Abstract
The ability to antagonize competing strains and species is often important for bacterial fitness in microbial communities. The extent to which intra-species antagonism drives phenotypic diversity of bacterial species is rarely examined in a comprehensive manner at both the genetic and phenotypic levels. Here we show that for nine abundant human gut Bacteroides species examined, there are only a few LPS glycan genetic types. We show that for a given Bacteroides species, there is a predominant lipopolysaccharide (LPS) glycan locus present in the majority of strains. However, other strains have replacements of glycosyltransferase-encoding genes, in most cases, adjacent to a membrane attack/perforin (MACPF) domain-encoding gene not present in the predominant type. We show that the MACPF genes present in LPS glycan biosynthesis loci of four Bacteroides species encode antimicrobial proteins and in Bacteroides vulgatus and Bacteroides dorei, we show the MACPF toxin targets the LPS of strains with the predominant LPS glycan locus. By a combination of gene deletion and replacement, we converted a MACPF toxin-producing strain into a sensitive strain. Genetic diversity of LPS glycan biosynthesis regions in Bacteroides is similar to phage serotype conversion whereby the receptor is altered to render the strain immune to infection/toxicity, and is a rare example in bacteria of toxin immunity conferred to the toxin-producing strain by replacement of genetic material to modify the receptor rather than by a cognate immunity protein.
Introduction
Bacteria have evolved properties that allow them to survive and compete in their ecosystems. The ability of bacteria to directly attack competing organisms provides an important competitive advantage in microbial communities. As the human colon is one of the densest microbial ecosystems on earth and is colonized by numerous closely related strains and species, the production of factors that antagonize competing organisms is likely an important fitness determinant for gut symbionts. We are beginning to identify some of the antagonistic systems/factors produced by the human gut Bacteroidales, the most abundant Gram-negative bacteria of the human gut. These bacteria produce Type VI secretion systems of three different genetic architectures [1]. The genetic architecture 3 (GA3) T6SS loci are present exclusively in Bacteroides fragilis [1] and three different variants of the GA3 T6SSs, encoding different effector and immunity proteins, have been shown to antagonize diverse gut Bacteroidales strains and species [2–4]. Gut Bacteroidales also secrete diffusible molecules that antagonize closely related strains and species. To date, three diffusible antimicrobial proteins of gut Bacteroidales have been identified, each with eukaryotic-like features [5–7].
Bacteroidales-secreted antimicrobial proteins 1 and 2 (BSAP-1 and BSAP-2) both contain membrane attack complex/perforin (MACPF) domains that are present in various eukaryotic molecules including components of the complement system and perforin, which lyse target cells by pore formation. BSAP-1 is produced by a subset of B. fragilis strains [5] and targets a β-barrel outer membrane protein (OMP) of sensitive strains, whereas BSAP-2 is produced by a subset of B. uniformis strains and targets the short O-antigen of lipopolysaccharide (LPS) of sensitive strains [6]. Binding to these molecules places the MACPF toxin at the cell surface where they likely oligomerize to create membrane pores. We previously showed that these BSAP-encoding genes were acquired along with gene(s) encoding an ortholog of their target in sensitive cells and integrated into the respective genomes so that the BSAP target gene(s) are replaced [6]. In the case of BSAP-1, the MACPF gene was acquired with an adjacent gene encoding an orthologous β-barrel OMP rendering the strain resistant to BSAP-1. For BSAP-2, the gene was acquired with a new set of glycosyltransferase (GT)-encoding genes, replacing genes of the predominant LPS glycan locus, thereby altering the glycan and rendering the strain resistant to the toxin [6]. Using a gnotobiotic mouse colonization model, we showed that the targets of both BSAP-1 and BSAP-2 are essential for competitive colonization of the mammalian gut, accounting for the replacement rather than deletion of the target genes with the incoming MACPF gene [6].
The LPS of Gram-negative bacteria is comprised of three major components: lipid A, core glycan, and O-antigen. The O-antigen component of the LPS of most bacteria comprises different numbers of identical multi-sugar subunits that are added to the core glycan giving a laddering appearance when analyzed by gel electrophoresis. In contrast, Bacteroides species generally synthesize relatively short O-antigens comprised of only one or a few repeat units with LPS sizes < 10 kDa, with the major LPS forms in the 2.5–5 kDa range [8]. This published study also showed that among the Bacteroides, B. vulgatus tends to have the most O-antigen repeat additions displaying a laddering appearance; however, the majority of the LPS of B. vulgatus was shown to be a low molecular weight (MW) molecule.
BSAP-1 and BSAP-2 are the first bacterially produced MACPF domain proteins shown to kill other bacteria. We showed that genes encoding proteins with MACPF domains are rarely encoded in bacterial genomes, with the exception of members of the phylum Bacteroidetes, whose collective genomes contain hundreds of such genes [5]. In an attempt to identify other gut Bacteroidales MACPF proteins with antibacterial toxin activity, we previously tested five additional MACPF proteins for their ability to target strains of the same species [5]. These analyses did not demonstrate toxin activity for any of these proteins under the conditions of our assay and for the strains tested for sensitivity [5]. In this study, we sought to identify additional MACPF domain proteins of gut Bacteroidales with antimicrobial activity using gene neighboring analyses. Using this comprehensive approach, we found that nearly all Bacteroides species analyzed have a representative strain with a MACPF gene located in their predicted LPS glycan biosynthesis locus, surrounded by distinct GT gene(s) compared with other strains of the species. We show that a MACPF protein encoded by a subset of B. vulgatus and B. dorei strains is a toxin that targets the LPS glycan of sensitive strains providing an ecological rationale for the LPS antigenic switch. These data show that acquisition of MACPF-encoding genes drives LPS diversity in the gut Bacteroides.
