Abstract
Aims
Excessive consumption of alcoholic beverages is associated with cardiac remodeling and cardiomyopathy. We examined the possible association of alcohol use, common Asian genetic variants in genes involved in alcohol metabolism, and cardiac structures/functions alterations.
Methods
A prospective, community-dwelling survey among individuals with available complete echocardiography examined the associations of alcohol use, cardiac structure/functions, and three common alcohol metabolizing genetic variants, including aldehyde dehydrogenase 2 (ALDH2), alcohol dehydrogenase 1B (ADH1B) and cytochrome P450 (CYP) isoform 2E1 (CYP2E1).
Results
Among 1577 participants (mean age: 53 ± 9, 59.7% female), we observed that in subjects with more frequent weekly ethanol intake showed greater left ventricle (LV) mass, more impaired diastolic functions, and reduced global longitudinal strain (GLS), systolic (SRs) and early diastolic strain rates (SRe) (P<0.05). After propensity matching for clinical confounders (n = 330:30 for frequent users and non-users), frequent alcohol use and subjects carrying ALDH2 (A/G or A/A), ADH1B (A/A) or CYP2E1(T/C or T/T) polymorphisms were all associated with worse GLSRs and GLSRe, with combined alcohol use and any given genetic variant aggravated these associations (all P < 0.05). Finally, we observed Gene–Gene synergistic effects on LV functional decline in frequent alcohol users by using linear mixed effect model (all interaction P < 0.05).
Conclusions
Among East Asians, even moderate alcohol consumption can confer subclinical adverse effects on cardiac systolic functions, which was most pronounced in subjects carrying common variants in alcohol metabolizing genes. These findings challenge the notion of beneficial influences of less heavy ethanol consumption on the heart, especially among East Asians.
Short summary
This study evaluated the association of level of alcohol consumption and genetic variants in genes involved in alcohol metabolism with changes in cardiac function in East Asians. Even moderate alcohol use conferred subclinical adverse effects on cardiac systolic functions, which were most pronounced in subjects carrying common alcohol metabolizing genes.
INTRODUCTION
Over the past decades, it has been suggested that light to moderate ethanol use was protective against cardiovascular morbidities and mortality (Di Castelnuovo et al., 2006). According to the AHA guideline (Goldberg et al., 2001), it is safest for both men and women not to regularly drink >14 or > 7 alcoholic drinks, respectively, on weekly basis, and to spread the drinks evenly over 3 or more days (Goncalves et al., 2015a). However, it has also been proposed that chronic excessive alcohol consumption may alter cardiac structures and function (Lazarevic et al., 2000; Spies et al., 2001). Changes may include myofilament protein reduction (Mathews et al., 1981; Vary and Deiter, 2005), fibrosis (Wang et al., 2005), and subclinical myocardial contractility (Djousse et al., 2004; Laonigro et al., 2009). Left ventrical (LV) function might start to deteriorate prior to clinical onset of symptoms (Lazarevic et al., 2000; Mahmoud et al., 2007). Therefore, early recognition of pre-clinical LV systolic dysfunction may benefit functional recovery if complete alcohol withdrawal or abstinence can be achieved before overt mechanical heart failure occurs (Nicolas et al., 1998; Gavazzi et al., 2000). Several Asian-specific genetic polymorphisms in the enzymes that metabolize ethanol (i.e. alcohol dehydrogenase 1B [ADH1B], aldehyde dehydrogenase 2 [ALDH2] and cytochrome P4502E1 [CYP2E1]) have been identified (Choi et al., 2003; Yang et al., 2009). ADH1B and ALDH2 constitute the major metabolic detoxifying pathway of ethanol in the liver (Zakhari, 2006; Zakhari and Li, 2007). The most widely recognized ethanol-metabolizing enzyme variant is the ALDH2 ∗ 2 allele (rs671, A), which confers a nearly complete loss of the ALDH2 enzymatic activity and results in acetaldehyde accumulation (Crabb et al., 1989; Brooks et al., 2009). The ADH1B ∗ 2 allele, which contains a single nucleotide polymorphism (SNP) (rs1229984, A), metabolizes ethanol to acetaldehyde >8 times higher than the reference ADH1B ∗ 1 allele (Lee et al., 2006). CYP2E1, which can be induced by chronic ethanol consumption, also acts to metabolize ethanol into acetaldehyde, as well as to generate reactive oxygen species (Zakhari, 2006). It has been proposed that differences among individuals in their ability to metabolize alcohol are modulated by behavior of alcohol consumption superimposed on genetic polymorphisms. The higher prevalence of the genetic polymorphisms in the ADH1B, ALDH2 and CYP2E1 genes in East Asians warrants a more comprehensive survey on the impact of alcohol and genetic background on cardiac structure and function. Therefore, we sought to investigate the relationships among habitual alcohol intake, genetic polymorphisms and changes in cardiac function from an Asian community-dwelling population.
MATERIALS AND METHODS
Study participants
The study participants comprised a community-based population from the Mitochondria-Aging in Northern Taiwan (MAGNET) study (September 2010 to May 2012) (Hung et al., 2012; Chen et al., 2015). The MAGNET study recruited residents, aged 40–74 years, from the northern coastal areas of New Taipei City, Taiwan. Written informed consents were obtained from all participants prior to study start. The protocol was reviewed and approved by the Institution Review Board of Mackay Medical College (No. P990001).
Baseline demographics including age, gender, body height, weight, waist, and hip circumferences were recorded. Body surface area (BSA) and blood pressure (systolic and diastolic blood pressure) were assessed in the morning, and an average of three measurements performed by well-trained nurses was determined. After a 10-min rest, blood pressure was measured from all subjects on their upper right arms in the sitting position.
Genetic association studies to identify the potential links of several selected SNPs, life styles, clinical risk factors and cardiovascular disease phenotypes were also performed. Biochemical data from venous blood samplings were collected after at least 8 h of fasting and total cholesterol, high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), fasting triglycerides (FTG) and fasting sugar levels (FPG) were measured.
