Abstract
Background:
The retinal homeobox (rx/rax) gene is a transcription factor expressed in the developing eye field that is necessary for normal eye development. rax is necessary for retinal specification and stem cell development. The genetic program of early retinal development, including rax expression, can be induced in naïve ectoderm by activation of IGF signaling. We have undertaken a microarray-based approach to identify rax-dependent IGF-induced genes.
Results:
We identified 21 IGF-induced genes that exhibit at least a 2-fold decrease in expression when rax expression is knocked down. 10 of these genes were expressed in the developing eye, 8 were expressed in the ciliary marginal zone of the mature tadpole retina, and 4 could significantly rescue the rax knockdown phenotype. One of these, the nei endonuclease VIII-like 3 (neil3) gene, rescued the rax knockdown phenotype to a remarkable degree. We found that neil3 is necessary for normal retinal lamination and retinal neuron differentiation.
Conclusions:
We have identified neil3 as a component of the rax genetic pathway necessary for normal retinal progenitor cell development. neil3 is involved in the base excision DNA repair pathway, suggesting that this pathway is essential for normal rax-dependent progenitor cell development in the mature retina.
Keywords: retinal homeobox gene, rax, Xenopus, retinal progenitor cells, neil3, microarray
INTRODUCTION
The vertebrate retina develops from the eye field, a region of the anterior neural plate adjacent to the prospective forebrain. This region is characterized by the intersection of several signaling pathways: the wnt and BMP pathways must be inhibited and IGF and FGF signaling must be active (Messina et al. 2015; Esteve and Bovolenta 2006; Atkinson-Leadbeater et al. 2014; Richard-Parpaillon et al. 2002; Pera et al. 2001; Andreazzoli 2009). The eye field splits into the two developing eyes and undergoes stereotypical morphological changes and movements to yield the developing retina. At first, this specific region of neuroepithelium can be considered to be comprised of only retinal stem cells. These cells are pluripotent and give rise to all types of retinal neurons and glia. In cold-blooded vertebrates and young birds, retinal progenitor cells, including a population of retinal stem cells, persist at the peripheral margin of the retina, known as the ciliary marginal zone (CMZ) and contribute to the development of the peripheral retina after development of the central retina is complete (Amato et al. 2004; Perron and Harris 2000; Perron et al. 1998; Fischer et al. 2013).
Retinal development is governed by a web of transcription factors, referred to as eye field transcription factors (EFTFs) conserved across phyla (Zuber et al. 2003). EFTFs cross-regulate each other to specify retinal progenitor cells. One such factor, the retinal homeobox (rax) gene, is near the apex of the EFTF hierarchy (Zuber et al. 2003; Mathers et al. 1997; Bailey et al. 2004). IGF signaling plays a necessary and sufficient role in retinal specification and development. In Xenopus, lack of IGF signaling results in a lack of eye development, whereas ectopic activation of the pathway results in the development of ectopic eyes (Pera et al. 2001; Richard-Parpaillon et al. 2002). Activation of IGF signaling in naïve Xenopus ectoderm results in the activation of the genetic program of retinal development, including expression of many EFTFs, including rax (Pera et al. 2001; Richard-Parpaillon et al. 2002).
rax is expressed in all retinal progenitor cells in the neuroepithilium of the developing retina and in retinal progenitor cells in the mature retina (Bailey et al. 2004; Mathers et al. 1997; Pan et al. 2006; Pan et al. 2010). rax is essential for retinal development. rax loss of function, through naturally occurring mutations, engineered mutations, or gene product knockdown, results in deficiencies in retinal development [reviewed in (Bailey et al. 2004)]. Further, rax is necessary for differentiating the eye field from surrounding neuroectoderm and for the cell and tissue movements necessary for early eye development (Giannaccini et al. 2013; Chuang and Raymond 2001; Rembold et al. 2006; Kenyon et al. 2001).
rax is thought to function as a transcriptional regulator. It contains a conserved paired-type homeodomain that functions in DNA binding (Mathers et al. 1997; Furukawa et al. 1997; Casarosa et al. 1997; Voronina et al. 2004; Pan et al. 2006), has been shown to interact physically with promoters of genes containing the conserved PCE-1 site (Pan et al. 2010; Kimura et al. 2000), and can activate reporter genes that include PCE-1 site-containing promoters (Pan et al. 2010; Pan et al. 2006; Chen and Cepko 2002; Wang et al. 2004; Irie et al. 2015; Reks et al. 2014). However, as is the case for many of EFTFs, not much is known regarding the identity of the genes regulated by rax. Recently, Giudetti and colleagues used a microarray approach to identify a collection of genes that are up-regulated in rax gain-of-function conditions and down-regulated in loss-of-function conditions (Giudetti et al. 2014). Simultaneously and independently, we have undertaken a microarray-based approach to identifying genes expressed downstream of rax; specifically, genes that are down-regulated in the absence of rax expression.
In this paper, we describe the identification of several genes that exhibit rax-dependent expression. We report that one of these, the nei endonuclease VIII-like 3 (neil3) gene, can rescue the rax knockdown phenotype to a remarkable extent. neil3 belongs to a class of DNA glycosylase homologous to the bacterial Fpg/Nei family (Takao et al. 2002; Rosenquist et al. 2003; Bandaru et al. 2002; Morland et al. 2002; Liu et al. 2010). These glycosylases initiate the first step in base excision repair by cleaving bases damaged by reactive oxygen species and introduce a DNA strand break via the associated lyase reaction (Liu et al. 2013; Wallace 2013). neil3 is expressed in proliferating cells in the brain, hematopoetic system, and some cancers (Torisu et al. 2005; Rolseth et al. 2008; Hildrestrand et al. 2009). In this paper we demonstrate that neil3 is genetically downstream of rax and that neil3 involved in normal retinal development.
RESULTS
The genetic sequence of early retinal development can be induced in Xenopus embryos and naïve ectoderm (animal cap explants) by activation of the insulin-like growth factor (IGF) signaling pathway (Pera et al. 2001; Richard-Parpaillon et al. 2002). Misexpression of IGF in intact embryos results in the development of ectopic eyes. Activation of IGF signaling in animal cap explants results in the expression of a set of anterior genes including the eye field transcription factors (EFTFs) pax6, rax, and six3. In our hands, animal caps explanted from embryos injected with RNA encoding IGF expressed rax and pax6 but not six6 (also known as optx2) (data not show), indicating that expression of early EFTFs (rax, pax6) was induced, but not that of a later EFTF (six6). We then performed a microarray screen to identify IGF-induced genes that exhibit expression dependent on rax expression level. RNA was isolated from animal caps explants prepared from embryos injected with RNA encoding IGF and either rax antisense or control morpholino oligonucleotides (ASMO or COMO, respectively) and used to screen a Xenopus laevis microarray. We identified 21 genes expressed at least 2.5 fold lower in rax knockdown animal caps as compared to control explants (not shown). As a first validation step, we compared the expression patterns of these genes to that of rax by wholemount in situ hybridization. We determined that 10 of these genes exhibited an expression pattern overlapping that of rax; that is, expression in developing eyes of neurula and tailbud embryos (Table 1 and Figure 1). Most of these genes were expressed in the developing brain and branchial arches as well as the eye: abcf2 (Figure 1A), cbx5 (Figure 1B), e2f7 (Figure 1F), fam50a.S (Figure 1G), bnip3 (Figure 1H), sox11 (Figure 1I), and fam50a.L (Figure 1J). One gene, cox4i1, was also expressed in the developing pronephros (Figure 1D). One gene, neil3 were expressed predominantly in the eye (Figure 1E). In summary, the expression of these 10 genes overlapped with rax expression in the developing eye.
