Abstract
Sturgeon species are imperiled world-wide by a variety of anthropogenic stressors including chemical contaminants. Atlantic sturgeon, Acipenser oxyrinchus, and shortnose sturgeon, Acipenser brevirostrum, are largely sympatric acipenserids whose young life-stages are often exposed to high levels of benthic-borne PCBs and PCDD/Fs in large estuaries along the Atlantic Coast of North America. In previous laboratory studies, we demonstrated that both sturgeon species are sensitive to early life-stage toxicities from exposure to environmentally relevant concentrations of coplanar PCBs and TCDD. The sensitivity of young life-stages of fishes to these contaminants varies among species by three orders of magnitude and often is due to variation in the structure and function of the aryl hydrocarbon receptor (AHR) pathway. Unlike mammals, fishes have two forms of AHR (AHR1 and AHR2) with AHR2 usually being more highly expressed across tissues and functional in mediating toxicities. Based on previous studies in white sturgeon, A. transmontanus, we hypothesized that sturgeon taxa are unusually sensitive to these contaminants because of higher levels of expression and functional activity of AHR1 than in other fish taxa. To address this possibility, we characterized AHR1 in both Atlantic Coast sturgeon species, evaluated its’ in vivo expression in young life-stages and in multiple tissues of shortnose sturgeon, and tested its ability to drive reporter gene expression in AHR-deficient cells treated with graded doses of PCB126 and TCDD. Similar to white sturgeon and lake sturgeon, AHR1 amino acid sequences in Atlantic sturgeon and shortnose sturgeon were more similar to mammalian AHRs and avian AHR1s than to AHR1 in other fishes, suggesting their greater functionality in sturgeon species than in other fishes. Exposure to graded doses of coplanar PCBs and TCDD usually failed to significantly induce AHR1 expression in young life-stages or most tissues of shortnose sturgeon. However, in reporter gene assays, AHR1 drove higher levels of gene expression than AHR2 alone, but their binary combination failed to drive higher levels of expression than either AHR alone. In total, our results suggest that AHR1 may be more functional in sturgeon species than in other fishes, but probably does not explain their heightened sensitivity to these contaminants.
Introduction
Atlantic sturgeon, Acipenser oxyrinchus, and shortnose sturgeon, Acipenser brevirostrum, are sympatric acipenserids that spawn in large estuaries along the Atlantic Coast of North America from the St. Lawrence and Saint John rivers in Canada, respectively, to at least the Altamaha River, Georgia. Atlantic sturgeon were listed under the U.S. Endangered Species Act (ESA) since 2012 as having five Distinct Populations Segments (DPS), four of which are designated as endangered and the fifth as threatened. Canadian Maritime and St. Lawrence River populations were also listed as threatened in 2011 by the Committee on the Status of Endangered Wildlife (COSEWIC). Shortnose sturgeon were listed in 1973 under the ESA as endangered and as a species of special concern under COSEWIC in 2012.
Atlantic sturgeon are anadromous with juvenile life-stage restricted to natal estuaries for 2–6 years, while subadults and adults exhibit long distance and prolonged migrations in coastal waters, only returning to natal rivers to spawn at advanced ages and in intermittent years (Wirgin et al. 2015). In contrast, shortnose sturgeon are usually restricted to natal rivers throughout its life history (Wirgin et al. 2010). Both species are long-lived, late maturing, and bottom-dwelling (Bain 1997), and are vulnerable to a variety of anthropogenic threats in their natal estuaries including overharvest, vessel strikes, habitat alteration, and compromised water quality (Shortnose Sturgeon Status Review Team 2010; Federal Register 2012,a,b).
An additional threat often mentioned for sturgeon species, but the effects of which are rarely empirically evaluated, is exposure of early life-stages to toxic chemicals such as polychlorinated biphenyls (PCBs), PCDDs (polychlorinated dibenzo-p-dioxins), and PCDFs (polychlorinated dibenzofurans). These chemicals are structurally related, lipophilic, sediment-bound, environmentally persistent, and biomagnify in aquatic food chains (Van der Oost et al. 2003). In other fishes, they are known to impair normal development (Wirgin and Chambers 2006), reduce survivorship (Elonen et al. 1998), and likely curtail recruitment to adult populations. Exposure to contaminants may be particularly problematic for sturgeon species because of their highly adhesive benthic embryos which tend to bind to contaminated sediments. They are found at high concentrations in sediments of several major estuaries along the Atlantic Coast of the U.S. that host reproducing populations of both sturgeon species. Prominent among these is the Hudson River, New York, with some of the largest populations of both sturgeon species coastwide co-occurring with alarmingly high sediment concentrations of PCBs, PCDD/Fs and PAHs (Wirgin et al. 2006). The Hudson River Estuary contains two prominent U.S. federal Superfund sites; 320 km of main stem river because of PCB contamination (Hudson River PCBs site) and the Passaic River-upper Newark Bay in the western Estuary because of world-record levels of the most toxic polychlorinated dibenzo-p-dioxin congener, 2,3,7,8 tetrachlorodibenzo-p-dioxin (TCDD) (Diamond Alkali site). Thus, young life-stages of both sturgeon species are chronically exposed to mixtures of these and other contaminants in the Hudson River estuary.
Coplanar PCBs and PCDD/Fs are known to be especially toxic to the early lifestages of fishes through impaired development of the heart (Antkiewicz et al. 2005; King-Heiden et al. 2012; Lanham et al. 2014) and compromised cardiovascular system function. However, variation among fishes in vulnerability to PCB and PCDD/Fs-induced early life-stage toxicities is great, with LD50s among species to TCDD spanning at least three orders of magnitude with salmonids being among the most sensitive of taxa and zebrafish among the least (Elonen et al. 1998; King-Heiden et al. 2012). Apparently, sturgeon species are among the more sensitive of fishes on this continuum based on earlier in vitro studies with white sturgeon and lake sturgeon Acipenser fulvescens (Doering et al. 2014ab) and our in vivo studies with Atlantic sturgeon and shortnose sturgeon (Roy et al. 2011; Chambers et al. 2012).
Studies in a variety of vertebrate taxa have implicated variation in structure and function of the aryl hydrocarbon receptor (AHR) pathway as the basis of differing sensitivities to these toxicants among species and even populations within species (Head et al. 2008; Wirgin et al. 2011; Reid et al. 2016). In the canonical pathway, AHR, a cytoplasmic transcription factor, activates downstream gene expression and toxicity by binding toxicants in the cytoplasm, shedding its cytoplasmic chaperones, Hsp90s and the aryl hydrocarbon receptor interacting protein (AIP), and translocating to the nucleus. Once in the nucleus, AHR dimerizes with the aryl receptor nuclear translocator (ARNT), AHR-ARNT heterodimer binds to dioxin responsive elements (DREs) in the regulatory regions of xenobiotic responsive genes in the AHR battery, and recruits additional factors needed for transcription. Unlike mammals, most fishes have at least two forms of AHR, AHR1 and AHR2 (Hahn et al. 1997), and in elasmobranchs three forms of AHR (including AHR3) (Hahn et al. 2017) with most evidence indicating that AHR2 is most highly expressed (Powell et al. 2000), better binds these toxicants, and is likely more functional in mediating toxicities. Multiple forms of AHR exist in fishes. AHR1 and AHR2, are paralogues that arose from a tandem duplication that occurred prior to the divergence of cartilaginous and bony fishes and was followed by a whole genome duplication in teleost fishes. AHR1 I in most fishes was believed to be orthologous to mammalian AHR but the most recent evidence suggests that AHR and AHR1 represent different lineages (Hahn et al. 2017).