Questionnaire surveys and alcohol consumption dose
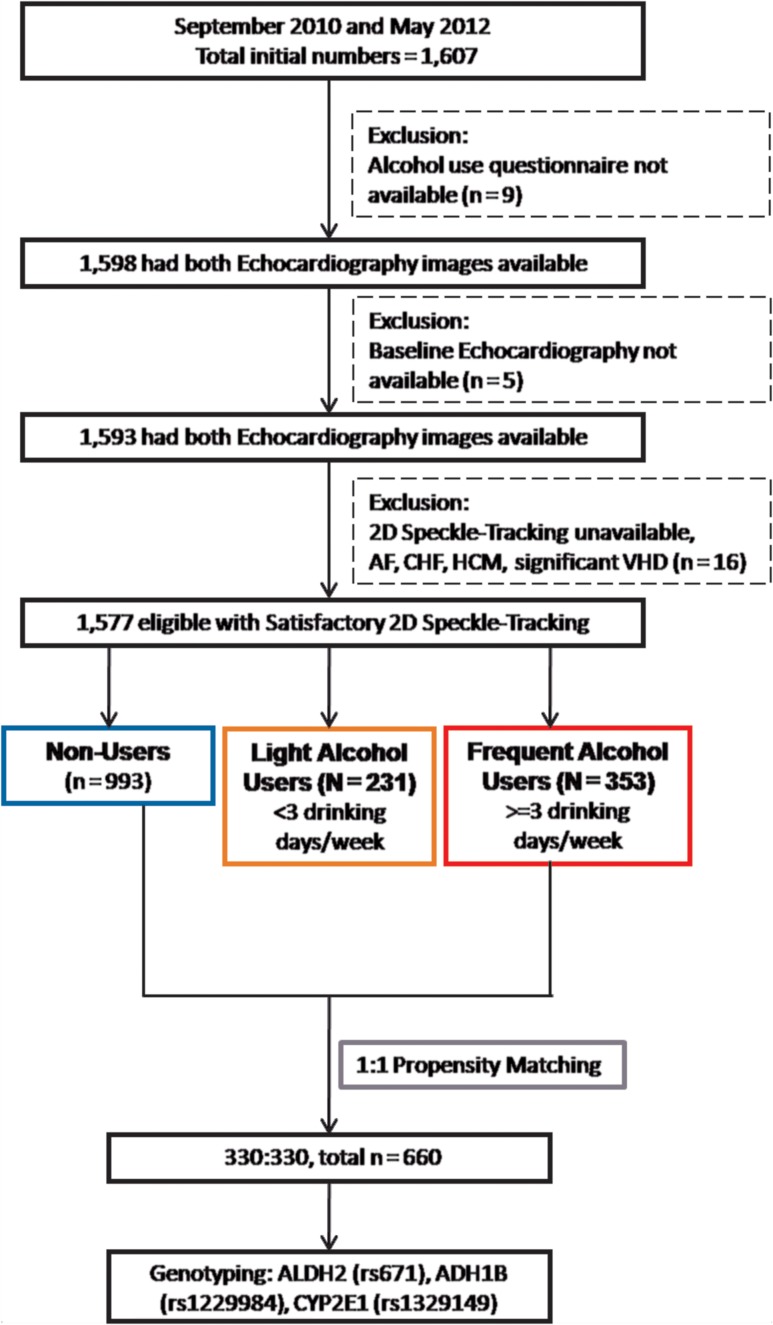
The average weekly consumption of alcohol including different types of alcoholic beverages was self-reported. Subjects were subsequently categorized as frequent alcohol users (defined as ≥3 drinking days per week, n = 351), light-users (defined as ≥1 and < 3 drinking days per week, n = 231), and non-users (defined as no alcohol intake, n = 993). Among all study participants, nine (0.73%) had no questionnaire data available for alcohol consumption behavior and were excluded. Participants were asked to indicate how often they consumed beer (alcohol by volume, 3.5–4.5%), wine, strong wine and liquor, as well as the amount of beer, wine and liquor consumed on a single occasion. For the frequency of alcohol consumption, six pre-defined categories, ranging from never to almost every day per week were queried. We calculated weekly alcohol consumption for each participant in gram units by multiplying the frequency of each consumed alcoholic beverage and the amount consumed from those subjects. This analysis of weekly consumption has been validated previously (Hung et al., 2016). From a small part of the responses where bottle or can quantities were unavailable, we estimated that one standard drinking cup or glass in Taiwan contains 75 ml of the beverage and one drink is equal to 12 g of pure alcohol. The measurement of total weekly alcohol intake was calculated based on the most recent alcohol consumption behavior (within the last 3 months). Medical histories including hypertension, diabetes, hyperlipidemia and associated medications use were obtained from the self-report questionnaires. The flow-chart showing the final study participants are detailed in Fig. 1.
Fig. 1.
The schematic flow-chart of our current recruited study participants.
Assessment of conventional echocardiography and Doppler-related imaging techniques
Among all 1607 study subjects enrolled in the study, 1593 had trans-thoracic echocardiography data available. Conventional echocardiography parameters including left ventricular (LV) wall thickness (both septal [IVS] and posterior walls [LVPW]), LV diameter (both end-diastolic [LVIDd] and end-systolic [LVIDs]), and derived LV mass index (indexed to BSA) were obtained. Among them, 1577 had good 2D imaging quality for speckle-tracking based strain analysis in all three LV apical planes and no evidence of clinical heart failure. Tissue Doppler imaging (TDI) was performed by pulsed-wave Doppler. Mitral inflow early (E) and late diastolic (A) LV filling velocities were obtained at the tip of the mitral leaflets from the apical 4-chamber view. Mitral annulus contraction (S′) and relaxation velocities (E′) were both assessed at lateral mitral annulus position with high frame rates using TDI, with LV filling pressure estimated as E/E′ ratio.
Speckle-tracking analysis and assessment of global LV deformation
We performed dedicated LV deformation imaging analysis using baseline 2D images from three LV apical views (4, 2 and 3-chamber views) for global longitudinal strains, three short-axis views (mitral, papillary and apex views) for LV circumferential strains, and LV twist (version 10.8, EchoPAC, GE Vingmed Ultrasound, Norway). Baseline 2D images were analyzed by off-line endocardial border manual tracing by the same experienced technicians, using novel proprietary software (version 10.8, EchoPAC, GE Vingmed Ultrasound, Norway). The mean frame rate for all participants was between 60 and 80 frames per second (fps) (averaged: 71 ± 10.4). Peak myocardial deformation percentage, St(t) = [L(t) – L0]/L0, from end-diastolic [L(t0)] to end-systolic phase [L(t1)] in longitudinal direction was defined as longitudinal strain (S). We also calculated the first derivative of longitudinal strain as the rate of deformation changes over time into strain rate (SR, where SR(t) = L−1(t)dL/dt), with totally three phases (peak systolic [SRs], early diastolic [SRe] and late diastolic [SRa]) measured in a single cardiac cycle. Based on automated speckle-tracking algorithms, the global LV longitudinal strain (GLS) and each global LV SR (GLSR) value were then averaged from three individual LV apical views (including 2-chamber [CH], 4CH and 3CH views). Global circumferential strain (GCS) curves were similarly obtained by averaging values from three individual short-axis levels (including mitral [MV], papillary [PM] and apical layer [AP]). The lower the absolute values of these deformational indices the worse the cardiac function.