Table 1 –
Summary of Candidate rax Downstream Genes Expressed in the Developing Eye.
| GENE | RAX KD | CMZ EXP | RESCUE | REFERENCES (XENOPUS) | UNIGENE ID |
|---|---|---|---|---|---|
| abcf2 | 162.0 | + | + | 15223337 | XI.4203 |
| cbx5 | 2.5 | + | − | 12805230 | XI.22796 |
| loc1 | 50.0 | − | ND | 16916602 | XI.44108 |
| cox4i1 | 16.0 | +* | ND | 11943455 | XI.48853 |
| neil3 | 2.5 | + | + | 26751644 | XI.60649 |
| e2f7 | 2.0 | + | + | XI.16369 | |
| fam50a.S | 2.0 | +* | ND | 27762356 | XI.19192 |
| bnip3 | 1.8 | − | ND | 24360906 | XI.59322 |
| sox11 | 4.0 | + | + | 8765746 | XI.891, XI.782582 |
| fam50a.L | 1.9 | + | − | 27762356 | XI.3766 |
GENE – gene name according to Xenbase.org; RAX KD – fold decrease in expression of gene upon knockdown of rax expression; CMZ EXP – expression in the ciliary marginal zone (CMZ) visualized by in situ hybridization on sectioned material as shown in Figure 2 (+ denotes CMZ expression, − denotes lack of CMZ expression, * denotes ubiquitous expression within the neural retina); RESCUE – significant rescue of the rax knockdown phenotype by microinjection of RNA encoding the gene (significance is stated in the text); REFERENCES (XENOPUS) – PubMed ID (PMID) of literature reference to the Xenopus ortholog of the gene, where it exists; UNIGENE ID – UniGene ID of Xenopus laevis ortholog of the gene.
Notes:
“loc” denotes loc100498624 and is also known as MGC86492
sox11.L and sox11.S have individual Unigene IDs.
Figure 1 – Ten of the genes identified as exhibiting rax-dependent expression are expressed in the developing eye.
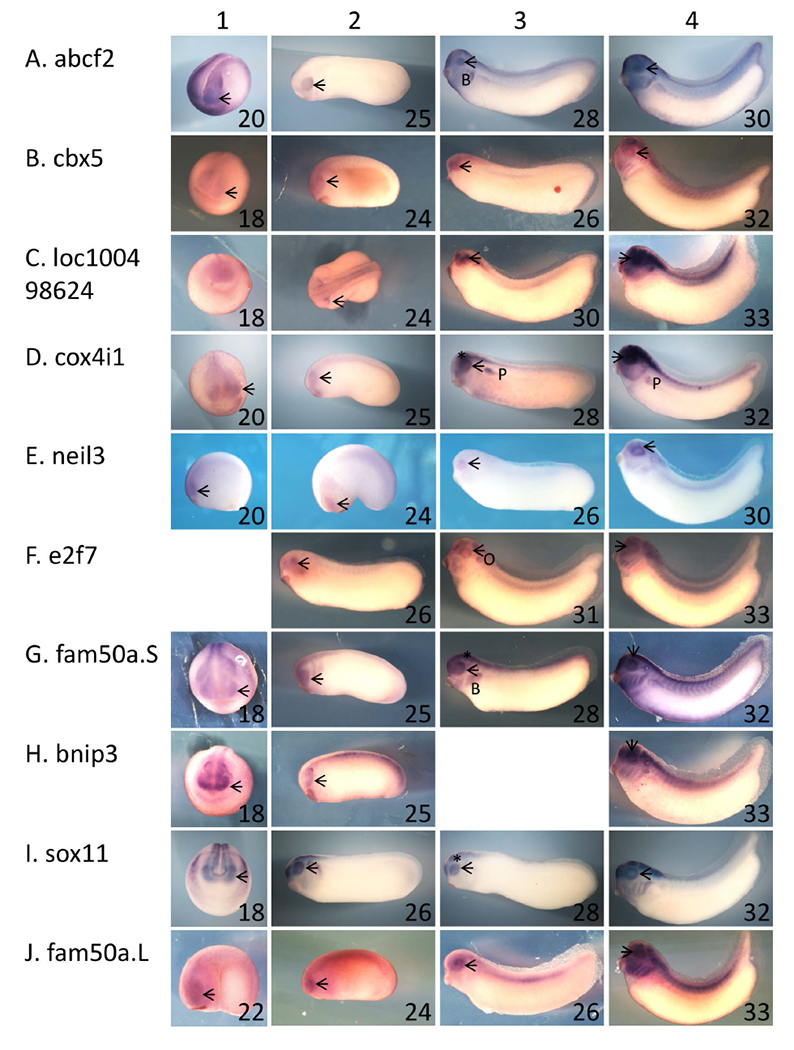
A – J: Xenopus laevis embryos were fixed and subjected to in situ hybridization using antisense riboprobes specific for 10 of the genes identified as exhibiting rax-dependent expression in the microarray screen described in the text. Genes/riboprobes are identified to the left of each row. Embryos used were late neurula (st 18 – 20, column 1), with the exception of J1 (st 22), early tailbud (st 24 and 25, column 2), mid tailbud (st 25 – 31, column 3), and mid-late tailbud (st 30 – 33, column 4). Lateral views are shown for all embryos except for A1, B1, C1, D1, G1, H1, I1, where anterior views are shown. In all cases, dorsal is towards the top of the figure. Abbreviations and marks: arrowhead – eye; * - brain; B – branchial arches; P – pronephros.
Next, we repeated IGF treatment of control or rax knockdown animal caps and analyzed expression of these 10 genes by quantitative RT-PCR (Table 1). We found that expression of these genes was reduced 1.8 – 162 fold in rax knockdown animal caps. Thus, all 10 of these genes exhibited expression overlapping with that of rax and were dependent on rax expression for induction by IGF and were therefore considered candidate rax downstream genes.
Of these 10 genes, 8 were expressed in the ciliary marginal zone (CMZ), the region of the tadpole retina containing retinal progenitor cells: abcf2, cbx5, cox4i1, e2f7, sox11, fam50a.L, fam50a.S, and neil3 (Figure 2, circled). cbx5, e2f7, fam50a.L, and neil3 were expressed exclusively in the CMZ (Figure 2B, F, J, and E, respectively) while abcf2 and sox11 were expressed in both the inner and outer nuclear layers (Figure 2A,I). cox4i1 and fam50a.S appeared to be expressed ubiquitously in the retina (Figure 2D,G), and were not studied further. We continued our investigations using this set of 6 genes: abcf2, cbx5, e2f7, sox11, fam50a.L, and neil3.
Figure 2 – Six of the genes identified in the microarray screen are expressed in the ciliary marginal zone.
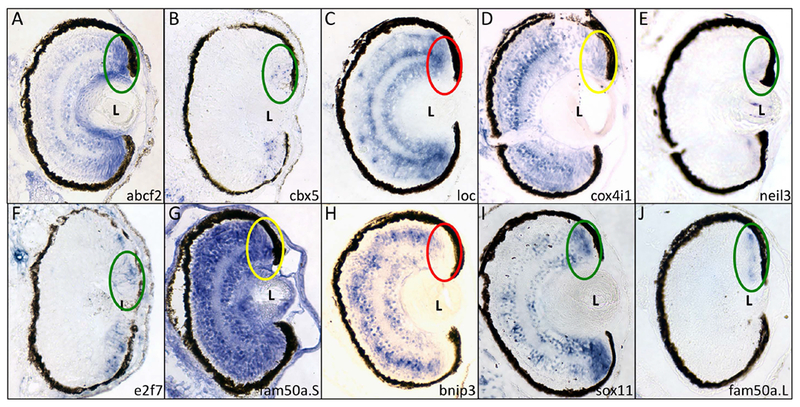
Fixed X. laevis tadpoles (st 41) were paraffinized, sectioned and subjected to in situ hybridization using antisense riboprobes for the 10 genes expressed in the eye (see Figure 1). Genes/riboprobes are identified in the lower left of each panel. Sections are presented with dorsal side towards the top of the figure. The dorsal CMZ is indicated with an oval. Key: green – positive expression in CMZ; red – negative expression in CMZ; yellow – positive expression in CMZ and ubiquitous expression in retina. L – lens.
To further analyze the genetic relationship between rax and the set of validated genes, we tested the ability of each gene to rescue the rax knockdown phenotype. As reported previously (Bailey et al. 2004; Chuang and Raymond 2001), reduction of rax expression by antisense morpholino oligonucleotides (ASMOs) results in small or no eyes (microphthalmia or anophthalmia, respectively) in approximately 80% of injected embryos (Figure 3A). Injection of a 4-mismatch control morpholino oligonucleotide typically resulted in approximately 60 - 80% of injected embryos exhibiting no detectable phenotype (data not shown). For each gene, we determined the amount of RNA that could be microinjected with no obvious phenotypic effect on eye development, typically 10 pg (not shown). Co-injection of abcf2, sox11, e2f7, and neil3 RNAs significantly diminished the severity of the rax knockdown phenotype (Figure 3A), while co-injection of cbx5 and fam50a.L did not. neil3 rescued the rax knockdown phenotype to a remarkable degree (10 fold), compared to abcf2, sox11, and e2f7 (2 – 3 fold). Examples of phenotypic rescue by neil3 are shown in Figure 3B. Our results to this point suggest that: four genes discovered through this microarray screen (abcf2, sox11, e2f7, and neil3) are genetically downstream of rax. All four genes are dependent on rax expression for expression, are expressed in the developing eye in general, are expressed in the CMZ in particular, and can rescue the phenotypic effects of rax knockdown to a significant degree. One gene, neil3, could rescue the rax knockdown phenotype to a remarkable degree; the remainder of this paper investigates the genetic relationship between rax and neil3.
