Abstract
Bovine mammary epithelial cells (MAC-Ts) are a common cell line for the study of mammary epithelial inflammation; these cells are used to mechanistically elucidate molecular underpinnings that contribute to bovine mastitis. Bovine mastitis is the most prevalent form of disease in dairy cattle that culminates in annual losses of 2 billion dollars for the U.S. dairy industry. Thus, there is an urgent need for improved therapeutic strategies. Histone deacetylase (HDAC) inhibitors are efficacious in rodent models of inflammation, yet their role in bovine mammary cells remain unclear. HDACs have traditionally been studied in the regulation of nucleosomal DNA, in which deacetylation of histones impacts chromatin accessibility and gene expression. Using MAC-T cells stimulated with tumor necrosis factor α (TNFα) as a model for mammary cell inflammation, we report that inhibition of HDACs 1 and 2 (HDAC1/2) attenuated TNFα-mediated inflammatory gene expression. Of note, we report that HDAC1/2-mediated inflammatory gene expression was partly regulated by c-Jun N terminal kinase (JNK) and extracellular signal-regulated kinase (ERK) phosphorylation. Here, we report that HDAC1/2 inhibition attenuated JNK and ERK activation and thus inflammatory gene expression. These data suggest that HDACs 1 and 2 regulate inflammatory gene expression via canonical (i.e. gene expression) and non-canonical (e.g. signaling-dependent) mechanisms. While further studies using primary cell lines and animal models are needed, our combined data suggest that HDAC1/2-specific inhibitors may prove efficacious for the treatment of bovine mastitis.
Keywords: HDAC inhibitors, HDACs, histone deacetylases, phosphorylation, bovine inflammation, bovine mammary epithelial cells, MAC-T
Introduction
Mastitis, inflammation of the mammary gland and udder tissue, is the most prevalent form of disease in dairy cattle that contributes to significant monetary loss for the U.S. dairy industry with annual estimates of 2 billion dollars (Akers and Nickerson, 2011; Erskine et al., 2003; Hogeveen et al., 2011). Mastitis is an inflammatory reaction characterized by tissue damage to the milk-producing cells in the udder; this is typically initiated by bacterial infection, the most common pathogen in mastitis is Escherichia coli (E. coli). Bovine mammary epithelial cells are milk producing cells that represent the first line of defense against infection (Rinaldi et al., 2010; Strandberg et al., 2005). Following infection, fibroblasts within the mammary stroma secrete tumor necrosis factor α (TNFα), which stimulates mammary epithelial cells to release chemoattractant proteins that recruit neutrophils and macrophages to the alveoli to amplify inflammation (Chen et al., 2016; Rinaldi et al., 2010; Zhao and Lacasse, 2008).
Inflammatory effects of TNFα within mammary epithelial cells are mediated through mitogen-activated protein kinase (MAPK) signaling pathways that involve c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK) and p38 (Suman et al., 2016). Phosphorylation of all three of these MAPKs has been shown to stimulate release of chemoattractant proteins that trigger migration of immune cells from the blood to the mammary gland (Suman et al., 2016; Zhao and Lacasse, 2008). Collectively, immune cell migration and epithelial cell inflammation contribute to tissue damage during mastitis.
Small molecule inhibitors of histone deacetylases (HDACs) have been shown efficacious in various inflammatory models including pulmonary hypertension and rheumatoid arthritis (Angiolilli et al., 2016; Cavasin et al., 2012; Stenmark et al., 2012; Zhao et al., 2012). Recent reports demonstrate that HDAC inhibitors attenuate signaling of the MAP kinase, ERK (Ferguson et al., 2013; Williams et al., 2014). HDACs catalyze the removal of acetyl groups from lysine residues of proteins, but have traditionally been studied as regulators of nucleosomal DNA. HDACs are grouped into four classes: class I HDACs (1, 2, 3 & 8), class II HDACs (4, 5, 6, 7, 9 & 10), class III HDACs (Sirt1–7) and the lone class IV HDAC, HDAC11 (Bradner et al., 2010; Gregoretti et al., 2004). Class I, II and IV HDACs require zinc for catalytic activity, while class III HDACs are nicotinamide adenine dinucleotide (NAD +)-dependent. Inhibition of zinc-dependent HDACs blocks inflammation (Das Gupta et al., 2016); yet the role for specific HDAC isoforms in the control of MAPK phosphorylation and bovine mammary epithelial cell inflammation have yet to be elucidated.
Based on previous reports, we postulated that small molecule HDAC inhibitors would inhibit bovine mammary epithelial cell MAPK signaling. Furthermore, we hypothesized that attenuation of MAP kinases partly contribute to anti-inflammatory actions attributed to HDAC inhibitors. To test these postulates, we relied on the commonly used MAC-T cell line (Huynh et al., 1991); this is an immortalized bovine mammary epithelial cell line that affords the opportunity to elucidate molecular mechanisms underpinning the inflammatory response. In this report, MAC-T cells were stimulated with TNFα as a model of bovine mammary inflammation. Using MAC-T cells, we showed that HDACs 1 and 2 regulate ERK and JNK phosphorylation in response to TNFα signaling. Moreover, we demonstrated that inhibition of MAPK signaling partially explained the anti-inflammatory actions attributed to HDAC inhibitors. Lastly, we showed that treatment with class I-selective or HDAC1/2-selective HDAC inhibitors normalized inflammatory gene profiles in MAC-T cells in response to TNFα. While further studies using class I- and HDAC1/2-selective inhibitors in primary bovine mammary epithelial cells and animal models of mammary inflammation are needed, these data suggest efficacy for HDAC inhibitors as additional therapeutics for bovine mastitis.
Materials and Methods
Cells and Cell Culture
MAC-T cells were kindly provided by Dr. Laura Hernandez (University of Wisconsin-Madison). MAC-T cells were used for all experiments (<12 passages). MAC-T cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Caisson Labs), containing 10 % fetal bovine serum (FBS; Atlanta Biologicals), 10 μg/mL insulin (Sigma-Aldrich), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Caisson Labs) at 37 °C in 5% CO2. MAC-T cells were cultured under basal conditions. At 80% confluence, MAC-T cells were pre-treated for 24 hours with HDAC inhibitors as indicated, trichostatin A (TSA; Sigma #T8552; 200 nM), MGCD0103 (MGCD; Selleckchem #S1122; 1 μM), Apicidin (Api; Enzo Life Sciences #BML-GR340; 1 μM), Romidepsin (HDAC1/2i; Selleckchem #S3020; 1 μM), RGFP966 (HDAC3i; Selleckchem #S7229; 1 μM), RG2833 (HDAC1/3i; Selleckchem #S7292; 1 μM), Diphenylacetohydroxamic acid (DPAH; Sigma #D6071; 10 μM), Tubastatin A (TubA; Selleckchem #S8049; 1 μM) prior to stimulation with tumor necrosis factor α (TNFα; Invitrogen #PMC3014; 300 pM). At 60% confluence, MAC-T cells were transiently transfected with Dicer-substrate siRNAs (DsiRNAs) for HDAC1#1 (IDT; rArGrU rGrArU rGrArC rUrArC rArUrU rArArA rUrUrC rUrUA C and rGrUrA rArGrA rArUrU rUrArA rUrGrU rArGrU rCrArU rCrArC rUrGrU), HDAC1#2 (IDT; rArUrG rArCrU rArCrA rUrUrA rArArU rUrCrU rUrArC rGrCT C and rGrArG rCrGrU rArArG rArArU rUrUrA rArUrG rUrArG rUrCrA rUrCrA) HDAC2 #1 (IDT; rArArU rUrUrG rArCrA rGrUrU rArArA rGrGrU rCrArU rGrCT A and rUrArG rCrArU rGrArC rCrUrU rUrArA rCrUrG rUrCrA rArArU rUrGrA), HDAC2 #2 (IDT; rGrArA rUrUrU rCrUrA rUrUrC rGrArG rCrArU rCrArG rArCA A and rUrUrG rUrCrU rGrArU rGrCrU rCrGrA rArUrA rGrArA rArUrU rCrUrC) or non-targeting scrambled Universal Negative DsiRNA control (siControl; IDT; 51–01–19–09) for 48 hrs, prior to stimulation with 300 pM TNFα. Gene knockdown was performed individually and in combination. IDT = Integrated DNA Technologies
HDAC Activity Assays
HDAC activity assays were performed as previously described (Lemon et al., 2011). MAC-T cells were lysed in PBS (pH 7.4) containing 0.5% Triton X-100, 300 mM NaCl and protease/phosphatase inhibitor cocktail (Thermo Fisher). Cell lysate was clarified by centrifugation prior to determination of protein concentration via BCA Protein Assay Kit (Pierce). Protein lysate (15 μg protein/well) was diluted into PBS buffer (100 μL/well) in a 96-well plate. Vehicle (DMSO) or HDAC inhibitors, TSA, Apicidin and MGCD0103 was added to cell lysate and incubated at 37 °C for 2 hrs. Class-specific HDAC substrates were then added (5 μL of 1 mM stock solution), and plates returned to the 37 °C incubator for 2 hrs. Developer/stop solution was added (50 μL per well of PBS with 1.5% Triton X-100, 3 μM TSA, and 0.75 mg/mL trypsin) and plates incubated at 37 °C for 20 min. Subsequent to deacetylation, trypsin was used to release AMC, resulting in increased fluorescence. AMC fluorescence was measured via BioTek Synergy2 plate reader, with excitation and emission filters of 360 nm and 460 nm, respectively.
