Abstract
Acquiring sufficient quantities of iron to support survival is often a critical limitation for pathogenic bacteria. To meet this demand, bacteria have evolved unique strategies to scavenge iron and circumvent the nutritional immunity exerted by their hosts. One common strategy, which is often a key virulence factor for bacterial pathogens, involves the synthesis, secretion, and re-uptake of iron chelators known as siderophores. In vitro and in vivo studies have demonstrated that the siderophore aerobactin is critical for virulence in the hypervirulent pathotype of Klebsiella pneumoniae (hvKP). Given the high rate of multi-drug resistance in Klebsiella pneumoniae, and in light of the ever-increasing demand for novel Gram-negative therapeutic targets, we identified aerobactin production as a promising antivirulence target in hvKP. Herein, we describe the development of a high-throughput biochemical assay for identifying inhibitors of the aerobactin synthetase IucA. The assay was employed to screen ~110,000 compounds across several commercially-available small-molecule libraries. IucA inhibitors with activity at micromolar concentrations were identified in our screening campaigns and confirmed using secondary orthogonal assays. However, the most potent compounds also exhibited some properties commonly observed with promiscuous/non-specific inhibitors, including incubation time and target enzyme concentration dependence, as well as the potential to antagonize unrelated enzymes.
INTRODUCTION
Iron is a critical nutrient that plays a variety of structural and functional roles in biological systems. It is involved in redox chemistry in multiple enzyme systems, sometimes as a lone cofactor, as in the iron and α-ketoglutarate dependent oxygenases, or in the context of larger molecular complexes, such as those found in heme-dependent enzymes and iron-sulfur clusters. Due to the low solubility of iron oxidized to the ferric state, bioavailable iron is limiting in many physiologic environments encountered by microorganisms. Microbes have therefore evolved to produce a wide array of siderophores, small-molecule iron chelators that are secreted into the environment where they form high-affinity chelation complexes with ferric ions.1 These iron-siderophore complexes are then assimilated back into the cell through specialized uptake systems, whose expression is often co-regulated with the siderophore biosynthetic genes.
A limited number of conserved iron-chelating functional groups are incorporated into a variety of structurally-diverse siderophores. Iron chelation is most commonly mediated by the oxygen atoms of carboxylate, catechol/phenol, and hydroxamate moieties that decorate different types of scaffolding. Many bacteria use nonribosomal peptide synthetases (NRPSs) to produce siderophores. NRPSs use a fascinating modular enzyme architecture to produce peptides without the use of mRNA or ribosomes.2 Further, the ability of the NRPSs to utilize non-proteinogenic amino acids and aryl acids enables the incorporation of these iron-binding chemical functionalities. Common NRPS-derived siderophores include enterobactin, which is present in many enteric Gram-negative species, pyoverdine and pyochelin from Pseudomonas, mycobactin from mycobacteria, as well as bacillibactin from Bacillus species.1
The NRPS-independent siderophores (NIS) constitute a second major class of siderophores.3 These compounds are most commonly composed of hydroxamate constituents linked together by polyanionic citrate, α-ketoglutarate, or succinate molecules. A shared feature of the NIS biosynthetic pathways is the presence of ATP-dependent ligases that couple the hydroxamate-bearing building block to a carboxylate of the di-/tri-acid backbone.4 As with NRPS siderophores, NISs are found in both Gram-positive bacteria, including staphyloferrin of Staphylococcus and desferrioxime of Streptomyces, and Gram-negative bacteria, including achromobactin of Pectobacterium, and aerobactin, which is produced by a number of Enterobacteriaceae species. Interestingly, some Bacillus species produce petrobactin, a hybrid NRPS-NIS stealth siderophore.1
The necessity of micromolar concentrations of iron to support microbial growth makes siderophore production an important phenotype of many bacteria. Targeting siderophore systems has therefore been examined from many angles for potential antivirulence treatment approaches, including obstructing ferric siderophore import systems,5 blocking siderophore uptake and cycling with rigidified analogues and mimetics,6 neutralization via siderophore-based immunization strategies,7, 8 increasing drug potency and specificity through siderophore-antibiotic “Trojan Horse” conjugates,9 as well as antagonizing siderophore biosynthetic enzymes.10 Multiple studies support the strategy of targeting siderophore systems by demonstrating that genetic or chemical disruption can result in the attenuation of microbial growth and virulence under iron limiting conditions.10, 11 We have previously used a high-throughput biochemical screening approach to identify inhibitors of pyoverdine production by blocking the fatty acid hydrolase critical for maturation of the pyoverdine chromophore. Inhibitors with Ki values in the sub-micromolar range were capable of blocking pyoverdine production in iron-depleted culture conditions.12
While K. pneumoniae has long been recognized as a common nosocomial pathogen, hypervirulent strains of K. pneumoniae (hvKP), which have the ability to infect otherwise healthy people in the community, are receiving renewed attention by the medical community.13 These hvKP strains were initially described in southeast Asia, but have now been reported all around the globe. Phylogenetic analysis revealed nearly all hvKP isolates harbor a ≈224 kB plasmid containing multiple virulence-related genes.14 Of significant concern, a contemporary report from China described the transfer of this virulence-conferring plasmid to a carbapenem-resistant K. pneumoniae strain.15 The coalescence of drug-resistant and hypervirulent phenotypes offers the possibility of an untreatable and highly virulent pathogen.
Analysis of the genetic markers of hvKP that contribute significantly to its virulence revealed that iron acquisition plays a major role.16, 17 Most hvKP strains are capable of producing four siderophores, the NRPS siderophores yersiniabactin, enterobactin, and the glycosylated enterobactin derivative salmochelin, as well as the NIS aerobactin. Genetic studies demonstrate that mutant hvKP strains unable to produce aerobactin showed significantly reduced virulence in subcutaneous and intra-peritoneal mouse infection models compared to either the parental wild-type organism, or to strains mutated in any or all of the other siderophore biosynthetic pathways.16, 17 The critical role that aerobactin plays in virulence has prompted our effort to understand its biosynthesis and develop tools that could further validate aerobactin production and/or utilization as a novel antivirulence target.
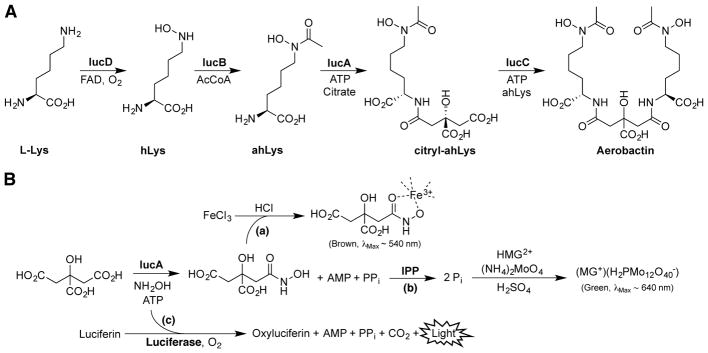
The aerobactin operon encodes four biosynthetic enzymes (IucABCD) and a transmembrane transporter (IutA) involved in aerobactin uptake.18 IucD and IucB hydroxylate and acetylate L-lysine to produce N6-acetyl-N6-hydroxy-L-lysine (ahLys). Our recent biochemical and structural studies demonstrate that two equivalents of this iron-chelating amino acid derivative are linked together via sequential ligation to a single citrate molecule through the stereoselective activity of two NIS synthetases, IucA and IucC (Fig. 1A).19, 20 These ATP-dependent ligases initially produce an activated citryl-adenylate intermediate that reacts with an incoming nucleophile to complete the ligation (Fig. S1).