Figure 3 – Four of the genes identified in the microarray screen can significantly rescue the rax knockdown phenotype.
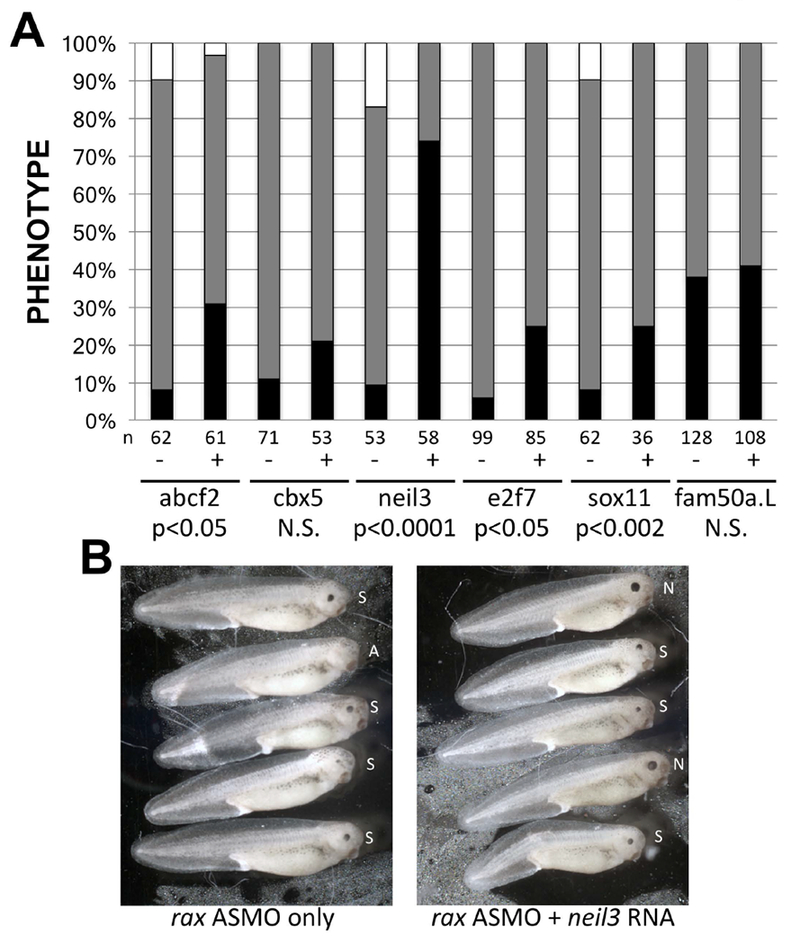
X. laevis embryos were injected with rax antisense morpholino oligonucleotides (ASMO) with or without mRNA encoding each of the 6 microarray screen genes expressed in the CMZ (Figure 2). RNAs and lissamine-tagged MOs were injected into a dorsal blastomere at the 4-cell stage and embryos were screened for one-sided injection and normal, small, or absent eyes at st 41. A. Comparison of frequency of normal (black bar portion), small (gray bar portion), and absent (white bar portion) eye phenotypes in embryos injected with rax ASMO with or without RNAs encoding microarray genes (marked “+” or “−”, respectively, and identified at bottom of graph). Statistical significance of rescue was determined (student’s t-test performed using frequency of normal eyes in RNA injected vs uninjected samples); p-values are shown at the bottom of the graph. N.S. = not significant (p>0.05). Number of embryos for each condition (n) is shown below each column. B. Sampling of embryos injected with rax ASMO with or without neil3 RNA (right and left panels, respectively). Embryos were injected with lissamine-tagged rax ASMO with or without neil3 RNA. Shown are injected sides of st 41 embryos scored as having normal (N), small (S), or an absent (A) eye.
neil3 is genetically downstream of rax.
To further investigate the genetic relationship between rax and neil3, we performed overexpression and co-knockdown studies. First, we investigated the ability of rax to activate neil3 expression in naïve ectoderm. RNA encoding rax was injected into embryos; animal caps explants were prepared and assayed for neil3 expression by real time RT-PCR. We found that expression of rax induced expression of neil3 in a dose-dependent manner Figure 4A). Thus, even in a non-neural explant system, rax can induce expression of neil3.
Figure 4 – neil3 and rax are in a common genetic pathway.
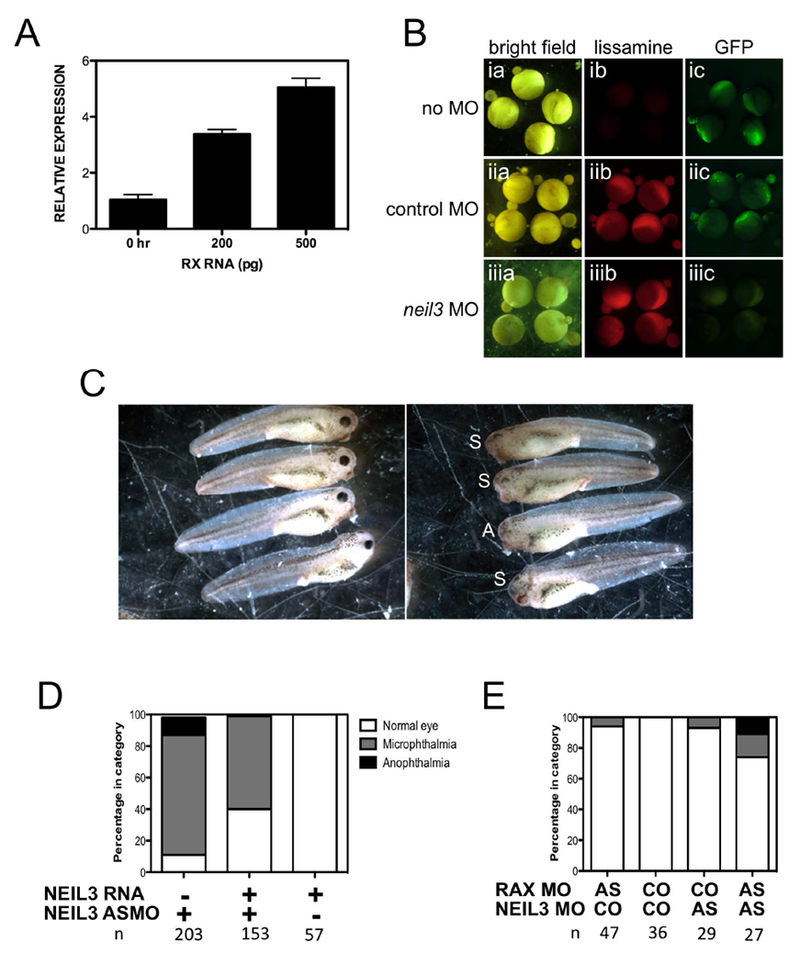
A. neil3 is expressed in animal caps prepared from rax-injected embryos in a dose-dependent manner. X. laevis embryos were injected with rax RNA (amount shown in graph). Animal caps were explanted when embryos reached early blastula stage and cultured until early gastrula stage. RNA was extracted and neil3 expression was determined by quantitative RT-PCR. Results are normalized to an internal control and then to the neil3 expression level in caps prepared from uninjected embryos. B. The neil3 antisense morpholino oligonucleotide (ASMO) can inhibit expression of a neil3-gfp reporter. The reporter contains a portion of neil3 5’-UTR and coding region, including the ASMO target, fused in-frame with the GFP coding region downstream of the CMV promoter. The reporter was injected alone (top row, ia-c) or with lissamine-tagged COMO (middle row, iia-c) or lissamine-tagged neil3 ASMO (bottom row, iiia-c). Embryos were imaged under bright field (left column, ia-iiia), red fluorescence to detect the lissamine-tagged MOs (middle column, ib-iiib), or green fluorescence to detect GFP (right column, iiia-iiic). C. Injection of a neil3 ASMO results in embryos with small or no eyes. X. laevis embryos were injected unilaterally (one dorsal blastomere at the 4-cell stage) with neil3 ASMO. Left panel: normal eye development on uninjected side. Right panel: small (S) or absent (A) eyes on the injected side. D. Co-injection of neil3 RNA partially rescues knockdown phenotype. X. laevis embryos were injected with neil3 ASMO, RNA, or a mixture of both. Embryos were cultured to st 41 and scored for eye phenotype on the injected side. E. Co-injection of neil3 and rax ASMOs results in a more severe phenotype than injection of either ASMO alone. X. laevis embryos were injected with a low dose of either rax or neil3 ASMO alone or a mixture of both. Embryos were cultured to st 41 and scored for eye phenotype on the injected side. Number of embryos for each condition (n) is shown below each column in D and E.
