygen concentrations. If Ca. Nitrotoga are indeed motile, they may migrate towards areas of greater reducing potential, and likely lose external motile structures when growing in biofilms or in culture, as the presence of flagella or pili have not been noted in previous studies [2, 13, 33–35].
Summary comparison of genomic features with other NOB
Overall, the genomic inventory for Ca. Nitrotoga suggests an expansive metabolic potential including nitrogen, sulfur, hydrogen, and organic carbon metabolism. Many of these metabolisms are also predicted in other groups of NOB, including Nitrospira, Nitrobacter, Nitrolancea, Nitrospina, Nitrococcus, and Ca. Nitromaritima (Supplementary Table S2). Additional features such as motility, quorum sensing, and siderophore production contribute to the physiology and ecology of NOB. These findings extend the overall theme that NOB are highly versatile [10] and that NOB should not only be associated with nitrite oxidation. In fact, the primary function of NOB in the environment may be a metabolism other than nitrite oxidation. Horizontal gene transfer of the key protein involved in nitrite oxidation suggests that Ca. Nitrotoga (as well as Nitrospira and Nitrospina [49, 50]) may not have originally been a nitrite oxidizer, further drawing into question what its primary metabolism is. Niche specificity of each NOB species is likely a complex scenario based on genetic repertoire, substrate availability, interactions with other organisms, enzyme kinetics, and physiological limits (e.g., temperature, pH). Multiple NOB groups can co-occur within an environment, but occupy separate niches to avoid competition (e.g., Nitrospira growing chemoorganoheterotrophically with formate and Ca. Nitrotoga growing chemolithoautotrophically with nitrite).
Environmental distribution
Ca. Nitrotoga-like sequences in Colorado rivers
Ca. Nitrotoga distribution was evaluated across 80 water column and sediment samples from three rivers in Colorado (Bear Creek, Cherry Creek, and South Platte River), based on amplification with general 16S rRNA gene primers targeting the total bacterial community (Supplemental Figure S6). Ca. Nitrotoga-like OTUs were identified in 85% of the samples, and Nitrospira-like OTUs were identified in 98% of the samples. Both groups co-occurred in 85% of the samples. No other NOB sequences were identified. Within each individual sample, the summed relative abundance ranged from 0 to 4.5% of the total bacterial community for Ca. Nitrotoga-like OTUs and 0–6.4% for Nitrospira-like OTUs. The summed relative abundance of Ca. Nitrotoga-like OTUs was greater than that of Nitrospira-like OTUs in 21% of the samples. Nitrobacter and Nitrospira are typically the more commonly studied NOB in freshwater systems [10, 103], but future efforts should now also consider Ca. Nitrotoga as potential important players in freshwater nitrite oxidation.
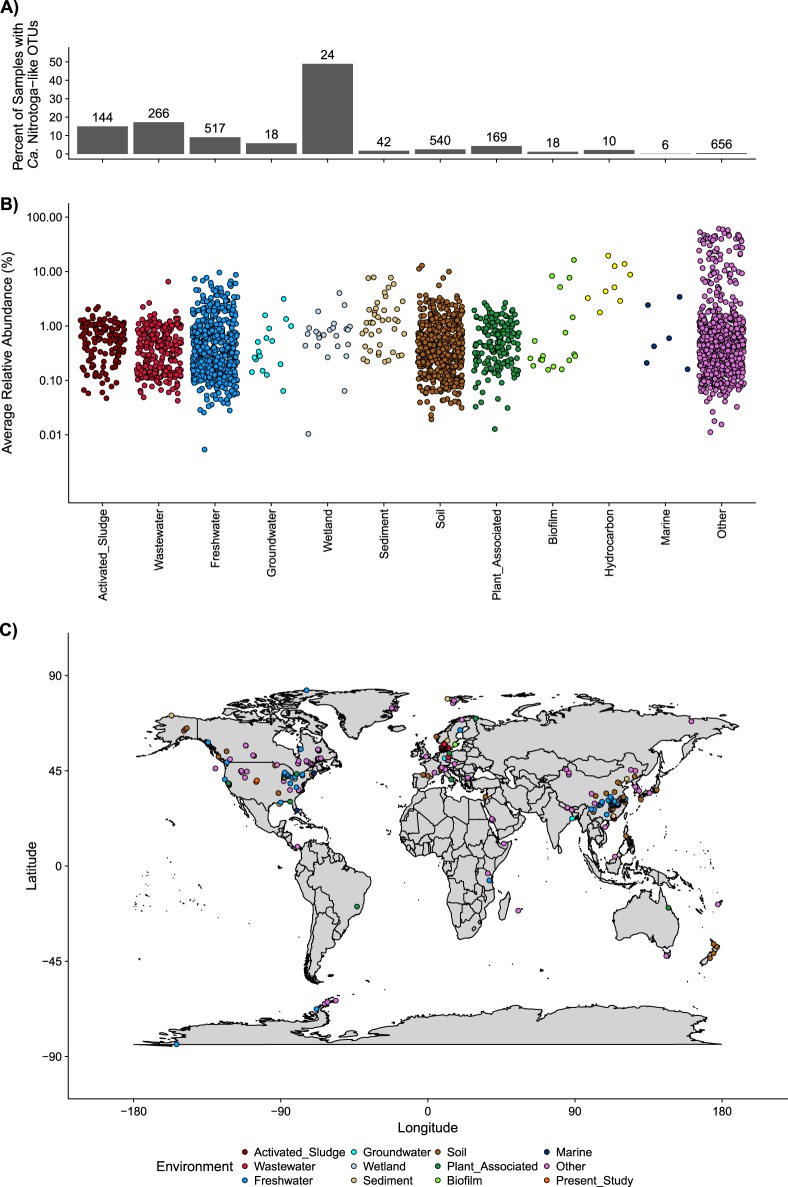
Global distribution of Ca. Nitrotoga-like 16S rRNA sequences