Materials and methods
All primers used in this study are shown in Table S5.
Identification of MACPF domain proteins encoded in human gut Bacteroidales genomes
The 2813 genomes identified in NCBI’s taxonomy database [9] as of 9 February 2018 as belonging to the division “CFB group bacteria” and that provided gene translation data were included. The profile hidden Markov model (HMM) of the MAC/Perforin (MACPF) domain (Pfam v. 31 accession PF01823.18 [10]) was used (along with hmmsearch v. 3.1b2, http://hmmer.org) to search all 8,637,350 proteins encoded by these genomes for matches having a full sequence bit score that equaled or exceeded the gathering threshold cutoff (21.50) set for the model. The initial set recovered (536 proteins from 365 genomes) under these criteria was reduced by retaining only MACPF proteins derived from Bacteroidetes genomes of human gut origin.
If a sequenced isolate was not identified to the species level, its species was predicted based on 16S rRNA gene sequence(s) included with the genome submission (by searching NCBI’s 16S ribosomal database using targeted loci BLAST and/or the Ribosome Database Project’s sequence match facility [11] RDP release 11), or by using the BBSketch tools included in BBMap (v. 37.90, https://sourceforge.net/projects/bbmap) to calculate the average nucleotide identity of the genome in question to genome sketches maintained at the Joint Genome Institute’s (https://jgi.doe.gov) non-redundant sketch server (https://nt-sketch.jgi-psf.org/sketch). MACPF proteins detected in genome sequences that could not be identified to the species level or that were detected in species determined not to be of human gut origin were excluded. Genomes where species-level identification was deduced by either or both of these methods are shown in parenthesis in the supplemental tables.
The 282 MACPF domain containing proteins from human gut Bacteroidales clustered into 68 groups using the blastclust program (v. 2.2.2 [12]) at the 95% amino-acid identity level over 85% of their length. A member of each cluster was selected as a representative, and these 68 protein sequences were aligned using Muscle [13]. The multiple alignment was analyzed using the Maximum Likelihood method based on the LG model [14] using MEGA X [15] to create the bootstrap consensus tree ([16] Fig. S2) based on 500 replicates. Branches reproduced in < 50% of the bootstrap replicates are collapsed, and the percentage of replicate trees in which the remaining bootstrap branches were reproduced are shown. The initial tree was produced using the Neighbor-Join and BioNJ algorithms with a pairwise distance matrix estimated using a JTT model, and the topology with the best log likelihood value was selected. Rate differences among sites were modeled using a discrete gamma distribution (five categories (+G, parameter = 2.6640). All positions with < 95% site coverage were eliminated.
Gene neighborhood analysis
Up to 12 genes flanking both sides of each of the 68 MACPF cluster representative genes were retrieved and their protein sequences were analyzed using hmmsearch for matches where the full sequence bit score met or exceeded the gathering threshold cutoff indicated in the profile HMM models contained in v. 31 of the Pfam database [10]. The best scoring Pfam (lowest full sequence e value) was retained. Each protein sequence retrieved was also scanned for the presence of a signal sequence using LipoP version 1.0a [17] and for transmembrane helices using TMHMM version 2.0c [18]. The results of these analyses, including the direction of transcription and the size of the protein in amino acids, is shown in Table S2.
Bacterial strains and growth conditions
Bacteroides stains used experimentally in this study are shown in Table S3. Bacteroides strains were grown in supplemented basal medium [19] or on supplemented brain heart infusion plates. Erythromycin (5 µg/ml) and gentamicin (200 µg/ml) were added where appropriate. Escherichia coli strains were grown in L broth or on L agar plates with antibiotics (carbenicillin 100 µg/ml, trimethoprim 100 µg/ml, kanamycin 50 µg/ml) added where appropriate.
Agar spot assay
Analysis of the ability of one strain to inhibit the growth of another by the secretion of diffusible inhibitory molecules was assayed using the agar spot test [20] modified as described in ref. [6].
Creation of deletion mutants
Internal non-polar deletion mutants were constructed by amplifying DNA upstream and downstream of the gene or region to be deleted using the primers listed in Table S5. PCR products were digested and cloned by three-way ligation into pNJR6 [21]. The resulting plasmids were conjugally transferred into Bacteroides spp. using helper plasmid R751and cointegrates were selected on erythromycin and gentamicin plates. Erythromycin-sensitive double cross outs were screened by PCR for the mutant genotype.
Cloning and heterologous expression of genes intrans
Genes expressed in trans were PCR amplified and cloned into expression vector pFD340 [22]. Resulting plasmids were verified by sequencing and transferred to Bacteroides strains by conjugation using helper plasmid RK231.