Genotyping of selected SNPs
Genomic DNA of each selected study participant was extracted from EDTA-containing whole blood samples by a semi-automated extraction system (Smart LabAssist, Taiwan Advanced Nanotech Inc., Tau-Yuan County, Taiwan) with TANBead Blood DNA plate (Taiwan Advanced Nanotech Inc.). TaqMan probes were then used to examine the target polymorphisms. The DNA samples were genotyped for the relevant SNPs for each candidate gene using an established commercial service (Genomics BioSci & Tech. Co., Ltd, Taiwan). The designated SNPs were determined by real-time polymerase chain reaction (PCR) allelic discrimination using reagents for the TaqMans SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The detailed assay information and genetic sequencing for each target alcohol-metabolizing enzyme coding genes are listed in Supplementary Table 1. PCR amplification and allelic discrimination were carried out using a ViiA7 Real-Time PCR System (Applied Biosystems). The quantitative PCR program was 60°C for 30 s, then initial denaturation for 10 min at 95°C, followed by 40 cycles of denaturation at 95°C for 15 s and annealing at 60°C for 1 min. The allele-detection process and allelic discrimination were performed for 1 min at 60°C. The raw data was then analyzed by ViiA7 Software v1.2.4
The following three known SNPs were examined: ADH1B Arg47His (rs1229984, G/A), ALDH2 Glu487Lys (rs671, G/A) and CYP2E1 (rs1329149, C/T). The success rates of genotyping was >96.6% with a mismatch rate of 0.0% for the analysis of the above mentioned SNPs.
Statistical analysis
Continuous data are presented as mean and standard deviation (SD), with categorical variables presented as proportions or percentages in distributions and compared by using Χ2. Data including baseline characters and cardiac structures/functions were displayed across three alcohol use groups (non-, light- and frequent alcohol users) and compared using oneway ANOVA test with post hoc paired comparisons made between groups (Tables 1 and 2).
Table 1.
Baseline demographics of all study participants across three alcohol user groups
| Non-users (n = 993) | Light alcohol users (n = 231) | Frequent alcohol users (n = 353) | P value | |
|---|---|---|---|---|
| Baseline demographics | ||||
| Age (years) | 53.4 ± 9.1 | 50.9 ± 8.8* | 53.1 ± 8.5# | 0.726 |
| Sex, female (%) | 719 (72.4%) | 128 (55.4%) | 95 (26.9%) | <0.001 |
| Body height (cm) | 159.0 ± 7.5 | 161.8 ± 8.3* | 164.3 ± 7.2*# | <0.001 |
| Body weight (kg) | 61.7 ± 10.8 | 64.2 ± 11.9* | 68.2 ± 10.8*# | <0.001 |
| BMI (kg/m2) | 24.4 ± 3.6 | 24.3 ± 3.5 | 25.3 ± 3.5*# | <0.001 |
| SBP (mmHg) | 127.1 ± 19.3 | 126.1 ± 19.7 | 129.9 ± 19.4 | 0.029 |
| DBP (mmHg) | 78.3 ± 13.4 | 78.5 ± 13.7 | 81.4 ± 13.8*# | 0.001 |
| Pulse pressure (mmHg) | 48.8 ± 13.6 | 47.6 ± 12.8 | 48.6 ± 14.2 | 0.49 |
| HR (beat/min) | 69.1 ± 10.3 | 68.6 ± 10.7 | 68.2 ± 11.7 | 0.4 |
| Laboratory data | ||||
| Fasting glucose (mg/dl) | 99.5 ± 27.8 | 96.6 ± 18.6 | 100.5 ± 27.5 | 0.21 |
| Total cholesterol (mg/dl) | 207.6 ± 37.4 | 208.3 ± 35.3 | 208.2 ± 38.5 | 0.96 |
| FTG (mg/dl) | 108.2 ± 72.6 | 113.5 ± 98.6 | 138.3 ± 120.9*# | <0.001 |
| LDL-C (mg/dl) | 123.7 ± 33.2 | 125.1 ± 31.3 | 123.7 ± 34.3 | 0.84 |
| HDL-C (mg/dl) | 57.1 ± 15.7 | 56.6 ± 14.9 | 53.8 ± 14.4* | 0.002 |
| eGFR (ml/min/1.73 m2) | 87.9 ± 22.5 | 85.7 ± 20.9 | 84.1 ± 20.9* | 0.015 |
| Medical histories | ||||
| Active smokers, n (%) | 83 (8.3%) | 31 (13.2%) | 85 (24.1%) | <0.001 |
| HTN, n (%) | 193 (19.4%) | 33 (14.3%) | 73 (20.7%) | 0.128 |
| DM, n (%) | 95 (9.6%) | 10 (4.3%) | 40 (11.3%) | 0.013 |
| Hyperlipidemia, n (%) | 307 (30.9%) | 72 (31.2%) | 126 (35.7%) | 0.244 |
| CVD, n (%) | 79 (8.0%) | 14 (6.1%) | 28 (7.9%) | 0.609 |
BMI, body mass index; CVD, cardiovascular diseases; DBP, diastolic blood pressure; DM, diabetes; FTG, fasting triglyceride level; HDL-C, high-density lipoprotein cholesterol; HR, heart rate; HTN, hypertension; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure. *P < 0.05 vs non-user; #P < 0.05 vs light users.
Table 2.