Ca. Nitrotoga-like 16S rRNA sequences were present in 2410 different publically available SRA samples deposited in GenBank (from 183,154 total SRA runs analyzed with IMNGS) (Fig. 5). Ca. Nitrotoga containing samples comprised 70 different user-defined environments with a global distribution across seven continents spanning from the tropics to the poles (Fig. 5c). About 9% of all freshwater environments (517/5678) had Ca. Nitrotoga-like OTUs, with some communities consisting of nearly 10% relative abundance of Ca. Nitrotoga-like OTUs. Nearly half of all wetland samples (24/49) had Ca. Nitrotoga-like OTUs with an average relative abundance of 0.89%, however most of these samples were part of the same BioProject and sampling location. A high proportion of activated sludge (15%) and wastewater (17%) samples also harbored relevant populations of Ca. Nitrotoga-like OTUs (0.55% average relative abundance), similar to previous reports [13]. The relative abundance of Ca. Nitrotoga-like OTUs ranged from 0.02–13.03% across 540 soil samples. Of the few (6/6233) marine samples that had Ca. Nitrotoga-like OTUs, most were found near freshwater rivers or wetlands.
Fig. 5.
Global distribution of Ca. Nitrotoga-like sequences. OTUs from 16S rRNA gene amplicon studies deposited as SRA runs were clustered by IMNGS. All runs with OTUs ≥97% identity to cultured Ca. Nitrotoga 16S rRNA gene sequences (from this study), an alignment of at least 200 bp, and at least 100 total reads were kept. a The percent of SRA runs with Ca. Nitrotoga-like OTUs within respective environments are shown with the number of SRA runs with Ca. Nitrotoga-like OTUs displayed above each bar. b The relative abundance of all Ca. Nitrotoga-like OTUs was averaged across all four queried Ca. Nitrotoga 16S rRNA gene sequences and plotted by environment. c Global distribution of SRA runs with Ca. Nitrotoga-like OTUs. 484 of the 2410 SRA runs with Nitrotoga-like OTUs did not have geographic information available, including all of the “Hydrocarbon” environmental samples. Orange points represent sampling locations for the enrichment cultures presented in this study
A few samples in the SRA dataset had very high relative abundance of Ca. Nitrotoga-like OTUs compared to other samples. Several of these samples were associated with enrichment cultures from their respective environments (e.g., biofilm, hydrocarbon, and other). For example, a series of samples from a single study which involved enrichment from waterworks sand filters in the Netherlands had Ca. Nitrotoga-like OTUs comprising as much as 61% of the microbial community.
These findings further corroborate the notion that the major NOB groups are unequally distributed in the environment (reviewed in the ref. [10]). For instance, Ca. Nitrotoga-like sequences are rarely found in marine systems while sequences from other NOB groups seem to only occur in marine settings (including Ca. Nitromaritima, Nitrospina, and Nitrospira lineage IV). Of the seven major NOB groups, only three are typically found in freshwater settings (Ca. Nitrotoga, Nitrospira, and Nitrobacter). NOB found in engineered settings include Ca. Nitrotoga, Nitrospira, Nitrobacter, Nitrolancea, and Nitrococcus ([10] and references within). Nitrospira are the only known thermophilic NOB; whereas, Ca. Nitrotoga, Nitrospina, and Nitrospira have been detected in polar settings [104–106]. Overall, Ca. Nitrotoga, Nitrobacter, and Nitrospira lineage II seem to have the broadest environmental distribution among the major NOB groups spanning five of the six habitat types defined by Daims et al. [10]. When all of the individual Nitrospira lineages are evaluated together, they represent the most diverse and broadly distributed NOB genus (present in all six habitat types) [10].