Western immunoblot blot analyses and silver staining
Antiserum to whole-cell B. vulgatus CL10T00C06 was prepared in rabbits by Lampire Biologicals (Pipersville, PA) using the EXPRESS-LINE polyclonal antiserum protocol. To create an antibody fraction specific to the molecule lost in B. vulgatus CL10T00C06Δ2GT mutant, we performed an antibody adsorption [23] to remove antibodies to surface molecules common to both the wild-type (WT) and deletion mutant strains, leaving only antibodies to surface molecules lost in the mutant. For polyacrylamide gel electrophoresis analysis, bacteria or purified LPS was boiled in lithium dodecyl sulfate sample buffer and subjected to electrophoresis using NuPAGE 12% or 4–12% Bis-Tris polyacrylamide gels with 2-(N-morpholino)ethanesulfonic buffer (Life Technologies). In some cases, the gel was silver stained (Peirce Silver Stain Kit, Thermo Scientific). For western immunoblot analysis, the contents of the gels were transferred to polyvinylidene difluoride membranes and probed with the adsorbed serum. Alkaline phosphatase-labeled anti-rabbit IgG (Pierce) was the secondary antibody, and the membranes were developed with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (KPL, Gaithersburg, MD).
LPS extraction
LPS was purified by a modified aqueous-phenol extraction as described [24]. Cultures were grown to an OD600 = 0.5 and 1.5 ml was centrifuged and resuspended in 200 µl of buffer containing 2% β-mercaptoethanol, 2% sodium dodecyl sulphate (SDS), 10% glycerol, bromophenol blue and 0.1 M Tris-HCl pH 6.8. To lyse the cells, samples were boiled for 15 minutes and incubated overnight at 37°C with 10 µl of 10 mg/ml Proteinase K (Promega Corp.). To extract LPS, 200 µl of Tris-saturated phenol was added and samples were incubated at 65°C for 15 minutes. 1 ml of diethyl ether was added and the samples were vortexed then centrifuged. After centrifugation, the bottom layer was retained, and this extraction was repeated three times. The final extraction volume was doubled using a buffer containing 4% β-mercaptoethanol, 4% SDS, 20% glycerol, bromophenol blue and 0.1 M Tris-HCl pH 6.8.
Genome sequence of B. vulgatus CL10T00C06
The genome sequence of B. vulgatus CL10T00C06 from DNA isolation and library construction to assembly and annotation proceeded by the same pipeline as recently described [7]. This whole-genome shotgun project was deposited in DDBJ/ENA/GenBank under BioProject ID PRJNA400641 and under BioSample ID SAMN07572298.
Results
Gene neighboring analyses of MACPF domain-encoding genes of human gut Bacteroidales
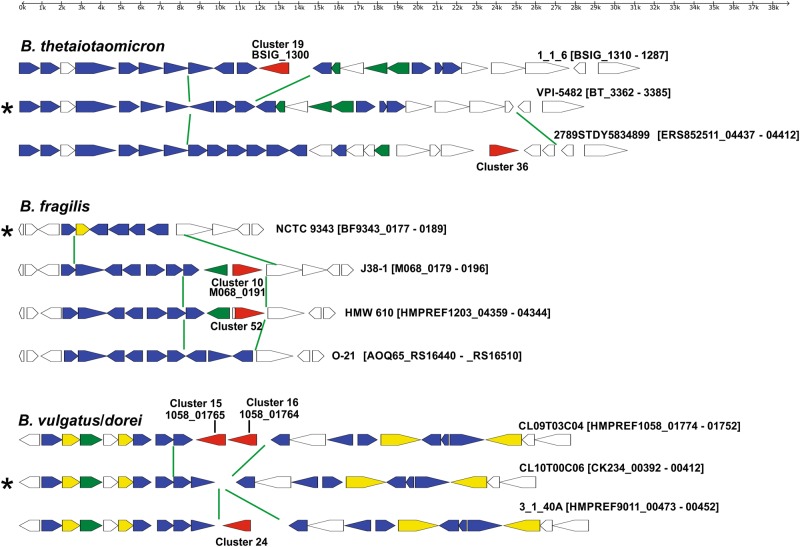
To identify potential antimicrobial toxins among the large number of MACPF domain proteins of human gut Bacteroidales, we took advantage of the fact that BSAP-1- and BSAP-2-encoding genes are in the same locus in producing strains as are their target genes in sensitive strains. We therefore searched for a pattern of replacement of OMP or GT-encoding genes adjacent to MACPF genes. The MACPF domain proteins of human gut Bacteroidales species group into 68 clusters at ≥ 95% identity, 85% length (Table S1). In most cases, MACPF proteins clustered strictly by species with the exception of B. ovatus/B. xylanisolvens and B. vulgatus/B. dorei, both sets of species that are very similar to each other [25]. In addition, the cluster 4 MACPF protein is encoded by the genomes of 12 different gut Bacteroidales species (Table S1). We previously showed that this MACPF gene was transferred to diverse Bacteroidales species on an integrative conjugative element [26]. Of the 68 clusters, the cluster 1 MACPF protein of B. fragilis is BSAP-1 [5] and the cluster 14 MACPF of B. uniformis is BSAP-2 [6]. A representative member of each of the 68 clusters was chosen (corresponding to the first protein listed in Table S1) and we analyzed the genetic region surrounding these MACPF genes. DNA comprising 12 genes on each side of the MACPF-encoding gene was retrieved (where available) and motifs were identified, as were predicted signal sequences (cleaved by SpI or SpII), and predicted transmembrane domains (Table S2). These analyses revealed that many of these MACPF genes are present in the vicinity of genes encoding putative GTs (19 of 68 clusters) or genes encoding fimbriae components Mfa-1 or FimA (10 of 68). We performed a comprehensive analysis of the regions containing the 19 MACPF genes in the vicinity of GT-encoding genes and analyzed these corresponding regions from all sequenced strains of these species including those without the corresponding MACPF gene. The genetic composition of these regions suggests that the genes