To further explore the genetic relationship between rax and neil3, we investigated the effects of knocking down expression of both genes. We obtained ASMOs designed to knock down neil3 expression by inhibiting its translation. To verify ASMO function, we prepared a neil3-gfp reporter and co-injected it with lissamine-tagged control morpholino oligonucleotide (COMO) or neil3 ASMO (Figure 4B). We found that the reporter was expressed in control and in COMO-injected embryos (Figure 4Bic and Figure 4Biic, respectively) but expressed at a reduced level in neil3 ASMO-injected embryos (Figure 4Biiic), demonstrating that the neil3 ASMO can effectively and specifically reduce expression of the reporter. Injection of the neil3 ASMO results in microphthalmic and anophthalmic embryos (Figure 4C,D), similar to the Rx knockdown phenotype. Injection of the neil3 ASMO resulted in approximately 90% of embryos exhibiting a small or absent eye. Co-injection of a neil3 RNA containing silent mutations that render it non-targetable partially rescued the phenotype – approximately 3-fold more embryos with normal eyes (Figure 4D). These results demonstrate that neil3 expression is specifically necessary for normal eye development. We further found that co-injection of low doses of the rax and neil3 ASMOs result in more severe eye phenotypes than either ASMO alone (Figure 4E). Injection of the ASMO mixture resulted in approximately 30% of embryos having either small or absent eyes. This represents a higher frequency of affected embryos, since injection of either ASMO alone resulted in approximately 5% of embryos expressing a phenotype. Further, injection of the ASMO mixture results in a more severe phenotype since injection of each AMSO alone resulted in only microphthalmia. Taken together, these results strongly indicate that rax and neil3 are part of a common genetic pathway and that rax is genetically upstream of neil3.
neil3 is necessary for normal retinal development.
As discussed above, knockdown of neil3 expression results in microphthalmia or anophthalmia. We further analyzed small eyes in neil3 knockdown embryos. We found that the retinas of neil3 knockdown embryos do not exhibit normal lamination or cellular differentiation. Histological analysis of neil3 knockdown embryos revealed that retina is severely disorganized (Figure 5B,C). There is little evidence of lamination and the retina has several apparent photoreceptor rosettes. Further, neil3 knockdown retinas have fewer differentiated retinal neurons (Figure 5E,G). Predictably, these neurons are not arranged into discernable lamina. Surprisingly, the eyes from the uninjected sides of these embryos were not completely normal. The ventral portions of these eyes were not histologically normal – cells in the INL and ONL exhibited abnormally elongated and spindle shaped nuclei (Fig 5A) and lacked normal expression of markers of differentiated retinal neurons (Fig 5D,F). Lineage tracing experiments have demonstrated that the retina develops primarily from ipsilateral dorsal ectodermal blastomeres but that there is contribution from contralateral blastomeres as well (Moody 1987a, b). These results demonstrate that NEIL3 is necessary for normal development and differentiation of the neural retina.
Figure 5 – neil3 knockdown embryos exhibit deficits in retinal development.
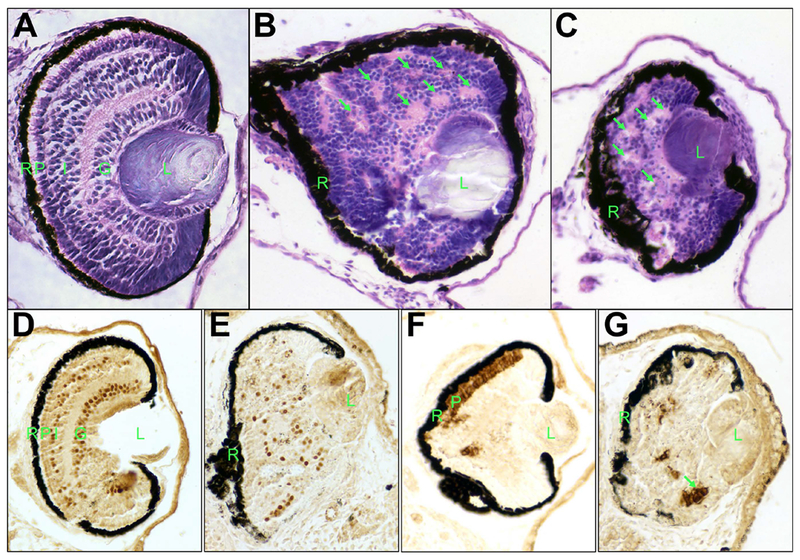
A – C. Knockdown of neil3 results in a disorganized retina. Histological staining of retinas from uninjected (A) or injected (B,C) sides of embryos injected with neil3 antisense morpholino oligonucleotide (ASMO), sectioned, and stained with hemotoxylin/eosin. D – F. Knockdown of neil3 results in aberrant retinal cell differentiation. Embryos injected with neil3 ASMO were fixed and subjected to immunohistochemistry using antibodies against Islet-1 (D,E) or rhodopsin (F,G). Panels D and F show uninjected side and E and G show injected sides. R – retinal pigmented epithelium (RPE); P – photoreceptor layer; I – inner nuclear layer (INL), G – ganglion cell layer; L – lens. Arrows indicate examples of putative photoreceptor rosettes.
neil3 is expressed in the developing eye from neural tube to tadpole stages and is necessary for normal retinal development. To further investigate the basis for the knockdown phenotype, we analyzed the expression of RPC markers in neil3 knockdown embryos prior to retinas maturation, at neural tube and tailbud stages. We found that expression of early pan-progenitor markers, rax and pax6, was weaker in embryos injected with neil3 ASMO (Figure 6A-D). Additionally, these markers are expressed in a smaller domain, consistent with the micro-/an-ophthalmia phenotype observed. These results indicate that there may be abnormalities in induction or maintenance of retinal progenitor cells. We next analyzed expression of neurod1, a marker of a subset of RPCs that are proliferating and undifferentiated, but are no longer uncommitted retinal progenitor cells (Perron et al. 1998). neurod1 expression was essentially absent in the developing eyes of neil3 knockdown embryos but largely unaffected in other expression domains, such as the brain and cranial ganglia (Figure 6E,F). Similar results were observed for expression of another late RPC marker, notch (not shown). These results suggest that neil3 expression is important for the development of later RPCs.
Figure 6 – Molecular markers of retinal progenitor cells (RPCs) are expressed abnormally in neil3 knockdown embryos.