Temperature range of habitats containing Ca. Nitrotoga-like sequences
All previously reported Ca. Nitrotoga cultures were enriched at low temperatures (4–17 °C) [2, 33–35]. Sampling for the Ca. Nitrotoga enrichment cultures presented here occurred in winter and spring when in situ river water temperatures are typically <15 °C, and cultures were incubated at room temperature (~25 °C). Ca. Nitrotoga were originally shown to outcompete Nitrospira and Nitrobacter spp. at lower temperatures [33] and have appeared in colder WWTPs [13]. However, Ca. Nitrotoga were also shown to outcompete Nitrospira at 23–25 °C and higher nitrite loads [21], suggesting additional competitive advantages besides tolerance of low temperatures in some settings.
In our global SRA database analysis, a small number of samples containing Ca. Nitrotoga-like sequences (101 BioSamples from 17 unique BioProjects) reported temperature metadata, which ranged from 0 to 33 °C across freshwater, marine, sediment, soil, and “other” environmental categories (Supplemental Figure S5). The minimum reported temperature (0 °C) was from permafrost samples in Alaska [107] and the maximum reported temperature (33 °C) was from a bioreactor sample in China (SAMN02438470). There were no trends observed between temperature and relative abundance of Ca. Nitrotoga-like OTUs (Supplemental Figure S5). Other samples in this analysis (without reported temperature data) were found at latitudes ranging from near the equator to the north and south poles, which may reflect a similar temperature range (though many other factors would need to be considered including depth, elevation, slope, etc.). Overall, this small dataset suggests that Ca. Nitrotoga may not be restricted to cold temperature habitats. Individual phylotypes may show distinct temperature ranges and temperature optima. Physiology experiments will be necessary to confirm nitrite oxidation by Ca. Nitrotoga over the reported range of temperatures.
Interactions with other organisms
Though not directly observed here, Ca. Nitrotoga cells likely interact with other organisms in the environment similar to patterns observed with other NOB ([10] and references within). Ca. Nitrotoga have been documented to grow in flocs and were shown to co-aggregate with AOB in WWTPs [2, 13]. Ca. Nitrotoga likely forms a mutualistic symbiosis with ammonia oxidizers similar to other NOB, where the NOB consume nitrite produced by ammonia oxidizers, which benefit from nitrite detoxification by NOB. Nitric oxide produced by Ca. Nitrotoga via NirK (see above) might facilitate biofilm formation with ammonia oxidizers, which can be triggered by nitric oxide [108]. Extracellular polymeric substances (EPS) production by Ca. Nitrotoga (observed via microscopy in [35] and in genomic predictions presented here) and ammonia oxidizers [109] likely supports nitrification aggregation formation. The Ca. Nitrotoga genomes do not encode urease or cyanase genes, and thus are not likely to participate in “reciprocal feeding” reported in other NOB where urea/cyanate are broken down into ammonium, which is oxidized by ammonia oxidizers to produce nitrite for the NOB (Palatinszky et al. [110, 62]). However, ammonia produced during nitrile degradation by Ca. Nitrotoga (Supplemental Note) may result in similar “reciprocal feeding” interactions. Organic carbon degradation by Ca. Nitrotoga would produce carbon dioxide for uptake by autotrophic ammonia oxidizers. The presence of genes for a single acyl homoserine lactone synthase (LuxI homolog) in each genome, as well as several LuxR family transcriptional receptors and multiple homoserine efflux transporters (RhtA), indicate that Ca. Nitrotoga may use quorum sensing to communicate. Other NOB have been documented to use quorum sensing at high cell densities and the Ca. Nitrotoga LuxI homolog grouped phylogenetically with other sequenced Betaproteobacteria [111, 112]. Ca. Nitrotoga may use quorum sensing at high cell densities in nitrification aggregates enabling communication among Ca. Nitrotoga cells or possibly with other nitrifiers.
Based on genomic predictions, there are several interactions that might occur between Ca. Nitrotoga cells and heterotrophs. Organic compounds produced by Ca. Nitrotoga cells growing autotrophically could be consumed by heterotrophic microorganisms. When growing heterotrophically, Ca. Nitrotoga might compete with heterotrophs for organic carbon compounds under oxic or anoxic conditions (with oxygen or nitrate as the terminal electron acceptor). At the boundary of oxic/anoxic layers, aerobic nitrite oxidation might be supported by anaerobic nitrate reduction with organic carbon by Ca. Nitrotoga, other NOB, or other anaerobic heterotrophs.