Cytochrome P4501A (CYP1A) is a prominent member of the AHR battery because of its wide window of inducibility in fishes resulting in its use as a biomarker and its function in the metabolism of PAHs. The non-canonical AHR pathway is also known to interact with an ever-increasing number of signaling pathways in immunological, cellular stress, and hormonal responses (Tappenden et al. 2013). Studies have demonstrated that genetic variation within and immediately downstream of the ligand binding domain of AHR among strains of mice (Poland et al. 1994; Ema et al. 1994), fishes (Wirgin et al. 2011; Doering et al. 2015) and birds (Karchner et al. 2006; Head et al. 2008; Farmahin et al. 2013) alter the vulnerability of species to aromatic hydrocarbon-induced toxicities. Furthermore, genetic variants in the transactivation domain of AHR in rats (Okey et al. 2005), and in multiple components of the AHR2 pathway among populations of Atlantic killifish Fundulus heteroclitus (Reid et al. 2016) alter the ability of AHR to drive gene expression (usually CYP1A) and toxicity.
We have previously demonstrated that CYP1A mRNA expression in both Atlantic sturgeon and shortnose sturgeon was dose-responsive and significantly inducible at environmentally relevant concentrations of four coplanar PCBs (Roy et al. 2018) and TCDD (Roy et al. 2011). For example, significant CYP1A induction was observed in Atlantic sturgeon and shortnose sturgeon larvae that were waterborne exposed to TCDD at the lowest level tested, 0.001 parts per billion (ppb). This level of TCDD has been reported in the livers of the sympatric finfish, Atlantic tomcod, from the Hudson River Estuary (Fernandez et al. 2004). Furthermore, we showed that graded doses of PCB126 and TCDD induced a variety of early life-stage toxicities in both Atlantic Coast sturgeon species; including altered hatching rate, reduced growth, pericardial and yolk sac edema, impaired eye development and reduced duration of life-span of starved larvae at environmentally relevant concentrations of these chemicals (Chambers et al. 2012). We concluded that both Atlantic Coast sturgeon species were among the more sensitive of fishes to toxicities induced by these compounds, however the mechanistic basis of these toxic responses was unknown.
Studies in other North American sturgeon taxa similarly suggested that they were among the more sensitive of fishes to these toxicants and possibly provided a mechanistic explanation for these findings. In transient transfection assays with AHR-deficient cells transfected with white sturgeon A. transmontanus AHR1 and AHR2 and treated with PCDD/Fs and PCBs and, Doering et al. (2014a) observed that the combination of two AHRs activated greater reporter gene expression than either AHR alone. They also reported that EC50s for both AHRs were lower than for any vertebrate AHR similarly tested. Additionally, they determined that both AHRs had similar basal and induced expression levels in the livers of beta-naphthoflavone (β-NF) treated juvenile white sturgeon leading them to hypothesize that cooperative expression of the two AHRs increased the sensitivity of sturgeon species to aromatic hydrocarbons contaminants.
In our study, we sought to characterize AHR1 structure and functionality in both Atlantic Coast sturgeon species and determine whether their sensitivities to PCB and TCDD-induced toxicities result from the joint activity of the two forms of AHR as reported for white sturgeon. This involved cloning of AHR1, quantifying its expression and comparing it to that of AHR2 in TCDD-and PCB- treated young life-stages and tissues of shortnose sturgeon, and evaluating the efficacy of AHR1 alone, AHR2 alone, and their binary combination in driving reporter gene expression in transient transfection assays in AHR-deficient mammalian cells treated with TCDD or coplanar PCB126.
Methods
Characterization of AHR1 cDNA
Total RNA was isolated from Atlantic sturgeon and shortnose sturgeon embryos of Saint John River, New Brunswick, Canada, ancestry using Ultraspec reagent (Biotexc, Houston, TX) as per the manufacturer’s recommendations. To generate first strand cDNA, 500 ng of random hexamers (Integrated DNA Technologies Inc., Coralville, IA) were added to 100 ng of total RNAs in 15 μl volumes and incubated at 75° C for 5 min. The mixture was chilled and 10 μl of reverse transcriptase (RT) mix was added so that the final reaction contained 1X MMLV reaction buffer (Promega Life Science, Madison, WI), 20 U MMLV reverse transcriptase (Promega), 10 U RNasin RNase inhibitor (Promega), and 0.5 mM dNTPs (GE Healthcare Bio-Sciences Corp. Piscataway, NJ). Reactions were incubated at 420C for 1 h, denatured at 980 C for 5 min, chilled on ice, and used in PCR to amplify AHR1 cDNAs.
Initially, multiple sequence alignments were performed to identify conserved areas among four AHR1 sequences (white sturgeon, quail, albatross and cormorant) in order to develop primers to amplify AHR1 from the two Atlantic Coast sturgeon taxa (Table 1). Amplification reactions were in 30 μl total volumes that contained 0.5 μM of forward and reverse primers, 0.2 mM dNTPs (GE Healthcare), 1X PCR buffer (Roche Molecular Systems, Indianapolis, IN), 1 U Taq DNA polymerase (Roche), 4 μl of first strand cDNA product, and H20 to volume. Cycling parameters were 950 C for 5 min, 35 cycles at 950 C for 15 s, 500 C for 15 s, 720 C for 60 s, and a final extension at 720 C for 7 min. RT-PCR products were purified with Qiagen MinElute PCR purification kits (Qiagen, Valencia, CA), sequenced, and compared with white sturgeon sequences. Derived sequences were used to develop additional Atlantic sturgeon and shortnose sturgeon specific primers that were paired with white sturgeon AHR1 primers for subsequent use in RT-PCR (Table 1). Using this approach, we were able to obtain the complete AHR1 coding and some 5’ and 3’ noncoding sequences for both species.
Table 1.
Primers used to amplify and sequence Atlantic sturgeon and shortnose sturgeon AHR1
| Forward primers for RT-PCR | |
| STAHR1_23F | GAAAGCCAGTGCAGAAA |
| STAHR1_151F | GTTATCTCCAAACTGG |
| STAHR1_423F | TGTCATCCACCAGAGTG |
| STAHR1_592F | CTTCCTCCAGAGAATTC |
| STAHR1_1189F | GCTACTGGGGAGGCTGT |
| STAHR1_1595F | TTCAGCAAGATGAGGAG |
| STAHR1_1852F | CAGCAGCAGCTCTGCCA |
| STAHR1_2146F | GAATTGGAGGATTTCCT |
| STAHR1_2254F | GTGTCTATGTACCAGTG |
| STAHR1_354F | GGTTAGTGCCGATGGTTCAG |
| STAHR1_484F | GCGTCAGCTTCACTGGG |
| STAHR1_−48F | CGAGACGGGATGATGAAC |
| STAHR1_−99F | CTGGCTTTTTAAATATAA |
| STAHR1_−88F | AATATAAAAAGCGTCTTC |
| Reverse Primers for RT-PCR | |
| STAHR1_476R | AATTCTGCTCTGTCTTC |
| STAHR1_608R | GAATTCTCTGGAGGAAG |
| STAHR1_885R | TTTTCCTTTGGCATCAC |
| STAHR1_1205R | ACAGCCTCCCCAGTAGC |
| STAHR1_1611R | CTCCTCATCTTGCTGAA |
| STAHR1_1612R | ACTCTTCATCTTGCTGA |
| STAHR1_1868R | TGGCAGAGCTGCTGCTG |
| STAHR1_2165R | TCCAGGAAATCCTCCAA |
| STAHR1_2383R | TATCCATTTTGGAACTT |
| STAHR1_2502R | TGGAAAGCCACTGGATG |
| STAHR1_2568R | CTTCAGAAGGCAGCACTC |
| STAHR1_2656R | CTGCACAGTATAACCAGC |
| STAHR1_219R | GAAGCTTTTGGCTCTCAG |
| STAHR11268R | CCTTTGGCCTTGGCATGC |
| STAHR11311R | CTCTTGATCTCTTGCTC |
| Primers used for Real Time PCR | |
| STRT_2213F | GCCCTCAGGATAGTATGATTACTTCAC |
| STSNRT_2284R | CCGGCAGACACTGGTACATAG |
| Sturact60F | CATTGTCACCAACTGGGATGAC |
| Sturact125R | ACACGCAGCTCATTGTAGAAGGT |
Phylogenetic analysis of AHR1 in sturgeon species
A UPGMA dendrogram was generated using MEGA7 (Kumar et al. 2016) containing AHR amino acid sequences from mammals, AHR1 sequences from fishes and birds, and AHR2 sequences from fishes and birds. Sequences used and their Genbank accession numbers were red seabream Pagrus major AHR1 (BAE02824), Fugu rubripes AHR1b (NM_001037959), Japanese medaka Oryzias latipes AHR1a (BAB62012), Japanese medaka AHR1b (BAB62011), fathead minnow Pimephales promelas AHR1 (KX912260), Atlantic salmon Salmo salar AHR1a (NM_001123686), Atlantic salmon AHR1b (AY456091), northern pike Esox lucius AHR1b (KX912265), Atlantic killifish Fundulus heteroclitus AHR1 (AF024591), spiny dogfish Squalus acanthias AHR1 (AFR24092), lake sturgeon Acipenser fulvescens AHR1 (KM236089), white sturgeon Acipenser transmontanus AHR1 (KJ420394), Atlantic sturgeon Acipenser oxyrinchus oxyrinchus AHR1 (MH925108), shortnose sturgeon Acipenser brevirostrum AHR1 (MH925109), Japanese quail Coturnix japonica AHR1 (NM_001323184), chicken Gallus gallus domesticus AHR1 (NP_989449), human Homo sapiens AHR (L19872), mouse Mus musculus AHR (NP_038492), spotted gar Lepisosteus oculatus AHR1a (XP_015213432), black-footed albatross Phoebastria nigripes AHR2 (BAC87796), common cormorant Phalacrocorax carbo AHR2 (BAF64245), rainbow trout Oncorhynchus mykiss AHR2a (NP_001117723), rainbow trout AHR2b (NP_001117724), Atlantic tomcod Microgadus tomcod AHR2 (FJ215752), red seabream AHR2 (AB197788), Atlantic Killifish AHR2 (FHU29679), goldfish Carassius auratus AHR2 (FJ554572), zebrafish AHR2 (AF063446), Atlantic sturgeon Acipenser oxyrinchus AHR2 (MH223597), shortnose sturgeon Acipenser brevirostrum AHR2 (MH223598), white sturgeon AHR2 (KJ420395), and lake sturgeon AHR2 (AIW39681).