Figure 1.
(A) Aerobactin biosynthesis. The citryl-hydroxamate NIS is biosynthesized from L-lysine and citrate building-blocks via the combined activity of four Iu-cABCD enzymes. (B) Summary of assay chemistries. IucA catalyzes the ATP-dependent ligation of citrate with the surrogate substrate hydroxylamine. (a) Detecting the citryl-hydroxamate product via chelation with ferric iron. (b) A malachite green (MG) assay in which the pyrophosphate by-product is cleaved into two equivalents of phosphate (Pi) and detected by complexation with ammonium molybdate and MG. (c) Measuring residual ATP by conversion to light with luciferase.
While there are multiple potential avenues to block the aerobactin system, we sought to leverage our recent structural and functional studies of IucA and target this critical enzyme for inhibition. A similar strategy was adopted to identify natural product inhibitors of the related NIS synthetases SbnE and AsbB of staphyloferrin B and petrobactin biosynthesis, respectively, identifying the baulamycins as micromolar inhibitors of the two target enzymes.21 Bioactivity assays against both Gram-positive and Gram-negative species harboring NIS pathways were promising, showing IC50 values that ranged from 20 to 150 μM. However, the baulamycins were surprisingly equally effective at inhibiting growth in both iron-rich and iron-limiting conditions, suggesting that an off-target effect may predominate under the conditions investigated. Indeed, the general toxicity of the baulamycins was recently reported to stem from damage to bacterial membranes.22 Toward the goal identifying a small-molecule probe capable of inhibiting IucA, we describe our high-throughput primary assay development, initial screening studies, and the secondary orthogonal assays used to evaluate the activity of inhibitors identified in our screening. Our studies identified several compounds that were active in the primary and secondary orthogonal assays at μM concentrations. However, initial follow-up analysis of the most potent compounds revealed some in vitro characteristics consistent with non-specific enzyme interference mechanisms.
MATERIALS & EXPERIMENTAL METHODS
Materials
All commercial reagents were used as provided unless otherwise indicated. Inorganic pyrophosphatase (IPP, S. cerevisiae), myokinase (MK, rabbit muscle), pyruvate kinase/lactate dehydrogenase enzymes (PK/LDH, rabbit muscle), adenosine triphosphate disodium salt hydrate (ATP), sodium citrate tribasic dihydrate, hydroxylamine hydrochloride, malachite green oxalate, ammonium molybdate, Tween 20, sulfuric acid, ferric chloride hexahydrate, acetohydroxamic acid, anhydrous DMSO, and SYPRO Orange were purchased from Sigma-Aldrich (St. Louis, MO). TCEP was purchased from Hampton Research, Inc. (Aliso Viejo, CA). The Kinase-Glo Plus assay kit was purchased from Promega Corp. (Madison, WI) and used according to the manufacturer’s instructions. Multiple commercial compound collections were screened for inhibitors of IucA. All libraries were sourced as 10 mM solutions in DMSO. An initial pilot screen of 4,400 compounds was conducted using bioactive compounds from the Library of Pharmacologically Active Compounds (LOPAC, Sigma-Aldrich), Spectrum Collection (MicroSource Discovery Systems, Inc., Gaylordsville, CT), and Tocriscreen Total (Tocris Bioscience, Bristol, UK) chemical libraries. Larger-scale screening campaigns included the EXPRESS-Pick library (ChemBridge (CB) Corp., San Diego, CA) and the HitDiscover collection (Maybridge (MB), Thermo Fisher Scientific, Waltham, MA). Potential leads identified from the larger screening campaigns were re-ordered as solids directly from ChemBridge and Maybridge, dissolved in DMSO to yield 10–20 mM stock solutions, and used without any further analysis or purification.
IucA Expression & Purification
The target enzyme in this study was aerobactin synthetase IucA from hypervirulent K. pneumoniae hvKP1 (GenBank EMB09144.1). The iucA gene was amplified from hvKP1 genomic DNA using primers to incorporate restriction sites at the 5′ and 3′ ends of the gene. The gene was subcloned into a modified pET15b vector containing an N-terminal 5xHis tag and a TEV protease recognition site. The expression vector was transformed into E. coli BL21(DE3) for protein production. Cells were grown in LB media at 37 °C (250 RPM) for approximately three hours to an OD600 of ≈0.65. IucA expression was induced with the addition of 500 μM IPTG, followed by incubation at 16 °C (250 RPM) for ≈18 hours. Cells were harvested by centrifugation at 6 × 103g for 15 min at 4 °C. After decanting the supernatant media, the cell pellet was flash frozen in liquid N2 and stored at −80 °C for later use.
Frozen cell pellet was re-suspended in a lysis buffer containing 50 mM HEPES, 250 mM NaCl, 10 mM imidazole, 0.2 mM TCEP, and 10% glycerol, at pH 7.5. Cell lysis was carried out by sonication and the resulting slurry was separated by ultracentrifugation at 185 × 103g. The supernatant was filtered through a 0.45 μm polysulfone membrane before being subjected to immobilized metal affinity chromatography. The lysate supernatant was passed over a 5 mL Ni2+-NTA column and bound proteins were eluted from the column using lysis buffer containing 300 mM imidazole. Fractions that were shown to contain His-tagged IucA by SDS-PAGE were combined and dialyzed overnight at 4 °C with TEV protease in dialysis buffer (50 mM HEPES, 250 mM NaCl, 0.2 mM TCEP, 0.5 mM EDTA, 10% glycerol, pH 7.5). After spiking with imidazole to 20 mM, the dialyzed sample was passed over the Ni2+-NTA column for a second time. The flow through fractions containing IucA without the His tag were combined and concentrated using a centrifugal filter before being subjected to size exclusion chromatography (SEC). The concentrated protein solution was eluted over the SEC column (HiLoad 16/60 Superdex 200, GE Healthcare Life Sciences, Marlborough, MA) using a buffer of 50 mM HEPES, 150 mM NaCl, 0.2 mM TCEP, pH 7.5. The desired fractions, identified by chromatographic absorbance and confirmed by SDS-PAGE, were combined, concentrated, flash frozen in liquid N2, and stored at −80 °C.
High-throughput Assay Optimization
Assay conditions were optimized in 100 μL reaction volumes in 96-well clear polystyrene microplates (NUNC Brand) and absorbance measurements were performed using a BioTek (Winooski, VT) Synergy 4 microplate reader. Initially, a ferric-hydroxamate assay was explored in which ferric iron was added to the citryl-hydroxamate product of the IucA-catalyzed reaction (Fig. S1), yielding a brown-colored chelation complex. A standard curve was constructed by combining 90 μL of acetohydroxamic acid standards with 10 μL of 10% (w/v) FeCl3·6H2O in 0.7M HCl and measuring absorbance at 540 nm. After observing that this assay had insufficient sensitivity, an alternative malachite green (MG) assay was investigated (Fig. S1).23 This assay is based on colorimetrically detecting inorganic phosphate (Pi) using an acidic solution of MG dye and ammonium molybdate. Inorganic pyrophosphatase (IPP) was included in the reaction mixture to cleave the pyrophosphate (PPi) by-product into two equivalents of Pi, and by doing so, coupling the assay readout to IucA activity. The developing solution containing 1.0 mg/mL MG oxalate, 1.5% (w/v) ammonium molybdate, 0.15% (v/v) Tween 20, and 4.7 N sulfuric acid, was prepared based on previously-described methods.24 A standard curve was constructed by combining 25 μL of this developing solution with 100 μL of sodium phosphate standards (1:4 ratio) and measuring absorbance at 637 nm. Numerous reaction parameters including IucA concentration, substrate concentration, reaction time, and development time were optimized in parallel using this general MG assay. The final assay conditions used in high-throughput screening are outlined below.