Conclusions
The enrichment of four putative Ca. Nitrotoga species, characterization of the first reported Ca. Nitrotoga genomes, and analysis of the distribution of Ca. Nitrotoga-like 16S rRNA sequences has expanded our knowledge of NOB ecology. The divergent NXR enzyme may indicate a novel evolution of nitrite oxidation in Ca. Nitrotoga separate from other known NOB. Ca. Nitrotoga are predicted to have a diverse metabolic profile, likely allowing proliferation under variable nutrient and oxygen conditions. The putative Ca. Nitrotoga species presented here were enriched from the water column and sediments of urban- and agriculturally-impacted rivers, suggesting a broad habitat range within freshwater systems and a greater role in freshwater nitrification than previously expected. The prevalence of Ca. Nitrotoga-like sequences found in globally distributed habitats (e.g., freshwater, soil, wetland, and wastewater) over a range of reported temperatures (0–33 °C) suggests that Ca. Nitrotoga likely play a previously underappreciated functional role on a global scale.
Future work should determine Ca. Nitrotoga ecophysiology, including comparative nitrite oxidation kinetics and optimal growth conditions of each putative species. Determining the functional redundancy and niche differentiation of Ca. Nitrotoga compared to other NOB will be important for understanding their role in nitrite oxidation under varying environmental conditions. Understanding the sensitivity or resilience of Ca. Nitrotoga to disturbances will be important for predicting their response to environmental change.
Data availability
The datasets generated during the current study are available in the NCBI GenBank repository under BioProject PRJNA437159 and PRJNA464356.
Electronic supplementary material
Acknowledgements
We thank Adrienne Narrowe and Christopher Miller for guidance on bioinformatic data analyses. We would also like to thank Hannah Clark, Bhargavi Ramanathan, Nicklaus Deevers, Colin Beacom, Michael Kain, and Munira Lantz for their assistance in cultivation and preliminary physiology experiments and Sladjana Subotic and Anna Scopp for their help with environmental distribution sample collection and processing. Portions of this manuscript were previously published as a part of University of Colorado Denver Master’s thesis submission (AB, 2017). Funding was provided by the University of Colorado, Denver and the City and County of Denver.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0240-8) contains supplementary material, which is available to authorized users.
References
- 1.Lebedeva EV, Alawi M, Jozsa PG, Daims H, Spieck E. Physiological and phylogenetic characterization of a novel lithoautotrophic nitrite-oxidizing bacterium, ‘Candidatus Nitrospira bockiana’. Int J Syst Evol Microbiol. 2008;58:242–50. doi: 10.1099/ijs.0.65379-0. [DOI] [PubMed] [Google Scholar]
- 2.Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E. Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J. 2007;1:256–64. doi: 10.1038/ismej.2007.34. [DOI] [PubMed] [Google Scholar]
- 3.Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U. Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium. Arch Microbiol. 1986;144:1–7. [Google Scholar]
- 4.Watson SW, Waterbury JB. Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Mikrobiol. 1971;77:203–30. [Google Scholar]
- 5.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–9. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorokin DY, Lücker S, Vejmelkova D, Kostrikina Na, Kleerebezem R, Rijpstra WIC, et al. Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012;6:2245–56. doi: 10.1038/ismej.2012.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngugi DK, Blom J, Stepanauskas R, Stingl U. Diversification and niche adaptations of Nitrospina-like bacteria in the polyextreme interfaces of Red Sea brines. ISME J. 2015;10:1383–99. doi: 10.1038/ismej.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinto AJ, Marcus DN, Ijaz Z, Bautista-de los Santos QM, Dick GJ, Raskin L. Metagenomic evidence for the presence of comammox nitrospira-like bacteria in a drinking water system. mSphere. 2015;1:e00054–15. doi: 10.1128/mSphere.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Füssel J, Lücker S, Yilmaz P, Nowka B, van Kessel MAHJ, Bourceau P, et al. Adaptability as the key to success for the ubiquitous marine nitrite oxidizer Nitrococcus. Sci Adv. 2017;3:e1700807. doi: 10.1126/sciadv.1700807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch H, Galushko A, Albertsen M, Schintlmeister A, Spieck E, Richter A, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Res Rep. 2014;1696:761–3. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 13.Lücker S, Schwarz J, Gruber-Dorninger C, Spieck E, Wagner M, Daims H. Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J. 2015;9:708–20. doi: 10.1038/ismej.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pradhan S, Srinivas TNR, Pindi PK, Kishore HH, Begum Z, Singh PK, et al. Bacterial biodiversity from Roopkund Glacier, Himalayan mountain ranges, India. Extremophiles. 2010;14:377–95. doi: 10.1007/s00792-010-0318-3. [DOI] [PubMed] [Google Scholar]