Amplification and insertion of AHR1 and AHR2 cDNAs into expression vectors
First strand cDNAs were prepared from Atlantic and shortnose sturgeon as described above and subjected to Extra-Long PCR amplification using a LongAmp® Taq PCR Kit (New England Biolabs, Ipswich, MA) to separately amplify the 5’ and 3’ halves of both AHR1 and AHR2 coding sequences. Random hexamers (200 ng) were added to 50 ng of total RNAs in 6 μl volumes, reactions were incubated at 680 C for 5 min, and chilled on ice. Reverse transcriptase mixture was added to the RNA-hexamer mix so that it contained 500 μM dNTPs (GE Healthcare), 1X RT buffer (Promega), 10 U of RNasin ribonuclease inhibitor (Promega) and 80 U of M-MLV Reverse transcriptase (Promega). Reactions were incubated at 420 C for 30 min after which 40 μl of LongAmp PCR mix was added to the first strand cDNA mixtures. Reactants included 1X LongAmp buffer (New England Biolabs), 300 μM dNTPs (GE Healthcare), 1 μM of forward and reverse primers (Integrated DNA Technologies) and 0.5 μl of LongAmp Taq polymerase (New England Biolabs). PCR conditions were 940 C for 2 min, 10 cycles at 940 C for 10 s, 440 C for 30 s and 680 C for 180 s, and then 25 cycles at 940 C for 10 s, 500 C for 30 s, and 680 C for 180 s with addition of 5 s in each successive cycle, and a final extension at 680 C for 7 min.
The 5’ half of the AHR2 coding sequence from both sturgeon species was amplified using primers XRTPCREB and XRTPCRBE (Table 2), digested with EcoRI and BamHI, and cloned into pUC19 vector. The 3’ half of the AHR2 coding sequence from both species was amplified with XRTPCRBK and XRTPCRKB, digested with BamHI and KpnI, and cloned into pUC19. EcoRI–EcoRV fragments from 5’ AHR2 and EcoRV-KpnI fragments from 3’ AHR2 were cloned into the linearized expression vector pcDNA3.1/Zeo(−). The cloned fragments in pAT-AHR2 and pSS-AHR2 were sequenced to ensure that they were identical to the Atlantic sturgeon and shortnose sturgeon AHR2 sequences previously determined (Roy et al. 2018).
Table 2.
Primers used for cloning Atlantic sturgeon and shortnose sturgeon AHR1 and AHR2 coding sequences into the pcDNA3.1(-Zeo) expression vector
| Primers used to clone AHR1 | |
| STAHR1–75F | TTTTTCTAGAAGTGAATTGCG |
| STAHR11268R | CCTTTGGCCTTGGCATGC |
| STAHR12544RA | CAGGCATGGTACCAGAGTTC |
| STAHR12544RS | CAGGCATGGTACCAGGGTTT |
| Primers used to clone AHR2 | |
| XRTPCREB | GAGAAGGAATTCTACTAGCTACAC |
| XRTPCRBE | AACAAGGGATCCTCAACATAGGAG |
| XRTPCRBK | CAAGATGGATCCTAACAACAGTGA |
| XRTPCRKB | TTTTTTGGTACCCTAAAATACAGT |
The 5’ half of AHR1 coding sequence from both sturgeon taxa was amplified with the primers STAHR1–75F and STAHR11268R. The 3’ half of AHR1 coding sequence was amplified using STAHR11251F as forward primer and the reverse primer was STAHR12544RA for Atlantic sturgeon and STAHR12544RS for shortnose sturgeon. Amplification conditions for LongAmp PCR were identical to those for AHR2. The 1,295 bp amplicon from the 3’ end was digested with KpnI (within the primer sequence) and BamHI (within the cDNA sequence) and inserted into the corresponding restriction sites of pBluescript II SK (+) cloning vector. This resultant recombinant (3’ AHR1-BS) was then used to clone the 5’ end of AHR1 cDNA. A 1,343 bp amplicon from the 5’ half of cDNA was digested with XbaI (within the primer sequence) and BamHI (within the cDNA sequence) and inserted into 3’AHR1-BS. Because there were two BamHI sites within the 1,295 bp fragment product, the insert was missing a 75 bp BamHI fragment of the AHR1 cDNA. A 679 bp AHR1 sequence, containing the 75 bp BamHI fragment from both species, was amplified using primers STAHR1_1189F and STAHR1_1868R, digested with BamHI, and inserted into the vector that contained both the 5’ and 3’ end of the AHR1 cDNA minus the 75 bp BamHI fragment. The clones were sequenced to determine the orientation of the 75 bp fragment. Full length Atlantic sturgeon and shortnose sturgeon AHR1 cDNAs were isolated by digesting the plasmids with XbaI and KpnI and cloned into the linearized expression vector pcDNA3.1/Zeo(−) to generate the pAT-AHR1 and pSS-AHR1 plasmids.