High-Throughput Screening Assay
For high-throughput screening, the assay volumes were cut in half for use in 384-well format (50 μL reaction solution, 13 μL developing solution). An “enzyme-initiated” protocol was carried out in which test compounds were first added to the reaction mixture before the reaction was initiated by the addition of IucA (Fig. S4A). A solution containing 55.6 mM HEPES pH 7.5, 0.11% Tween 20, 16.7 mM MgCl2, 55.6 mM hydroxylamine, 55.6 μM ATP, 55.6 μM citrate, and 0.28 U/mL IPP was dispensed (45 μL) into clear polystyrene microplates (Corning, Inc.) using a BioTek MicroFlo dispenser. Next, 40 nL of test compounds (10 mM in DMSO, 8 μM final concentration) were transferred from deep-well blocks to the reaction solution using a stainless-steel pin tool operated by a robotic workstation (JANUS, PerkinElmer, Waltham, MA). The IucA-catalyzed reaction was initiated by adding 5 μL of 3 μM IucA in 25 mM HEPES, 75 mM NaCl, and 0.1 mM TCEP at pH 7.5. The reactions were allowed to proceed for 30 min at room temperature before being quenched by dispensing (μFill, BioTek) 13 μL of MG developing solution. After allowing the assay color to develop/stabilize for 30 min, the absorbance at 620 nm was measured (EnVision 2103 Multilabel Microplate Reader, PerkinElmer). A second, “substrate-initiated” variation of this assay was also employed in which the test compounds were allowed to pre-incubate with IucA in 40 μL of reaction solution (62.5 mM HEPES pH 7.5, 0.13 % Tween 20, 18.8 mM MgCl2, 93.8 mM NaCl, 0.25 mM TCEP, 0.3 U/mL IPP, and 380 nM IucA) for 15 min prior to reaction initiation with the addition of 10 μL of a substrate solution (250 μM ATP, 250 μM citrate, 250 mM hydroxylamine, and 50 mM HEPES pH 7.5) (Fig. S4B).
Each microplate contained positive (+) controls (DMSO vehicle only) in columns 1, 2, and 23, as well as negative (−) controls (either no IucA or no substrates) in column 24. Using these controls, assay performance was continually assessed by statistics including dynamic range (R) and screening window coefficient (Z′) described by Zhang and colleagues:25
| Eq. (1) |
| Eq. (2) |
where Ā is mean absorbance and σ is standard deviation (SD) of positive and negative controls. Hits in the primary screen were generally defined by >20 % inhibition. Some compounds that marginally missed this threshold, but still appeared to be outliers from the baseline were also conservatively included as primary hits for inclusion in follow-up testing. Hits were substantiated by looking for dose-dependent inhibition in duplicate experiments using serial dilutions of test compounds aliquoted from the screening stocks. Percent inhibition was defined as:
| Eq. (3) |
Orthogonal Luminescence Assay
The substrate-initiated IucA reaction was carried out as described above on 100 μL scale in 96-well solid white microplates (Greiner Bio-one). Test compounds (1 μL) were added as 100× stock solutions in DMSO and allowed to pre-incubate with IucA in the reaction solution (80 μL) for 30 min before the reaction was initiated with the solution of substrates (20 μL). After 30 min of reaction time at room temperature, 50 μL of Kinase-Glo Plus reagent (Promega Corp.) containing luciferase and luciferin was added to the reaction mixture. The luminescence signal was allowed to stabilize for 30 min before being measured on a BioTek Synergy 4 plate reader in luminescence mode. Negative (−) controls (DMSO vehicle, minimum signal) and positive (+) controls (no IucA, maximum signal) were used to calculate % inhibition based on light intensity in a manner analogous to the method described above. All dose-response curves of purchased compounds were fit with non-linear regression (dose-response inhibition, variable slope) curves using GraphPad Prism 6 software. IC50 values are reported ± standard error (SE) of the fitted curve.
Promiscuity AK/PK/LDH Assay
To evaluate the potential off-target effects (promiscuity) of inhibitors, a coupled AMP-NADH oxidation assay was employed (Fig. S1).26 This assay utilizes myokinase (MK), pyruvate kinase (PK), and lactate dehydrogenase (LDH) to couple the production of AMP to the oxidation of NADH, which can be followed by measuring absorbance at 340 nm. A reaction mixture containing 50 mM HEPES pH 7.5, 15 mM MgCl2, 50 mM NaCl, 0.2 mM TCEP, 0.1% Tween 20, 3 mM PEP, 500 μM NADH, and 10 U/mL MK, PK, and LDH was incubated with test compounds at room temperature for 90 min. The reaction was initiated by the addition of 500 μM AMP and 50 μM ATP. Initial reaction velocities were determined by measuring absorbance at 340 nm every 10 s for 10 min. Negative (−) (no AMP/ATP) and positive (+) controls (DMSO vehicle) allowed percent inhibition to be calculated based on initial reaction velocity in a manner analogous to the method described above.
Thermal Shift Assay
A florescence-based thermal shift assay was carried out to determine if inhibitors were able to bind and thermally stabilize IucA.27 Samples (30 μL, 25 mM HEPES, 75 mM NaCl, 0.1 mM TCEP, pH 7.5) were prepared containing 10 μg IucA, 5× SYPRO Orange dye, and various concentrations of inhibitors in 96-well PCR plates (Stratagene, San Diego, CA) sealed with polyolefin film. The plate was placed in a Stratagene Mx3005P real-time PCR instrument and the temperature was increased from 25 to 99 °C over 45 min while the fluorescence intensity (λEX = 545 nm, λEM = 568 nm) was measured every 0.5 °C. Melting temperatures (TM) were determined by calculating the maximum value of first derivative of the melting curves. When evaluating the effect of the substrates ATP (1.5 mM) and citrate (1.5 mM) on IucA thermal denaturation, a more concentrated buffer system (50 mM HEPES, 150 mM NaCl, 0.2 mM TCEP, pH 7.5) was employed.
RESULTS
Assay Development & Optimization
An in vitro reaction system was developed and optimized to be employed in high-throughput screening for antagonists of aerobactin synthetase IucA. Because ahLys, the native nucleophilic substrate of IucA, was not commercially available, hydroxylamine was used as a surrogate nucleophile (Fig. 1B). Prior studies indicated that hydroxylamine is able to react with the activated citryl-adenylate intermediate to yield a simple citryl-hydroxamate product. A reporter system was required to monitor reaction progress because none of the reaction products were conveniently directly measurable. A number of continuous and non-continuous assay chemistries were considered for detecting each of the three products, citryl-hydroxamate, AMP, or pyrophosphate (PPi) (Fig. S1).