- 15.Sattin SR, Cleveland CC, Hood E, Reed SC, King AJ, Schmidt SK, et al. Functional shifts in unvegetated, perhumid, recently-deglaciated soils do not correlate with shifts in soil bacterial community composition. J Microbiol. 2009;47:673–81. doi: 10.1007/s12275-009-0194-7. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt SK, Nemergut DR, Miller AE, Freeman KR, King AJ, Seimon A. Microbial activity and diversity during extreme freeze-thaw cycles in periglacial soils, 5400 m elevation, Cordillera Vilcanota, Peru. Extremophiles. 2009;13:807–16. doi: 10.1007/s00792-009-0268-9. [DOI] [PubMed] [Google Scholar]
- 17.Srinivas TNR, Singh SM, Pradhan S, Pratibha MS, Kishore KH, Singh AK, et al. Comparison of bacterial diversity in proglacial soil from Kafni Glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles. 2011;15:673–90. doi: 10.1007/s00792-011-0398-8. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Wu L, Boden R, Hillebrand A, Kumaresan D, Moussard H, et al. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J. 2009;3:1093–104. doi: 10.1038/ismej.2009.57. [DOI] [PubMed] [Google Scholar]
- 19.Roden EE, McBeth JM, Blöthe M, Percak-Dennett EM, Fleming EJ, Holyoke RR, et al. The microbial ferrous wheel in a neutral pH groundwater seep. Front Microbiol. 2012;3:1–18. doi: 10.3389/fmicb.2012.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White CP, DeBry RW, Lytle DA. Microbial survey of a full-scale, biologically active filter for treatment of drinking water. Appl Environ Microbiol. 2012;78:6390–4. doi: 10.1128/AEM.00308-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinnunen M, Gulay A, Albrechtsen HJ, Dechesne A, Smets BF. Nitrotoga is selected over Nitrospira in newly assembled biofilm communities from a tap water source community at increased nitrite loading. Environ Microbiol. 2017;0:1–41. doi: 10.1111/1462-2920.13792. [DOI] [PubMed] [Google Scholar]
- 22.Christner BC, Priscu JC, Achberger AM, Barbante C, Carter SP, Christianson K, et al. A microbial ecosystem beneath the West Antarctic ice sheet. Nature. 2014;512:310–3. doi: 10.1038/nature13667. [DOI] [PubMed] [Google Scholar]
- 23.Na H, Kim OS, Yoon SH, Kim Y, Chun J. Comparative approach to capture bacterial diversity of coastal waters. J Microbiol. 2011;49:729–40. doi: 10.1007/s12275-011-1205-z. [DOI] [PubMed] [Google Scholar]
- 24.Martiny JBH, Eisen JA, Penn K, Allison SD, Horner-Devine MC. Drivers of bacterial Beta-diversity depend on spatial scale. Proc Natl Acad Sci. 2011;108:7850–4. doi: 10.1073/pnas.1016308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan L, Song C, Meng S, Qiu L, Zheng Y, Wu W, et al. Spatial distribution of planktonic bacterial and archaeal communities in the upper section of the tidal reach in Yangtze River. Sci Rep. 2016;6:39147. doi: 10.1038/srep39147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Qi R, Yang M, Zhang Y, Yu T. Bacterial community characteristics under long-term antibiotic selection pressures. Water Res. 2011;45:6063–73. doi: 10.1016/j.watres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Chen S, Li H. (2016). Dewatering sewage sludge by a combination of hydrogen peroxide, jute fiber wastes and cationic polyacrylamide. Int Biodeterior Biodegrad. 2016;128:78–84.
- 28.Ziegler AS, McIlroy SJ, Larsen P, Albertsen M, Hansen AA, Heinen N, et al. Dynamics of the fouling layer microbial community in a membrane bioreactor. PLoS One. 2016;11:1–14. doi: 10.1371/journal.pone.0158811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figdore BA, Stensel HD, Winkler MKH. Comparison of different aerobic granular sludge types for activated sludge nitrification bioaugmentation potential. Bioresour Technol. 2017;251:189–96. doi: 10.1016/j.biortech.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Karkman A, Mattila K, Tamminen M, Virta M. Cold temperature decreases bacterial species richness in nitrogen-removing bioreactors treating inorganic mine waters. Biotechnol Bioeng. 2011;108:2876–83. doi: 10.1002/bit.23267. [DOI] [PubMed] [Google Scholar]
- 31.Bereschenko LA, Stams AJM, Euverink GJW, Van Loosdrecht MCM. Biofilm formation on reverse osmosis membranes is initiated and dominated by Sphingomonas spp. Appl Environ Microbiol. 2010;76:2623–32. doi: 10.1128/AEM.01998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders AM, Albertsen M, Vollesen J, Nielsen PH. The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 2015;10:11–20. doi: 10.1038/ismej.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alawi M, Off S, Kaya M, Spieck E. Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep. 2009;1:184–90. doi: 10.1111/j.1758-2229.2009.00029.x. [DOI] [PubMed] [Google Scholar]
- 34.Hüpeden J, Wegen S, Off S, Lücker S, Bedarf Y, Daims H, et al. Relative abundance of nitrotoga spp. in a biofilter of a cold freshwater aquaculture plant appears to be stimulated by a slightly acidic pH-value. Appl Environ Microbiol. 2016;82:1838–45. doi: 10.1128/AEM.03163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishii K, Fujitani H, Soh K, Nakagawa T, Takahashi R, Tsuneda S. Enrichment and physiological characterization of a cold-adapted nitrite oxidizer nitrotoga sp. from Eelgrass sediments. Appl Environ Microbiol. 2017;83:1–14. doi: 10.1128/AEM.00549-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–72. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowka B, Daims H, Spieck E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol. 2015;81:745–53. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griess-Romijn van Eck E (1966). Physiological and chemical tests for drinking water. NEN 1056 IV-2 Nederlands Normalisatie Instituut: Rijswijk, The Netherlands 10.1038/ismej.2012.70.