Immunoblotting
Cells were treated as indicated and cell lysate collected in lysis buffer (PBS, 300 mM NaCl, 0.5% Triton-X, pH 7.4) containing protease and phosphatase inhibitors (HALT™, Thermo Scientific). Cells were sonicated prior to clarification by centrifugation (at 16,000 × g for 5 min). Protein concentration was measured via BCA Assay Kit (Pierce) and proteins resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membranes (BioRad) and probed with antibodies against phospho-JNK (Cell Signaling Technology; 4668), phospho-p38 (Cell Signaling Technology; 4511), phospho-ERK (Cell Signaling Technology; 4370), total JNK (Santa Cruz Biotechnology; sc-571), total p38 (Santa Cruz Biotechnology; sc-7149), total ERK (Santa Cruz Biotechnology; sc-1647) HDAC1 (Cells Signaling Techology; 5356), or HDAC2 (Cell Signaling Technology; 5113). Horseradish Peroxidase (HRP)-conjugated secondary antibodies (Southern Biotech) were used at a concentration of 1:2000. SuperSignal West Pico chemiluminescence system (Thermo Scientific) and a ChemiDoc XRS+ imager (BioRad) were used to detect protein.
Real time quantitative PCR
To assess pro-inflammatory gene expression, a panel of 83 genes was analyzed using a quantitative polymerase chain reaction (qPCR) array (Qiagen; PABT-011ZA). MAC-T cells were pretreated with Vehicle, MGCD, or SP600125 (JNKi) prior to stimulation with TNFα (300 pM) or Vehicle (distilled H2O). Following TNFα (1h post) treatment, total RNA was harvested using TRI Reagent (Life Technologies). RNA samples were diluted to 100 ng/μL (500 ng) and then converted to cDNA using the RT2 First Strand cDNA Synthesis Kit (Qiagen; 330401). SABioscence RT2 Profiler Cow Inflammatory Cytokines & Receptors PCR Array (Qiagen; PABT-011ZA) was used to complete the PCR array analysis. PCR array data was verified with quantitative PCR (qPCR); biological triplicates were used for each treatment.
For qPCR, 500 ng of RNA was converted to cDNA via the Verso cDNA Synthesis Kit (Thermo Scientific; AB-1453) and qPCR performed using Apex qPCR GREEN Master mix (Genesee Scientific; 42–120) on a BioRad CF96X qPCR instrument (BioRad). PCR primers for chemokine (C-C motif) ligand 26 (CCL26) (Fwd: gtt cct gac aag aca gag gaa t) and (Rev: ccg aga tcg aca ctc aag ata ag), colony stimulating factor 2 (CSF2) (Fwd: ct acc cac aaa gag cca aa) and (Rev: tca aca tgg ccc tga aca a), tumor necrosis factor α (TNFα) (Fwd: gcc aac tcc ctc tgt tta tgt) and (Rev: gac acc ttg acc tcc tga ata a), lymphotoxin beta (LTB) (Fwd: cga gag ggt gta cgt caa tat c) and (Rev: cat cag gca gta gct gtt ctt a), and 18S ribosomal RNA (Fwd: gcc gct aga ggt gaa att ctt a) and (Rev: ctt tcg ctc tgg tcc gtc tt) were designed using Integrated DNA Technologies (IDT) Primer Quest Tool.
Statistical analyses
Statistical analysis was completed by ANOVA followed by Tukey’s post-hoc analysis unless otherwise noted using GraphPad Prism software. Statistical significance (defined as p<0.05) is reported as applicable.
Results
TNFα-induced phosphorylation of JNK and p38 in MAC-T cells.
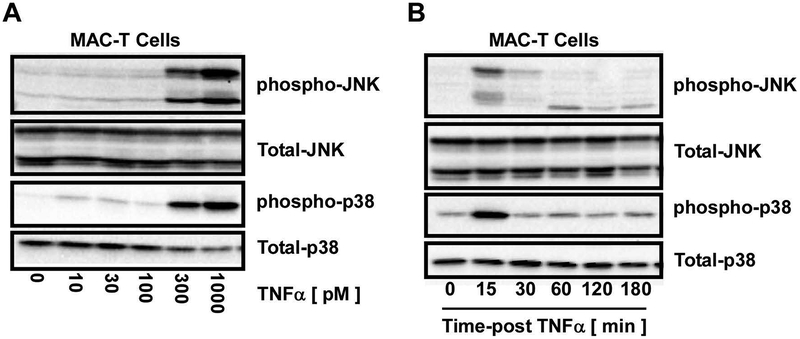
Dose-response curves and time course experiments were initially designed to elucidate TNFα-mediated induction of JNK and p38 signaling (i.e. phosphorylation) in MAC-T cells. Phosphorylation of JNK and p38 was assessed via immunoblotting with phospho-specific antibodies that recognize JNK and p38 when bisphosphorylated on the constitutive threonine-X-tyrosine (T-X-Y) motif that is known to be essential and sufficient for catalytic activity (Pearson et al., 2001). At 80% confluence, MAC-T cells were stimulated with increasing concentrations of TNFα for 15 minutes prior to immunoblot analysis for phosphorylated and total JNK and p38. Immunoblot analysis demonstrated dose-dependent increases in JNK and p38 phosphorylation, with 300 pM TNFα identified for further analysis (Fig.1A). Time course experiments were next designed to elucidate peak phosphorylation of JNK and p38 in response to 300 pM TNFα. JNK and p38 phosphorylation was robust and transient with peak activation observed at 15 minutes post-TNFα stimulation (Fig.1B). Based on these data, MAC-T cells were lysed 15 minutes post-TNFα (300 pM) stimulation for studies outlined below unless otherwise noted.
Figure 1. Increased MAPK phosphorylation in MAC-T cells in response to TNFα.
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were A) stimulated with increasing doses of TNFα and cell lysates harvested at 15 min post-TNFα stimulation. Phospho-JNK and phospho-p38 as well as total-JNK, and p38 were examined by immunoblotting. B) MAC-T cells were stimulated with TNFα (300 pM). Cell lysates were collected over time post-TNFα stimulation and immunoblotted for phospho-JNK, phospho-p38 as well as total-JNK and p38.