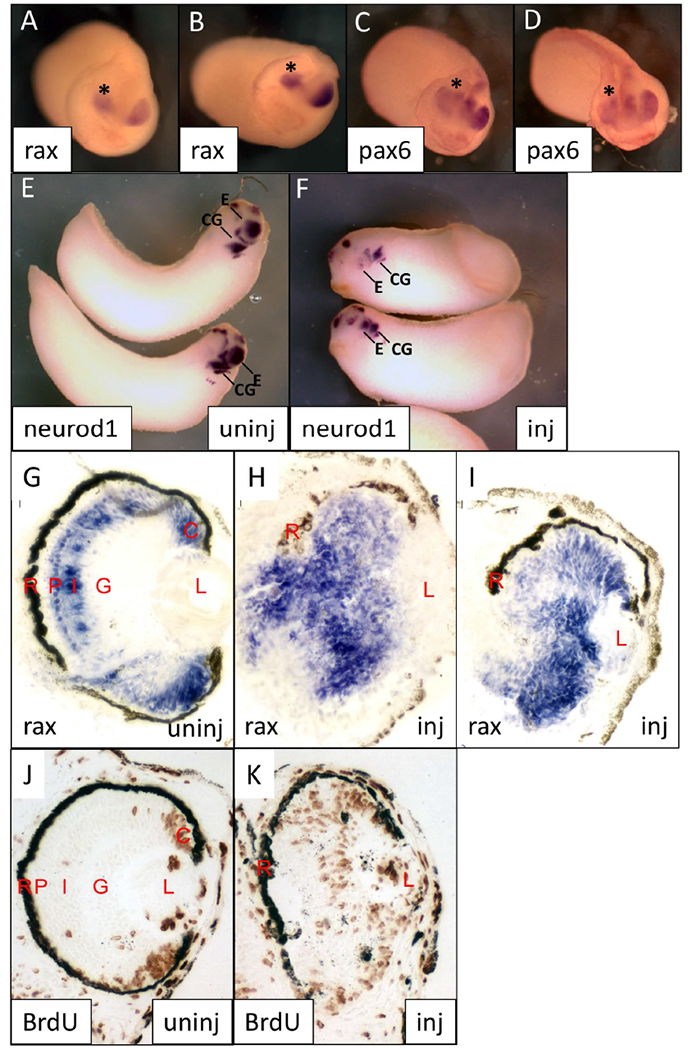
Embryos were injected with neil3 antisense morpholino oligonucleotides (one dorsal blastomere at 4-cell stage), cultured, and subjected to wholemount (A-F) or section (G-I) in situ hybridization using the indicated riboprobes. A-D. Early tailbud stage embryos stained for rax (A,B) or pax6 (C,D). Embryos or oriented with dorsal towards the top of the panel and anterior facing the viewer. Eye on uninjected side is indicated with an asterisk (*). E,F. Early tailbud embryos stained for neurod1 expression. Uninjected side is shown in E and injected side is shown in F. G-K. St 38 embryos stained for rax expression (G-I) or BrdU incorporation (J,K). Examples of eyes from uninjected (G,J) or uninjected (H,I,K) embryo sides are shown. Abbreviations: C – ciliary marginal zone; CG – cranial ganglia; E – eye; G – ganglion cell layer; I – inner nuclear layer; L – lens; P – photoreceptor layer; R – retinal pigmented epithelium.
To further understand the differentiation state of the NEIL3 knockdown retina, we analyzed the expression of rax by in situ hybridization and cell proliferation by BrdU incorporation (Figure 6 G-K). At st 38, we found that the retina of neil3 knockdown embryos had widespread rax expression (Figure 6H,I) and BrdU incorporation (Figure 6K). At this stage, rax expression is normally primarily observed in the CMZ and photoreceptor layer (Figure 6G) and BrdU incorporation is generally limited to the CMZ (Figure 6J). Thus, in addition to the lack of lamination and expression of markers of differentiated retinal neurons (Figure 5), neil3 KD retinas contain persistent RPCs. Taken together, these results suggest that NEIL3 may be necessary for retinal progenitor cell progression from uncommitted progenitor cell to neuronal progenitor cell.
NEIL3 function in retinal development requires DNA binding and DNA glycosylase activity.
As a DNA glycosylase, Neil3 is similar to other members of the Type VIII endonuclease family, including a nearly invariant aspartate residue that is part of the DNA glycosylase active site, D166 (Bandaru et al. 2002). Mutation of this aspartate impairs DNA glycosylase function in Type VIII endoglycosylases from yeast to man. To investigate the involvement of NEIL3 activity as a DNA glycosylase in its function in the retina, we mutated D166 to asparagine (D166N) and assayed its ability to rescue the rax knockdown phenotype. We found that D166N was impaired in its ability to rescue the rax KD phenotype as compared to the wild-type neil3 (Figure 7B). These results demonstrate that DNA glycosylase function is necessary for Neil3 function downstream of rax in retinal development.
Figure 7 – Stereotypical peptide domains are required for neil3 function.
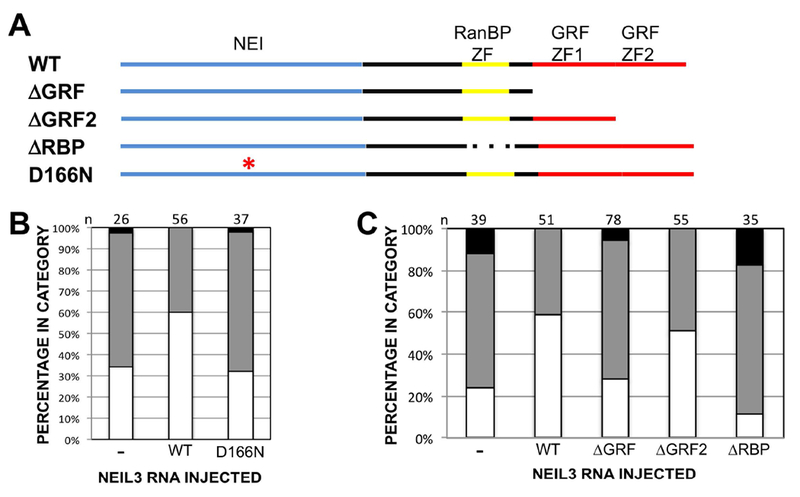
A. Diagram of neil3 protein highlighting stereotypical peptide domains: NEI domain (blue), RanBP-type zinc finger (yellow), and GRF-type zinc fingers (red). Wild type (top line) and mutated versions are depicted. ΔGRF: both GRF zinc fingers deleted; ΔGRF2: second GRF zinc finger deleted; ΔRBP: RanBP zinc finger deleted; D166N: point mutation resulting in aspartate 166 substitution by asparagine. B,C. RNA encoding neil3 containing the D166N mutation (B) or deletions of RanBP- or GRF-type zinc fingers (C) does not rescue the rax knockdown phenotype to the same extent as wild type neil3. Open/white portion of graph bar: normal eye; gray: microphthalmia; black: anophthalmia. X. laevis embryos were injected with rax antisense morpholino oligonucleotides (ASMO) alone or with mRNA encoding wild type or nutated neil3. RNAs and lissamine-tagged MOs were injected into a dorsal blastomere at the 4-cell stage and embryos were screened for one-sided injection and normal, small, or absent eyes at st 41. Number of embryos for each condition (n) is shown below each column in B and C.
Neil3 requires zinc fingers for function and sequence-specific DNA binding.
Neil3 is an atypical member of the fpg/Nei family of DNA glycosylases since it encodes zinc fingers not found in other family members (Bandaru et al. 2002; Rosenquist et al. 2003; Takao et al. 2002; Morland et al. 2002). Two of these zinc fingers are related to GRF zinc fingers, thought to function in DNA binding. Neil3 also contains an additional zinc finger that is related to one found in RanBP, thought to function in protein-protein interaction. We found that these zinc fingers are involved in the function of Neil3 in retinal development. Deletion of both GRF zinc fingers abrogates the ability of Neil3 to rescue the rax knockdown phenotype (Figure 7C). Further, deletion of only the C-terminal GRF zinc finger results in an intermediate level of rescue, indicating that both GRF zinc fingers are involved in Neil3 function. Deletion of the RanBP-like zinc finger results in enhancement of the rax knockdown phenotype (Figure 7C), indicating that this construct may act in a dominant negative manner.
DISCUSSION
We identified 4 genes expressed downstream of rax: abcf2, e2f7, neil3, and sox11. These genes could rescue the rax knockdown phenotype to a significant extent. Of these genes, only abcf2 is not overtly associated with progenitor cell development. While e2f7 has not been studied in Xenopus, e2f7 is an atypical E2F and may be involved in regulating proliferation by repressing transcription of traditional E2F target genes [reviewed in (Lammens et al. 2009)]. Interestingly, sox11 has been found to be involved in Xenopus retinal development (Cizelsky et al. 2013); knockdown of sox11 results in a phenotype reminiscent of the one we observed for knockdown of neil3. Overexpression of neil3 rescued the rax knockdown phenotype to a surprising degree. It was expected that any single rax downstream gene product would only partially rescue the rax knockdown phenotype and that co-expression of several downstream genes might be required for near-full rescue. The high degree of rescue exhibited by neil3 suggests that it may be a primary target of rax and that it may play a major role in mediating the expression of other rax downstream genes. Additional studies are required to determine if neil3 is a direct target of rax.