- 39.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2014;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 40.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new Genome Assembly Algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed]
- 44.Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Palaniappan K, et al. The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v 4) Stand Genom Sci. 2015;10:1–6. doi: 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eren AM, Esen OumlC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-R LM, Konstantinidis KT. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ. 10.7287/peerj.preprints.1900v1
- 48.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lücker S, Nowka B, Rattei T, Spieck E, Daims H. The genome of nitrospina gracilis Illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol. 2013;4:1–19. doi: 10.3389/fmicb.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, et al. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:1–9. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parada AE, Needham DM, Fuhrman JA. (2015). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 10.1111/1462-2920.13023. [DOI] [PubMed]
- 54.Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinform. 10.1128/JVI.02271-09. [DOI] [PMC free article] [PubMed]
- 55.Ramanathan B, Boddicker AM, Roane TM, Mosier AC. Nitrifier gene abundance and diversity in sediments impacted by acid mine drainage. Front Microbiol. 2017;8:1–16. doi: 10.3389/fmicb.2017.02136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF, Pace N, et al. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 2011;12:R44. doi: 10.1186/gb-2011-12-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caro-Quintero A, Konstantinidis KT. Bacterial species may exist, metagenomics reveal. Environ Microbiol. 2012;14:347–55. doi: 10.1111/j.1462-2920.2011.02668.x. [DOI] [PubMed] [Google Scholar]
- 59.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S (2017) High-throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. bioRxiv 225342 10.1101/225342. [DOI] [PMC free article] [PubMed]
- 60.Konstantinidis KT, Rosselló-móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-R LM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe Mag. 2014;9:111–8. [Google Scholar]
- 62.Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci. 2015;112:11371–6. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spieck E, Aamand J, Bartosch S, Bock E. Immunocytochemical detection and location of the membrane-bound nitrite oxidoreductase in cells of Nitrobacter and Nitrospira. FEMS Microbiol Lett. 1996;139:71–6. [Google Scholar]
- 64.Spieck E, Ehrich S, Aamand J, Bock E. Isolation and immunocytochemical location of the nitrite-oxidizing system in Nitrospira moscoviensis. Arch Microbiol. 1998;169:225–30. doi: 10.1007/s002030050565. [DOI] [PubMed] [Google Scholar]
- 65.Starkenburg SR, Chain PSG, Sayavedra-Soto LA, Hauser L, Land ML, Larimer FW, et al. Genome sequence of the chemolithoautotrophic nitrite-oxidizing bacterium Nitrobacter winogradskyi Nb-255. Appl Environ Microbiol. 2006;72:2050–63. doi: 10.1128/AEM.72.3.2050-2063.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hemp J, Lücker S, Schott J, Pace LA, Johnson JE, Schink B, et al. Genomics of a phototrophic nitrite oxidizer: insights into the evolution of photosynthesis and nitrification. ISME J. 2016;10:1–10. doi: 10.1038/ismej.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sargent F. The twin-arginine transport system: moving folded proteins across membranes. Biochem Soc Trans. 2007;35:835–47. doi: 10.1042/BST0350835. [DOI] [PubMed] [Google Scholar]
- 68.Martinez-espinosa RM, Dridge EJ, Bonete MJ, Butt JN, Butler CS, Sargent F, et al. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol Lett. 2007;276:129–39. doi: 10.1111/j.1574-6968.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 69.Bartosch S, Wolgast I, Spieck E, Bock E. Identification of nitrite-oxidizing bacteria with monoclonal antibodies recognizing the nitrite oxidoreductase. Appl Environ Microbiol. 1999;65:4126–33. doi: 10.1128/aem.65.9.4126-4133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kloer DP, Hagel C, Heider J, Schulz GE. Crystal structure of ethylbenzene dehydrogenase from aromatoleum aromaticum. Structure. 2006;14:1377–88. doi: 10.1016/j.str.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 71.Wrage N, Velthof GL, Van Beusichem ML, Oenema O. Role of nitrifier denitrification in the production of nitrous oxide. Soil Biol Biochem. 2001;33:1723–32. [Google Scholar]