Class I HDAC inhibitors attenuate TNFα-mediated JNK and ERK phosphorylation in MAC-T cells.
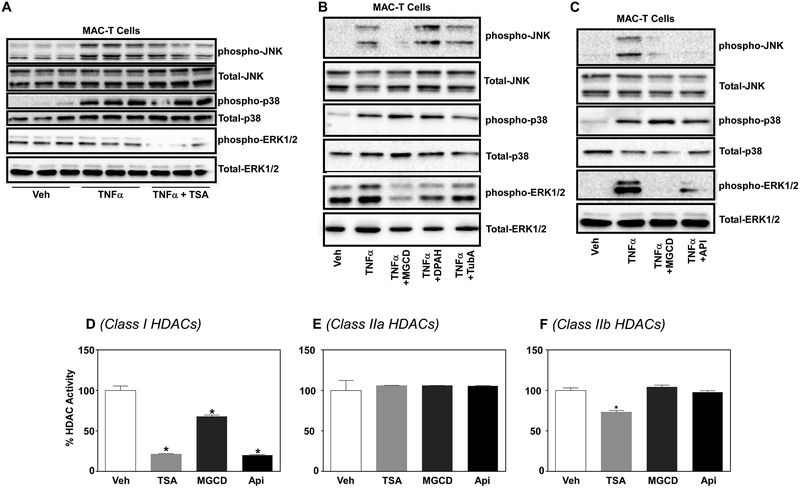
Small molecule HDAC inhibitors have recently been shown to regulate phosphorylation cascades including that of the MAPK, ERK (Barter et al., 2010; Ferguson et al., 2013; Williams et al., 2014). Thus, we postulated that inhibition of HDACs would block phosphorylation of the stress-activated protein kinases, JNK and p38. JNK and p38 phosphorylation increased in MAC-T cells stimulated for 15 min with 300 pM TNFα; phosphorylation of JNK not p38 was attenuated with broad spectrum HDAC inhibition via 200 nM trichostatin A (TSA) (Fig.2A). In addition, TSA attenuated ERK phosphorylation in MAC-T cells (Fig.2A), consistent with previous studies (Ferguson et al., 2013). TSA acts as a ‘pan’ HDAC inhibitor to target class I and II HDACs. As such, we treated MAC-T cells with class-selective HDAC inhibitors; MGCD0103 (MGCD; class I HDACi), Diphenylacetohydroxamic acid (DPAH; class IIa HDACi) and Tubastatin A (TubA; class IIb HDACi) for 24 hours prior to TNFα (300 pM) stimulation for 15 min. JNK, ERK and p38 phosphorylation increased with TNFα stimulation. JNK and ERK phosphorylation, but not p38 phosphorylation, was completely blocked with the class I-selective HDAC inhibitor, MGCD (Fig.2B). As pharmacological inhibitors can have off target effects, we next examined JNK and ERK phosphorylation in MAC-T cells treated with two independent class I-selective HDAC inhibitors, MGCD and Apicidin (Api; class I HDACi). Treatment with MGCD or Apicidin completely blocked TNFα-stimulated JNK and ERK phosphorylation (Fig.2C). No inhibition of p38 phosphorylation was observed (Fig.2C). In addition, there were no changes in total JNK or ERK protein expression, suggesting that class I HDACs regulate either the phosphorylation or dephosphorylation of these two MAP kinases. Quantitation for these experiments was performed, as each experiment was performed a minimum of three times (Supplemental Fig.1). Quantitative results for JNK can be observed with the graphs on the left and results for ERK observed with graphs on the right (Supplemental Fig.1). Moreover, figure letters A-C for Supplemental Figure 1 correspond to Figure 2A-C.
Figure 2. Class I HDACs regulate JNK and ERK phosphorylation in MAC-T cells in response to TNFα.
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were A) treated with trichostatin A (TSA) for 24 hrs prior to TNFα stimulation (300pM). Cell lysates were harvested at 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phospho-p38, phosphor-ERK, total-JNK, total-ERK and total-p38. B) MAC-T cells were treated with class selective HDAC inhibitors MGCD0103 (MGCD; class I), DPAH (Class IIa) and Tubastating A (TubA; class IIb) prior to TNFα (300 pM) stimulation. Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phospho-p38, phosphor-ERK as well as total-JNK, total-ERK and total-p38. C) MAC-T cells were treated with class I selective HDAC inhibitors MGCD and apicidin (Api) 24 hrs prior to TNFα (300 pM) stimulation. Cells lysates were harvested 15 min post-TNFα stimulation and immunoblotted as above. D) Protein was harvested from MAC-T cells treated with basal (10% FBS, 10 mg/ml insulin) media. Protein lysate (30 μg) was subsequently incubated with vehicle (DMSO), TSA, MGCD, and apicidin for 2 hrs prior to addition of class-specific HDAC substrates. Developer solution was added for an additional 20 minutes and fluorescent activity measured via BioTek plate reader. One-way ANOVA with Dunnett’s post-hoc analysis was used to assess statistical significance when p<0.05.
To demonstrate class selectivity for Apicidin and MGCD, substrate-specific HDAC assays were performed to examine class I, IIa and IIb HDAC activity. MAC-T protein lysate was incubated with TSA (pan-HDACi), MGCD, or Apicidin for 2 hrs prior to the addition of class-specific HDAC substrates. This assay has been outlined in detail (Heltweg et al., 2004; Lemon et al., 2011). HDAC activity demonstrated that MGCD and Apicidin only target class I HDACs for inhibition, while TSA was capable of inhibiting class I and IIb HDAC substrates in MAC-T cells (Fig.2D). It should be noted that no visible signs of cell death or stress were observed in MAC-T cell culture. Moreover the doses used for HDAC inhibition in our studies are typical for cell culture experiments, with no signs of toxicity reported in those studies not using cancer cell lines (Angiolilli et al., 2016; Antos et al., 2003; McKinsey, 2012; Schmitz and de la Vega, 2015; Wagner et al., 2013; Williams et al., 2014).
HDACs 1 and 2 are essential for JNK and ERK phosphorylation in TNFα-stimulated MAC-T cells.