An independent study characterized the rax transcriptome and identified several direct rax targets ( ). The set of rax downstream genes identified in that study differs from that found in our study. Our analysis of the data from this study (GEO accession number GSE35918) revealed that the rax downstream genes identified in our study were not expressed at consistent levels in the replicate samples from the Guidetti et al study (not shown). This likely resulted in exclusion of these genes from consideration. The differences in genes identified in the two studies could also be due to differences in study design. The study by Giudetti et al focused on genes with changed expression levels in both rax gain-of-function (RNA overexpression) and loss-of-function (gene knockdown) in whole embryos while our study identified rax—dependent genes in animal caps overexpressing IGF. Even though overexpression of IGF in animal caps recapitulates the genetic pathway of early eye development ( ), it may differ substantially from the natural course of eye development in the context of an intact embryo. Finally, expression of neil3 was generally lower in rax lof embryos than in control embryos in the Giudetti et al dataset, but was not among the genes with the most highly significant change in expression.
The precise roles of neil3 in retinal progenitor cells are uncertain, but likely involve DNA repair. neil3 is a DNA glycosylase involved in the base excision repair (BER) pathway that is also necessary for neural stem cell biology (Sejersted et al. 2011; Reis and Hermanson 2012; Regnell et al. 2012; Hildrestrand et al. 2009). First, neil3 is activated upon induction of proliferation or release from quiescence (Neurauter et al. 2012). Second, it is necessary for neural stem cells to exhibit normal differentiation potential and essential for adult neurogenesis in the mouse hippocampus (Reis and Hermanson 2012; Regnell et al. 2012; Sejersted et al. 2011). In the latter of these studies, neil3 knockout mice exhibited deficits in learning and memory (Regnell et al. 2012). These mice did not have overt abnormalities in eye morphology or function (Sejersted et al. 2011; Torisu et al. 2005). In our case, we observed gross abnormalities in retinal development upon knockdown of neil3 expression in Xenopus. We found that knockdown of Xenopus neil3 resulted in small eyes containing poorly differentiated retinas. These results are reminiscent of results obtained by Reis and Hermanson demonstrating that knockdown of neil3 expression results inhibited differentiation of neural stem cells (Reis and Hermanson 2012). These cells survived but exhibited decreased rates of proliferation and signs of senescence. Knockdown neil3 neural stem cells exhibited elevated accumulation of damaged DNA. Further studies are required to determine the prevalence of DNA damage in normal and neil3 knockdown frog retinas.
Notably, knockout of neil3 in mice did not show abnormalities in embryonic neurogenesis; instead, defects in adult neurogenesis were observed (Regnell et al. 2012; Sejersted et al. 2011). Retinal development in cold-blooded animals occurs in two phases – early development of the retina from embryonic neuroepithelium and later development from the ciliary marginal zone (CMZ) and its precursors (Straznicky and Gaze 1971; Johns 1977; Perron and Harris 2000; Harris and Perron 1998; Fischer et al. 2013; Hollyfield 1971, 1968). Almost all of the Xenopus tadpole retina is derived from the CMZ – only the most central portion of the retina is derived from the initial embryonic neuroepithelium (Straznicky and Gaze 1971). Perhaps CMZ retinal progenitor cells are more like neural stem cells found in the adult mammalian brain than embryonic neuroepithelial stem cells and cannot develop and differentiate properly without normal levels of neil3 expression.
We found that each of the stereotypical neil3 peptide domains was essential for its ability to rescue the rax knockdown phenotype. Deletion of one or both GRF-type or the RanBP2-like zinc fingers resulted in more severe lack of rescue of the rax knockdown phenotype. These zinc fingers are thought to mediate the DNA interactions necessary for neil3 function (Wallace et al. 2017; Wallace 2013). We also found that mutation of a conserved aspartate in the Nei domain impaired the ability of NEIL3 to rescue the Rx knockdown phenotype, suggesting that the DNA glycosylase function of NEIL3 was necessary for its function in retinal development in the context of the rax genetic pathway.
Our results suggest that neil3 may be a major mediator of rax function as a regulator of retinal progenitor cell gene transcription. neil3 has been linked to gene regulation through its ability to remove oxidized guanine lesions from quadruplex DNA structures found in gene regulatory regions (Zhou et al. 2013). G-quadruplex DNA structures are found throughout the genome and are estimated to be contained in the promoter regions of approximately 40% of transcribed genes potentially including genes that harbor multiple G-rich transcriptional regulatory elements, such as response elements for members of the KLF family that includes SP1 (Rigo et al. 2017; Rhodes and Lipps 2015). KLF transcription factors occur relatively frequently in promoters, adding to the notion that G-quadruplexes may be involved in transcriptional regulation of many genes. C-myc is a notable G-quadruplex-containing gene that is important for retinal progenitor cell biology, although it is not clear if neil3 plays a role in its regulation (Zhou et al. 2015). Interestingly, repair of DNA oxidation is also linked to epigenetic regulation of gene expression. DNA oxidation has been shown to be a driver of estrogen receptor-induced gene regulation involving histone demethylation (Perillo et al. 2008). In plants, a zinc finger-containing DNA glycosylase, DEMETER, is involved in DNA demethylation (Gehring et al. 2006; Morales-Ruiz et al. 2006). Interestingly, neil1 and neil2, but not neil3, have been implicated in enzymatic DNA demethylation involving removal of 5-methylcytosine oxidized by Tet enzymes (Schomacher et al. 2016). Taken together, these findings paint an intriguing picture of a potential role for neil3 in regulating gene expression downstream of rax in progenitor cells of the maturing retina.
EXPERIMENTAL PROCEDURES
Embryos.
Embryos were obtained by in vitro fertilization, staged, and microinjected with RNAs or morpholino oligonucleotides as described previously (Sive et al. 2000; Nieuwkoop 1994). All work with live animals was performed in compliance with an animal use protocol approved by the Nationwide Children’s Hospital Research Institute Institutional Animal Care and Use Committee.
Genes.
Names of genes identified by microarray screening are as listed in Xenbase (http://www.xenbase.org/, RRID:SCR_003280).
Plasmids.
The neil3 EST (IMAGE:5155774) was obtained from Open Biosystems (http://www.openbiosystems.com/). To prepare pCS2-NEIL3, the coding region of NEIL3 was amplified from the EST using primers 5’-GATCGAATTCAATTATGGTGGAGGGTCC-3’ and 5’-GATCCTCGAGCTACTCTGTTTTTGCCCA-3’ and subcloned into pCS2 using EcoRI and XhoI (included in primers – underlined). To prepare Neil3 lacking both GRF zinc fingers pCS2-NEIL3ΔGRF12), pCS2-NEIL3 was digested by AflII and XbaI. Overhangs were filled in using Klenow and self-ligated. To delete only the second GRF zinc finger, pCS2-NEIL3 was digested using NcoI and XhoI. Overhangs were filled in using Klenow and self-ligated.
To delete the RanBP zinc finger, site direct mutagenesis was performed using the QuickChangeXL kit (Stratagene) according to the manufacturer’s instructions and the following primers 5’-AACATGTAGCAACAAGTGATATCTCGTCTCTTGCAGTT-3’ 5’-AACTGCAAGAGACGAGATATCACTTGTTGCTACATGTT-3’. Successful mutations were identified using an EcoRV site included in the primers (underlined). The glycosylase domain mutation, pCS2-NEIL3D166N, was generated by site directed mutagenesis of pCS2-NEIL3 using the following mutagenic primer and its reverse complement (changed nucleotides are in lower case): 5’-CTATGCGACATTCTGCTTaATCAGATGATTCTACCTG-3’.
To prepare the reporter for testing efficacy of the antisense morpholino oligonucleotide (ASMO), pCS2-N’-NEIL3-SGP, the N-terminal portion of NEIL3 cDNA, including the 5’ UTR and the ASMO target, was amplified using 5’-CTAGAAGCTTCGCGAGGGGCAATTAT-3, and 5’-GATCTCTAGAGCTGCATCCAGTAAGTGA-3’, and subcloned into the HindIII and XbaI sites of pCS2-SGP. This plasmid expresses a fusion protein containing the N-terminal portion of Neil3, including the ASMO target, and GFP under the control of the CMV promoter.