- 72.Kozlowski JA, Kits KD, Stein LY. Comparison of nitrogen oxide metabolism among diverse ammonia-oxidizing bacteria. Front Microbiol. 2016;7:1090. doi: 10.3389/fmicb.2016.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stein LY. Heterotrophic nitrification and nitrifier denitrification. In: Ward B, Arp D, Klotz M, editors. Nitrification. Washington, DC: ASM Press; 2011. pp. 95–114. [Google Scholar]
- 74.Starkenburg SR, Larimer FW, Stein LY, Klotz MG, Chain PSG, Sayavedra-Soto LA, et al. Complete genome sequence of Nitrobacter hamburgensis X14 and comparative genomic analysis of species within the genus Nitrobacter. Appl Environ Microbiol. 2008;74:2852–63. doi: 10.1128/AEM.02311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zorz JK, Kozlowski JA, Stein LY, Strous M, Kleiner M. Comparative proteomics of three species of ammonia-oxidizing bacteria. Front Microbiol. 2018;9:139. doi: 10.3389/fmicb.2018.00938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kozlowski JA, Price J, Stein LY. Revision of N2O-producing pathways in the ammonia-oxidizing bacterium Nitrosomonas europaea ATCC 19718. Appl Environ Microbiol. 2014;80:4930–5. doi: 10.1128/AEM.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carini, P, Dupont, CL, Santoro, AE (2018). Patterns of thaumarchaeal gene expression in culture and diverse marine environments. Environ Microbiol. 10.1111/1462-2920.14107 [DOI] [PubMed]
- 78.Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J. 2016;10:1836–45. doi: 10.1038/ismej.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qin W, Amin SA, Lundeen RA, Heal KR, Martens-Habbena W, Turkarslan S, et al. Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 2018;12:508–19. doi: 10.1038/ismej.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci USA. 1998;95:10378–83. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morris RL, Schmidt TM. Shallow breathing: bacterial life at low O2. Nat Rev Microbiol. 2013;11:205–12. doi: 10.1038/nrmicro2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han H, Hemp J, Pace LA, Ouyang H, Ganesan K, Roh JH, et al. Adaptation of aerobic respiration to low O2 environments. Proc Natl Acad Sci. 2011;108:14109–14. doi: 10.1073/pnas.1018958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, Günter M, et al. Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 2012;6:1200–9. doi: 10.1038/ismej.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pachiadaki MG, Sintes E, Bergauer K, Brown JM, Record NR, Swan BK, et al. Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science. 2017;358:1046–51. doi: 10.1126/science.aan8260. [DOI] [PubMed] [Google Scholar]
- 85.Emerson D, Field EK, Chertkov O, Davenport KW, Goodwin L, Munk C, et al. Comparative genomics of freshwater Fe-oxidizing bacteria: implications for physiology, ecology, and systematics. Front Microbiol. 2013;4:1–17. doi: 10.3389/fmicb.2013.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI (2011). The cyotchrome bd respiratory oxygen reductases. Biochim Biophys Acta. 2011;1807:1398–413. [DOI] [PMC free article] [PubMed]
- 87.Refojo PN, Teixeira M, Pereira MM. The alternative complex III: properties and possible mechanisms for electron transfer and energy conservation. BBA. 2012;1817:1852–9. doi: 10.1016/j.bbabio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 88.Bock E, Koops HP, Möller UC, Rudert M. A new facultatively nitrite oxidizing bacterium, Nitrobacter vulgaris sp. nov. Arch Microbiol. 1990;153:105–10. [Google Scholar]
- 89.Sundermeyer-Klinger H, Meyer W, Warninghoff B, Bock E. Membrane-bound nitrite oxidoreductase of Nitrobacter: evidence for a nitrate reductase system. Arch Microbiol. 1984;140:153–8. [Google Scholar]
- 90.Søndergaard D, Pedersen CNS, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:1–8. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, et al. [FeFe]- and [NiFe]-hydrogenase diversity, mechanism, and maturation. Biochim Biophys Acta. 2015;1853:1350–69. doi: 10.1016/j.bbamcr.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 92.Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol. 2003;185:2759–73. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PSG, et al. Complete genome sequence of nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microbiol. 2008;74:3559–72. doi: 10.1128/AEM.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei X, Sayavedra-Soto LA, Arp DJ. The transcription of the cbb operon in Nitrosomonas europaea. Microbiology. 2004;150:1869–79. doi: 10.1099/mic.0.26785-0. [DOI] [PubMed] [Google Scholar]