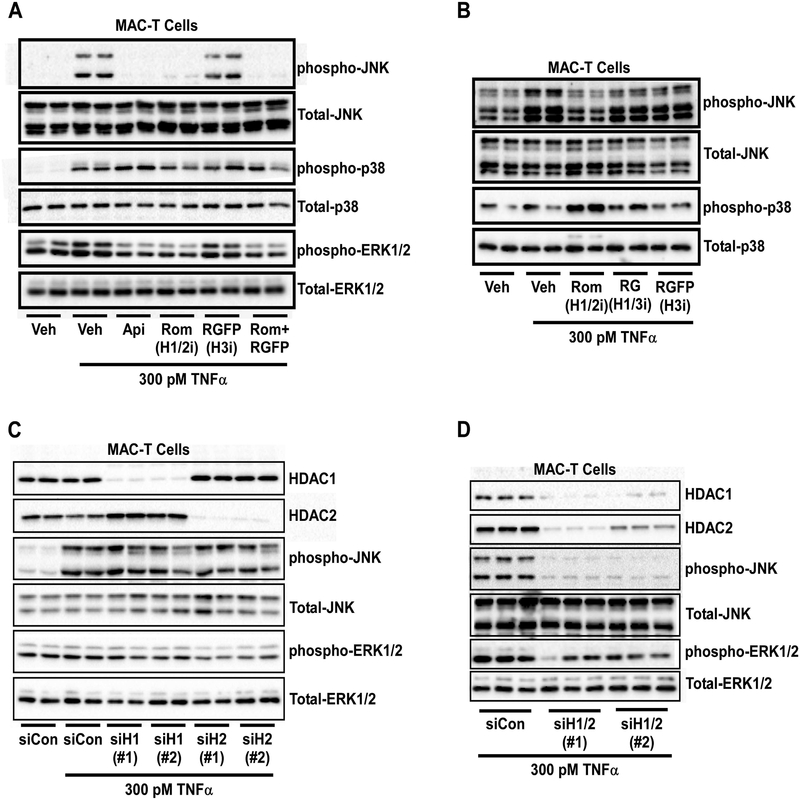
Data above demonstrated that class I HDACs regulate TNFα-mediated JNK and ERK signaling in MAC-T cells, yet the HDAC isoform(s) responsible remain unknown. To address this question, MAC-T cells were pretreated for 24 hrs with the class I HDACi Apicidin or isoform-selective HDAC inhibitors Romidepsin (Rom; HDAC1/2i), RGFP966 (RGFP; HDAC3i) or Rom and RGFP in combination prior to TNFα stimulation at 300 pM for 15 min. It should be noted that MGCD0103 does not target HDAC8 for inhibition (Fournel et al., 2008), as such inhibitors for HDAC8 were not used in these studies. TNFα-mediated phosphorylation of JNK and ERK was attenuated by the class I HDAC inhibitor (Api) and HDAC1/2 inhibitor (Rom) but not the HDAC3 inhibitor (RGFP) (Fig.3A). Again, p38 phosphorylation remained unchanged and no inhibition was observed for total MAPK protein expression. Additional pharmacological tools were used to distinguish HDAC1- or HDAC2-specific actions on JNK signaling. For these studies, MAC-T cells were pretreated for 24 hrs with Romidepsin (Rom; HDAC1/2i), RG2833 (RG; HDAC1/3i), or RGFP966 (RGFP; HDAC3i) prior to TNFα stimulation. Again, inhibition of HDACs 1 and 2 (Rom) attenuated TNFα-induced JNK phosphorylation to basal (i.e. unstimulated) levels (Fig.3B). However, inhibition of HDACs 1 and 3 (RG) or HDAC3 alone was not sufficient to attenuate JNK signaling in response to TNFα (Fig.3B), suggesting that HDAC2 or HDAC1 and 2 in combination are needed for JNK phosphorylation.
Figure 3. HDACs 1 and 2 regulate JNK and ERK phosphorylation in MAC-T cells in response to TNFα.
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were A) treated with vehicle (DMSO), apicidin (Api), Romidepsin (Rom), RGFP966 (RGFP), or a combination of Rom + RGFP for 24 hrs prior to TNFα (300 pM) stimulation. Cell lysates were harvested at 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phospho-p38, phosphor-ERK, total-JNK, total-ERK and total-p38. B) MAC-T cells were treated with vehicle (DMSO), Romidepsin (Rom), RG2833 (RG) or RGFP966 (RGFP) for 24 hrs prior to TNFα (300 pM) stimulation. Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phospho-p38 as well as total-JNK and p38. C) MAC-T cells were transiently transfected with dicer-substrate siRNA for two different regions of HDAC1 (siH1#1 or siH1#2), HDAC2 (siH2#1 or siH2#2), or siControl (siCon) using lipofectamine 3000 reagent for 48 hrs prior to TNFα stimulation (300 pM). Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for HDAC1, HDAC2, phospho-JNK, phopho-ERK, total-JNK and total ERK. D) MAC-T cells were transiently transfected with independent siRNAs targeted to knockdown HDACs 1 and 2 in combination (siH1/2#1 or siH1/2#2) or siControl (siCon) using lipofectamine 3000 reagent for 48 hrs prior to TNFα stimulation (300 pM). Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for HDAC1, HDAC2, phospho-JNK, phopho-ERK, total-JNK and total ERK.
To determine which HDAC isoform, HDAC1 or 2, regulates MAPK phosphorylation, we next used dicer-substrate small interfering RNA (siRNA) to genetically knockdown HDAC1 or 2 individually. MAC-T cells were transiently transfected with siRNA for two independent regions of the HDAC1 gene (siH1#1 or siH1#2), HDAC2 gene (siH2#1 or siH2#2) or non-targeting control (siCon) for 48 hrs prior to stimulation with TNFα. Small interfering RNAs for HDAC1, targeted HDAC1 but not HDAC2 for knockdown (Fig.3C). Likewise, siRNAs targeted against HDAC2, attenuated HDAC2 protein expression but not HDAC1 (Fig.3C). Surprisingly, loss of HDAC2 did not attenuate JNK phosphorylation as postulated (Fig.3C), suggesting both HDACs 1 and 2 are necessary for JNK signaling. However, loss of HDAC2 did inhibit ERK phosphorylation (Fig.3C); these data were quantified (Supplemental Fig.2A).
Given that individual knockdown of HDAC1 or HDAC2 did not inhibit JNK phosphorylation, we next used siRNA targeted against HDACs 1 and 2 in combination. Combination knockdown with two different siRNAs directed against HDACs 1 and 2 (siH1/2#1 or siH1/2#2) was sufficient to inhibit HDAC1 and 2 protein expression (Fig.3D). Moreover, loss of HDACs 1 and 2 in combination attenuated JNK and ERK phosphorylation (Fig.3D); these data were quantified (Supplemental Fig.2B). Based on quantitative analysis, knockdown of HDAC2 or combination knockdown of HDACs1 and 2 attenuated ERK phosphorylation to the same extent (~50%), suggesting that HDAC2 is the main regulator of ERK signaling while HDACs1 and 2 are both required for JNK phosphorylation.
Class I HDACs regulate inflammatory gene expression in MAC-T cells.
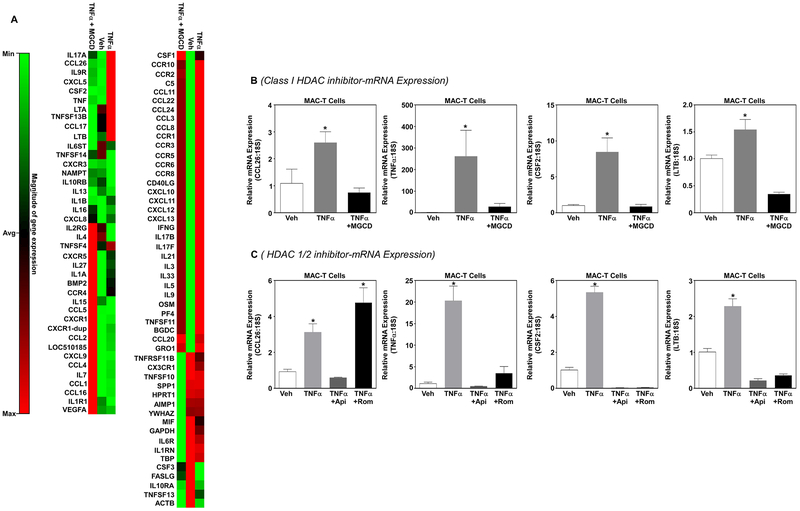
Small molecule inhibitors of HDACs have previously been shown to block inflammation (Das Gupta et al., 2016). We postulated that class I selective HDAC inhibitors would block TNFα-induced inflammatory gene expression. To test this postulate, MAC-T cells were pretreated with MGCD for 24 hrs prior to TNFα stimulation (300 pM). RNA was harvested 1hr post-TNFα stimulation and PCR arrays used to assess bovine inflammatory gene expression. PCR array analysis demonstrated that inhibition of class I HDACs normalized TNFα-mediated changes in inflammatory gene expression in MAC-T cells (Fig.4A); these data are visualized via heatmap (Fig.4A). Quantitative PCR (qPCR) further demonstrated that inhibition of class I HDACs significantly attenuated TNFα-induced expression of c-c motif chemokine ligand 26 (CCL26), tumor necrosis factor α (TNFα), granulocyte-macrophage colony-stimulating factor 2 (CSF2) and lymphotoxin beta (LTB) (Fig.4B). While these data demonstrated that class I-selective HDAC inhibitors block TNFα-induced inflammatory gene expression, it remains unclear what role HDACs 1 and 2 play in the regulation of these genes. Therefore, MAC-T cells were pretreated with Apicidin (Api) or Romidepsin (Rom; HDAC1/2 inhibitor) prior to stimulation with TNFα. Similar to the class I selective HDAC inhibitor MGCD, the class I-selective HDAC inhibitor Apicidin significantly inhibited TNFα-induced expression of CCL26, TNFα, CSF2 and LTB (Fig.4C). Of note, the HDAC1/2 selective inhibitor Rom only attenuated expression of TNFα, CSF2 and LTB but not CCL26 (Fig.4C). These data would suggest that HDAC3 regulates CCL26 gene expression.