are not involved in capsular polysaccharide biosynthesis [27, 28] or protein glycan synthesis [29, 30] and therefore are likely involved in LPS glycan (core glycan or O-antigen) synthesis. As further support of the involvement of these regions in LPS glycan synthesis, most of these regions contain additional genes involved in LPS synthesis (Fig. 1, S1). In addition, the B. uniformis ATCC 8492 and CL03T00C23 regions and the B. thetaiotaomicron VPI 5482 region were previously shown to encode O-antigens [6, 31]. We also found that these putative LPS glycan biosynthesis loci are present in the same genetic regions of Bacteroides chromosomes flanked by conserved genes often including a gene encoding a nitroreductase on one side and a gene encoding a protein of the PP-binding superfamily on the other side. Intra-species comparisons revealed that each Bacteroides species that we identified as having a MACPF gene in the LPS glycan genetic region has between two and four LPS glycan genetic types. Where genome sequences of numerous strains of a given Bacteroides species were available, there was typically one LPS glycan genetic type that predominated, which in most cases does not contain a MACPF-encoding gene (Fig. 1, S1).
Fig. 1.
Maps of LPS glycan genetic regions of four Bacteroides species. Green lines delineate the area of divergence between LPS glycan genetic types within a species. MACPF genes are shown in red, putative glycosyltransferase-encoding genes are shown in blue, genes encoding other products involved in glycan synthesis are colored green and other putative lipid A/core glycan synthesis genes are shown in yellow. Asterisks indicate the predominant LPS glycan genetic type of a species
Identification of toxin activity of three MACPF proteins
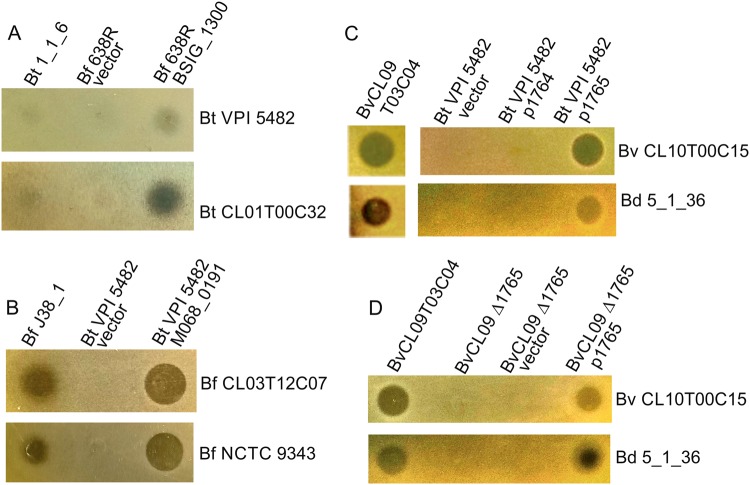
We predicted that some of the MACPF proteins encoded in these regions are toxins that target the LPS of species-matched strains without the MACPF gene. The replacement of GT-encoding genes would allow the producing strain to synthesize a distinct LPS glycan so as not to self-intoxicate. To determine whether we could detect toxin activity by MACPF proteins encoded in these regions, we selected three to test for activity based on the availability of both the MACPF producing and predicted sensitive strains in our collection (B. thetaiotaomicron 1_1_6 (BSIG_1300, cluster 19), B. fragilis J38-1 (M068_0191, cluster 10) and B. vulgatus CL09T03C04 adjacent MACPF genes (HMPREF1058_01765 and HMPREF1058_01764, clusters 16 and 17, respectively) Table S1 and Fig. 1. To determine whether any detected toxin activity was due to the respective MACPF proteins, we also cloned each of the four MACPF genes individually into a Bacteroides expression vector for constitutive expression and placed them in a heterologous Bacteroides species to assay for acquisition of a secreted toxin phenotype. Little secreted antimicrobial activity was detected from B. thetaiotaomicron 1_1_6 against two strains with the predominant LPS genetic type under the conditions of our assay, but inhibitory zones were detected when the gene was constitutively expressed in B. fragilis 638 R (Fig. 2a). The MACPF gene present in the B. fragilis J38-1 LPS glycan region encodes potent antimicrobial activity against two B. fragilis strains with the predominant LPS glycan genetic locus (Fig. 2b). B. vulgatus and B. dorei strains share the same putative LPS glycan genetic loci (Fig. 1). Of the two adjacent MACPF genes of B. vulgatus CL09T03C04 (BvCL09), HMPREF1058_01765 (1765) but not HMPREF1058_01764 (1764) antagonized both B. vulgatus and B. dorei strains with the predominant LPS genetic type (Fig. 2c). We broadened our analysis to include additional B. fragilis and B. vulgauts/dorei strains, checking for sensitivity to the toxin produced by the cluster 10 or 16 MACPF proteins, respectively. As shown in Table S3, all B. fragilis strains with the predominant, non-MACPF, LPS glycan locus were sensitive to the cluster 10 MACPF (M068_0191), and similarly, all B. vulgatus/dorei strains with the predominant LPS glycan genetic region were sensitive to the cluster 16 MACPF toxin (HMPREF1058_01765). The two resistant B. fragilis strains both had the same LPS glycan genetic regions containing the gene encoding the cluster 10 MACPF toxin and the two resistant B. vulgauts/dorei strains similarly has the same LPS glycan genetic regions containing the gene encoding the cluster 16 MACPF toxin. We also tested the ability of these toxins to target other Bacteroides species and found no inter-strain killing as expected owing to their distinct LPS glycan genetic regions.