A simple and low-cost assay based on chelating ferric iron to the citryl-hydroxamate product was initially evaluated (Fig. 1B). In this assay, addition of ferric iron to the reaction mixture containing the IucA-synthesized citryl-hydroxamate product led to the formation of a ferric citryl-hydroxamate chelation complex exhibiting a characteristic brown color (Fig. S2A). Initial trials suggested a relatively high concentration of substrates were required to observe a significant absorbance signal. Substituting acetohydroxamic acid as a proxy for the citryl-hydroxamate product, a standard curve demonstrated that the ferric-hydroxamate species had a relatively low molar absorptivity (≈1,100 M−1cm−1) (Fig. S2A). In order to maximize the dynamic range of the assay, it was estimated that the reaction would require the substrates ATP and citrate in the low mM range, well above the KM values reported for IucA with these substrates (ATP, 130 ± 30; citrate, 180 ± 30 μM).20 Because performing the reaction with saturating substrates would likely hamper our ability to identify enzyme inhibitors competitive with these substrates, we explored alternative assays with greater sensitivity.
A malachite green (MG) assay was evaluated for its potential to be a more sensitive alternative.23, 24 In this assay, inorganic pyrophosphatase (IPP) was used to cleave the PPi byproduct into two equivalents of inorganic phosphate (Pi) (Fig. 1B). The phosphate in the reaction mixture was then reacted with MG and ammonium molybdate under acidic conditions to form a green-colored MG-phosphomolybdate species, which was measured colorimetrically at 620–640 nm (Fig. S2B). A standard curve prepared using sodium phosphate standards indicated that the MG-phosphomolybdate complex had a much higher molar absorptivity (≈94,000 M−1cm−1) than the ferric citryl-hydroxamate complex (Fig. S2C).
With the MG-based assay shown to have superior sensitivity, the in vitro IucA reaction system was optimized for subsequent use in high-throughput screening (Fig. S3). A number of variables were evaluated, including reaction time, IucA concentration, substrate concentration, and development time. Due to the increased sensitivity of the MG assay, the reaction could be carried out with the concentration of ATP and citrate below their KM values, making the assay sensitive to inhibitors competitive with these substrates (Fig. S3A). Concentrations greater than 50 μM exhibited a tendency for the MG-phosphomolybdate complex to precipitate and were therefore avoided in subsequent assay optimization. The reaction progress curve plateaued around 30 min (Fig. S3B). To balance maximizing assay signal while maintaining sensitivity toward marginal enzyme inhibitors, a reaction time of 30 min was selected. An IucA concentration of 200–300 nM achieved a good dynamic range, while remaining insignificant compared to the test compound concentration (8 μM) (Fig. S3B–C). Finally, following the addition of the development solution containing MG and ammonium molybdate, the time required for absorbance signal stabilization was dependent on IucA concentration and the amount of reaction product produced (Fig. S3C). Thorough mixing of the assay solution was observed to expedite signal stabilization.
High-Throughput Screening
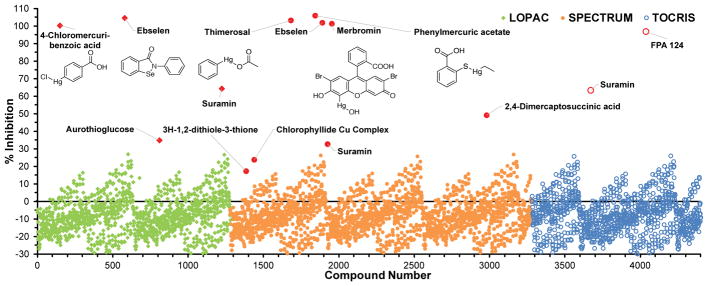
After optimizing assay parameters in low-throughput format, an initial automated high-throughput pilot screen was carried out. First, the assay volumes were cut in half for use in 384-well microplates (Fig S4). Next, test plates of positive (no test compounds) and negative (no IucA) controls were set-up to ensure adequate dynamic range and reproducibility when utilizing automated dispensing. Finally, a pilot screen consisting of three commercially-available collections totaling 4,400 bioactive compounds was performed (Fig. 2, Table 1). Systematic fluctuation in the background signal periodic over two consecutive microplates was observed during the pilot screen. The fluctuation was traced to very minor dispensing oscillations, which were only appreciable due to the high sensitivity of the assay. In spite of these oscillations, assay performance appeared sufficient (avg. Z′ = 0.6 ± 0.1) to confidently identify assay inhibitors. By inspecting the scatter plot in Fig. 2, 14 compounds exhibiting assay inhibition were identified and selected for follow-up testing to verify their observed activity. Of the 14, five compounds displayed a significant level of dose-dependent inhibition starting at 10 μM (Fig. S5). Unfortunately, each of the five significantly active compounds contained a selenium or mercury atom (Fig. 2 & S5). In this context, we surmised that these organometallic compounds were most likely inhibiting IucA via non-specific protein reactivity. Despite only turning-up suspicious organometallic compounds, the pilot screen did prove the assay capable of identifying inhibitors.
Figure 2.
Scatter plot of the initial pilot screen of 4,400 bioactive molecules from three smaller chemical libraries. Hits are highlighted in red and labeled with compound name. The chemical structure of the five most potent compounds in follow-up testing are also included.
Table 1.
Statistical summary of screening campaigns.
| Statistic | Pilot | ChemBridge (CB) EXPRESS-Pick | Maybridge (MB) HitDis-cover |
|---|---|---|---|
| No. of Compounds | 4,400 | 55,230 | 52,160 |
| Mean (+) Control ± SD (AU) | 1.00 ± 0.01 | 0.98 ± 0.03 | 0.94 ± 0.04 |
| Mean (−) Control ± SD (AU) | 0.438 ± 0.004 | 0.28 ± 0.01 | 0.28 ± 0.01 |
| Mean R (AU) ± SD | 0.56 ± 0.01 | 0.70 ± 0.03 | 0.66 ± 0.04 |
| Mean Z′ ± SD | 0.6 ± 0.1 | 0.7 ± 0.1 | 0.7 ± 0.1 |
| No. of Hits (%) | 14 (0.318%) | 36 (0.065%) | 101 (0.194%) |
| No. Replicated & Active* (%) | 5 (0.114%) | 16 (0.029%) | 12 (0.023%) |
>20% inhibition at 20 μM
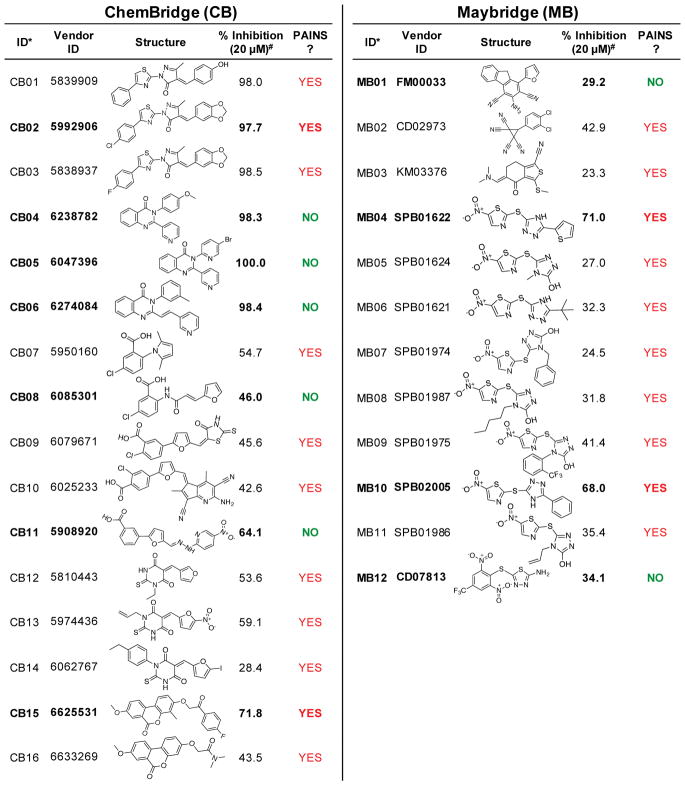
We expanded our screening to encompass two larger diversity-oriented commercial libraries totaling ≈110,000 compounds, seeking to discover a suitable lead for probe development (Table 1). When screening the ChemBridge (CB) EXPRESS-Pick library, we used the “enzyme initiated” assay protocol identical to the pilot screen. From this library, 36 (0.065%) hits were identified and rescreened; 16 compounds reproduced the inhibitory activity in follow-up assessment (Fig. 3). The relatively low number of hits in this library led us to seek ways to adjust the assay protocol in a manner that would increase the hit rate. The protocol was modified so that test compounds were allowed to pre-incubate with IucA for 15 min prior to reaction initiation with a solution of substrates (Fig. S4B). In this “substrate-initiated” protocol, slow-binding inhibitors had the opportunity to interact with IucA before the reaction was initiated. When employing this protocol for the Maybridge (MB) HitDiscover library, the number of hits increased (101, 0.194%), although it is unclear if this was directly the result of the protocol change. Significant dose-dependent inhibition was replicated for 12 of the 101 hits (Fig. 3). Across both libraries, a total of 28 compounds were identified with reproducible >20% inhibition at 20 μM.
