Abstract
Most soil bacterial taxa are thought to be dormant, or inactive, yet the extent to which they synthetize new rRNA is poorly understood. We analyzed 18O composition of RNA extracted from soil incubated with H218O and used quantitative stable isotope probing to characterize rRNA synthesis among microbial taxa. RNA was not fully labeled with 18O, peaking at a mean of 23.6 ± 6.8 atom percent excess (APE) 18O after eight days of incubation, suggesting some ribonucleotides in soil were more than eight days old. Microbial taxa varied in the degree they incorporated 18O into their rRNA over time and there was no correlation between the APE 18O of bacterial rRNA and their rRNA to DNA ratios, suggesting that the ratios were not appropriate to measure ribonucleotide synthesis. Our study indicates that, on average, 94% of soil taxa produced new rRNA and therefore were metabolically active.
Introduction
Most bacteria in soil are thought to be dormant [1, 2], while only small active fractions control ecosystem processes [3]. Active bacterial cells have higher metabolic rates than dormant cells, leading to higher protein and rRNA synthesis. In contrast, dormant bacteria have very-low metabolic activity [4]. RNA concentrations likely decrease as most metabolic processes, including RNA synthesis, are halted, while DNA concentrations may remain stable because dormant cells are alive.
The relative abundances of ribosomal RNA (rRNA) and DNA extracted from environmental samples are commonly used as indicators of microbial metabolic activity [5]. However, rRNA to DNA ratios among taxa in communities substantially vary, often unrelated to metabolic activity, suggesting RNA alone may not be reliable indicator of active populations [6].
Stable isotope probing (SIP) can assess microbial activity independent of rRNA to DNA ratios. SIP with 18O-labeled water is powerful for assessing growth and activity of microbial communities because water is a universal substrate for nucleic acid synthesis [7]. In this study, we incubated 2 grams of soil with 400 µl of sterile 95 atom % H218O or with 400 µl of sterile, natural abundance H218O, for 1, 4, and 8 days (N = 18), and extracted total RNA following each incubation. Newly synthetized 18O-containing RNA has higher buoyant density than old RNA, and can be separated through isopycnic ultracentrifugation on a CsTFA density gradient. We fractionated the ultracentrifuged RNA, purified the fractions and sequenced a fragment of the 16S rRNA gene from complementary DNA (cDNA) as described in Supplement S1. Sequencing data were analyzed using a QIIME 1.7 based [8] chained workflow: https://github.com/alk224/akutils-v1.2 [9]. To assess rRNA synthesis of individual taxa, we measured the incorporation of 18O into rRNA by calculating taxon-specific shift in rRNA density and by converting it to atom percent excess (APE) 18O using a freely available R code (https://bitbucket.org/QuantitativeSIP/qsip_repo). APE 18O indicated the excess of 18O in microbial rRNA relative to natural abundance of the isotope, and was used to estimate rRNA synthesis rate, a measure of microbial activity. We assessed temporal patterns and variation in rRNA synthesis rates among soil microbial populations using qSIP, and compared our results to RNA to DNA ratios.
All taxa contained 18O-labeled rRNA after four days of incubation with H218O. Densities of rRNA in non-labeled incubations varied slightly around the mean (1.7808 ± 0.0011 g/ml), whereas densities of labeled rRNA significantly differed on each day (Fig. 1), which likely reflects taxonomic variation in the rate of metabolic activity [10] or differential reliance among taxa on de novo ribonucleotide synthesis [11] vs. ribonucleotide salvaging. If ribonucleotides are synthesized de novo, 18O will be assimilated throughout the ribonucleotide in addition to its assimilation into phosphodiester bonds [12], which will increase the 18O composition of rRNA more than recycling alone.
Fig. 1.