To prepare the neil3 rescue RNA construct, containing 5 silent mutations, decreasing its ability to be targeted by the NEIL3 antisense morpholino oligonucleotide, the neil3 coding region was amplified from pCS2-NEIL3 and inserted into pCS2. Amplification of the neil3 coding region was performed using the following the same reverse primer as before and the following forward primer (changed nucleotides are in lower case, EcoRI site used for subcloning is underlined): GATCGAATTCGATGGTtGAaGGTCCaGGcTGcACGCTGAATGG.
RNAs.
To make neil3 antisense riboprobe, the coding region of neil3 was amplified and TA-cloned into pCRII (TOPO TA Kit, Invitrogen). This plasmid was linearized using BamHI and digoxygenin-labelled antisense probe was transcribed by using T7 RNA polymerase. Riboprobes for rax, pax6, and neurod were prepared as described previously (Martinez-De Luna et al. 2011; Pan et al. 2006).
Capped RNAs for microinjection were prepared using mMessage mMachine kits (Ambion) and purified using RNA Mini Columns (Roche), and stored at −80°C (Krieg and Melton 1984). pCS2-NEIL3 and different neil3 truncations were linearized using NotI and transcribed using Sp6 RNA polymerase. pCS2-NSL-RanBP-GRFs-VP16/Engrailed and pCS2-NSL-GRFs-VP16/Engrailed were linearized by KpnI and transcribed using Sp6 RNA polymerase.
Morpholino oligonucleotides.
Morpholino oligonucleotides (MOs) with 5’-lissamine tags were designed by and obtained from Gene Tools, LLC, to inhibit translation of both X. laevis rax.S and rax.L (also known as Rx1A and Rx2A). rax antisense MO (rax ASMO): 5’-CTGTGCAGGTGCATTGAGGACCCTT-3’. rax control MO (COMO), containing five mismatches (in lower case): 5’-CTcTGCAcGTGgATTGAGcACCgTT-3’. neil3 knockdown was achieved using neil3 ASMO: 5’-GCAGCCCGGACCCTCCACCATAATT-3’; standard COMO: 5’-CCTCTTACCTCAgTTACAATTTATA-3’. For single MO experiments, embryos were injected with 10 nl of 0.25 mM MO solution. For simultaneous knockdown of rax and neil3, embryos were injected with 10 nl of solution containing 0.05 mM each MO.
Microarray.
IGF RNA (4 ng) was injected near the animal pole of embryos at 1-cell stage. When embryos reached the 4-cell stage, 0.25 mM of either Rx ASMO or COMO was injected into all four cells. Animal caps were then cut at stage 9 and cultured in 1 X MMR until sibling embryos reached stage 13. Animal caps were collected and RNA extracted using Trizol (Invitrogen) following manufacturer’s protocol. RNA extracted from these animal caps (from embryos co-injected with IGF and Rx ASMO or COMO) was used to probe Affymetrix Xenopus laevis 1.0 microarrays following manufacturer’s recommendations by the Nationwide Children’s Hospital Genomics Core. In brief, GeneChips were scanned in a GeneChip Scanner 3000 (Affymetrix Inc.). CEL files were generated from DAT files by GeneChip Operating Software (GCOS, Affymetrix Inc.). The probe set signals were generated by the RMA algorithm in ArrayAssist 3.4 (Stratagene) and were used to determine differential gene expression by pairwise comparisons. The fold of gene expression alteration was used for further interpretation of the microarray data.
Histology, in situ hybridization, and immunohistochemistry.
Paraffin-embedding embryos were sectioned at 8 μM. R.T.U. Vector Staining kit was used to stain section with different antibody according to manufacturer’s instructions. Primary antibodies were used at the following dilutions: mouse anti-rhodopsin (RetP1; Biomeda, Foster City, CA) 1:50; mouse anti-islet 1, 1:50 (39.4D5; Developmental Studies Hybridoma Bank [DSHB], University of Iowa)._Whole mount in situ hybridization was performed as described previously (Sive 2000)._Section in situ hybridization was performed on 8 μM sections as described previously (Viczian et al. 2003).
Quantitative RT-PCR.
RNA extracted from animal caps or embryos was treated with DNase to eliminate genomic DNA contamination. cDNA was then prepared using qScripTM cDNA Supermix (Quanta).
Real-time PCR was performed in a 25-μL amplification mixture containing 1 μL cDNA, 12.5 μL of 2x PCR master mix (SYBR Green; Applied Biosystems, Foster City, CA), and 100 nM forward and reverse primers, respectively (NEIL3: 5’-AAAAGCTCCTTTCCCAGCG-3’, 5’-CCCTGCTTCCTCCCATAATGA-3’; L8: 5’-AAGCTTCGGGCTATCGACTTT-3’, 5’-ACGGCCTGGATCATGGATAA-3’). The PCR conditions included a polymerase activation step at 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds and run on a sequence detector (Model 7500; Applied Biosystems). The CT for Neil3 and Rax was normalized by an internal control gene L8. The statistical significance of relative differences in expression levels was determined by Student’s group t-test.
ACKNOWLEDGEMENTS
We thank Drs. Andy Fischer, Vidu Garg, Kathy Moore, and Chris Phiel for helpful discussions and critical suggestions. We also thank Bogna Brzezinska, Dr. Jessica Buescher, Annie Rorick, and Sydney Viox for performing some of the rescue experiments. This work was funded by a grant from NIH (NEI) EY015480 to HME.
Footnotes
The authors have no conflicts of interest to declare.
REFERENCES
- Amato MA, Arnault E, Perron M. 2004. Retinal stem cells in vertebrates: parallels and divergences. Int J Dev Biol 48 (8-9):993–1001. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M 2009. Molecular regulation of vertebrate retina cell fate. Birth Defects Res C Embryo Today 87 (3):284–295. [DOI] [PubMed] [Google Scholar]
- Atkinson-Leadbeater K, Hehr CL, McFarlane S. 2014. Fgfr signaling is required as the early eye field forms to promote later patterning and morphogenesis of the eye. Dev Dyn 243 (5):663–675. [DOI] [PubMed] [Google Scholar]
- Bailey TJ, El-Hodiri H, Zhang L, Shah R, Mathers PH, Jamrich M. 2004. Regulation of vertebrate eye development by Rx genes. Int J Dev Biol 48 (8-9):761–770. [DOI] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP. 2002. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 1 (7):517–529. [DOI] [PubMed] [Google Scholar]
- Casarosa S, Andreazzoli M, Simeone A, Barsacchi G. 1997. Xrx1, a novel Xenopus homeobox gene expressed during eye and pineal gland development. Mech Dev 61 (1-2):187–198. [DOI] [PubMed] [Google Scholar]
- Chen CM, Cepko CL. 2002. The chicken RaxL gene plays a role in the initiation of photoreceptor differentiation. Development 129 (23):5363–5375. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Raymond PA. 2001. Zebrafish genes rx1 and rx2 help define the region of forebrain that gives rise to retina. Dev Biol 231 (1):13–30. [DOI] [PubMed] [Google Scholar]
- Cizelsky W, Hempel A, Metzig M, Tao S, Hollemann T, Kuhl M, Kuhl SJ. 2013. sox4 and sox11 function during Xenopus laevis eye development. PLoS One 8 (7):e69372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve P, Bovolenta P. 2006. Secreted inducers in vertebrate eye development: more functions for old morphogens. Curr Opin Neurobiol 16 (1):13–19. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Bosse JL, El-Hodiri HM. 2013. The ciliary marginal zone (CMZ) in development and regeneration of the vertebrate eye. Exp Eye Res 116:199–204. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Kozak CA, Cepko CL. 1997. rax, a novel paired-type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci U S A 94 (7):3088–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. 2006. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 (3):495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannaccini M, Giudetti G, Biasci D, Mariotti S, Martini D, Barsacchi G, Andreazzoli M. 2013. Brief report: Rx1 defines retinal precursor identity by repressing alternative fates through the activation of TLE2 and Hes4. Stem Cells 31 (12):2842–2847. [DOI] [PubMed] [Google Scholar]
- Giudetti G, Giannaccini M, Biasci D, Mariotti S, Degl’innocenti A, Perrotta M, Barsacchi G, Andreazzoli M. 2014. Characterization of the Rx1-dependent transcriptome during early retinal development. Dev Dyn 243 (10):1352–1361. [DOI] [PubMed] [Google Scholar]
- Harris WA, Perron M. 1998. Molecular recapitulation: the growth of the vertebrate retina. Int J Dev Biol 42 (3):299–304. [PubMed] [Google Scholar]
- Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjoras M, Luna L. 2009. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollyfield JG. 1968. Differential addition of cells to the retina in Rana pipiens tadpoles. Dev Biol 18 (2):163–179. [DOI] [PubMed] [Google Scholar]
- Hollyfield JG. 1971. Differential growth of the neural retina in Xenopus laevis larvae. Dev Biol 24 (2):264–286. [DOI] [PubMed] [Google Scholar]