Figure 3.
Compounds with reproducible inhibition from the ChemBridge and Maybridge collections. Twenty-eight compounds were identified from the primary screen and showed reproducible inhibition in replicate measurements. #Percent inhibition is shown as mean of duplicate measurements. Purchased compounds are shown in bold and were used is dose-dependent analysis.
Preliminary in Vitro Assessment of Purchased Hit Compounds
The 28 compounds with reproducible assay activity were examined to better understand their in vitro behavior (Fig. 3). First, the chemical structure of the compounds was inspected for the presence of known pan-assay interference (PAINS) motifs. Twenty-two of the 28 compounds were flagged as potential PAINS by an automated PAINS sub-structure filter.28 Because these PAINS predictions are based on limited empiric data, we decided to proceed with further experimental evaluation against our target enzyme.29 However, we remained cautious of the potential for inhibitor activity resulting from a mechanism other than binding site engagement.
Eleven compounds were purchased for further evaluation based on availability and chemical relatedness. The activities of the 11 purchased compounds were first re-tested in a dose-dependent manner using the primary substrate-initiated MG assay employed for screening the MB library (Fig. 4A & S6). CB02 displayed the highest potency, while compounds CB04-06 also exhibited encouraging dose-dependent activity. CB08, CB11, and CB15 only showed significant inhibitory activity in the enzyme-initiated assay; very minimal inhibitory activity was observed when evaluated using the substrate-initiated protocol (Fig. S6).
Figure 4.
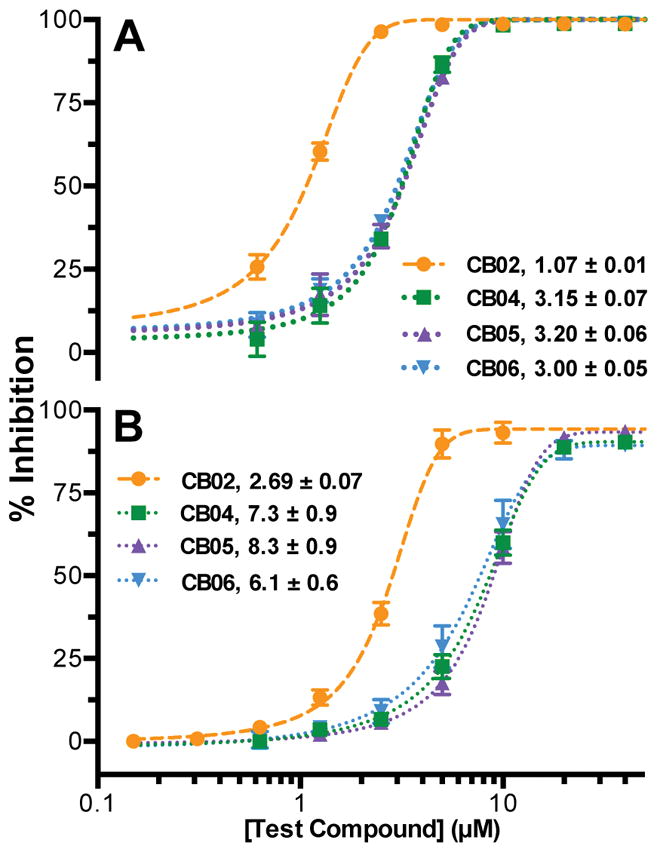
Dose-response of the most potent purchased compounds employing the (A) primary MG and (B) secondary luminescence assays. Data points represent the mean of triplicate measurements ± SD fitted with non-linear regression. Listed in the legend are IC50 values ± SE.
To validate IucA-specific inhibition observed with the primary MG assay chemistry, an orthogonal luminescence-based secondary assay was employed to re-evaluate eight of the most active purchased compounds (Fig. 4B & S7). In this assay, unreacted ATP was coupled to light production via the ATP-dependent conversion of luciferin to oxyluciferin by luciferase (Fig. 1B). The inhibition of all the tested compounds were closely replicated by the secondary orthogonal assay, exhibiting the same degree of inhibition whether we monitored product (PPi) production or substrate (ATP) utilization. This confirmed that activity was not an artifact of assay chemistry. In both primary and orthogonal assays, CB02 and CB04-06 exhibited the most potent dose-dependent activity.
Initial Investigation of Inhibitory Mechanism
A simple biophysical assay was then employed to determine if inhibitor-IucA binding could be detected. To this end, a fluorescence-based thermal shift assay was utilized to investigate if the purchased test compounds were able to influence the thermal stability of IucA.27 Addition of substrates (ATP, and the combination ATP and citrate) resulted in a reproducible +2–5 °C shift in the melting temperature (TM) of IucA (Fig. S8A). Knowing that ligand binding could thermally stabilize IucA, the assay was also carried out with the purchased hit compounds to probe if any could produce a similar positive thermal shift. The IucA melting curves with purchased hit compounds illustrated that none significantly shifted the melting temperature of IucA in a positive direction at concentrations up to 20 μM (Fig. S8B). However, a dose-dependent increase in baseline fluorescence was observed with CB04-6 and MB12, potentially indicating a direct hydrophobic interaction with the fluorescent dye or partial denaturation of IucA.