Comparisons of cardiac structure/function and myocardial deformations across three alcohol user groups
| Non-users (n = 993) | Light alcohol users (n = 231) | Frequent alcohol users (n = 353) | P value | |
|---|---|---|---|---|
| Conventional measures | ||||
| IVS (mm) | 8.9 ± 1.1 | 9.0 ± 1.1 | 9.5 ± 1.2*# | <0.001 |
| LVPW (mm) | 8.9 ± 1.0 | 9.0 ± 1.0 | 9.4 ± 1.0*# | <0.001 |
| LVIDd (mm) | 45.8 ± 3.3 | 45.9 ± 2.9 | 46.6 ± 3.2*# | <0.001 |
| LVIDd (mm) | 28.6 ± 2.7 | 28.7 ± 2.4 | 29.2 ± 2.8*# | <0.001 |
| LVEDV (ml) | 96.9 ± 16.1 | 97.4 ± 14.4 | 100.9 ± 16.2*# | <0.001 |
| LVESV (ml) | 31.6 ± 7.9 | 31.7 ± 6.6 | 33.3 ± 7.9*# | 0.0011 |
| LVEDVi (ml/m2) | 55.0 ± 8.4 | 53.8 ± 7.9 | 53.6 ± 8.6* | 0.013 |
| LVESVi (ml/m2) | 17.9 ± 4.1 | 17.5 ± 3.3 | 17.6 ± 4.0 | 0.33 |
| LVEF (%) | 67.4 ± 5.8 | 67.4 ± 5.0 | 67 ± 5.5 | 0.56 |
| LV mass (LVM) (g) | 137.3 ± 31.6 | 140.7 ± 29.2 | 156.0 ± 34.6*# | <0.001 |
| LV mass index (LVMi) (g/m2) | 77.4 ± 14.6 | 77.2 ± 12.8 | 82.4 ± 16.2*# | <0.001 |
| LV mass–volume ratio | 1.42 ± 0.23 | 1.45 ± 0.25 | 1.55 ± 0.25*# | <0.001 |
| Diastolic indices | ||||
| TDI E′ (mean) (cm/s) | 9.7 ± 2.4 | 10.0 ± 2.4 | 9.6 ± 2.2 | 0.12 |
| E/A ratio | 1.20 ± 0.42 | 1.23 ± 0.5 | 1.15 ± 0.39 | 0.085 |
| DT (ms) | 195.1 ± 36.5 | 197.1 ± 37.4 | 206.6 ± 37.6*# | <0.001 |
| IVRT (ms) | 89.3 ± 13.9 | 90.0 ± 14.1 | 93.7 ± 16.1*# | <0.001 |
| E/E′ (mean) ratio | 7.99 ± 2.63 | 7.26 ± 2.11* | 7.35 ± 2.00* | <0.001 |
| Myocardial deformations | ||||
| GLS (%) | −20.63 ± 2.08 | −20.73 ± 1.92 | −20.48 ± 2.02 | 0.32 |
| GCS (%) | −21.28 ± 2.91 | −21.37 ± 2.76 | −21.19 ± 2.66 | 0.76 |
| GLSRs (1/s) | −1.14 ± 0.17 | −1.15 ± 0.16 | −1.12 ± 0.16* | 0.014 |
| GLSRe (1/s) | 1.35 ± 0.34 | 1.37 ± 0.34 | 1.26 ± 0.34*# | <0.001 |
| GLSRa (1/s) | 1.07 ± 0.23 | 1.04 ± 0.20 | 1.13 ± 0.34*# | <0.001 |
E′, myocardial relaxation velocity by Tissue Doppler imaging; GCS, global circumferential strain; GLS, global longitudinal strain; GLSRs, GLSRe and GLSRa, global longitudinal strain rate for systolic (s), early diastolic (e) and late diastolic (a) ventricular phases; IVS, interventricular septum; LVPW, posterior wall thickness; LVIDd, left ventricular internal diastolic dimension; LVM, left ventricular mass; LVMi, left ventricular mass index; LVEF, left ventricular ejection fraction; E/A, early to late mitral inflow velocity ratio; DT, deceleration time; IVRT, isovolumetric relaxation time. *P < 0.05 vs non-user; #P < 0.05 vs light users.
To explore the associations between weekly alcohol use dose (as independent variable, per + 100 g/week) and each cardiac structure/deformation parameter (as separate, dependent variables: LV mass, GLS, GLSRs, GLSRe and GLSRa), linear regression models were conducted with coefficient values (Coef.) and 95% confidence interval (CI) reported, and adjusted for baseline clinical information (including age, sex, body size [BMI], blood pressures, heart rate and renal function in terms of estimated glomerular filtration rate [eGFR]), medical histories (hypertension, diabetes, cardiovascular diseases), and active smoking as potential confounders (Kishi et al., 2015). We further conducted nested case control study by matching those frequent alcohol users (>3 drinking days/week) with those individuals who did not consume alcohol for baseline clinical covariates, resulting after matching the inclusion of 330 subjects from each group (Fig. 1). The propensity matching methods and more detailed descriptions were further addressed in supplemental materials.
The three SNPs were compared between matched groups. Genotype frequencies were determined, and Hardy–Weinberg equilibrium was assessed for each SNP. The associations between cardiac structural/functional measures including deformations (as dependent variable), patterns of alcohol use (as independent variable) and three genetic polymorphisms (as independent variable) were tested using linear mixed effect model (LMM) (Table 3). The regression coefficient was estimated under a random effect model that allowed the baseline values of dependent variable to vary among any two specific matched samples who were clustered in a unique stratum in our 330 matched individuals in each group. That is, the regression coefficient was based on a subject-specific approach which was also called ‘conditional’ model. For the analysis of main effect of alcohol use/genetic polymorphisms, we examined the associations among various cardiac structures/functions or myocardial deformations (including LVMi, GLS, GCS, GLSRs, GLSRe and GLSRa) separately (as dependent variable) and regular alcohol use (as drinkers and controlled non-drinkers) with three genetic polymorphisms as independent variables in same model (Table 3, Models 1 and 2) after propensity matching procedure (n = 330 for each group). We also examined the two-way interaction effects (various genotypes on cardiac structural/functional values) by examining the main effect of frequent alcohol use and three genotypes and their interactions (Gene–Gene or Alcohol–Gene) introduced into the LMM simultaneously after testing no apparent multi-collinearity between these variables (Table 3, Model 3 for Gene–Gene and Model 4 for Alcohol–Gene). Finally, we further tested three-way interaction effects of frequent alcohol use and any combined two genetic variants and their interactions (Alcohol–Gene–Gene) introduced into the LMM simultaneously to see whether there may exist any genetic synergistic effects with frequent alcohol use on various cardiac structures/functions or myocardial deformations (Table 3, Model 5).
Table 3.