Figure 4. Class I HDACs regulate MAC-T inflammatory gene expression in response to TNFα.
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were treated with the class I selective HDAC inhibitor MGCD for 24 hrs prior to TNFα stimulation (300 pM) and RNA isolated at 1hr post-TNFα stimulation. A) Heatmap generated from a quantitative polymerase chain reaction (qPCR) array was used to examine inflammatory gene expression. B) Real-time qPCR was used to validate CCL26, TNFα, CSF2 and LTB gene expression from mRNA identified in the array. C) In addition, MAC-T cells were pretreated with an independent class I selective HDAC inhibitor apicidin (Api; 1 μM) or an HDAC1/2-specific inhibitor (Romidepsin, Rom 1 μM) for 24 hrs prior to TNFα stimulation (300 pM). RNA was isolated 1hr post-TNFα stimulation and qPCR used to examine CCL26, TNFα, CSF2 and LTB gene expression. One-way ANOVA with Tukey’s post-hoc analysis was used to assess statistical significance when p<0.05.
MAPK-mediated regulation of inflammatory gene expression in MAC-T cells in response to TNFα.
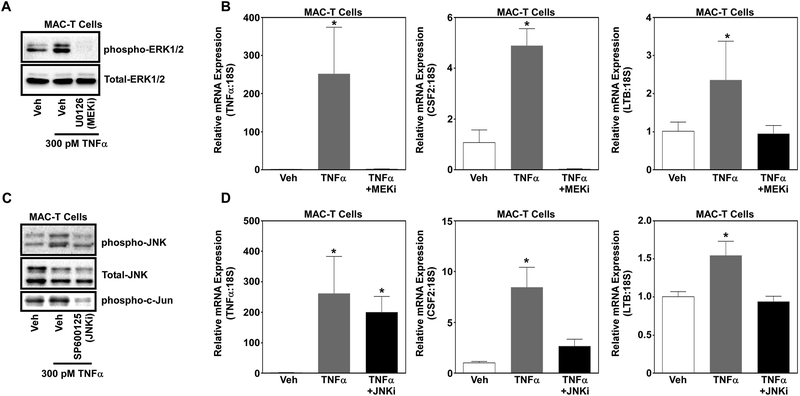
Data above demonstrated that HDACs 1 and 2 regulate TNFα-induced MAPK signaling and inflammatory gene expression. Thus, we postulated that HDACs regulate inflammatory gene expression in part via changes in ERK and JNK phosphorylation. To test this postulate, MAC-T cells were pretreated with inhibitors of MEK (U0126), the upstream kinase to ERK, or JNK (SP600125) prior to stimulation with TNFα. Western blot analysis of ERK phosphorylation demonstrated that U0126 inhibited TNFα-induced ERK signaling (Fig.5A). In addition, treatment with SP600125 attenuated JNK signaling in MAC-T cells stimulated with TNFα (Fig.5C). While SP600125 is an inhibitor JNK, others have shown similar results regarding JNK phosphorylation. However, we also observed that treatment with SP600125 attenuated phosphorylation of c-Jun, a transcription factor downstream of JNK (Fig.5C). In keeping with our postulate, inhibition of ERK and JNK significantly reduced expression of CSF2 and LTB gene expression (Fig.5B&D). However, treatment with the ERK inhibitor and not the JNK inhibitor blocked TNFα expression, demonstrating selectivity towards ERK (Fig.5B&D).
Figure 5. ERK and JNK signaling partly regulate inflammatory gene expression in MAC-T cells in response to TNFα.
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were treated with the MEK inhibitor (U0126; 10 μM) or JNK inhibitor (SP600125; 20 μM) for 30 min prior to TNFα stimulation (300 pM). Protein was isolated 15 minutes post-TNFα and immunoblotted for A) phosphor-ERK and total-ERK as well as B) phosphor-JNK, phosphor-c-Jun and total-JNK. RNA was isolated 1hr post-TNFα stimulation and qPCR used to assess inflammatory gene expression of TNFα, CSF2 and LTB in MAC-Ts treated with the C) MEK inhibitor or D) JNK inhibitor. One-way ANOVA with Tukey’s post-hoc analysis was used to assess statistical significance when p<0.05.
Discussion
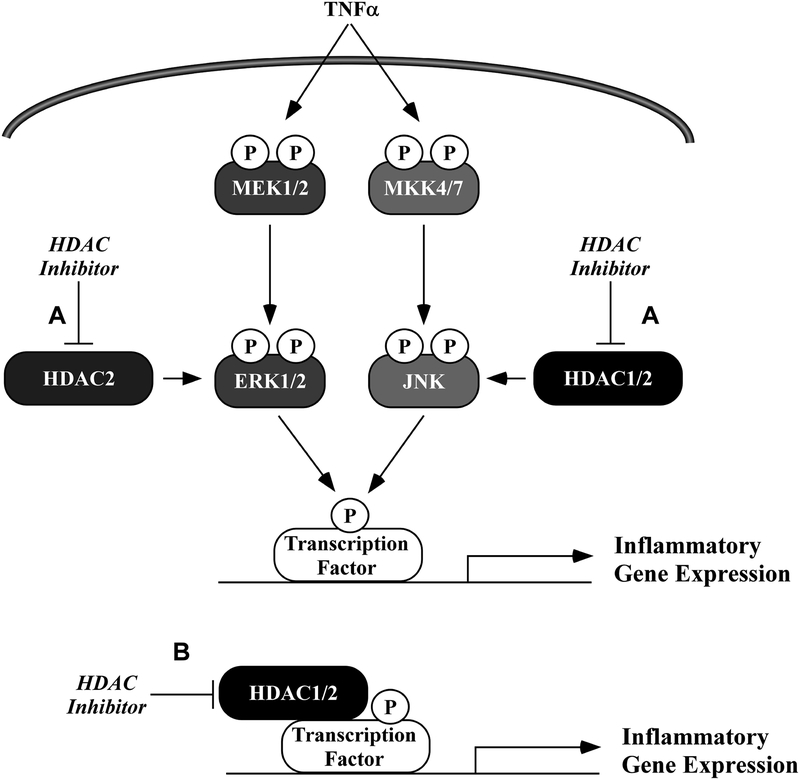
We present new findings that HDAC2 regulated ERK phosphorylation while HDACs 1 and 2 regulated JNK signaling in MAC-T bovine mammary epithelial cells. Based on our findings, we propose a model in which inhibition of HDACs 1 and 2 attenuate JNK and ERK phosphorylation in response to TNFα (Fig.6A). In addition, HDAC inhibitors normalized inflammatory gene expression in MAC-Ts in response to TNFα; this was likely mediated via MAPK-dependent (Fig.6A) regulation, e.g. MAPK-targeted phosphorylation of a transcription factor (e.g. c-Jun), as well as direct regulation of chromatin remodeling and gene expression (Fig.6B). These findings represent the first demonstration in bovine mammary epithelial cells for the protective actions of zinc-dependent HDAC inhibitors and suggest efficacy for class I- or HDAC1/2-selective HDAC inhibitors as therapeutics for mastitis.