Reporter gene assays
Mouse hepatoma c12 cells (B15Ciii2; CRL 2710) derived from Hepa-1c1c7 cells and expressing reduced levels of AHR mRNA and protein were maintained as recommended by ATCC (Manassas, VA). Cells (30,000/well) were plated into 48-well plates. Transfections were done in triplicate wells 24 h after plating. For each well, a total of 300 ng of DNA was complexed with 1 μl of Lipofectamine 2000 (Invitrogen Corp., Carlsbad, CA) in 50 μl of serum free DMEM. Amounts of DNA transfected were 50 ng of pAT-AHR1, pAT-AHR2, pSS-AHR1, pSS-AHR2 singly or a binary mixture pAT-AHR1 and pAT-AHR2 or pSS-AHR1and pSS-AHR2, 50 ng of pAT-ARNT1 DNA, 40 ng of pGudLuc 6.1, and 3 ng of pRL-TK (Promega). Amounts of transfected DNA were kept constant by the addition of pcDNA3.1/Zeo(−) DNA. We did not test lesser amounts of DNA (i.e. 25 ng of plasmids each) in exposures with binary mixtures. At 5 h post transfections, cells were treated with graded doses of either TCDD (0.1, 1.0, 10, 100 ppb) or PCB126 (1.0, 10, 100, 1000 ppb) in acetone or acetone alone at a 0.5% final concentration. At 17 hr after addition of chemicals, cells were lysed and subjected to both firefly and renilla luciferase activity assays using a Dual Luciferase Reporter Assay system (Promega) according to manufacturer’s instructions.
Sources of embryos and tissues for gene expression analysis
Shortnose sturgeon embryos were obtained from Acadian Sturgeon and Caviar, Inc., Saint John, New Brunswick, Canada. Shortnose sturgeon broodstock were captive for several years and were originally collected from the Saint John River. Embryos were transported at approximately 48 h post-fertilization to the Howard Marine Sciences Laboratory of NOAA Fisheries, Sandy Hook, New Jersey, where they were reared and chemically treated as described in Roy et al.(2011) and Chambers et al. (2012).
Chemical exposures of shortnose sturgeon embryos
Shortnose sturgeon embryos (n = 25/treatment group, 3 d post-fertilization) were water-borne exposed for 24 hr in 25 ml of 1-ppt artificial sea water in 100-ml glass beakers to graded doses of TCDD (nominal doses of 0.0005 parts per billion (ppb), 0.005 ppb, 0.05 ppb, 0.5 ppb, 5 ppb and 50 ppb (AccuStandard; 99.1% purity)); PCB77 (nominal doses of 0.1 ppb, 1.0 ppb, 10 ppb, 100 ppb, 1000 ppb, and 10,000 ppb (AccuStandard; 99.7% purity)); PCB81 (nominal doses of 0.1 ppb, 1.0 ppb, 10 ppb, 100 ppb, 1000 ppb, and 10,000 ppb (AccusStandard 99.8% purity)); PCB126 (nominal doses of 0.1 ppb, 1.0 ppb, 10 ppb, 100 ppb, 1000 ppb, and 10,000 ppb (AccuStandard; 99.7% purity)); PCB169 (nominal doses of 0.1 ppb, 1.0 ppb, 10 ppb, 100 ppb, 1000 ppb, and 10,000 ppb (AccuStandard; 99.0% purity)); and an Aroclor mixture of Aroclor 1248 (40%), Aroclor 1254 (40%), and Aroclor 1260 (20%) (Accustandard) in acetone vehicle or to acetone alone.
Shortnose sturgeon embryos were maintained in exposure water for 24 h at 12oC after which they were rinsed and transferred to 750-ml Pyrex dishes with 500 ml of clean 1 ppt seawater for rearing until hatch. Every 12 h, dishes were cleaned of dead embryos, newly hatched larvae were removed, and an 80% percent water change was performed. Hatchlings were transferred and held alive in beakers for 24 h, snap frozen in liquid nitrogen, and stored at −80° C until RNA isolations.
Chemical exposures of juvenile shortnose sturgeon
Juvenile shortnose sturgeon that were approximately one-year-old and of Connecticut River ancestry that were captive at the Conte Lab of the USGS (Turners Falls, MA) were i.p. injected with graded doses of PCB126 (0.00 ppb, 0.01 ppb, 0.1 ppb, 1.0 ppb, 10 ppb, and 50 ppb) in 25 μl of corn oil solvent and sacrificed 8 d after exposure. Five fish were i.p. injected per treatment group. Multiple tissues (gill, heart, intestine, liver, pectoral fin clip) were harvested and immediately flash frozen on dry ice.
RNA isolations
All RNA isolations for individual shortnose sturgeon larvae (n=6/exposure group) and tissues (n = 5/exposure group) were done with Ultraspec reagent (Biotex, Houston, TX) as per the manufacturer’s recommendations and modified in Yuan et al. (2006). RNA concentrations and purities were evaluated with a NanoDrop ND-1000 spectrophotometer (NanoDrop Products, Wilmington, DE). RNA concentrations of all samples were adjusted to 100 ng/μl for subsequent procedures.
Expression of AHR1 in early life-stages and tissues of shortnose sturgeon
We used real-time RT-PCR to quantitatively compare AHR1 expression among exposure groups of larval shortnose sturgeon and tissues from juvenile sturgeon with the primers STRT_2213F and STSNRT_2284R. β-actin was amplified as the endogenous control using the primers Sturact60F and Sturact125 (Roy et al. 2011). Reactions contained 1.5 μl of RT products, 1.5 μl of primer mix (100 nM final concentration of each primer) (IDT) and 3 μl of Power Syber Green PCR Master Mix (Applied Biosystems (ABI), Foster City, CA). PCR cycling parameters were 950 C for 10 min and 40 cycles of 950 C for 15 s and 600 C for 60 s for amplifications on an ABI 7900 real-time PCR instrument.
Statistical analysis of gene expression data
Mean fold induction of AHR1 and CYP1A was calculated using the relative comparison Ct method for real-time RT-PCR as described in Livak and Schmittgen (2001). This method was applied this to calculate ß-actin-normalized fold induction of AHR1 and CYP1A in RT-PCR assays. The mean values of ΔAHR1 and ΔCYP1A for each larval exposure group or among tissues were compared using a one-way ANOVA with a Tukey’s post hoc test to determine the significance of differences in response among toxicant groups and negative controls, and among different doses within the same chemical.
Results
We compared AHR1 sequences among white sturgeon and three avian species to develop species-specific primers that were successfully used to amplify complete AHR1 cDNA sequences in both Atlantic sturgeon and shortnose sturgeon. The complete AHR1 coding sequences in Atlantic sturgeon and shortnose sturgeon were 2,559 and 2,547 nucleotides, respectively. Full-length peptide sequences of AHR1 in Atlantic sturgeon and shortnose sturgeon were 852 and 848 amino acid residues, respectively. Nucleotide and amino acid similarity of AHR1 was high among the four North American sturgeon species characterized to date with a mean nucleotide similarity of 95.2% (Table 3). The mean amino acid similarity of AHR1 among the four sturgeon species was also high (96.3%). Similarity of AHR1 nucleotide and amino acid sequence was higher between both Atlantic Coast species and white sturgeon than with lake sturgeon.
Table 3:
Evaluation of nucleotide and amino acid sequence identity of Atlantic sturgeon and shortnose sturgeon AHR1 to that of four other fishes (white sturgeon, lake sturgeon, Atlantic killifish, and zebrafish) and two avian species (quail and albatross). Genbank Accession No. KJ420394, KM236089, NM_001323184 (quail), AB106109 (albatross), AF024591 (killifish), and AF258854 (zebrafish, AHR1B) respectively. The LALIGN program in http://www.ebi.ac.uk/Tools/psa/lalign/nucleotide.html was used for this analysis.