Fig. 2.
Agar spot assays to assess antimicrobial activity of MACPF domain proteins. For each panel, the strains listed at the top are those dotted and tested for toxin activity and the strains listed along the side are those tested for sensitivity. a Assays of WT B. thetaiotaomicron (Bt) 1_1_6 and B. fragilis (Bf) 638R with vector alone or expressing the MACPF gene BSIG_1300 overlaid with two Bt strains without the MACPF gene. b Analysis of WT Bf J38-1 and Bt VPI 5482 with vector alone or expressing the MACPF gene M068_0191 overlaid with two Bf strains without the MACPF gene. c Analysis of WT B. vulgatus (Bv) CL09T03C04 or Bt VPI 5482 with vector alone or constitutively expressing each MACPF gene individually when overlaid with a Bv and B. dorei (Bd) strain of the predominant LPS genetic type. d Agar spot assay showing zones of inhibition of sensitive strains from WT, ∆1765 and the deletion mutant with empty vector or the gene restored in trans
As the cluster 16 MACPF toxin of B. vulgatus/dorei had potent activity, and the LPS glycan genetic region of these species had not been studied, we chose to further analyze the MACPF toxin and LPS genetic regions in B. vulgatus/dorei. An internal, non-polar deletion mutant of 1765 rendered B. vulgatus CL09 unable to inhibit the growth of sensitive B. vulgatus and B. dorei strains, and placement of the 1765 gene in trans restored activity (Fig. 2d). Based on these analyses, we named the cluster 16 MACPF protein of B. vulgatus and B. dorei, BSAP-3. A MACPF protein cladogram (from human gut Bacteroidales strains) based on global similarity is shown in Fig S2 with confirmed antimicrobial toxins highlighted in red.
BSAP-3 targets the LPS of sensitive strains
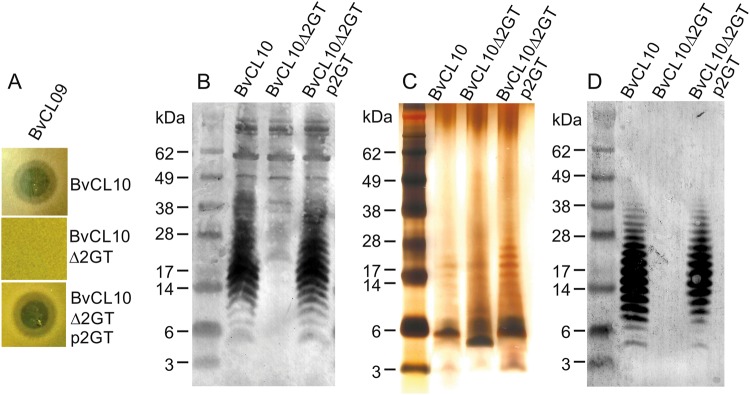
We predicted that BSAP-3 targets the LPS of sensitive B. vulgatus/B. dorei strains, and that replacement of GT genes along with acquisition of the BSAP-3-encoding gene results in a structurally distinct LPS glycan that does not serve as a target. A deletion mutant in sensitive strain B. vulgatus CL10T00C06 (BvCL10) of genes CK234_00400 - CK234_00401, the two GT-encoding genes unique to the sensitive strains (Fig. 1, Table S4) rendered the strain resistant to killing by BSAP-3 and sensitivity was restored to this mutant when these two genes were returned in trans (Fig. 3a). To determine whether this target molecule is a component of the LPS, we raised antiserum to the WT BvCL10 strain and adsorbed it using the CK234_00400-1 deletion, leaving only antibodies to surface molecules absent in the mutant strain. Western immunoblot analysis of whole-cell lysates showed that this adsorbed antiserum reacted with a laddering LPS of the WT BvCL10 strain not present in the deletion mutant (Fig. 3b). To confirm that these immunoreactive molecules are LPS, we purified LPS from the WT, deletion mutant, and complemented strain and analyzed by silver stain and western immunoblot (Fig. 3c, d). These analyses confirmed that the mutant was deficient in a laddering LPS that was restored to the mutant with replacement of CK234_00400-1 (p2GT) (Fig. 3d). The silver strain revealed that the majority of the LPS of the WT strain is not modified with extensive O-antigen repeats, rather most of the LPS likely contains only a single O-antigen repeat with a total apparent MW of ~ 5.5 kDa (Fig. 3c). The deletion of the two GT-encoding genes (∆2GT) resulted in an even lower MW form of the LPS that does not react with the adsorbed antiserum (Fig. 3b, c).