Figure 6. Working model of HDAC inhibitor-mediated suppression of ERK and JNK phosphorylation and inflammatory gene expression.
A) HDAC 2 regulates TNFα-induced ERK signaling. HDACs 1 and 2 in combination regulate TNFα-induced JNK phosphorylation. Inhibition of ERK and JNK partly explain anti-inflammatory mechanisms linked to class I selective HDAC inhibitors in bovine mammary epithelial cells (MAC-Ts). B) In addition, inhibition of HDACs attenuates pro-inflammatory gene expression through mechanisms associated with canonical regulation of gene expression, independent of MAPK signaling.
Bovine mastitis is often caused by bacterial infection or trauma that results in immune activation, in which mammary epithelial cells secrete pro-inflammatory chemokines to recruit immune cells such as neutrophils and macrophages to the mammary tissue (Akers and Nickerson, 2011; Chen et al., 2016; Erskine et al., 2003; Hoeben et al., 2000). Immune cells respond by secreting pro-inflammatory molecules like TNFα (Hueber et al., 2000). Circulating TNFα functions in macrophages to enhance microbicidal capacity or its killing activity for microorganisms such as bacteria (Hueber et al., 2000). However, TNFα also functions to activate inflammatory expression within the mammary epithelium (Hoeben et al., 2000; Hueber et al., 2000). As TNFα plays a key role in mammary epithelial cell inflammation after initial bacterial infection, we relied on TNFα as our inflammatory agonists in an immortalized bovine mammary epithelial cell line. As expected, TNFα significantly increased inflammatory gene expression in MAC-T cells. Of note, we report that TNFα stimulation significantly increased expression of the TNFα gene within mammary epithelial cells, suggesting a feedforward mechanism that perpetuates the inflammatory environment. However, protein expression of TNFα was not examined. While combined the immune/inflammatory response is designed to rid the host of infection or remediate and repair trauma to the mammary tissue, chronic inflammation contributes to mammary tissue damage.
Due to the prevalence and costs associate with bovine mastitis, the dairy industry is interested in therapeutics that can prevent or reduce mammary inflammation. HDAC inhibitors possess anti-inflammatory actions (Angiolilli et al., 2016; Bradner et al., 2010; Zwergel et al., 2015). HDAC inhibitors have traditionally been studied in the context of nucleosomal histones, in which changes in histone acetylation impact chromatin accessibility and thus alter gene expression (Bradner et al., 2010; Gregoretti et al., 2004). This epigenetic modulation would be expected to contribute to genome wide changes in gene expression, and thus could be postulated to have deleterious consequences for cell function. However, many reports have shown efficacy in vitro and in vivo for the use of small molecule HDAC inhibitors for the treatment of cardiovascular disease (Ferguson and McKinsey, 2015; Jeong et al., 2018), rheumatoid arthritis (Angiolilli et al., 2016), pulmonary hypertension (Cavasin et al., 2012; Stenmark et al., 2012) as well as diabetes and metabolic disease (Christensen et al., 2011; Dali-Youcef et al., 2007). Further, four HDAC inhibitors have been approved by the FDA for the treatment of cancer (Yoon and Eom, 2016). Combined, these reports would suggest potential efficacy for HDAC inhibitors as a therapeutic for bovine mastitis. Consistent with this, we demonstrate that treatment with HDAC inhibitors blocked pro-inflammatory gene expression in bovine mammary epithelial cells. While further studies using primary bovine mammary cell lines in addition to in vivo testing are needed, these data provide the first step towards acknowledging HDAC inhibitors as anti-inflammatory therapeutics for bovine mammary epithelial cell inflammation.
In addition, this report highlights class I- and HDAC1/2-selective inhibitors as efficacious anti-inflammatory therapeutics in the MAC-T cell line. Treatment with class- or isoform-selective HDAC inhibitors would be expected to limit widespread epigenetic changes in gene expression and thus limit off-target actions associated with more traditional pan-HDAC inhibitors, such as the FDA-approved HDAC inhibitor, SAHA (vorinostat). Future research examining HDAC1/2 inhibitors in vitro and in vivo for the treatment of bovine mastitis will add significantly to the field.
Intracellular signaling cascades link inflammatory external stimuli such as TNFα to internal actions such as pro-inflammatory gene expression. MAPKs and NF-κB represent two major signaling cascades that link inflammatory stimuli to nuclear induction of pro-inflammatory gene expression (Gil et al., 2007; Solinas and Becattini, 2017; Wajant et al., 2003). HDACs have recently been shown to regulate inflammatory signaling cascades such as the MAPKs as well as NF-κB (Barter et al., 2010; Chen et al., 2018; Ferguson et al., 2013). For instance, HDAC inhibitor treatment was reported to reduce neuro-inflammation via inhibited HDAC3 activity that contributed to acetylation of the NF-κB subunit p62, which altered inflammatory gene expression in a spinal cord injury model (Chen et al., 2018). In addition, this group reported that HDAC inhibition led to increased STAT1 acetylation, suggesting that HDAC inhibitors can regulate activity of many intracellular signaling cascades. We report that HDAC1/2-selective inhibitors attenuated JNK and ERK phosphorylation in bovine mammary epithelial cells. While inhibition of JNK and ERK was sufficient to attenuate gene expression of select genes, our data do not infer that anti-inflammatory actions were solely the result of ERK and JNK signaling. Regarding the literature, in which HDACs have been shown to regulate NF-κB (Chen et al., 2018), JAK/STAT (Chen et al., 2018; Yang and Seto, 2008), MAPKs (Blakeslee et al., 2017; Ferguson et al., 2013) and PI3K/AKT (Yang and Seto, 2008) among others, we would postulate that anti-inflammatory actions mediated by HDAC inhibitors are a result of MAPK inhibition in combination with changes in other signaling cascades. Assessment of these intracellular pathways in bovine mammary epithelial cells would be interesting to explore.
Future research into how HDACs 1 and 2 regulate JNK and ERK signaling is sure to yield interesting, mechanistic actions for these compounds. Current evidence demonstrate that HDACs work to regulate gene expression (Gregoretti et al., 2004; Yang and Seto, 2007), suggesting that HDAC inhibition potentially regulates ‘inducible’ phosphatases that act as feedback regulators for JNK and ERK dephosphorylation in bovine mammary epithelial cells. Indeed, we previously reported that HDAC3 regulated the phosphatase, dual-specificity phosphatase 5 (DUSP5), as a feedback inhibitor for ERK dephosphorylation in cardiac muscle cells (Ferguson et al., 2013). However, it should be noted that others have reported that HDACs directly interact with and deacetylate phosphatases, such as dual-specificity phosphatase 1 (DUSP1), in order regulate phosphatase activity or stability. For instance, HDACs 1, 2 and 3 were shown to deacetylate DUSP1, which led to inhibition of DUSP1 activity and increased p38 phosphorylation and subsequent inflammation in macrophages (Jeong et al., 2014). Lastly, direct HDAC-MAPK regulation has also been shown. In this report, treatment with HDAC inhibitors was shown to increase p38 phosphorylation (Pillai et al., 2011), opposite our findings for ERK and JNK. Moreover, it was reported that treatment with HDAC inhibitors increased p38 acetylation and that acetylation of p38 on lysine 53 (K53) enhanced p38 activity and ATP binding affinity (Pillai et al., 2011). Combined, these data would suggest that HDAC inhibitors potentially regulate ERK and JNK signaling through canonical epigenetic actions via induction of phosphatase gene expression or non-canonical actions directed against changes in MAPK or phosphatase acetylation, which could change protein activity or stability.