- Irie S, Sanuki R, Muranishi Y, Kato K, Chaya T, Furukawa T. 2015. Rax Homeoprotein Regulates Photoreceptor Cell Maturation and Survival in Association with Crx in the Postnatal Mouse Retina. Mol Cell Biol 35 (15):2583–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns PR. 1977. Growth of the adult goldfish eye. III. Source of the new retinal cells. J Comp Neurol 176 (3):343–357. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Zaghloul N, Moody SA. 2001. Transcription factors of the anterior neural plate alter cell movements of epidermal progenitors to specify a retinal fate. Dev Biol 240 (1):77–91. [DOI] [PubMed] [Google Scholar]
- Kimura A, Singh D, Wawrousek EF, Kikuchi M, Nakamura M, Shinohara T. 2000. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J Biol Chem 275 (2):1152–1160. [DOI] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. 1984. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res 12 (18):7057–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens T, Li J, Leone G, De Veylder L. 2009. Atypical E2Fs: new players in the E2F transcription factor family. Trends Cell Biol 19 (3):111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. 2010. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A 107 (11):4925–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Doublie S, Wallace SS. 2013. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat Res 743-744:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-De Luna RI, Kelly LE, El-Hodiri HM. 2011. The Retinal Homeobox (Rx) gene is necessary for retinal regeneration. Dev Biol 353 (1):10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathers PH, Grinberg A, Mahon KA, Jamrich M. 1997. The Rx homeobox gene is essential for vertebrate eye development. Nature 387 (6633):603–607. [DOI] [PubMed] [Google Scholar]
- Messina A, Lan L, Incitti T, Bozza A, Andreazzoli M, Vignali R, Cremisi F, Bozzi Y, Casarosa S. 2015. Noggin-Mediated Retinal Induction Reveals a Novel Interplay Between Bone Morphogenetic Protein Inhibition, Transforming Growth Factor beta, and Sonic Hedgehog Signaling. Stem Cells 33 (8):2496–2508. [DOI] [PubMed] [Google Scholar]
- Moody SA. 1987a. Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev Biol 119 (2):560–578. [DOI] [PubMed] [Google Scholar]
- Moody SA. 1987b. Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev Biol 122 (2):300–319. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, Martinez-Macias MI, Ariza RR, Roldan-Arjona T. 2006. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci U S A 103 (18):6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. 2002. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res 30 (22):4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurauter CG, Luna L, Bjoras M. 2012. Release from quiescence stimulates the expression of human NEIL3 under the control of the Ras dependent ERK-MAP kinase pathway. DNA Repair (Amst) 11 (4):401–409. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. 1994. Normal Table of Xenopus Laevis (Daudin): A Systematical & Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis. Garland Science. [Google Scholar]
- Pan Y, Martinez-De Luna RI, Lou CH, Nekkalapudi S, Kelly LE, Sater AK, El-Hodiri HM. 2010. Regulation of photoreceptor gene expression by the retinal homeobox (Rx) gene product. Dev Biol 339 (2):494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Nekkalapudi S, Kelly LE, El-Hodiri HM. 2006. The Rx-like homeobox gene (Rx-L) is necessary for normal photoreceptor development. Invest Ophthalmol Vis Sci 47 (10):4245–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera EM, Wessely O, Li SY, De Robertis EM. 2001. Neural and head induction by insulin-like growth factor signals. Dev Cell 1 (5):655–665. [DOI] [PubMed] [Google Scholar]
- Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. 2008. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science 319 (5860):202–206. [DOI] [PubMed] [Google Scholar]
- Perron M, Harris WA. 2000. Retinal stem cells in vertebrates. Bioessays 22 (8):685–688. [DOI] [PubMed] [Google Scholar]
- Perron M, Kanekar S, Vetter ML, Harris WA. 1998. The genetic sequence of retinal development in the ciliary margin of the Xenopus eye. Dev Biol 199 (2):185–200. [DOI] [PubMed] [Google Scholar]
- Regnell CE, Hildrestrand GA, Sejersted Y, Medin T, Moldestad O, Rolseth V, Krokeide SZ, Suganthan R, Luna L, Bjoras M, Bergersen LH. 2012. Hippocampal adult neurogenesis is maintained by Neil3-dependent repair of oxidative DNA lesions in neural progenitor cells. Cell Rep 2 (3):503–510. [DOI] [PubMed] [Google Scholar]
- Reis A, Hermanson O. 2012. The DNA glycosylases OGG1 and NEIL3 influence differentiation potential, proliferation, and senescence-associated signs in neural stem cells. Biochem Biophys Res Commun 423 (4):621–626. [DOI] [PubMed] [Google Scholar]
- Reks SE, McIlvain V, Zhuo X, Knox BE. 2014. Cooperative activation of Xenopus rhodopsin transcription by paired-like transcription factors. BMC Mol Biol 15:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rembold M, Loosli F, Adams RJ, Wittbrodt J. 2006. Individual cell migration serves as the driving force for optic vesicle evagination. Science 313 (5790):1130–1134. [DOI] [PubMed] [Google Scholar]
- Rhodes D, Lipps HJ. 2015. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res 43 (18):8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Parpaillon L, Heligon C, Chesnel F, Boujard D, Philpott A. 2002. The IGF pathway regulates head formation by inhibiting Wnt signaling in Xenopus. Dev Biol 244 (2):407–417. [DOI] [PubMed] [Google Scholar]
- Rigo R, Palumbo M, Sissi C. 2017. G-quadruplexes in human promoters: A challenge for therapeutic applications. Biochim Biophys Acta 1861 (5 Pt B):1399–1413. [DOI] [PubMed] [Google Scholar]
- Rolseth V, Runden-Pran E, Luna L, McMurray C, Bjoras M, Ottersen OP. 2008. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair (Amst) 7 (9):1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. 2003. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2 (5):581–591. [DOI] [PubMed] [Google Scholar]
- Schomacher L, Han D, Musheev MU, Arab K, Kienhofer S, von Seggern A, Niehrs C. 2016. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat Struct Mol Biol 23 (2):116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, Burrows CJ, Luna L, Bjoras M. 2011. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Natl Acad Sci U S A 108 (46):18802–18807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. 2000. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sive HLG RM ; Harland RM 2000. Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Straznicky K, Gaze RM. 1971. The growth of the retina in Xenopus laevis: an autoradiographic study. J Embryol Exp Morphol 26 (1):67–79. [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. 2002. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem 277 (44):42205–42213. [DOI] [PubMed] [Google Scholar]
- Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. 2005. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem 138 (6):763–772. [DOI] [PubMed] [Google Scholar]
- Viczian AS, Vignali R, Zuber ME, Barsacchi G, Harris WA. 2003. XOtx5b and XOtx2 regulate photoreceptor and bipolar fates in the Xenopus retina. Development 130 (7):1281–1294. [DOI] [PubMed] [Google Scholar]
- Voronina VA, Kozhemyakina EA, O’Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. 2004. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet 13 (3):315–322. [DOI] [PubMed] [Google Scholar]
- Wallace BD, Berman Z, Mueller GA, Lin Y, Chang T, Andres SN, Wojtaszek JL, DeRose EF, Appel CD, London RE, Yan S, Williams RS. 2017. APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proc Natl Acad Sci U S A 114 (2):304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS. 2013. DNA glycosylases search for and remove oxidized DNA bases. Environ Mol Mutagen 54 (9):691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG, Stone EM, Swaroop A, Zack DJ. 2004. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum Mol Genet 13 (10):1025–1040. [DOI] [PubMed] [Google Scholar]
- Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. 2015. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res 43 (8):4039–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. 2013. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem 288 (38):27263–27272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. 2003. Specification of the vertebrate eye by a network of eye field transcription factors. Development 130 (21):5155–5167. [DOI] [PubMed] [Google Scholar]