Fig. 3.
BSAP-3 targets the LPS of sensitive strains. a Agar spot assays showing the sensitivities of WT B. vulgatus CL10, a GT deletion mutant of CL10 and the mutants with the deleted genes restored in trans. (Δ2GT denotes deletion of the two glycosytransferase unique to sensitive B. vulgatus and B. dorei strains, Fig. 1). b Western immunoblot of whole-cell lysates probed with antiserum raised to WT BvCL10T00C06 adsorbed with the ∆2GT mutant. c Silver stained SDS-PAGE of purified LPS preparations. d Western immunoblot of purified LPS probed with the same adsorbed serum as (b)
Gene replacement converts a BSAP-3-producing strain to a BSAP-3 sensitive strain
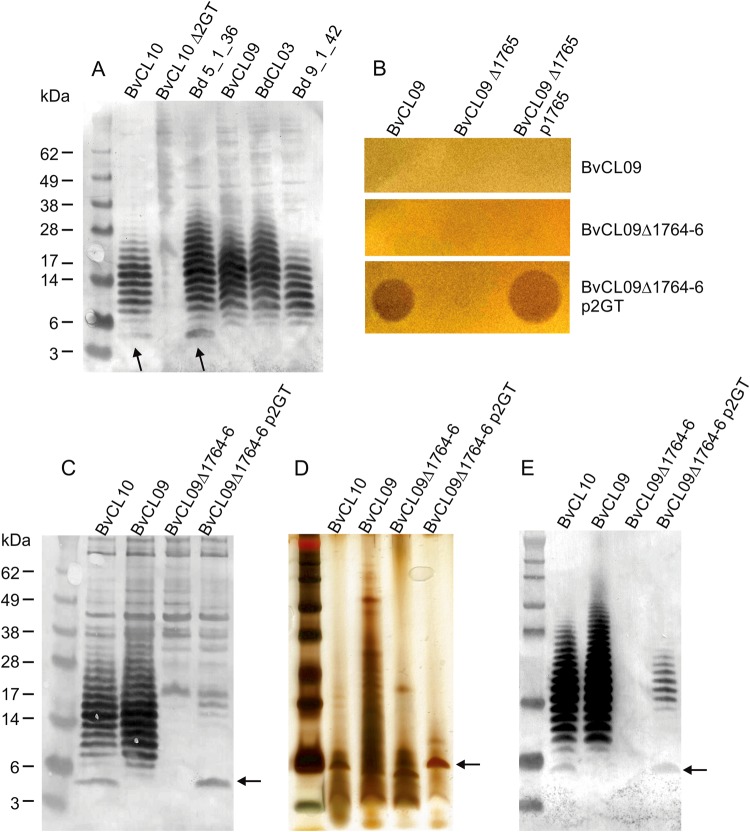
We predicted that the BSAP-3 encoding strains synthesize a structurally distinct LPS glycan. Western immunoblot analysis of whole-cell lysates showed that BSAP-3-encoding strains synthesized an O-antigen that is at least immunologically similar to that of BvCL10 as it is recognized by the antiserum to the LPS of strain BvCL10 (Fig. 4a). However, the predominant ~ 5.5 kDa LPS form of BvCL10, that is also present in BSAP-3 sensitive strain Bd 5_1_36, is absent in BSAP-3-encoding strains. This LPS form is also not present when the two GT-encoding genes of BvCL10 are deleted (Fig. 4a).
Fig. 4.
A small LPS glycoform correlates with BSAP-3 sensitivity. a Western immunoblot of whole-cell lysates of WT strains probed the adsorbed antiserum of Fig. 3. The arrow indicates a low MW LPS form only present in sensitive strains. b Overlay assays showing the conversion of a BSAP-3-producing strain to a sensitive strain by deletion of the three unique genes in the BvCL09 LPS region (1764-66) and addition of the two glycosyltransferase genes from BvCL10. c, d, e. Silver strain or western immunblot analyses of c, cell lysates or d, e, purified LPS preparations showing the shift in the low MW LPS form when the two BvCL10 GT genes are added to the BvCL09 Δ1764-66 mutant
As we showed that the two GT of BvCL10 are required for BSAP-3 sensitivity, deletion of these genes during acquisition of the BSAP-3 gene would have rendered BSAP-3 encoding strains resistant to the toxin. We sought to determine whether we could confer BSAP-3 sensitivity to BvCL09 by introduction of these two GT-encoding genes. We deleted HMPREF1058_1764-1766, the three genes unique to the BSAP-3 strains. Deletion of the three genes rendered BvCL09 unable to synthesize its O-antigen (Fig. 4c, e) demonstrating that the unique GT gene of BSAP-3 strains is required for O-antigen synthesis or its addition to the LPS. To this BvCL09 deletion mutant, we added the two GT genes of BvCL10 (CK234_00400-401). Addition of these genes rendered the BvCL09 recombinant strain sensitive to BSAP-3 (Fig. 2b). The major change in this strain was the acquisition of the abundant ~ 5.5 kDa LPS form unique to sensitive strains (Fig. 4c, d, e).