- 95.Yoo JG, Bowien B. Analysis of the cbbF genes from Alcaligenes eutrophus that encode fructose-1,6-/sedoheptulose-1,7-bisphosphatase. Curr Microbiol. 1995;31:55–61. doi: 10.1007/BF00294635. [DOI] [PubMed] [Google Scholar]
- 96.Sit B, Crowley SM, Bhullar K, Lai CCL, Tang C, Hooda Y, et al. Active transport of phosphorylated carbohydrates promotes intestinal colonization and transmission of a bacterial pathogen. PLoS Pathog. 2015;11:1–22. doi: 10.1371/journal.ppat.1005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Porcheron G, Garénaux A, Proulx J, Sabri M, Dozois CM. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations between strains, site of infection and the relative importance of the different metal transport systems for virulence. Front Cell Infect Microbiol. 2013;3:1–24. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Braun V, Mahren S, Ogierman M. Regulation of the Fecl-type ECF sigma factor by transmembrane signalling. Curr Opin Microbiol. 2003;6:173–80. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 99.Fenske D, Gnida M, Schneider K, Meyer-Klaucke W, Schemberg J, Henschel V, et al. A new type of metalloprotein: the Mo storage protein from Azotobacter vinelandii contains a polynuclear molybdenum-oxide cluster. ChemBioChem. 2005;6:405–13. doi: 10.1002/cbic.200400263. [DOI] [PubMed] [Google Scholar]
- 100.Thatoi H, Das S, Mishra J, Rath BP, Das N. Bacterial chromate reducatase, a potential enzyme for bioremediation of hexavalent chromium: a review. J Environ Manage. 2014;146:383–99. doi: 10.1016/j.jenvman.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 101.Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–41. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang Y, Xu G, Liang J, He Y, Xiong L, Li H, et al. DNA backbone sulfur-modification expands microbial growth range under multiple stresses by its anti-oxidation function. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-02445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cai M, Ng SK, Lim CK, Lu H, Jia Y, Lee PKH. Physiological and metagenomic characterizations of the synergistic relationships between ammonia- and nitrite-oxidizing bacteria in freshwater nitrification. Front Microbiol. 2018;9:1–13. doi: 10.3389/fmicb.2018.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ, et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012;6:1883–900. doi: 10.1038/ismej.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geyer KM, Altrichter AE, Takacs-Vesbach CD, Van Horn DJ, Gooseff MN, Barrett JE. Bacterial community composition of divergent soil habitats in a polar desert. FEMS Microbiol Ecol. 2014;89:490–4. doi: 10.1111/1574-6941.12306. [DOI] [PubMed] [Google Scholar]
- 106.Rani S, Koh HW, Rhee SK, Fujitani H, Park SJ. Detection and diversity of the nitrite oxidoreductase alpha subunit (nxrA) gene of nitrospina in marine sediments. Microb Ecol. 2017;73:111–22. doi: 10.1007/s00248-016-0897-3. [DOI] [PubMed] [Google Scholar]
- 107.Johnston ER, Rodriguez-R LM, Luo C, Yuan MM, Wu L, He Z, et al. Metagenomics reveals pervasive bacterial populations and reduced community diversity across the Alaska tundra ecosystem. Front Microbiol. 2016;7:1–16. doi: 10.3389/fmicb.2016.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmidt I, et al. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J Bacteriol. 2004;186:2781–8. doi: 10.1128/JB.186.9.2781-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arp D, Bottomley PJ. Nitrifiers: more than 100 years from isolation to genome sequences. Microbe. 2006;1:229–34. [Google Scholar]
- 110.Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M, et al. (2015). Cyanate as an energy source for nitrifiers. Nature 524: 105–108. [DOI] [PMC free article] [PubMed]
- 111.Mellbye BL, Spieck E, Bottomley PJ, Sayavedra-Soto LA. Acyl-homoserine lactone production in nitrifying bacteria of the genera nitrosopira, nitrobacter, and nitrospria identified via a survey of putative quorum-sensing genes. Appl Environ Microbiol. 2017;83:1–13. doi: 10.1128/AEM.01540-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sayavedra-Soto L, Ferrell R, Dobie M, Mellbye B, Chaplen F, Buchanan A, et al. Nitrobacter winogradskyi transcriptomic response to low and high ammonium concentrations. FEMS Microbiol Lett. 2015;362:1–7. doi: 10.1093/femsle/fnu040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available in the NCBI GenBank repository under BioProject PRJNA437159 and PRJNA464356.