Cognizant of the concerns raised by the PAINS analysis, further experiments were carried out to determine if these compounds were inhibiting IucA via a mechanism independent of target binding site engagement. Common characteristics of undesirable inhibitory mechanisms in biochemical assays include time dependence, sensitivity to detergent, sensitivity to enzyme concentration, and promiscuity toward unrelated enzymes.30, 31 Because detergent was already included in the primary screening assay, this characteristic was not re-evaluated. Only the purchased compounds with a relatively high level of dose-dependent activity, namely CB02 and CB04-6, were evaluated further. Also, because CB04-06 shared the same quinazolinone scaffold and exhibited equivalent potency, for the sake of efficiency and conserving limited quantities of CB04 and CB05, conclusions from experiments employing CB06 were presumed applicable to all three.
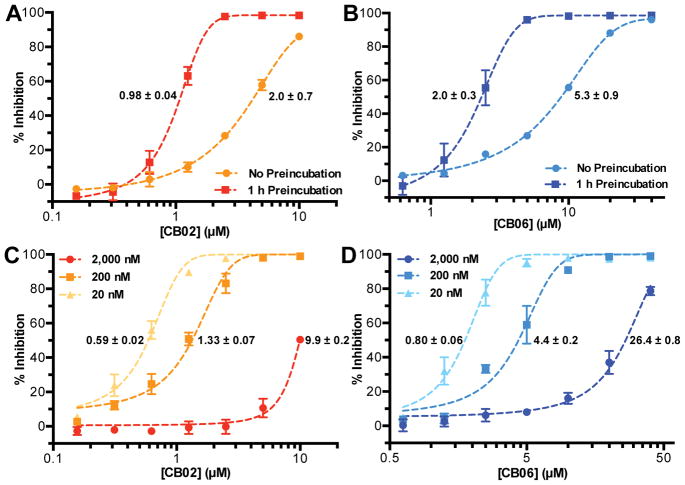
When CB02 and CB06 were allowed to pre-incubate with IucA for 1 h prior to reaction initiation, they showed increased apparent potency compared to reactions performed without pre-incubation (Fig. 5A–B). Moreover, when these compounds were added to reactions containing increasing concentrations of IucA (20, 200, or 2,000 nM), their apparent potency was reduced (Fig. 5C–D). Finally, the potential of CB02 and CB06 to inhibit unrelated enzymes was assessed using the coupling enzymes from the AMP-NADH oxidation assay (Fig. S1).26 This assay contained three enzymes (myokinase, pyruvate kinase, and lactate dehydrogenase) that could theoretically be targeted by the test compounds. Following pre-incubation with CB02 and CB06 for 90 min, a dose dependent reduction in the overall reaction velocity was observed for concentrations above 6.25 μM and 12.5 μM, respectively (Fig. S9).
Figure 5.
(A,B) The effect of pre-incubation time on ap-parent potency. Dose-response of CB02 and CB06 comparing 1 h pre-incubation with IucA to no pre-incubation (initiating reaction with enzyme) employing the MG assay. (C,D) The effect of target enzyme (IucA) concentration on apparent potency. Dose-response of CB02 and CB06 with three concentrations of IucA (2 μM, 200 nM, and 20 nM) in the MG assay. Plotted is the mean of triplicate reactions ± SD fitted with non-linear regression. IC50 values (μM) ± SE are listed next to each curve.
DISCUSSION
We report the development of a high-throughput screening assay for inhibitors of the NIS synthetase IucA from a clinically-isolated strain of hvKP. The assay was optimized for substrate and enzyme concentrations, reaction and development times, and employing a surrogate nucleophile in place of the native ahLys substrate. Following assay development and optimization, we report here the screening of multiple small-molecule libraries, as well as preliminary biochemical and informatic analyses of identified active compounds.
The assay was robust when miniaturized to 384-well plate format, enabling the screening of initial pilot libraries of a few thousand compounds. In the pilot screen, the assay displayed satisfactory performance with an average Z′ factor of 0.6, despite the average negative control (0.44 AU) displaying higher background signal than expected (Table 1). The assay proved capable of identifying enzyme inhibitors, with five compounds displaying significant dose-dependent inhibition. Ebselen was identified as an inhibitor from both the LOPAC and Spectrum collection. Known as a cysteine-modifier, ebselen is a potent covalent inhibitor of a number of thiol-dependent enzymes.32 While IucA is not predicted to rely on a cysteine thiol for its catalytic activity, general non-specific electrophilic modification of the enzyme could be responsible for the apparent activity of ebselen. In addition, four organomercury compounds were also identified in the initial screen. Merbromin, thiomersal, and phenylmercuric acetate were once widely employed in antiseptics, preservatives, or disinfectants, while 4-chloromercuribenzoic acid is a well-known thiol-reactive enzyme inhibitor.33, 34 Like ebselen, these organomercury compounds likely inhibited IucA via non-specific electrophilic reactivity. Despite uncovering inhibitors with probable non-ideal mechanisms of action, this limited preliminary screen proved that the assay was capable of picking out inhibitors.
Two larger diversity-oriented collections were screened with the goal of identifying a more suitable lead for probe development. First, screening the 55,230-compound CB Express-Pick library (avg. Z′ = 0.7 ± 0.1) turned up 36 (0.07%) compounds with apparent inhibitory activity. Pre-incubation of test compounds with the target is reported to increase the chances of identifying slow-binding inhibitors.35 Therefore, the 52,160-compound MB HitDiscover library was screened (avg. Z′ = 0.7 ± 0.1) using a second, substrate-initiated protocol in which IucA and test compounds were pre-incubated for 15 min prior to reaction initiation with substrates. The use of this experimental protocol resulted in a significantly increased hit rate (101, 0.19%), but the majority (89 out of 101) were not replicated in follow-up testing. The underlying mechanism behind the increased noise of the substrate-initiated protocol is unknown. In the end, a similar number of inhibitors with significant replicated activity were identified in both libraries (CB = 16 and MB = 12).
Many (22/28) of the active compounds were flagged for containing predicted PAINS motifs. In order to evaluate experimentally the empiric predictions, 11 compounds were purchased for further assessment. Compounds CB08, CB11, and CB15 only displayed significant inhibitory activity when tested using the original enzyme-initiated protocol. Given the resemblance of CB11 to the antibiotic nitrofurantoin, we hypothesized that CB11 may rely on redox activity to inhibit IucA. Its differential activity could be due to differences in TCEP exposure between the two protocols. Furthermore, this compound has also been reported as an inhibitor in a number of other reported high-throughput biochemical screens.36–39 The underlying mechanism behind the differential activity of CB08 and CB15 remains unclear.
Of the 11 purchased compounds, four displayed substantial dose-dependent inhibition of IucA in both the primary MG and the secondary luminescence assays. The most potent compound, pyrrolone-containing CB02, displayed IC50 values of 1–3 μM. CB04, CB05, and CB06, which all shared a quinazolinone scaffold, also displayed promising dose-dependent inhibition with IC50 values of 3–8 μM. Comparable inhibitory activity in the primary and secondary assays, which utilized distinctive chemistries coupled to unique products of the IucA-catalyzed condensation, suggested the compounds were acting as target enzyme inhibitors. However, we remained cautious, particularly towards CB02, which was highlighted for containing a PAINS substructure motif.