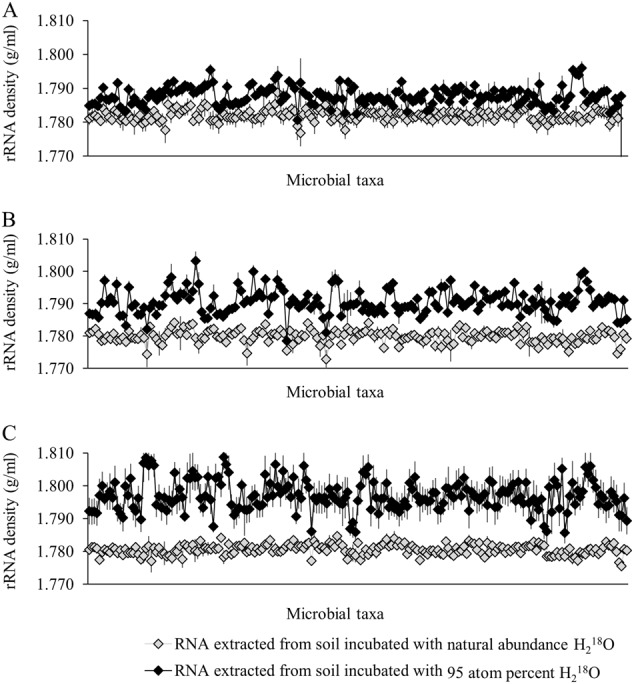
Densities of rRNA extracted from soil incubated with 95 atom % H218O (♦) or natural abundance H218O ( ) on three time points. a rRNA densities of taxa detected on day 1, b, c rRNA densities of taxa detected on day 4 and 8, respectively. Taxa are ranked by the same alphabetical order in each panel. Symbols represent means ± standard deviations
) on three time points. a rRNA densities of taxa detected on day 1, b, c rRNA densities of taxa detected on day 4 and 8, respectively. Taxa are ranked by the same alphabetical order in each panel. Symbols represent means ± standard deviations
Entirely dormant soil taxa were absent in our study, which challenges the widely accepted idea that dormancy is widespread among microbial taxa in the environment [13, 14]. We would observe many populations with non-labeled rRNA (i.e., containing 18O only at the natural abundance level) if dormancy were common under the incubation conditions. However, the average, positive 18O labeling we observed for all populations detected does not rule out individual cells within these populations that are dormant—non-labeled with 18O and exhibiting no rRNA synthesis. Positive APE 18O could also result from potential interactions between RNA molecules during ultracentrifugation [15], which possibly influenced our results and deserve additional research. Our observation of a weak correlation between the rRNA to DNA ratio and the APE 18O of rRNA across taxa (Spearman’s rank-order correlation, ρ(574) = −0.082, p = 0.051, Fig. 2) suggests that the ratio may be a poor proxy for metabolic activity, despite its positive correlation with microbial growth rate in some pure culture studies [16]. We expected that taxa with high rRNA to DNA ratios would have highly labeled rRNA, but this was not observed.
Fig. 2.

Relationship between rRNA to DNA ratios and APE 18O of rRNA among soil taxa on three time points: open symbols—day 1, black symbols—day 4, and gray symbols—day 8
We observed a significant temporal increase in 18O content for total RNA (F2,4 = 15.404, p = 0.013, Figure S1 and S2) and for RNA of phyla (Figure S3). RNA is thought to turn over rapidly [17], with estimates ranging from 20% per day [18] to 25% per hour [19]. In our experiment, we modeled rRNA turnover varied between 9 and 18% per day, which was slower than previously reported. The labeled RNA had ~23% of its oxygen atoms replaced with 18O, indicating that either some of the rRNA that was formed prior to H218O addition remained intact, and that the rRNA was newly synthesized but partly made with ribonucleotides that were more than 8 days old, or that newly synthesized ribonucleotides obtain part of their oxygen from organic substrates. Assuming that 50% of oxygen atoms come from H218O and 50% come from organic substrates [20], the isotopic composition of rRNA would be 50% at the fast modeled turnover rate and would have increased only minimally over time. The observed increase in 18O composition of RNA over time suggested that increasingly more ribonucleotides were synthesized and that the turnover rate of ribonucleotides in soil is ~23% per week.
Our knowledge of ribosome biosynthesis and degradation derives mostly from pure culture experiments, but, based on the soil we assessed, bacterial rRNA dynamics in soil may differ from those observed in pure cultures. Specifically, synthesis of new rRNA was slower than expected and unrelated to rRNA to DNA ratios in a soil microbial community, which is thought to have many dormant members. Yet, we found that all detected taxa synthesized new RNA during the 8-day incubation. Further research, conducted over longer incubations, will help determining maximum rRNA labeling and time required to reach it. Our work illustrates how RNA-qSIP can quantify taxon-specific activity relating to synthesis of new nucleic acids, opening doors to broader tests about microbial dormancy and metabolic activity across a range of soils and environments.
Accession numbers
All sequences have been deposited in NCBI SRA (accession numbers SAMN07960499 to SAMN07960874, SAMN07965143 to SAMN07965605, and SAMN07968111 to SAMN07968486). Data can directly be accessed at https://www.ncbi.nlm.nih.gov/Traces/study/?acc=SRP123236.