Discussion
In contrast to the loci encoding the multiple capsular polysaccharides of Bacteroides species [27, 28], few LPS glycan (core or O-antigen) biosynthesis regions have been identified in Bacteroides. Patrick et al. [32] studied the LPS of B. fragilis, and LPS glycan biosynthesis regions have been identified in B. uniformis [6] and B. thetaiotaomicron [31]. In this study, we identify putative LPS glycan biosynthesis loci of numerous other Bacteroides species and show that they are in a conserved region of the Bacteroides genome flanked by the same conserved genes in each species. A recent study also predicted the LPS or LOS glycan biosynthesis loci of one strain of several Bacteroides species [33], most of which are in agreement with the predominant LPS glycan genetic regions identified here.
We show that human gut Bacteroides have a MACPF-encoding gene in the LPS glycan biosynthesis locus of some strains of nine of the predominant gut species analyzed. This acquisition is accompanied by a switch to a new LPS glycan genetic variant of the species. The acquisition of MACPF domain-encoding genes likely co-occurred with the acquisition and replacement of distinct GT-encoding genes, resulting in an altered LPS glycan structure. We show here that in B. vulgatus/dorei, and previously showed in B. uniformis [6] that the MACPF genes in these regions target the predominant LPS glycan molecule of these species, providing a selective pressure for the glycan switch. Many Gram-negative bacteria such as E. coli, Salmonella enterica, and Vibrio cholerae have extensive diversity in their O-antigen locus, leading in some species to > 100 O-serotypes. The selective pressures contributing to the diversification of O-antigens differs for each species. For S. enterica and other pathogens, the host immune response is predicted to select new variants [34] as the O-antigen of LPS is typically highly immunogenic. The O-antigen of Salmonella is also under diversifying selection due to protozoan predation as different O-antigens allow for prey discrimination [35]. One of the best characterized factors leading to intra-species O-antigen diversity is that of serotype conversion mediated by temperate bacteriophage. Serotype conversion has been well studied in Shigella flexneri, a species in which most serotypes have a common O-antigen tetrasaccharide repeat backbone that is modified by the products of phage genes (reviewed in ref. [36]). These include genes encoding glycosyltransferases, acetyltransferases, and other products that alter the glycan sufficiently so that it is recognized differently by the host immune system and no longer serves as a receptor for the phage. LPS glycan diversity mediated by acquisition of new MACPF- and GT-encoding genes is most analogous to serotype conversion. In both cases, the glycan target is modified by products encoded by the incoming DNA so that the recipient strain is not sensitive to the toxin or phage.
The fact that each Bacteroides species analyzed typically has only a few LPS glycan genetic variants suggests that MACPF gene acquisition is the main driver of O-glycan diversity in these species, rather than host immunity. In fact, Peterson et al. [31] showed that an antibody to the O-antigen did not lead to a reduction in the numbers of B. thetaiotaomicron in a mouse colonization model, further supporting that diversifying selection of Bacteroides LPS glycans is not due to host immunity. The role of O-antigen in gut Bacteroides and the factors that govern selection for short O-antigens with only a few repeat units are unclear. In both B. uniformis and B. thetaiotaomicron, the O-antigen was shown to be important for gut colonization [6, 31]. In both studies, mutants with deletions or insertions affecting genes in their respective O-antigen loci were severely outcompeted by the isogenic wild-type strain in a gnotobiotic mouse colonization model. The importance of O-antigen for gut colonization is consistent with our finding that in each Bacteroides LPS glycan region, GT-encoding genes are replaced—rather than lost—along with acquisition of the incoming MACPF gene.
The antimicrobial protein Colicin N is a pore-forming toxin that binds the core glycan of E. coli LPS [37]. Unlike the MACPF toxins of Bacteroides, the Colicin N gene is adjacent to a gene encoding an immunity protein [38], and therefore, resistance of the producing strain does not require LPS core glycan diversity. In fact, most described bacterially produced toxins with bacterial targets, whether bacteriocins or toxic effectors secreted by Type VI secretion systems, have cognate immunity proteins. MACPF toxins are a rare example of immunity acquired by replacement of target genes in producing strains.
We do not predict that all of the remaining 63 distinct MACPF domain proteins of gut Bacteroidales have toxin activity. Like the cluster 17 MACPF analyzed in this study, we have not been able to demonstrate toxin activity for proteins of several other MACPF clusters [5, 6]. We do predict, however, that there are likely several other toxins within the remaining MACPF protein repertoire. It is likely that acquisition of MACPF genes encoding antibacterial toxins also contributes to the diversification of other surface molecules of Bacteroides species that serve as targets. Consistent with this prediction, we previously showed that a MACPF toxin produced by B. fragilis [5] targets an OMP important for gut colonization and that the MACPF gene is adjacent to a gene encoding an orthologous OMP that renders the producing strain resistant [6]. Therefore, the acquisition of MACPF-encoding genes likely impacts the diversity of other surface molecules important for gut colonization in human gut Bacteroides species.