Subsequent experiments were carried out to better characterize the in vitro behavior of the pyrrolone and quinazolinone compounds. The substrate ATP, and the combination of ATP and citrate, induced a positive shift in the thermal denaturation of IucA (Fig. S8A). In contrast, none of the quinazolinones were observed stabilize IucA by the thermal shift assay (Fig. S8B). However, melting curves with these compounds exhibited a dose-dependent increase in the baseline fluorescence, potentially a consequence of direct hydrophobic interaction with the fluorescent dye, or the compounds inducing partial unfolding of IucA. Given the recent attention that promiscuous aggregating inhibitors have garnered in the context of biochemical screening, we wanted to evaluate compound aggregation as a potential mechanism at an early stage in the discovery process.28, 30, 31 Testing of CB02 and CB06 revealed potential evidence of aggregation-based inhibition, including time dependence, enzyme concentration sensitivity, and promiscuity toward other enzymes. In this last assay, the promiscuity of the inhibitors was evaluated with an extended pre-incubation time (90 min) and with three possible enzyme targets. Inhibition was observed in this assay; however, a cumulative IC50 (50% inhibition against the three coupling enzymes) was approximate 20× higher for both CB02 and CB06 when compared to the pre-incubated inhibition against IucA.
To provide additional context to the apparent antagonism of CB02 and CB04-06 against our target enzyme IucA, we also examined their reported activity in publicly-available screening results. The pyrrolone compound CB02, (CID 5345903) was reported active in 2 out of 8 unique bioassays in the PubChem database, including against HIV-1 RNase H and CDC25B-CDK2/Cyclin A interaction. In contrast, no activity was reported for the quinazolinones CB04-06 (CID 5049702, 4359529, & 5729808), which were employed in 7, 2, or 1 unique assay(s) deposited in PubChem, respectively.
Further examination of the literature revealed the pyrrolone and quinazolinone compounds have also been previously reported as inhibitors in high-throughput biochemical screening projects. CB04 and CB05 were both identified as potent inhibitors of bacterial IspE kinase and P. falciparum glucose-6-phosphate dehydrogenase 6-phosphogluconalactonase in two disparate biochemical screening campaigns.36, 40 They were characterized as irreversible inhibitors with apparent potency dependent on target pre-incubation time. Furthermore, structure-activity relationship (SAR) investigation of the original lead was ambiguous and failed to identify a more potent analogue. The quinazolinone CB06 was identified in a third unrelated high-throughput biochemical screen as an inhibitor of human RAD51 recombinase in double-strand DNA repair.41 In contrast to CB04 and CB05, SAR analysis of CB06 revealed that RAD51 inhibition was sensitive to minor structural modifications, and it did not inhibit the E. coli homologue RecA. The pyrrolone compounds CB01-03 were flagged for containing a PAINS motif and all were previously identified as inhibitors in high-throughput biochemical screens against human and Yersinia fatty acid synthases.39, 42 Thus, while the prior reports suggest these compounds can antagonize other target enzymes in alternate assay environments, they do not rule out their potential utility as a lead for blocking aerobactin biosynthesis.
With aerobactin shown to play a critical role in the evolution of a novel hypervirulent pathogen (hvKP), we sought to develop a platform for identifying inhibitors of this critical virulence factor. Multiple lead compounds were identified against IucA following the development of a high-throughput biochemical screening assay. Inhibitors with activity against IucA by two orthogonal assays exhibited in vitro potency in the low-micromolar range. Initial follow-up experiments on the most potent inhibitors revealed some concerning in vitro characteristics consistent with non-specific enzyme interference mechanisms. Furthermore, any intra-cellular inhibitors will also face the challenge of cellular penetration, which is a particularly challenging problem for targeting Gram-negative bacteria. The use of more diverse libraries, including natural products, or those that may incorporate chemical features known to enable antibiotic accumulation,43 may help exploit this novel antivirulence target in an increasingly problematic pathogen.
Supplementary Material
Acknowledgments
Funding
This research was supported by the National Institutes of Health (AI-116998 to AMG) and pilot studies support from the Buffalo Clinical and Translational Research Center (NIH Grant UL1TR001412 (Timothy F. Murphy)). DCB was supported by NIH Training Grant T32-AI007614 (Laurie K. Read).
We thank Dr. Courtney Aldrich for helpful insights regarding hit compound evaluation. We thank Mr. Eric Drake for helpful discussions concerning assay design and development. Finally, we thank Dr. Thomas Russo for valuable dialogs and insights on hvKP.
Footnotes
Notes
The authors declare no competing financial interests.
References
- 1.Hider RC, Kong X. Chemistry and Biology of Siderophores. Nat Prod Rep. 2010;27(5):637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 2.Gulick AM. Structural Insight into the Necessary Conformational Changes of Modular Nonribosomal Peptide Synthetases. Curr Opin Chem Biol. 2016;35:89–96. doi: 10.1016/j.cbpa.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Challis GL. A Widely Distributed Bacterial Pathway for Siderophore Biosynthesis Independent of Nonribosomal Peptide Synthetases. ChemBioChem. 2005;6(4):601–11. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 4.Oves-Costales D, Kadi N, Challis GL. The Long-Overlooked Enzymology of a Nonribosomal Peptide Synthetase-Independent Pathway for Virulence-Conferring Siderophore Biosynthesis. Chem Commun (Camb) 2009;(43):6530–41. doi: 10.1039/b913092f. [DOI] [PubMed] [Google Scholar]
- 5.Yep A, McQuade T, Kirchhoff P, et al. Inhibitors of TonB Function Identified by a High-Throughput Screen for Inhibitors of Iron Acquisition in Uropathogenic Escherichia coli Cft073. mBio. 2014;5(2):e01089–13. doi: 10.1128/mBio.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohac TJ, Shapiro JA, Wencewicz TA. Rigid Oxazole Acinetobactin Analog Blocks Siderophore Cycling in Acinetobacter baumannii. ACS Infect Dis. 2017;3(11):802–806. doi: 10.1021/acsinfecdis.7b00146. [DOI] [PubMed] [Google Scholar]
- 7.Sassone-Corsi M, Chairatana P, Zheng T, et al. Siderophore-Based Immunization Strategy to Inhibit Growth of Enteric Pathogens. Proc Natl Acad Sci USA. 2016;113(47):13462–13467. doi: 10.1073/pnas.1606290113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mike LA, Smith SN, Sumner CA, et al. Siderophore Vaccine Conjugates Protect against Uropathogenic Escherichia coli Urinary Tract Infection. Proc Natl Acad Sci USA. 2016;113(47):13468–13473. doi: 10.1073/pnas.1606324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wencewicz TA, Long TE, Mollmann U, et al. Trihydroxamate Siderophore-Fluoroquinolone Conjugates Are Selective Sideromycin Antibiotics That Target Staphylococcus Aureus. Bioconjug Chem. 2013;24(3):473–86. doi: 10.1021/bc300610f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamb AL. Breaking a Pathogen’s Iron Will: Inhibiting Siderophore Production as an Antimicrobial Strategy. Biochim Biophys Acta. 2015;1854(8):1054–1070. doi: 10.1016/j.bbapap.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bilitewski U, Blodgett JAV, Duhme-Klair AK, et al. Chemical and Biological Aspects of Nutritional Immunity-Perspectives for New Anti-Infectives That Target Iron Uptake Systems. Angew Chem Int Ed Engl. 2017;56(46):14360–14382. doi: 10.1002/anie.201701586. [DOI] [PubMed] [Google Scholar]