| Nucleotide Identity (%) | |||||||
|---|---|---|---|---|---|---|---|
| Shortnose sturgeon |
White sturgeon |
Lake sturgeon |
Quail | Albatross | Atlantic killifish |
Zebrafish | |
|
Atlantic sturgeon |
97.5 | 96.6 | 91.9 | 64.3 | 64.9 | 58.6 | 59.2 |
|
Shortnose sturgeon |
98.2 | 93.5 | 64.6 | 65.0 | 58.9 | 59.7 | |
| White sturgeon | 93.3 | 64.4 | 64.7 | 59.1 | 59.6 | ||
| Lake sturgeon | 63.1 | 63.0 | 60.7 | 59.8 | |||
| Quail | 91.1 | 59.7 | 58.2 | ||||
| Albatross | 59.1 | 58.5 | |||||
| Atlantic killifish | 55.6 | ||||||
| Amino Acid Identity (%) | |||||||
|
Shortnose sturgeon |
White sturgeon |
Lake sturgeon |
Quail | Albatross |
Atlantic killifish |
Zebrafish | |
|
Atlantic sturgeon |
98.2 | 98.2 | 93.3 | 78.9 | 79.0 | 73.6 | 71.1 |
|
Shortnose sturgeon |
99.3 | 94.4 | 78.4 | 78.9 | 71.9 | 70.9 | |
| White sturgeon | 94.5 | 79.0 | 79.3 | 74.5 | 70.9 | ||
| Lake sturgeon | 76.2 | 76.4 | 68.5 | 71.0 | |||
| Quail | 97.4 | 77.9 | 73.2 | ||||
| Albatross | 78.1 | 67.3 | |||||
| Atlantic killifish | 74.0 | ||||||
UPGMA analysis generated a dendrogram which was evaluated for phylogenetic relationships of the putative AHR1 amino acid sequences of both Atlantic Coast sturgeon taxa to AHR sequences (AHR, AHR1, and AHR2) in mammals, birds, and other fishes (Fig. 1). Three major clades were evident, the first contained AHR1 sequences from all the fishes with the exception of the four sturgeon species and spotter gar. The second grouping contained the two AHR sequences from mammals (mouse and human), AHR1 sequences from the two avian species (quail and chicken), and AHR1a and AHR1 from the four sturgeon species and spotted gar. AHR1 sequences from the two Atlantic Coast sturgeon species clustered tightly. The final cluster contained AHR2 sequences from the two marine avian species (albatross and cormorant) and all of the fishes, including the four sturgeon taxa. Once again, the four sturgeon sequences clustered tightly with the two Atlantic Coast sturgeon species forming one grouping and lake sturgeon and white sturgeon forming a second.
Figure. 1.
Phylogenetic analyses of 32 full-length AHR, AHR1 and AHR2 sequences in vertebrate species. An unrooted UPGMA tree was generated by the maximum parsimony method with bootstrap values in MEGA 7-macOS ( Kumar et al. 2016) (See text for Genbank accession numbers).
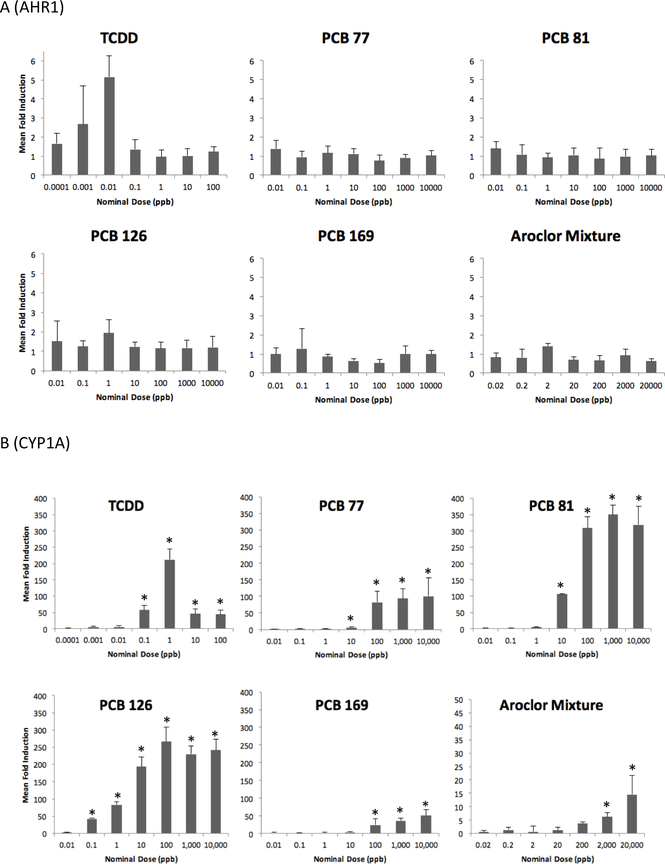
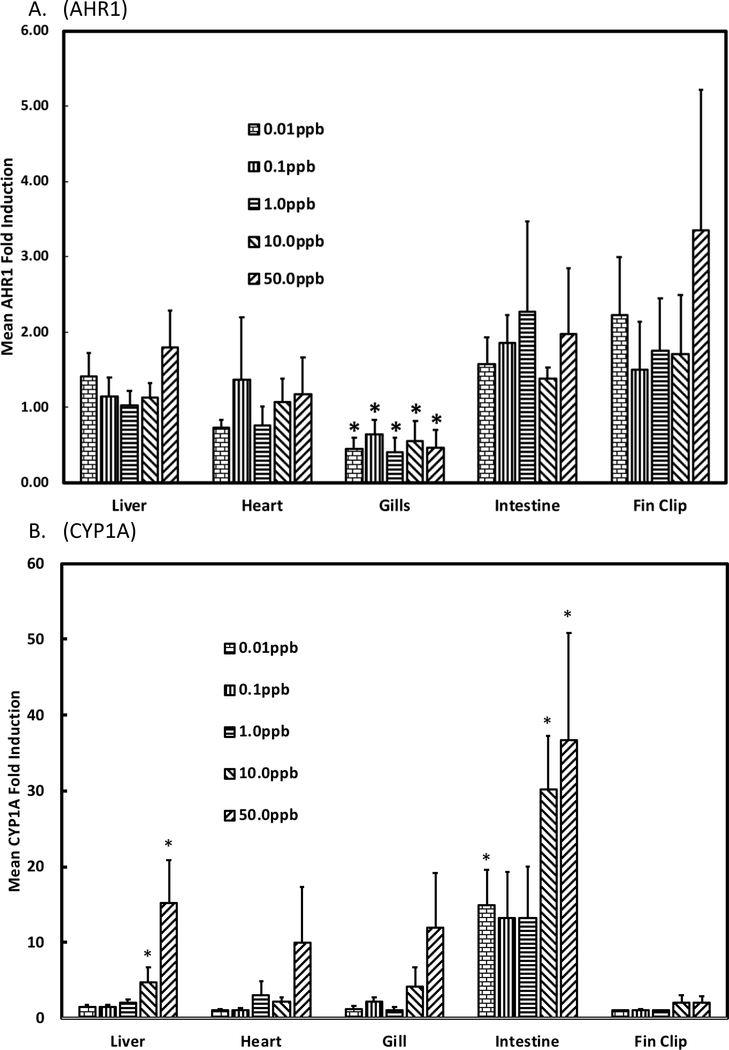
RT-PCR was used to evaluate expression of AHR1 in shortnose sturgeon larvae that were water-borne exposed for 24 h to graded doses of four coplanar PCBs, TCDD, and an environmentally relevant Aroclor mixture and compared its expression to that previously quantified for AHR2 in the same specimens (Roy et al. 2018). Unlike results for AHR2, we found no evidence of significantly induced AHR1 expression in shortnose sturgeon larvae for any of the six chemicals tested (Fig. 2a). This was in contrast to positive control CYP1A expression for which there was significant dose-responsive induction in larval shortnose sturgeon for graded doses of all six chemicals (Fig. 2b). Similarly, we found no evidence of significant induction of AHR1 in multiple tissues of juvenile shortnose sturgeon that had been i.p. injected with graded doses of PCB126 and sacrificed after 8 d (Fig. 3a). We did observe significant decreases in AHR1 expression in gills of specimens that had been treated with all five of the doses of PCB126. In contrast, there was significant induction of CYP1A in three of the six tissues tested: heart, liver, and intestine (Fig 3b). We also compared basal levels of AHR1 expression among the sturgeon tissues. Basal expression was determined in each tissue of specimens that were i.p. injected with corn oil. There was no significance difference in basal expression among liver, heart, gills, or intestine, but all of these tissues exhibited significantly higher levels of AHR1 expression than expression in fin clips. One caution to interpretation of our inter-tissue results is that we used the same housekeeping gene, ß-actin, for all tissues with unproven assumption that its expression was approximately equal across all tissues.