Electronic supplementary material
Acknowledgements
This research was supported by award 1142096 from the National Science Foundation, division of solar programs, NSF DEB-1241094, the Department of Energy’s Biological Systems Science Division, Program in Genomic Science (DE-SC0010579, and DE-SC0016207), and by the IGERT Fellowship. Additionally, the authors would like to thank Dr. Paul Dijkstra, Dr. Matthew Bowker and Lela Andrews from Northern Arizona University.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Subject Categories: Microbial population and community ecology, Microbial ecology and functional diversity of natural habitats
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0233-7) contains supplementary material, which is available to authorized users.
References
- 1.Khomutova TE, Demkina TS, Demkin VA. Estimation of the total and active microbial biomasses in buried subkurgan paleosoils of different age. Microbiology. 2004;73:196–201. doi: 10.1023/B:MICI.0000023989.04745.7b. [DOI] [PubMed] [Google Scholar]
- 2.Babiuk LA, Paul EA. The use of fluorescein isothiocyanate in the determination of the bacterial biomass of grassland soil. Can J Microbiol. 1970;16:57–62. doi: 10.1139/m70-011. [DOI] [PubMed] [Google Scholar]
- 3.Aanderud ZT, Jones SE, Fierer N, Lennon JT. Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front Microbiol. 2015;6:1–11. doi: 10.3389/fmicb.2015.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci USA. 2010;107:5881–6. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Větrovský T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–58. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7:2061–8. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz E. Characterization of growing microorganisms in soil by stable isotope probing with H218O. Appl Environ Microbiol. 2007;73:2541–6. doi: 10.1128/AEM.02021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krohn A. akutils-v12: facilitating analyses of microbial communities through QIIME. Zenodo. 2016;10:5281. [Google Scholar]
- 10.Campbell BJ, Kirchman DL. Bacterial diversity, community structure and potential growth rates along an estuarine salinity gradient. ISME J. 2012;7:210–20. doi: 10.1038/ismej.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebbole DJ, Zalkin H. Cloning and characterization of a 12-gene cluster from Bacillus subtilis encoding nine enzymes for de novo purine nucleotide synthesis. J Biol Chem. 1987;262:8274–87. [PubMed] [Google Scholar]
- 12.Richards OC, Boyer PD. 18O Labeling of deoxyribonucleic acid during synthesis and stability of the label during replication. J Mol Biol. 1966;19:109–19. doi: 10.1016/S0022-2836(66)80053-0. [DOI] [PubMed] [Google Scholar]
- 13.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 14.Cole JJ. Aquatic microbiology for ecosystem scientists: new and recycled paradigms in ecological microbiology. Ecosystems. 1999;2:215–25. doi: 10.1007/s100219900069. [DOI] [Google Scholar]
- 15.Lueders T, Manefield M, Friedrich MW. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol. 2004;6:73–8. doi: 10.1046/j.1462-2920.2003.00536.x. [DOI] [PubMed] [Google Scholar]
- 16.Muttray AF, Yu Z, Mohn WW. Population dynamics and metabolic activity of Pseudomonas abietaniphila BKME-9 within pulp mill wastewater microbial communities assayed by competitive PCR and RT-PCR. FEMS Microbiol Ecol. 2001;38:21–31. doi: 10.1111/j.1574-6941.2001.tb00878.x. [DOI] [Google Scholar]
- 17.Wellington EMH, Berry A, Krsek M. Resolving functional diversity in relation to microbial community structure in soil: exploiting genomics and stable isotope probing. Curr Opin Microbiol. 2003;6:295–301. doi: 10.1016/S1369-5274(03)00066-3. [DOI] [PubMed] [Google Scholar]
- 18.Ostle N, Whiteley AS, Bailey MJ, Sleep D, Ineson P, Manefield M. Active microbial RNA turnover in a grassland soil estimated using a 13CO2 spike. Soil Biol Biochem. 2003;35:877–85. doi: 10.1016/S0038-0717(03)00117-2. [DOI] [Google Scholar]
- 19.Yuan D, Shen V. Stability of ribosomal and transfer ribonucleic acid in Escherichia coli B/r after treatment with ethylenedinitrilotetraacetic acid and rifampicin. J Bacteriol. 1975;122:425–32. doi: 10.1128/jb.122.2.425-432.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaney SG, Duffy JJ, Boyer PD. Patterns of oxygen interchange between water, substrates, and phosphate compounds of Escherichia coli and Bacillus subtilis. J Biol Chem. 1972;247:2145–50. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