Electronic supplementary material
Acknowledgements
This work was supported by Public Health Service grant R01AI093771 from the NIH/National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. B. (thetaiotaomicron) 1_1_6, B. (dorei) 9_1_42FAA, B. (dorei) 5_1_36/D4, and B. fragilis 2_1_16, were obtained through BEI Resources as part of the Human Microbiome Project. We thank Kevin Roelofs and Leonor GarciaBayona for helpful discussions.
Compliance with ethical standards
Conflict of interest
The authors declare no conflicts of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0244-4) contains supplementary material, which is available to authorized users.
References
- 1.Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17:1–21. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubek Wadenburg J. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17:1281–91. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wexler AG, Bao Y, Whitney JC, Bobay LM, XAvier JB, Schofield WB, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–44. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–32. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol Microbiol. 2014;94:1361–74. doi: 10.1111/mmi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. Mbio. 2016;7: e01055-16. [DOI] [PMC free article] [PubMed]
- 7.Chatzidaki-Livanis, M., Coyne, M. J., Roelofs, K. G., Gentyala, R. R., Caldwell, J. M. & Comstock, L. E. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. Mbio. 2017;8:e01902-17 . [DOI] [PMC free article] [PubMed]
- 8.Maskell JP. Electrophoretic analysis of the lipopolysaccharides of Bacteroides spp. Antonie Van Leeuwenhoek. 1994;65:155–61. doi: 10.1007/BF00871756. [DOI] [PubMed] [Google Scholar]
- 9.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Punta M, Cogill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole JR, Wang Q, Fish JA, Chai B, Mcgarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye J, Mcginnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–9. doi: 10.1093/nar/gkl164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–20. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Juncker AS, Willenbrook H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–62. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnhammer EL, VON Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- 19.Pantosti A, Tzianobos AO, Onderdonk AB, Kasper DL. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun. 1991;59:2075–82. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avelar KE, Pinto LJ, Antunes LC, Lobo LA, Bastos MC, Domingues R, et al. Production of bacteriocin by Bacteriodes fragilis and partial characterization. Lett Appl Microbiol. 1999;29:264–8. doi: 10.1046/j.1365-2672.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- 21.Stevens AM, Shoemaker NB, Salyers AA. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol. 1990;172:4271–9. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CJ, Rogers MB, Mckee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–54. doi: 10.1016/0147-619X(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 23.Comstock LE, Coyne MJ, Tzianabos AO, Pantosti A, Onderdonk AB, Kasper DL. Analysis of a capsular polysaccharide biosynthesis locus of Bacteroides fragilis. Infect Immun. 1999;67:3525–32. doi: 10.1128/iai.67.7.3525-3532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis, M. R., Jr. & Goldberg, J. B. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J Vis Exp. 2012;28:pii: 3916 [DOI] [PMC free article] [PubMed]
- 25.Pedersen RM, Marmolin ES, Justesen US. Species differentiation of Bacteroides dorei from Bacteroides vulgatus and Bacteroides ovatus from Bacteroides xylanisolvens - back to basics. Anaerobe. 2013;24:1–3. doi: 10.1016/j.anaerobe.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 26.Coyne, M., Zitomersky, N., Mcguire, A., Earl, A. & Comstock, L. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio. 2014;5:e01305-14. [DOI] [PMC free article] [PubMed]
- 27.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–8. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 28.Coyne MJ, Comstock LE. Niche-specific features of the intestinal. Bacteroidales J Bacteriol. 2008;190:736–42. doi: 10.1128/JB.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coyne MJ, Fletcher CM, Chatzidaki-Livanis M, Posch G, Schaffer C, Comstock LE. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol Microbiol. 2013;88:772–83. doi: 10.1111/mmi.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–31. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson DA, Planer JD, Guruge JL, Xue L, Downey-virgin W, Goodman AL, et al. Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem. 2015;290:12630–49. doi: 10.1074/jbc.M114.633800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick S, Houston S, Thacker Z, Blakely GW. Mutational analysis of genes implicated in LPS and capsular polysaccharide biosynthesis in the opportunistic pathogen Bacteroides fragilis. Microbiology. 2009;155:1039–49. doi: 10.1099/mic.0.025361-0. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson AN, Choudhary BP, Fischbach MA. The biosynthesis of lipooligosaccharide from Bacteroides thetaiotaomicron. mBio 2018;9. [DOI] [PMC free article] [PubMed]
- 34.Wang L, Romana LK, Reeves PR. Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992;130:429–43. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wildschutte H, Wolfe DM, Tamewitz A, Lawrence JG. Protozoan predation, diversifying selection, and the evolution of antigenic diversity in Salmonella. Proc Natl Acad Sci USA. 2004;101:10644–9. doi: 10.1073/pnas.0404028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/S0966-842X(99)01646-7. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CL, Ridley H, Marchetti R, Silipo A, Griffin DC, Crawford L, et al. The antibacterial toxin colicin N binds to the inner core of lipopolysaccharide and close to its translocator protein. Mol Microbiol. 2014;92:440–52. doi: 10.1111/mmi.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugsley AP. The immunity and lysis genes of ColN plasmid pCHAP4. Mol Gen Genet. 1988;211:335–41. doi: 10.1007/BF00330613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.