- 12.Wurst JM, Drake EJ, Theriault JR, et al. Identification of Inhibitors of PvdQ, an Enzyme Involved in the Synthesis of the Siderophore Pyoverdine. ACS Chem Biol. 2014;9(7):1536–1544. doi: 10.1021/cb5001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shon AS, Russo TA. Hypervirulent Klebsiella pneumoniae: The Next Superbug? Future Microbiol. 2012;7(6):669–71. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- 14.Struve C, Roe CC, Stegger M, et al. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. mBio. 2015;6(4):e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu D, Dong N, Zheng Z, et al. A Fatal Outbreak of ST11 Carbapenem-Resistant Hypervirulent Klebsiella pneumoniae in a Chinese Hospital: A Molecular Epidemiological Study. Lancet Infect Dis. 2018;18(1):37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 16.Russo TA, Olson R, MacDonald U, et al. Aerobactin, but Not Yersiniabactin, Salmochelin and Enterobactin, Enables the Growth/Survival of Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae Ex Vivo and in Vivo. Infect Immun. 2015;83(8):3325–33. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russo TA, Olson R, Macdonald U, et al. Aerobactin Mediates Virulence and Accounts for Increased Siderophore Production under Iron-Limiting Conditions by Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae. Infect Immun. 2014;82(6):2356–67. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo V, Neilands JB. Characterization of iucA and iucC Genes of the Aerobactin System of Plasmid Colv-K30 in Escherichia coli. J Bacteriol. 1986;167(1):350–5. doi: 10.1128/jb.167.1.350-355.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey DC, Alexander E, Rice MR, et al. Structural and Functional Delineation of Aerobactin Biosynthesis in Hypervirulent Klebsiella pneumoniae. J Biol Chem. 2018;293(20):7841–7852. doi: 10.1074/jbc.RA118.002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey DC, Drake EJ, Grant TD, et al. Structural and Functional Characterization of Aerobactin Synthetase IucA from a Hypervirulent Pathotype of Klebsiella pneumoniae. Biochemistry. 2016;55(25):3559–3570. doi: 10.1021/acs.biochem.6b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tripathi A, Schofield MM, Chlipala GE, et al. Baulamycins A and B, Broad-Spectrum Antibiotics Identified as Inhibitors of Siderophore Biosynthesis in Staphylococcus aureus and Bacillus anthracis. J Am Chem Soc. 2014;136(4):1579–86. doi: 10.1021/ja4115924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steele AD, Ernouf G, Lee YE, et al. Diverted Total Synthesis of the Baulamycins and Analogues Reveals an Alternate Mechanism of Action. Org Lett. 2018;20(4):1126–1129. doi: 10.1021/acs.orglett.8b00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pegan SD, Tian Y, Sershon V, et al. A Universal, Fully Automated High Throughput Screening Assay for Pyrophosphate and Phosphate Release from Enzymatic Reactions. Comb Chem High Throughput Screen. 2010;13(1):27–38. doi: 10.2174/138620710790218203. [DOI] [PubMed] [Google Scholar]
- 24.Baykov AA, Evtushenko OA, Avaeva SM. A Malachite Green Procedure for Orthophosphate Determination and Its Use in Alkaline Phosphatase-Based Enzyme Immunoassay. Anal Biochem. 1988;171(2):266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 25.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 26.Wu MX, Hill KA. A Continuous Spectrophotometric Assay for the Aminoacylation of Transfer RNA by Alanyl-Transfer RNA Synthetase. Anal Biochem. 1993;211(2):320–3. doi: 10.1006/abio.1993.1276. [DOI] [PubMed] [Google Scholar]
- 27.Cummings MD, Farnum MA, Nelen MI. Universal Screening Methods and Applications of Thermofluor. J Biomol Screen. 2006;11(7):854–63. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 28.Baell JB, Holloway GA. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J Med Chem. 2010;53(7):2719–40. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 29.Capuzzi SJ, Muratov EN, Tropsha A. Phantom PAINS: Problems with the Utility of Alerts for Pan-Assay Interference Compounds. J Chem Inf Model. 2017;57(3):417–427. doi: 10.1021/acs.jcim.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorne N, Auld DS, Inglese J. Apparent Activity in High-Throughput Screening: Origins of Compound-Dependent Assay Interference. Curr Opin Chem Biol. 2010;14(3):315–24. doi: 10.1016/j.cbpa.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seidler J, McGovern SL, Doman TN, et al. Identification and Prediction of Promiscuous Aggregating Inhibitors among Known Drugs. J Med Chem. 2003;46(21):4477–86. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 32.Azad GK, Tomar RS. Ebselen, a Promising Antioxidant Drug: Mechanisms of Action and Targets of Biological Pathways. Mol Biol Rep. 2014;41(8):4865–79. doi: 10.1007/s11033-014-3417-x. [DOI] [PubMed] [Google Scholar]
- 33.Risher JF, Murray HE, Prince GR. Organic Mercury Compounds: Human Exposure and Its Relevance to Public Health. Toxicol Ind Health. 2002;18(3):109–60. doi: 10.1191/0748233702th138oa. [DOI] [PubMed] [Google Scholar]
- 34.Sheets RF, Hamilton HE, DeGowin EL. Hemolysis of Human Erythrocytes by a Sulfhydryl Inhibitor, p-Chloromercuribenzoic Acid. Proc Soc Exp Biol Med. 1956;91(3):423–427. doi: 10.3181/00379727-91-22283. [DOI] [PubMed] [Google Scholar]
- 35.Copeland RA. Mechanistic Considerations in High-Throughput Screening. Anal Biochem. 2003;320(1):1–12. doi: 10.1016/s0003-2697(03)00346-4. [DOI] [PubMed] [Google Scholar]
- 36.Preuss J, Hedrick M, Sergienko E, et al. High-Throughput Screening for Small-Molecule Inhibitors of Plasmodium falciparum Glucose-6-Phosphate Dehydrogenase 6-Phosphogluconolactonase. J Biomol Screen. 2012;17(6):738–51. doi: 10.1177/1087057112442382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S, Vossius S, Rahmouni S, et al. Multidentate Small-Molecule Inhibitors of Vaccinia H1-Related (Vhr) Phosphatase Decrease Proliferation of Cervix Cancer Cells. J Med Chem. 2009;52(21):6716–23. doi: 10.1021/jm901016k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed JC, Yip K, Sergienko E, et al. Methods and Compounds for Regulating Apoptosis. 0118135 A1. US. 2009
- 39.Smith J, Richardson R. Novel Antagonsits of the Human Fatty Acid Synthase Thioesterase. 0203236 A1. US. 2007 doi: 10.1158/1535-7163.MCT-07-0187. [DOI] [PubMed]
- 40.Tidten-Luksch N, Grimaldi R, Torrie LS, et al. IspE Inhibitors Identified by a Combination of in Silico and in Vitro High-Throughput Screening. PloS one. 2012;7(4):e35792. doi: 10.1371/journal.pone.0035792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang F, Motlekar NA, Burgwin CM, et al. Identification of Specific Inhibitors of Human Rad51 Recombinase Using High-Throughput Screening. ACS Chem Biol. 2011;6(6):628–35. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson RD, Smith JW. Novel Antagonists of the Thioesterase Domain of Human Fatty Acid Synthase. Mol Cancer Ther. 2007;6(7):2120–6. doi: 10.1158/1535-7163.MCT-07-0187. [DOI] [PubMed] [Google Scholar]
- 43.Richter MF, Drown BS, Riley AP, et al. Predictive Compound Accumulation Rules Yield a Broad-Spectrum Antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.