Joint interaction effects among alcohol use, cardiac structures/deformation measures and genetic variants in matched population
| Model/fixed effects | Regression coefficient (95% confidence interval)# | |||||
|---|---|---|---|---|---|---|
| LVMi (gm/m2) | GLS (%) | GCS (%) | GLSRs | GLSRe | GLSRa | |
| Model 1: Main effect of drink | ||||||
| Drinker/non-drinker | 2.35 (−0.12, 4.81) | 0.28 (−0.04, 0.59) | 0.04 (−0.44, 0.52) | 0.08 (0.06, 0.11)* | −0.12 (−0.17, −0.06)* | 0.01 (−0.02, 0.05) |
| Model 2: Main effect of genetic variant | ||||||
| rs671 | 2.27 (−0.33, 4.87) | 0.35 (0.01, 0.69)* | −0.05 (−0.56, 0.47) | 0.10 (0.07, 0.12)* | −0.18 (−0.24, −0.13)* | 0.03 (−0.01, 0.07) |
| rs1229984 | 0.25 (2.20, 2.71) | 0.31 (0.01, 0.63) | 0.69 (−0.34, 1.32) | 0.03 (0.01, 0.06)* | −0.07 (−0.12, −0.01)* | 0.02 (−0.02, 0.05) |
| rs1329149 | 3.16 (0.44, 5.89)* | 0.40 (0.04, 0.76)* | 0.20 (−0.34, 0.74) | 0.04 (0.01, 0.07)* | −0.11 (−0.17, −0.05)* | 0.00 (−0.04, 0.03) |
| Model 3: Gene–gene interaction | ||||||
| rs671 × rs1229984 | 2.86 (−2.09, 7.81) | −0.27 (−0.92, 0.38) | −0.03 (−1.02, 0.95) | 0.07 (0.02, 0.12)* | −0.12 (−0.23, −0.01)* | 0.07 (−0.001, 0.15) |
| rs671 × rs1329149 | 3.82 (−1.69, 9.34) | −0.15 (−0.87, 0.57) | 0.45 (−0.65, 1.54) | 0.06 (0.00004, 0.11)* | −0.13 (−0.25, −0.01)* | 0.01 (−0.06, 0.09) |
| rs1229984 × rs1329149 | −2.98 (−8.43, 2.47) | 0.15 (−0.57, 0.86) | −0.38 (−1.46, 0.71) | −0.01 (−0.06, 0.05) | −0.04 (−0.16, 0.07) | −0.08 (−0.16, 0.001) |
| Model 4: Drink–gene interaction | ||||||
| Drinker × rs671 | −3.08 (−8.29, 2.13) | 0.50 (−0.19, 1.18) | −0.28 (−1.33, 0.76) | 0.13 (0.08, 0.18)* | −0.17 (−0.28, −0.06)* | 0.02 (−0.05, 0.10) |
| Drinker × rs1229984 | 0.73 (−4.46, 5.91) | 0.66 (−0.02, 1.34) | −0.85 (−1.89, 0.18) | 0.08 (0.03, 0.13)* | −0.20 (−0.31, −0.09)* | 0.04 (−0.03, 0.12) |
| Drinker × rs1329149 | 2.41 (−3.32, 8.15) | 0.20 (−0.55, 0.95) | −0.29 (−1.43, 0.85) | 0.09 (0.04, 0.15)* | −0.16 (−0.28, −0.04)* | 0.04 (−0.04, 0.12) |
| Model 5: Drink–gene–gene interaction | ||||||
| Drink × rs671 × rs1229984 | −2.03 (−12.49, 8.42) | 1.58 (0.21, 2.94)* | −1.01 (−3.11, 1.08) | 0.25 (0.15, 0.35)* | −0.36 (−0.58, −0.15)* | 0.04 (−0.11, 0.18) |
| Drink × rs671 × rs1329149 | 6.51 (−5.16, 18.18) | 0.26 (−1.27, 1.78) | 0.67 (−1.65, 3.00) | 0.12 (0.01, 0.23)* | −0.18 (−0.34, 0.–14) * | −0.18 (−0.22, 0.07) |
| Drink × rs1229984 × rs1329149 | 0.48 (−10.49, 11.45) | −0.77 (−2.20, 0.65) | −0.01 (−2.19, 2.16) | 0.05 (−0.06, 0.15) | −0.26 (−0.49, −0.03)* | 0.02 (−0.13, 0.17) |
#The regression coefficient was estimated under a random effect model that allowed the baseline values of outcome to vary among any two specific matched samples who were clustered in a unique stratum (i.e. there were 330 matched IDs). That is, the regression coefficient was based on a subject-specific approach which was also called ‘conditional’ model.
For ADH1B (rs1229984), Non-Fast type (G/A or G/G) behaves as reference group; for ALDH (rs671), (G/G) behaves as reference group; for CYP2E1 (rs1329149), (C/C) serves as reference group; other abbreviations as Table 2.
*P < 0.05.
All calculations were carried out using Statistical Analysis System (SAS, version 9.0; SAS Institute, Inc.) and STATA software (version 11.0, Stata-Corp., College Station, TX, USA) with two-sided P value <0.05 considered to be significant.
RESULTS
Baseline clinical characters and echocardiography measurements in the study population
Baseline characteristics and echocardiography were available from a total of asymptomatic 1577 subjects (mean age: 53 ± 9 years, 942 females [59.7%]) included in this study (Tables 1 and 2). The light and frequent alcohol user groups were similar to the non-user group with respect to age (range, 50.9– 53.4 years), pulse pressure, heart rate, fasting glucose, total cholesterol, LDL-C, and prevalent hypertension, hyperlipidemia and cardiovascular disease (P > 0.05). Compared with non-users, light and frequent users were less likely to be females and had greater body height and weight values (P <0.05), diastolic blood pressure, fasting triglycerides, and lower eGFR than non-users (P <0.005) (Table 1).
Among 353 frequent alcohol users, baseline echocardiography showed greater wall thickness (IVS or LVPW), greater LV dimension (LVIDd), volume (LVEDV) and substantially greater LV mass and LV mass index (all P < 0.05). More prolonged DT and lower E/E’ were observed in frequent alcohol users (P <0.05), together with worse GLSRs, GLSRe, and augmented GLSRa compared with non-users (F-value: 4.28, 10.84 and 8.94, respectively, all P <0.05). In our linear regression models examining the associations between alcohol use and cardiac structure (LV mass) or deformation parameters (including GLS, GLSRs, GLSRe and GLSRa), increasing alcohol use (per 100gm ethanol intake/week) was associated with greater LV mass (Coef: 1.14, P = 0.047), lower GLS (Coef: 0.15, P < 0.001), borderline worse GLSRs (Coef: 0.006, P = 0.08) and reduced GLSRe (Coef: −0.02, P <0.001) after controlling for baseline clinical information, medical histories and active smoking status.
Baseline clinical characteristics, distribution of genetic polymorphisms and echocardiography measurements after matching
Propensity matching resulted in 330 matched subjects for frequent alcohol users and non-users with similar baseline demographic information including age, gender, body size (BMI), blood pressure, active smoking and medical histories.
Hardy–Weinberg equilibrium analysis showed that in the matched populations all tested loci were in Hardy–Weinberg equilibrium except greater prevalence of normal G/G in the ALDH2 (rs671) genotype in the frequent drinkers group (Supplementary Table 2). The alcohol consumption per week showed no major differences between target gene variants though a trend toward lower alcohol use for ALDH2 ∗ 2 carriers (Supplementary Figs 1 and 2).