Figure 2.
Expression of A) AHR1 and B) CYP1A in shortnose sturgeon larvae waterborne exposed to graded doses of TCDD, four coplanar PCBs, and an environmentally relevant Aroclor mixture. Asterisk indicates that expression is significantly induced compared to negative acetone vehicle control.
Figure 3.
Expression of A) AHR1 and B) CYP1A in multiple tissues of shortnose sturgeon i.p. injected with graded doses of PCB126 and sacrificed after 8 days. * denotes significantly decreased (AHR1) or increased (CYP1A) expression.
We proceeded to successfully clone full-length AHR1 and AHR2 coding sequences from both Atlantic sturgeon and shortnose sturgeon into the pcDNA3.1/Zeo(−) expression vector. This allowed evaluation of the efficacy of the two sturgeon AHR proteins in driving reporter gene expression in AHR-deficient mouse hepatoma c12 cells treated with graded doses of TCDD or PCB126. Our objectives were two-fold; to determine whether sturgeon AHR1, unlike in AHR1 in other fishes, had functional activity in driving gene expression and to determine whether the combination of AHR1 and AHR2 induced greater reporter gene expression than either AHR1 or AHR2 alone. Therefore, the c12 cells were transfected with Atlantic sturgeon and shortnose sturgeon AHR1 alone, Atlantic sturgeon and shortnose sturgeon AHR2 alone, and the binary combination of AHR1 and AHR2 for both species.
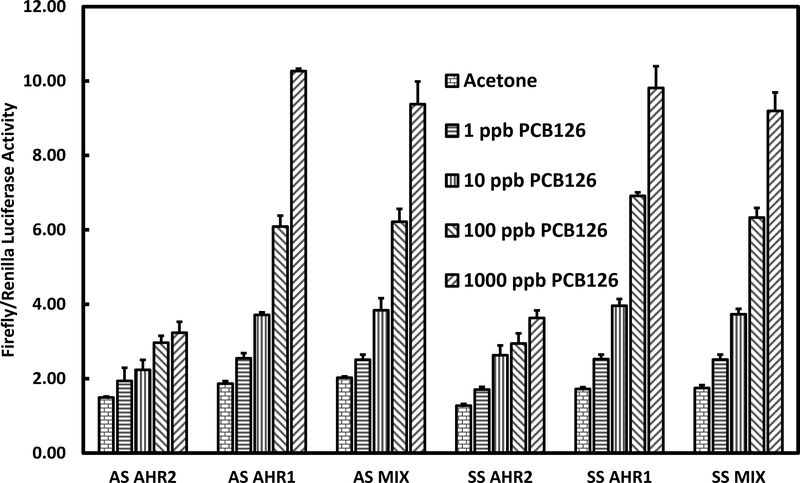
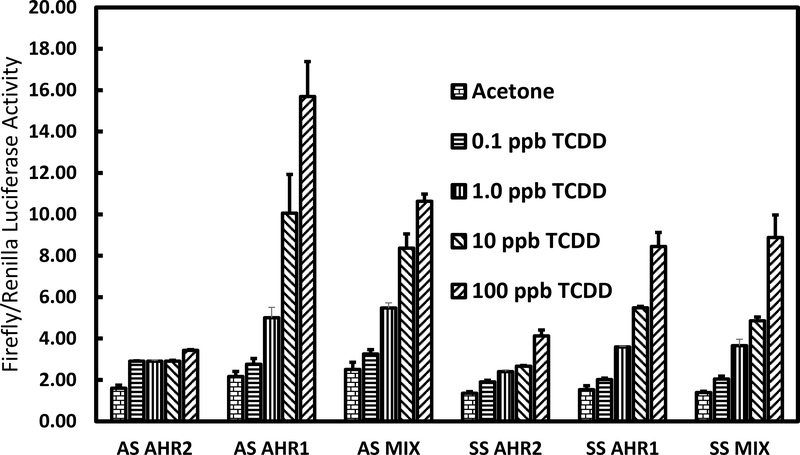
Only low levels of reporter gene expression occurred in transfections with either Atlantic sturgeon or shortnose sturgeon AHR2 alone for either TCDD or PCB126 treated-c12 cells (Fig. 4 and 5). Transfections with either Atlantic sturgeon or shortnose sturgeon AHR1 singly showed dose responsive and higher levels of reporter gene expression than seen with either AHR2 alone. Transfections of the binary combination of AHR1 and AHR2, however, did not show higher levels of gene expression than AHR1 in either species.
Figure 4.
Reporter gene expression in mouse hepatoma c12 cells treated with graded doses of PCB126 or acetone control and transfected with Atlantic sturgeon (AS) and shortnose sturgeon (SS) AHR1 singly, AHR2 singly, and their binary combination, pGudLuc 6.1 reporter gene plasmid and the control reporter vector pRL-TK.
Figure 5.
Results of reporter gene assay in mouse hepatoma c12 cells treated with graded doses of TCDD or acetone control and transfected with Atlantic sturgeon (AS) and shortnose sturgeon (SS) AHR1 singly, AHR2 singly, and their binary combination, pGudLuc 6.1 reporter gene plasmid and the control reporter vector pRL-TK.
Discussion
The major finding of our study is that functional activity of AHR1 is not enhanced in Atlantic Coast sturgeons and is probably not the mechanistic basis for the recently detected sensitivity of their young life-stages to TCDD and coplanar PCB toxicity. This result was unexpected given the recent finding of increased functional activity of AHR1 singly, and in binary concert with AHR2, in white sturgeon from the Pacific Coast.
The major hypothesis addressed in our study was that the previously reported sensitivity of Atlantic Coast sturgeon species to induced gene expression (Roy et al. 2011) and early life-stage toxicities (Chambers et al. 2012) from exposure to TCDD and coplanar PCBs was due to enhanced expression and functional activity of AHR1 in sturgeon species compared to other fishes and cooperativity between AHR1 and AHR2 in activating downstream gene expression and toxicities. This hypothesis emerged from prior results in white sturgeon where basal and β-NF induced levels of AHR1 were higher across multiple tissues than seen in other fishes and were sometimes similar to that of AHR2 (Doering et al. 2014a). Furthermore, in reporter gene assays in AHR-deficient cells, white sturgeon AHR1 was able to drive in vitro gene expression and the combination of AHR1 and AHR2 induced higher levels of gene expression than either AHR alone (Doering et al. 2014b).
To empirically address this hypothesis, we cloned and characterized AHR1 in both Atlantic Coast sturgeon species in this study and AHR2 previously (Roy et al. 2018). We demonstrated previously that AHR2 expression was significantly induced in larvae of both Atlantic Coast sturgeon species by exposure to TCDD, four different coplanar PCBs, and an Aroclor mixture (Roy et al. 2018) and in some tissues of shortnose sturgeon by PCB126 (Roy et al. 2011). Here, primers were also developed to quantify AHR1 expression in vivo in shortnose sturgeon larvae and tissues in RT-PCR assays. Furthermore, characterization of AHR1 here and AHR2 previously (Roy et al. 2018) allowed us to insert the full length AHR1 and AHR2 coding sequences of both sturgeon species into expression vectors to evaluate their in vitro functional activities.