In conclusion, we report that inhibition of HDACs 1 and 2 attenuate TNFα-activated JNK and ERK signaling; this partially contributes to inhibition of inflammatory gene expression. These data provide the first step towards examining class I- and HDAC1/2-selective HDAC inhibitors as potential anti-inflammatory therapeutics for bovine mastitis.
Supplementary Material
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were A) treated with trichostatin A (TSA) for 24 hrs prior to TNFα stimulation (300pM). Cell lysates were harvested at 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phosphor-ERK, total-JNK and total-ERK. Data were quantified B) MAC-T cells were treated with class selective HDAC inhibitors MGCD0103 (MGCD; class I), DPAH (Class IIa) and Tubastating A (TubA; class IIb) prior to TNFα (300 pM) stimulation. Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phosphor-ERK as well as total-JNK and total-ERK prior to quantifiication. C) MAC-T cells were treated with class I selective HDAC inhibitors MGCD and apicidin (Api) 24 hrs prior to TNFα (300 pM) stimulation. Cells lysates were harvested 15 min post-TNFα stimulation and immunoblotted as above. Data were quantified. All data were assessed via one-way ANOVA with Dunnett’s post-hoc analysis. Statistical significance was set at p<0.05.
A) MAC-T cells were transiently transfected with siRNA for two different regions of HDAC1 (siH1#1 or siH1#2), HDAC2 (siH2#1 or siH2#2), or siControl (siCon) using lipofectamine 3000 reagent for 48 hrs prior to TNFα stimulation (300 pM). Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for HDAC1, HDAC2, phospho-JNK, phopho-ERK, total-JNK and total ERK. Quantification was performed for ERK phosphorylation normalized to total-ERK. B) MAC-T cells were transiently transfected with independent siRNAs targeted to knockdown HDACs 1 and 2 in combination (siH1/2#1 or siH1/2#2) or siControl (siCon) using lipofectamine 3000 reagent for 48 hrs prior to TNFα stimulation (300 pM). Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for HDAC1, HDAC2, phospho-JNK, phopho-ERK, total-JNK and total ERK. Quantification was performed for JNK phosphorylation (Left Panel) and ERK phosphorylation (Right Panel) normalized to corresponding total MAPK protein. Significance was set at p<0.05. One-way ANOVA with Dunnett’s post-hoc analysis was performed for all data.
Acknowledgements:
This work is supported by the USDA National Institute of Food and Agriculture (Hatch-NEV00727) to B.S.F. Core facilities used for Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under grant number P20 GM103554.
Footnotes
Conflict of Interest:
The authors declare no conflicts of interest.
References
- Akers RM, Nickerson SC. 2011. Mastitis and its impact on structure and function in the ruminant mammary gland. J Mammary Gland Biol Neoplasia 16(4):275–289. [DOI] [PubMed] [Google Scholar]
- Angiolilli C, Kabala PA, Grabiec AM, Van Baarsen IM, Ferguson BS, Garcia S, Malvar Fernandez B, McKinsey TA, Tak PP, Fossati G, Mascagni P, Baeten DL, Reedquist KA. 2016. Histone deacetylase 3 regulates the inflammatory gene expression programme of rheumatoid arthritis fibroblast-like synoviocytes. Ann Rheum Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antos CL, McKinsey TA, Dreitz M, Hollingsworth LM, Zhang CL, Schreiber K, Rindt H, Gorczynski RJ, Olson EN. 2003. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J Biol Chem 278(31):28930–28937. [DOI] [PubMed] [Google Scholar]
- Barter MJ, Pybus L, Litherland GJ, Rowan AD, Clark IM, Edwards DR, Cawston TE, Young DA. 2010. HDAC-mediated control of ERK- and PI3K-dependent TGF-beta-induced extracellular matrix-regulating genes. Matrix Biol 29(7):602–612. [DOI] [PubMed] [Google Scholar]
- Blakeslee WW, Lin YH, Stratton MS, Tatman PD, Hu T, Ferguson BS, McKinsey TA. 2017. Class I HDACs control a JIP1-dependent pathway for kinesin-microtubule binding in cardiomyocytes. J Mol Cell Cardiol 112:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. 2010. Chemical phylogenetics of histone deacetylases. Nat Chem Biol 6(3):238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavasin MA, Demos-Davies K, Horn TR, Walker LA, Lemon DD, Birdsey N, Weiser-Evans MC, Harral J, Irwin DC, Anwar A, Yeager ME, Li M, Watson PA, Nemenoff RA, Buttrick PM, Stenmark KR, McKinsey TA. 2012. Selective class I histone deacetylase inhibition suppresses hypoxia-induced cardiopulmonary remodeling through an antiproliferative mechanism. Circ Res 110(5):739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, He G, Zhang W, Xu T, Qi H, Li J, Zhang Y, Gao MQ. 2016. Stromal fibroblasts derived from mammary gland of bovine with mastitis display inflammation-specific changes. Sci Rep 6:27462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ye J, Chen X, Shi J, Wu W, Lin W, Lin W, Li Y, Fu H, Li S. 2018. Valproic acid attenuates traumatic spinal cord injury-induced inflammation via STAT1 and NF-kappaB pathway dependent of HDAC3. J Neuroinflammation 15(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DP, Dahllof M, Lundh M, Rasmussen DN, Nielsen MD, Billestrup N, Grunnet LG, Mandrup-Poulsen T. 2011. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Mol Med 17(5–6):378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. 2007. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med 39(5):335–345. [DOI] [PubMed] [Google Scholar]
- Das Gupta K, Shakespear MR, Iyer A, Fairlie DP, Sweet MJ. 2016. Histone deacetylases in monocyte/macrophage development, activation and metabolism: refining HDAC targets for inflammatory and infectious diseases. Clin Transl Immunology 5(1):e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erskine RJ, Wagner S, DeGraves FJ. 2003. Mastitis therapy and pharmacology. Vet Clin North Am Food Anim Pract 19(1):109–138, vi. [DOI] [PubMed] [Google Scholar]
- Ferguson BS, Harrison BC, Jeong MY, Reid BG, Wempe MF, Wagner FF, Holson EB, McKinsey TA. 2013. Signal-dependent repression of DUSP5 by class I HDACs controls nuclear ERK activity and cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A 110(24):9806–9811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson BS, McKinsey TA. 2015. Non-sirtuin histone deacetylases in the control of cardiac aging. J Mol Cell Cardiol 83:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournel M, Bonfils C, Hou Y, Yan PT, Trachy-Bourget MC, Kalita A, Liu J, Lu AH, Zhou NZ, Robert MF, Gillespie J, Wang JJ, Ste-Croix H, Rahil J, Lefebvre S, Moradei O, Delorme D, Macleod AR, Besterman JM, Li Z. 2008. MGCD0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol Cancer Ther 7(4):759–768. [DOI] [PubMed] [Google Scholar]
- Gil A, Maria Aguilera C, Gil-Campos M, Canete R. 2007. Altered signalling and gene expression associated with the immune system and the inflammatory response in obesity. Br J Nutr 98 Suppl 1:S121–126. [DOI] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 338(1):17–31. [DOI] [PubMed] [Google Scholar]
- Heltweg B, Dequiedt F, Marshall BL, Brauch C, Yoshida M, Nishino N, Verdin E, Jung M. 2004. Subtype selective substrates for histone deacetylases. J Med Chem 47(21):5235–5243. [DOI] [PubMed] [Google Scholar]