We observed in frequent drinkers compared with non-drinkers slight increase in LV mass and LV mass index (82.4 vs 79.7gm/m2), greater prolonged IVRT (94 vs 90.9 ms), and substantially reduction in global longitudinal strain (GLS: −20.5 vs −20.9%), systolic SR (GLSRs: −1.12 vs −1.18 1/s), and early diastolic SR (GLSRe: 1.18 vs 1.30 1/s) after matching (all P <0.05, Supplementary Table 3). The standardized difference before and after matching was also provided to judge the clinical significance (Supplementary Table 4).
Associations among alcohol use, variants of alcohol metabolizing genes and cardiac structure/deformation measures
Table 3 showed the associations among patterns of alcohol use (frequent alcohol users vs non-users, n = 330 for each) and 3 genetic polymorphisms on cardiac structure/myocardial deformation indices (including LVMi, GLS, GCS, GLSRs, GLSRe and GLSRa, respectively) utilizing ‘conditional’ LMM after matching. Individuals with frequent alcohol use was associated with worse GLSRs (Coef: 0.08, 95% CI: 0.06 to 0.11) and lower GLSRe (Coef: −0.12, 95% CI: −0.17 to −0.06, all P < 0.05) when compared to non-users. Subjects carrying ALDH2 variants (A/G or A/A) were associated with worse GLS (Coef: 0.35, 95% CI: 0.01–0.69), with all three individual genetic polymorphism carriers (A/G or A/A for ALDH2, A/A for ADH1B, T/C or T/T for CYP2E1) associated with both worse GLSRs and GLSRe (all P < 0.05, Models 1 and 2 in Table 3). For the two-way interaction effects (various genotypes on cardiac structural/functional values) by examining the main effect of regular alcohol use and three genotypes and their interactions (Gene–Gene or Alcohol–Gene), we showed that combined ALDH2-ADH1B and ALDH2-CYP2E1, or combined alcohol use and any type of these three genetic polymorphism carriers were associated with both worse GLSRs and GLSRe (Models 3 and 4 in Table 3, all interaction P < 0.05). Finally, three-way interaction analysis showed genetic synergistic effects of ALDH2-ADH1B on GLS, GLSRs and GLSRe; also in ALDH2-CYP2E1 on GLSRs and GLSRe, as well as ADH1B-CYP2E1 on GLSRe in frequent alcohol users (Model 5 in Table 3, all interaction P < 0.05).
DISCUSSION
In this study, we demonstrated that light-to-moderate alcohol use was associated with cardiac structural and functional changes, including greater LV mass and worse diastolic indices, in a community-based population. A dose-dependent relationship between weekly alcohol consumption and systolic myocardial deformation decline was also observed, which remained unchanged even in a fully adjusted model. In addition, our genetic analysis revealed that ALDH2 ∗ 2 carriers tended to consume less alcohol. In individuals carrying the ALDH2 polymorphisms, either A/G or A/A, along with those carrying fast ADH1B alcohol-metabolizing genotypes (A/A) or CYP2E1 (C/T or T/T), habitual alcohol use was associated with the greatest impact on sublinical LV contractile functions. To our knowledge, this is the first study to evaluate the association between three well-defined genetic variants in the Asian population that affect alcohol metabolism and cardiac function.
Several mechanisms may contribute to the observed alcohol-dependent cardiac toxicity, including the accumulation of the ethanol metabolite, acetaldehyde, which may cause direct cardiac toxicity; possible nutritional deficiencies; or elevated blood pressures due to excessive alcohol use (Hajnoczky et al., 2005). Morphological features and functional alterations of the heart in excessive alcohol users have been characterized by LV wall thickening, chamber enlargement, reduced LV total mass, diastolic dysfunction and overtly depressive cardiac contractility (e.g. LV ejection fraction < 50%) (Askanas et al., 1980). Recently, a large population study has reported reduced longitudinal myocardial deformation in a sensitive, pre-clinical LV marker, which demonstrated an inverse relationship between the level of alcohol consumption and decline in cardiac deformation (Goncalves et al., 2015b). This relationship indicates the potential myocardial toxic effects of the long-term exposure to even a moderate dose of ethanol.
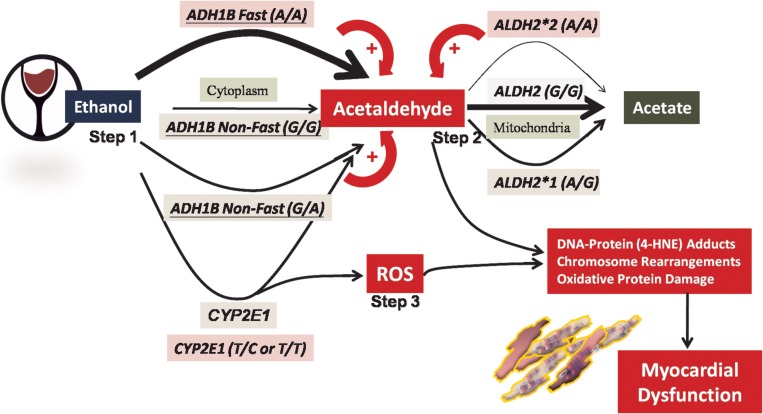
The three physiologically relevant alcohol-metabolizing genetic polymorphisms, ADH1B (rs1229984), ALDH2 (rs671) and CYP2E1 (rs1329149), have previously been shown to be associated with several malignant conditions of the upper aerodigestive tract cancers in East Asians (Whitfield, 2002; Badger et al., 2003; Choi et al., 2003; Seitz and Stickel, 2006; Yang et al., 2009). ADH constitutes a complex enzyme family, with the fast metabolizing ADH1B gene (rs1229984, A allele) observed to be the predominant variant in more than 80% of East Asians (Shen et al., 1997). Carriers of the A/A genotype are known to have a rapidly rising level of circulating acetaldehyde immediately after ethanol consumption, which is due to an efficient conversion of alcohol to its more toxic metabolite, acetaldehyde (Fig. 2; Step 1). Such a rapid rise has been associated with a greater risk of organ toxicity or malignancy with chronic alcohol use among East Asians (Seitz and Stickel, 2006). Furthermore, more than 35% of East Asians of the Chinese-Han descent, including the Taiwanese, Chinese, Korean, and Japanese, carry an ALDH2 ∗ 2 variant (rs671, A allele), which is ineffective if acetaldehyde is removed (Badger et al., 2003; Chang et al., 2012). Thus far, 560 million of East Asians are estimated to carry the ALDH2 ∗ 2-deficient allele (Brooks et al., 2009).