Phylogenetic analysis that included newly characterized AHR1 amino acid sequences in this study and previously characterized AHR, AHR1 and AHR2 sequences in other fishes (including two other North American sturgeon taxa), birds, and mammals, indicated that we had succeeded in cloning AHR1 from both Atlantic Coast sturgeon species. AHR, AHR1 and AHR2 sequences from all taxa depicted formed three distinct clades. One clade contained AHR2 from all the fishes and avian taxa, including the four previously characterized sturgeon AHR2 genes which formed a tight cluster. The second clade contained AHR1 from all fishes except the four sturgeon species and spotted gar. The third grouping contained AHR from mammals, AHR1 from avian taxa and AHR1 from the four sturgeon species and spotted gar. Interestingly, AHR1 from all four sturgeon species clustered tightly, but were within the avian AHR1 and mammalian AHR grouping rather than the grouping that contained all the other fish AHR1 sequences. This result was consistent with that of Doering et al. (2014a) in white sturgeon and suggests that expression and function of AHR1 in sturgeon species may be different from that of other fishes and more similar to that of AHR in mammals and AHR1 in birds. This result is consistent with Hahn et al.’s (2017) hypothesis that AHR1 seen in most fishes may have been lost in sturgeon taxa and gar, and that their second AHR is actually an AHR orthologue. Importantly, it also suggested that the AHR1 protein from sturgeon species may function more like mammalian AHRs and avian AHR1s than AHR1s in other fishes.
To initially evaluate the potential increased functional activity of AHR1 in sturgeon taxa compared to AHR1 in other fishes in activating gene expression and toxicity, we evaluated the expression of AHR1 in shortnose sturgeon larvae that were water-borne exposed to graded doses of TCDD, four coplanar PCBs, and an Aroclor mixture. We found no significant induction of AHR1 in shortnose sturgeon larvae at any dose of any of the chemicals. In contrast, CYP1A was significantly and dose-responsively induced in these same larval specimens by at least one dose of each chemical and often at the lowest dose tested. These AHR1 results contrast to our earlier results on AHR2 in these same specimens where AHR2 was significantly induced by TCDD, three of four coplanar PCBs and the Aroclor mixture and levels of AHR2 and CYP1A expression were significantly correlated in both shortnose and Atlantic sturgeon (Roy et al. 2018). This data indicates that AHR2, not AHR1, expression is the likely driver of induced CYP!A expression in these two sturgeon species. In total, these results do not support an increased role of AHR1 in mediating toxicity in the two Atlantic Coast sturgeon species.
We also compared basal and induced expression of AHR1 among five tissues of juvenile shortnose sturgeon that had been i.p. injected with graded doses of PCB126 or corn oil vehicle. We found no difference in basal levels of AHR1 expression among tissues of the specimens that had been i.p. injected with corn oil vehicle except that fin clips exhibited lower levels of expression than in all the other tissues tested. Furthermore, PCB126 failed to induce significant expression of AHR1 at any of the doses in any of the tissues tested. These results differ from those of Doering et al. (2014a) who previously evaluated basal expression among nine tissues of juvenile white sturgeon and found significant differences among tissues with liver having higher levels of basal expression than seven other tissues; the exceptions being heart and gill. Doering et al. (2014a) also investigated the effects of i.p. injection of two doses of β–NF (50 and 500 mg β–NF/kg bw) on AHR1 expression in liver, gill and intestine of juvenile white sturgeon. They found significant increases of AHR1 expression in all three tissues at the lowest dose of BNF used, but not at the highest dose in liver and gill. Thus, basal and inducible expression of AHR1 differed between the Atlantic Coast and Pacific coast sturgeon species despite their structural relatedness. These results do not support an increased functional role for AHR1 in shortnose sturgeon.
A reporter gene expression assay provides a strong empirical test of the functionality of variant forms of AHR (Wirgin et al. 2011). With it, mammalian cells that were inherently deficient in AHR expression, were transfected with the pGudluc 6.1 reporter gene vector that contained the firefly luciferase gene and four dioxin response elements (DREs) to which activated AHR-ARNT complex could bind, a plasmid expressing exogenous Atlantic sturgeon ARNT1, and the target AHR1 or AHR2 plasmid for which the functionality of its protein product was tested. Transfected cells were treated with graded doses of AHR agonists, TCDD or coplanar PCB126, and the amount of firefly luciferase expression product was quantified. The greater functionality of the target AHR, the more AHR-ARNT1 complex that forms and binds DREs, the greater the amount of luciferase gene product that is made.
Initially, we found little evidence of reporter gene expression in TCDD or PCB126 treated cells that were transfected with either sturgeon AHR1 or AHR2 and Atlantic tomcod ARNT1 (Genbank Accession Number ACX53265). In contrast, positive control transfections in the same system with Atlantic tomcod AHR2 yielded significantly increased reporter gene expression in either PCB126 or TCDD-treated cells (data not shown). This result prompted us to isolate ARNT1 from Atlantic sturgeon as described in the Methods of the Supplementary Information. As depicted in the UPGMA dendrogram in Figure 1 of the Supplementary Information, we succeeded in isolating a protein in Atlantic sturgeon that was genetically similar to ARNT1 from other fishes. Subsequently, we substituted Atlantic sturgeon ARNT1 for tomcod ARNT1 in the reporter gene assays. Surprisingly, transfections with both Atlantic sturgeon and shortnose sturgeon AHR1 exhibited dose-responsive and higher levels of reporter gene expression than with either sturgeon AHR2. Maximum reporter gene expression in cells transfected with Atlantic sturgeon AHR1 and treated with TCDD was approximately 16-fold and approximately 10-fold in cells treated with PCB126 compared to vehicle treated controls. Similarly, maximum reporter gene expression in cells transfected with shortnose sturgeon AHR1 and treated with either TCDD or PCB126 was dose-responsive and higher than in cells transfected with shortnose sturgeon AHR2. However, reporter gene expression in cells treated with binary combinations of either Atlantic sturgeon or shortnose sturgeon AHR1 and AHR2 was not greater than with AHR1 alone from either species. Thus, mixtures of the two AHRs from both species did not support greater reporter gene expression and likely not greater functional activity than either AHR1 alone.
Our results contrasted with those reported in white sturgeon in which the mixture of AHR1 and AHR2 supported higher levels of reporter gene activity than either AHR alone in cells treated with β-NF (a model PAH) and led the authors to conclude that both AHRs were functionally active in species of sturgeons and may have resulted in the hypersensitivity of sturgeon species compared to other fishes to aromatic hydrocarbon contaminants. It is possible that the difference in results between the reporter gene studies with white sturgeon versus Atlantic and shortnose sturgeon is the chemicals used in exposures; a non-halogenated PAH versus TCDD and a PCB. For example, we have previously suggested that in Atlantic tomcod, AHR2 mediates responses to halogenated aromatic hydrocarbons and that a second protein, perhaps AHR1, mediates responses to non-halogenated PAHs (Wirgin et al. 1992; Yuan et al. 2006).
At this point, the mechanistic basis of the heightened sensitivity of Atlantic Coast sturgeon species to TCDD or PCB induced toxicity remains to be determined. Phylogenetic analysis of their AHR1 suggested that its functional activity might be similar to that of mammalian AHR and avian AHR1. Consistent with that suggestion, Atlantic and shortnose sturgeon AHR1 drove higher levels of reporter gene expression than their AHR2. Yet, in vivo expression of AHR1 was not inducible in sensitive young life-stages nor in multiple tissues of juvenile shortnose sturgeon. Unlike AHR2, expression of AHR1, was not directly correlated or predictive of downstream expression of CYP1A. Most importantly, the combination of AHR1 and AHR2 did not drive higher levels of reporter gene expression than AHR1 alone. While our results do not support increased functional activity of AHR1 in Atlantic coast sturgeons compared to other fishes, knockdown or knockout experiments with AHR1 and AHR2, would provide a more definitive test of the functional role of these two proteins in mediating aromatic hydrocarbon toxicity in this primitive taxon.
Supplementary Material
Highlights.