- Hoeben D, Burvenich C, Trevisi E, Bertoni G, Hamann J, Bruckmaier RM, Blum JW. 2000. Role of endotoxin and TNF-alpha in the pathogenesis of experimentally induced coliform mastitis in periparturient cows. J Dairy Res 67(4):503–514. [DOI] [PubMed] [Google Scholar]
- Hogeveen H, Huijps K, Lam TJ. 2011. Economic aspects of mastitis: new developments. N Z Vet J 59(1):16–23. [DOI] [PubMed] [Google Scholar]
- Hueber AO, Zornig M, Bernard AM, Chautan M, Evan G. 2000. A dominant negative Fas-associated death domain protein mutant inhibits proliferation and leads to impaired calcium mobilization in both T-cells and fibroblasts. J Biol Chem 275(14):10453–10462. [DOI] [PubMed] [Google Scholar]
- Huynh HT, Robitaille G, Turner JD. 1991. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp Cell Res 197(2):191–199. [DOI] [PubMed] [Google Scholar]
- Jeong MY, Lin YH, Wennersten SA, Demos-Davies KM, Cavasin MA, Mahaffey JH, Monzani V, Saripalli C, Mascagni P, Reece TB, Ambardekar AV, Granzier HL, Dinarello CA, McKinsey TA. 2018. Histone deacetylase activity governs diastolic dysfunction through a nongenomic mechanism. Sci Transl Med 10(427). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y, Du R, Zhu X, Yin S, Wang J, Cui H, Cao W, Lowenstein CJ. 2014. Histone deacetylase isoforms regulate innate immune responses by deacetylating mitogen-activated protein kinase phosphatase-1. J Leukoc Biol 95(4):651–659. [DOI] [PubMed] [Google Scholar]
- Lemon DD, Horn TR, Cavasin MA, Jeong MY, Haubold KW, Long CS, Irwin DC, McCune SA, Chung E, Leinwand LA, McKinsey TA. 2011. Cardiac HDAC6 catalytic activity is induced in response to chronic hypertension. J Mol Cell Cardiol 51(1):41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA. 2012. Therapeutic potential for HDAC inhibitors in the heart. Annu Rev Pharmacol Toxicol 52:303–319. [DOI] [PubMed] [Google Scholar]
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev 22(2):153–183. [DOI] [PubMed] [Google Scholar]
- Pillai VB, Sundaresan NR, Samant SA, Wolfgeher D, Trivedi CM, Gupta MP. 2011. Acetylation of a conserved lysine residue in the ATP binding pocket of p38 augments its kinase activity during hypertrophy of cardiomyocytes. Mol Cell Biol 31(11):2349–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi M, Li RW, Bannerman DD, Daniels KM, Evock-Clover C, Silva MV, Paape MJ, Van Ryssen B, Burvenich C, Capuco AV. 2010. A sentinel function for teat tissues in dairy cows: dominant innate immune response elements define early response to E. coli mastitis. Funct Integr Genomics 10(1):21–38. [DOI] [PubMed] [Google Scholar]
- Schmitz ML, de la Vega L. 2015. New Insights into the Role of Histone Deacetylases as Coactivators of Inflammatory Gene Expression. Antioxid Redox Signal 23(1):85–98. [DOI] [PubMed] [Google Scholar]
- Solinas G, Becattini B. 2017. JNK at the crossroad of obesity, insulin resistance, and cell stress response. Mol Metab 6(2):174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark KR, Frid MG, Yeager M, Li M, Riddle S, McKinsey T, El Kasmi KC. 2012. Targeting the adventitial microenvironment in pulmonary hypertension: A potential approach to therapy that considers epigenetic change. Pulm Circ 2(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg Y, Gray C, Vuocolo T, Donaldson L, Broadway M, Tellam R. 2005. Lipopolysaccharide and lipoteichoic acid induce different innate immune responses in bovine mammary epithelial cells. Cytokine 31(1):72–86. [DOI] [PubMed] [Google Scholar]
- Suman S, Sharma PK, Rai G, Mishra S, Arora D, Gupta P, Shukla Y. 2016. Current perspectives of molecular pathways involved in chronic inflammation-mediated breast cancer. Biochem Biophys Res Commun 472(3):401–409. [DOI] [PubMed] [Google Scholar]
- Wagner FF, Wesmall yi UM, Lewis MC, Holson EB. 2013. Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics 10(4):589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H, Pfizenmaier K, Scheurich P. 2003. Tumor necrosis factor signaling. Cell Death Differ 10(1):45–65. [DOI] [PubMed] [Google Scholar]
- Williams SM, Golden-Mason L, Ferguson BS, Schuetze KB, Cavasin MA, Demos-Davies K, Yeager ME, Stenmark KR, McKinsey TA. 2014. Class I HDACs regulate angiotensin II-dependent cardiac fibrosis via fibroblasts and circulating fibrocytes. J Mol Cell Cardiol 67:112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. 2007. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26(37):5310–5318. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31(4):449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S, Eom GH. 2016. HDAC and HDAC Inhibitor: From Cancer to Cardiovascular Diseases. Chonnam Med J 52(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Chen CN, Hajji N, Oliver E, Cotroneo E, Wharton J, Wang D, Li M, McKinsey TA, Stenmark KR, Wilkins MR. 2012. Histone deacetylation inhibition in pulmonary hypertension: therapeutic potential of valproic acid and suberoylanilide hydroxamic acid. Circulation 126(4):455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Lacasse P. 2008. Mammary tissue damage during bovine mastitis: causes and control. J Anim Sci 86(13 Suppl):57–65. [DOI] [PubMed] [Google Scholar]
- Zwergel C, Valente S, Jacob C, Mai A. 2015. Emerging approaches for histone deacetylase inhibitor drug discovery. Expert Opin Drug Discov 10(6):599–613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MAC-T cells under basal (10% FBS, 10 mg/ml insulin) conditions were A) treated with trichostatin A (TSA) for 24 hrs prior to TNFα stimulation (300pM). Cell lysates were harvested at 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phosphor-ERK, total-JNK and total-ERK. Data were quantified B) MAC-T cells were treated with class selective HDAC inhibitors MGCD0103 (MGCD; class I), DPAH (Class IIa) and Tubastating A (TubA; class IIb) prior to TNFα (300 pM) stimulation. Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for phospho-JNK, phosphor-ERK as well as total-JNK and total-ERK prior to quantifiication. C) MAC-T cells were treated with class I selective HDAC inhibitors MGCD and apicidin (Api) 24 hrs prior to TNFα (300 pM) stimulation. Cells lysates were harvested 15 min post-TNFα stimulation and immunoblotted as above. Data were quantified. All data were assessed via one-way ANOVA with Dunnett’s post-hoc analysis. Statistical significance was set at p<0.05.
A) MAC-T cells were transiently transfected with siRNA for two different regions of HDAC1 (siH1#1 or siH1#2), HDAC2 (siH2#1 or siH2#2), or siControl (siCon) using lipofectamine 3000 reagent for 48 hrs prior to TNFα stimulation (300 pM). Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for HDAC1, HDAC2, phospho-JNK, phopho-ERK, total-JNK and total ERK. Quantification was performed for ERK phosphorylation normalized to total-ERK. B) MAC-T cells were transiently transfected with independent siRNAs targeted to knockdown HDACs 1 and 2 in combination (siH1/2#1 or siH1/2#2) or siControl (siCon) using lipofectamine 3000 reagent for 48 hrs prior to TNFα stimulation (300 pM). Cell lysates were collected 15 min post-TNFα stimulation and immunoblotted for HDAC1, HDAC2, phospho-JNK, phopho-ERK, total-JNK and total ERK. Quantification was performed for JNK phosphorylation (Left Panel) and ERK phosphorylation (Right Panel) normalized to corresponding total MAPK protein. Significance was set at p<0.05. One-way ANOVA with Dunnett’s post-hoc analysis was performed for all data.