Fig. 2.
Hypothetical model of mechanism of ethanol toxicity and cardiac injury through different pathways, including more rapid acetaldehyde conversion and accumulation by the activity of fast ADH1B (Step 1), delayed metabolism of toxic acetaldehyde due to reduced enzyme activity due to the variant ALDH2 (Step 2), and reactive oxygen species (ROS) generation (Step 3) by higher CYP2E1 activity due to genetic polymorphisms. CYP2E1 activity can also be induced by chronic alcohol use. Thicker arrow denotes faster metabolism, and thinner arrow denotes slower metabolism; + denotes facilitation of acetaldehyde accumulation. Arrows alone indicate enzyme functions, and arrows with a + mark denote the biological modulation resulting in acetaldehyde formation or accumulation.
Based on this and a previous study, Taiwan probably has the highest prevalence (45–47%) of ALDH2*2 carriers in the general population (Gouillon et al., 2000; Zhang et al., 2013). Consequently, individuals carrying one or two copies of the ALDH2*2 variant alleles have been observed to be more susceptible to alcohol-related toxicity due to acetaldehyde accumulation, which may explain the observed myocardial damage even with moderate but regular ethanol use (Fig. 2, Step 2). Therefore, it is not surprising that combined genetic polymorphisms of the fast metabolizing ADH1B and defective ALDH2*2 variants manifested a synergistic effect in compromised myocardial functions.
CYP2E1 is one of the cytochrome P450 isoenzymes predominantly present in the microsomes or vesicles, which contributes to an alternative and minor oxidative process of alcohol oxidation in the liver (Zakhari, 2006). Polymorphism of CYP2E1 (rs1329149), together with the variant rs671 in ALDH2, has been shown to be associated with a higher risk of colorectal cancer in chronic ethanol users (Yang et al., 2009). A substantial induction of hepatic CYP2E1 has been reported in a chronic alcohol user in a dose-dependent manner (Seitz and Stickel, 2006). Importantly, alcohol metabolism by CYP2E1 not only produces acetaldehyde but also generates reactive oxygen species, such as hydroxyethyl, superoxide anion or hydroxyl radicals, which can be induced by chronic alcohol intake and may further accentuate oxidative tissue injury, lipid peroxidation and greater cancer risks in long-term alcohol users (Gouillon et al., 2000; Badger et al., 2003; Zhang et al., 2013). Therefore, a trend toward a possible synergistic effect between CYE2E1 rs1329149 and ALDH2 rs671 polymorphisms in long-term alcohol users may result in aggravated acetaldehyde or peroxidized toxic metabolites accumulation, which may accentuate the degree of myocardial damage (Wu et al., 2012) (Fig. 2, Step 3).
Overall, our data suggest that habitual alcohol use, even short-term, is associated with LV remodeling and altered myocardial functions. ALDH2 as the major alcohol-metabolizing gene with prevalent polymorphism in the Taiwanese population together with ADH1B or CYP2E1 variants were associated with subclinical myocardial dysfunction in a population with moderate alcohol use, and further showed Gene–Gene synergistic effects on aggravated LV functional decline in frequent alcohol users.
LIMITATIONS
There are several limitations on our study. Firstly, our data is cross-sectional, and may not explain real casual-relationships and the true effect size of genetic variants on myocardial functional alterations or disturbances. Further, direct evidence from serum markers for alcoholism is lacking, though we followed the quantification method well validated from several important publications previously. In particular, the actual dose of alcohol consumption may vary between those with genetic polymorphisms or wild types participants among each genetic trait in the real world, especially those subjects relevant to ALDH2*2 (rs671) centered mutation who tended to have smaller dosage of alcohol use as compared to non-mutants. The tendency of lessened alcohol use in such population by nature to avoid discomfort clinical symptoms may possibly lead to underestimate of the true myocardial harmful effects of such genetic polymorphism population.
CONCLUSION
In conclusion, we demonstrated in a community-dwelling population that alcohol use may be associated with cardiac remodeling together with subclinical myocardial dysfunction assessed by deformation imaging. In addition, these functional abnormalities were related to greater amount of alcohol use. We also observed a synergistic, detrimental effect of ALDH2 polymorphisms with ADH1B or CYP2E1 on myocardial dysfunction, indicating combination of common East Asian-specific ethanol metabolizing genes may aggravate cardiac functional declines (Chien et al., 2016). These findings challenge the notion of the beneficial effects of mild to moderate ethanol use on the heart, and highlight the need for a larger scale prospective study to further evaluate the impact of alcohol on the heart, especially among East Asians.
Supplementary Material
SUPPLEMENTARY MATERIAL
Supplementary data are available at Alcohol and Alcoholism online.
FUNDING
This research supported by Ministry of Science and Technology (Taiwan) (NSC-101-2314-B-195-020, NSC103-2314-B-010-005-MY3, 103-2314-B-195-001-MY3, 101-2314-B-195 -020 –MY1, MOST 103-2314-B-195-006-MY3, NSC102-2314-B-002-046-MY3), and funds from Mackay Memorial Hospital (10271, 10248, 10220, 10253, 10375, 10358, E-102003).
CONFLICT OF INTEREST Statement
None declared.
REFERENCES
- Askanas A, Udoshi M, Sadjadi SA (1980) The heart in chronic alcoholism: a noninvasive study. Am Heart J 99:9–16. [DOI] [PubMed] [Google Scholar]
- Badger TM, Ronis MJ, Seitz HK, et al. (2003) Alcohol metabolism: role in toxicity and carcinogenesis. Alcohol Clin Exp Res 27:336–47. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Enoch MA, Goldman D, et al. (2009) The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med 6:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Chiu YF, Lee IT, et al. (2012) Common ALDH2 genetic variants predict development of hypertension in the SAPPHIRe prospective cohort: gene-environmental interaction with alcohol consumption. BMC Cardiovasc Disord 12:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Hsu KH, Chiou HY, et al. (2015) Associations of lipid-related genetic markers with elevated carotid intima-media thickness in middle-age adults and elders. Int J Cardiol 189:264–6. [DOI] [PubMed] [Google Scholar]
- Chien J, Liu J, Lee MH, et al. (2016) Risk and predictors of hepatocellular carcinoma for chronic hepatitis B patients with newly developed cirrhosis. J Gastroenterol Hepatol 31:1971–77. [DOI] [PubMed] [Google Scholar]
- Choi JY, Abel J, Neuhaus T, et al. (2003) Role of alcohol and genetic polymorphisms of CYP2E1 and ALDH2 in breast cancer development. Pharmacogenetics 13:67–72. [DOI] [PubMed] [Google Scholar]
- Crabb DW, Edenberg HJ, Bosron WF, et al. (1989) Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest 83:314–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Castelnuovo A, Costanzo S, Bagnardi V, et al. (2006) Alcohol dosing and total mortality in men and women: an updated meta-analysis of 34 prospective studies. Arch Intern Med 166:2437–45. [DOI] [PubMed] [Google Scholar]
- Djousse L, Levy D, Benjamin EJ, et al. (2004) Long-term alcohol consumption and the risk of atrial fibrillation in the Framingham Study. Am J Cardiol 93:710–3. [DOI] [PubMed] [Google Scholar]
- Gavazzi A, De Maria R, Parolini M, et al. (2000) Alcohol abuse and dilated cardiomyopathy in men. Am J Cardiol 85:1114–8. [DOI] [PubMed] [Google Scholar]
- Goldberg IJ, Mosca L, Piano MR, et al. (2001) AHA Science Advisory: wine and your heart: a science advisory for healthcare professionals from the Nutrition Committee, Council on Epidemiology and Prevention, and Council on Cardiovascular Nursing of the American Heart Association. Circulation 103:472–5. [DOI] [PubMed] [Google Scholar]
- Goncalves A, Claggett B, Jhund PS, et al. (2015. a) Alcohol consumption and risk of heart failure: the Atherosclerosis Risk in Communities Study. Eur Heart J 36:939–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves A, Jhund PS, Claggett B, et al. (2015. b) Relationship between alcohol consumption and cardiac structure and function in the elderly: the Atherosclerosis Risk In Communities Study. Circ Cardiovasc Imaging 8:e002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouillon Z, Lucas D, Li J, et al. (2000) Inhibition of ethanol-induced liver disease in the intragastric feeding rat model by chlormethiazole. Proc Soc Exp Biol Med 224:302–8. [DOI] [PubMed] [Google Scholar]
- Hajnoczky G, Buzas CJ, Pacher P, et al. (2005) Alcohol and mitochondria in cardiac apoptosis: mechanisms and visualization. Alcohol Clin Exp Res 29:693–701. [DOI] [PubMed] [Google Scholar]
- Hung CL, Wu YJ, Liu CC, et al. (2012) Rationale and design of MAGNET (Mitochondria-AGing in NorthErn Taiwan) study: a community-based cohort investigating mitochondria-related aging and cardiovascular diseases in suburban areas of Northern Taiwan. Int J Gerontol 6:122–26. [Google Scholar]
- Hung CL, Goncalves A, Lai YJ, et al. (2016) Light to moderate habitual alcohol consumption is associated with subclinical ventricular and left atrial mechanical dysfunction in an asymptomatic population: dose-response and propensity analysis. J Am Soc Echocardiogr 29:1043–51 e4. [DOI] [PubMed] [Google Scholar]
- Kishi S, Reis JP, Venkatesh BA, et al. (2015) Race-ethnic and sex differences in left ventricular structure and function: the Coronary Artery Risk Development in Young Adults (CARDIA) Study. J Am Heart Assoc 4:e001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laonigro I, Correale M, Di Biase M, et al. (2009) Alcohol abuse and heart failure. Eur J Heart Fail 11:453–62. [DOI] [PubMed] [Google Scholar]
- Lazarevic AM, Nakatani S, Neskovic AN, et al. (2000) Early changes in left ventricular function in chronic asymptomatic alcoholics: relation to the duration of heavy drinking. J Am Coll Cardiol 35:1599–606. [DOI] [PubMed] [Google Scholar]
- Lee SL, Chau GY, Yao CT, et al. (2006) Functional assessment of human alcohol dehydrogenase family in ethanol metabolism: significance of first-pass metabolism. Alcohol Clin Exp Res 30:1132–42. [DOI] [PubMed] [Google Scholar]
- Mahmoud S, Beauchesne LM, Davis DR, et al. (2007) Acute reversible left ventricular dysfunction secondary to alcohol. Can J Cardiol 23:475–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews EC Jr, Gardin JM, Henry WL, et al. (1981) Echocardiographic abnormalities in chronic alcoholics with and without overt congestive heart failure. Am J Cardiol 47:570–8. [DOI] [PubMed] [Google Scholar]
- Nicolas JM, Antunez E, Thomas AP, et al. (1998) Ethanol acutely decreases calcium transients in cultured human myotubes. Alcohol Clin Exp Res 22:1086–92. [DOI] [PubMed] [Google Scholar]
- Seitz HK, Stickel F (2006) Risk factors and mechanisms of hepatocarcinogenesis with special emphasis on alcohol and oxidative stress. Biol Chem 387:349–60. [DOI] [PubMed] [Google Scholar]
- Shen YC, Fan JH, Edenberg HJ, et al. (1997) Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res 21:1272–7. [PubMed] [Google Scholar]
- Spies CD, Sander M, Stangl K, et al. (2001) Effects of alcohol on the heart. Curr Opin Crit Care 7:337–43. [DOI] [PubMed] [Google Scholar]
- Vary TC, Deiter G (2005) Long-term alcohol administration inhibits synthesis of both myofibrillar and sarcoplasmic proteins in heart. Metabolism 54:212–9. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhou Z, Saari JT, et al. (2005) Alcohol-induced myocardial fibrosis in metallothionein-null mice: prevention by zinc supplementation. Am J Pathol 167:337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JB. (2002) Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet 71:1247–50. author reply 50-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YZ, Yang H, Zhang L, et al. (2012) Application of crossover analysis-logistic regression in the assessment of gene-environmental interactions for colorectal cancer. Asian Pac J Cancer Prev 13:2031–7. [DOI] [PubMed] [Google Scholar]
- Yang H, Zhou Y, Zhou Z, et al. (2009) A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol Biomarkers Prev 18:2522–7. [DOI] [PubMed] [Google Scholar]
- Zakhari S. (2006) Overview: how is alcohol metabolized by the body? Alcohol Res Health 29:245–54. [PMC free article] [PubMed] [Google Scholar]
- Zakhari S, Li TK (2007) Determinants of alcohol use and abuse: impact of quantity and frequency patterns on liver disease. Hepatology 46:2032–9. [DOI] [PubMed] [Google Scholar]
- Zhang RH, Gao JY, Guo HT, et al. (2013) Inhibition of CYP2E1 attenuates chronic alcohol intake-induced myocardial contractile dysfunction and apoptosis. Biochim Biophys Acta 1832:128–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.