Cloning and characterization of AHR1 cDNA in endangered Atlantic sturgeon and shortnose sturgeon
Amino acid sequence in both sturgeons was more similar to mammalian AHR than AHR1 in other fishes
AHR1 was not significantly induced by four coplanar PCBs nor TCDD in young lifestages of shortnose sturgeon
AHR1 was not significantly induced by PCB126 in multiple tissues of juvenile shortnose sturgeon
AHR1 expression vectors that were developed for both sturgeon species drove higher gene expression than AHR2 expression vectors in reporter gene assays
The binary combination of AHR1 and AHR2 expression vectors failed to drive higher gene expression than AHR1 alone in reporter gene assays
Acknowledgements
We acknowledge support of the Hudson River Foundation, NOAA’s Northeast Fisheries Science Center, use of the Molecular Facilities Core of NYU NIEHS Center Grant ES00260, and laboratory contributions of Kristin Habeck in conducting this research. We thank Cornel Ceapa of Acadian Sturgeon and Caviar, Inc. for the Atlantic sturgeon and shortnose sturgeon embryos used in these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Antkiewicz DS, Burns CG, Carney SA, Peterson RE, Heideman REW 2005. Heart malformation is an early response to TCDD in embryonic zebrafish. Toxicol. Sci 84:368–377. [DOI] [PubMed] [Google Scholar]
- Bain MD 1997. Atlantic and shortnose sturgeons of the Hudson River: common and divergent life history attributes. Environ Biol. Fishes 48:347–358. [Google Scholar]
- Chambers RC, Davis DD, Habeck EA, Roy NK, Wirgin I 2012. Early life-stage toxic effects of PCB126 and TCDD on shortnose sturgeon and Atlantic sturgeon. Environ. Toxicol. Chem 31:2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doering JA, Wiseman S, Beitel SC, Giesy JP, Hecker M 2014a. Identification and expression of aryl hydrocarbon receptors (AhR1 and AhR2) provide insight in an evolutionary context regarding sensitivity of white sturgeon (Acipenser transmontanus) to dioxin-like compounds. Aquat. Toxicol 150:27–35. [DOI] [PubMed] [Google Scholar]
- Doering JA, Farmahin R, Wiseman S, Kennedy SW, Giesy JP, Hecker M 2014b. Functionality of aryl hydrocarbon receptors (AhR1 and AhR2) of white sturgeon and implications for the risk assessment of dioxin-like compounds. Environ. Sci. Technol 15:8219–8226. [DOI] [PubMed] [Google Scholar]
- Doering JA, Farmahin R, Wiseman S, Beitel SC, Kennedy SW, Giesy JP, Hecker M 2015. Differences in activation of aryl hydrocarbon receptors of white sturgeon relative to lake sturgeon are predicated by identities of key amino acids in the ligand binding domain. Environ. Sci. Technol 49:4681–4690. [DOI] [PubMed] [Google Scholar]
- Elonen GE, Spehar RL, Holcombe GW, Johnson RD, Fernandez JD, Erickson RJ, Tietge JE, Cook PM 1998. Comparative toxicity of 2,3,7,8 tetrachlorodibenzo-p-dioxin to seven freshwater fish species during early life-stage development. Environ. Toxicol. Chem. 17:472–483. [Google Scholar]
- Ema M, Ohe N, Suzuki M, Mimura J, Sogawa K, Ikawa S, Fujii-Kuriyama Y 1994. Dioxin binding activities of polymorphic forms of mouse and human aryl hydrocarbon receptors. J. Biol. Chem 269:27337–27343. [PubMed] [Google Scholar]
- Farmahin R.Manning, Crump D, Wu D, Mundy LJ, Jones SP, Hahn ME, Karchner SI, Giesy JP, Bursian SJ, Zwiernik MJ, Fredericks T, Kennedy SW 2013. Amino acid sequence of the ligand-binding domain of the aryl hydrocarbon receptor 1 predicts sensitivity of wild birds to effects of dioxin-like compounds. Toxicol. Sci 131:139–152. [DOI] [PubMed] [Google Scholar]
- Federal Register. 2012a. Endangered and threatened wildlife and plants; threatened and endangered status for distinct population segments of Atlantic sturgeon in the Northeast region. 77, 5880–5912, February 6, 2012. [Google Scholar]
- Federal Register. 2012b. Endangered and threatened wildlife and plants; threatened and endangered status for two distinct population segments of Atlantic sturgeon Acipenser oxyrinchus oxyrinchus in the southeast. 77, 5914–5984, February 6, 2012. [Google Scholar]
- Fernandez MP, Ikonomou MG, Courtenay SC, Wirgin II 2004. Spatial variation in hepatic levels and patterns of PCBs and PCDD/Fs among young-of-the-year and adult Atlantic tomcod (Microgadus tomcod) in the Hudson River Estuary. Environ. Sci. Technol 38:976–983. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Merson RR 2017. Diversity as opportunity: insights from 600 million years of AHR evolution. Curr. Opin. Toxicol 2:58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Shapiro MA, Perera SA 1997. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc. Natl. Acad. Sci. USA 94:13743–13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head JA, Hahn ME, Kennedy SW 2008. Key amino acids in the aryl hydrocarbon receptor predict dioxin sensitivity in avian species. Environ. Sci. Technol 42:7535–7541. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Kennedy SW, Hahn ME 2006. The molecular basis for differential dioxin sensitivity in birds: role of the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 103:6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Heiden TC, Mehta V, Xiong KM, Lanham KA, Antkiewicz DS, Ganser A, Heideman W, Peterson RE 2012. Reproductive and development toxicity of dioxin in fish. Mol. Cell. Endocrinol 354:121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamur K 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanham KA, Plavicki J, Peterson RE, Heideman W 2014. Cardiac myocyte-specific AHR activation phenocopies TCDD-induced in zebrafish. Toxicol. Sci 141:141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using realtime quantitative PCR and the 2- CT method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Okey AB, Franc MA, Moffat ID, Tijet N, Boutros PC, Korkalainen M, Tuomisto J, and Pohjanvirta R 2005. Toxicological implications of polymorphisms in receptors for xenobiotic chemicals: The case of the aryl hydrocarbon receptor. Toxicol. Appl. Pharmacol 207:S43–S51. [DOI] [PubMed] [Google Scholar]
- Poland A, Palen D, Glover E 1994. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol. Pharmacol 46:915–921. [PubMed] [Google Scholar]
- Powell WH, Bright R, Bello SM, Hahn ME 2000. Developmental and tissue-specific expression of AHR1, AHR2, and ARNT2 in dioxin-sensitive and-resistant populations of the marine fish Fundulus heteroclitus. Toxicol. Sci 57:229–239. [DOI] [PubMed] [Google Scholar]
- Reid NM, Proestou DA, Clark BW, Warren WC, Colbourne JK, Shaw JR, Karchner SI, Hahn ME, Nacci D, Oleksiak MF, Crawford DL, Whitehead A 2016. The genomic landscape of rapid repeated evolutionary adaptation to toxic pollution in wild fish. Science. 354:1305–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NK, Candelmo A, DellaTorre MS, Chambers RC, Nadas A, Wirgin I 2018. Characterization of AHR2 and CYP1A expression in Atlantic sturgeon and shortnose sturgeon treated with coplanar PCBs and TCDD. Aquat. Toxicol 197:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NK, Walker N, Chambers RC, Wirgin I 2011. Characterization and expression of cytochrome P4501A in Atlantic sturgeon and shortnose sturgeon experimentally exposed to TCDD and coplanar PCB 126. Aquat. Toxicol 104:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortnose Sturgeon Status Review Team. 2010. A Biological Assessment of Shortnose Sturgeon (Acipenser brevirostrum). Report to National Marine Fisheries Service, Northeast Regional Office. November 1, 2010. 417 pp. [Google Scholar]
- Tappenden DM, Hwang HJ, Yang L, Thomas RS, LaPres JL 2013. The aryl-hydrocarbon receptor protein interactions network (AHR-PIN) as identified by tandem affinity purification (TAP) and mass spectrophotometry. J. Toxicol http://dx.doi.org/10.1155/2013/279829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Oost R, Beyer J, Vermeulen NPE 2003. Fish bioaccumulation and biomarkers in environmental risk assessment: a review. Environ. Toxicol. Pharmacol 13:57–149. [DOI] [PubMed] [Google Scholar]
- Wirgin II, Chambers RC 2006. Atlan tic tomcod (Microgadus tomcod): A model species for the response of Hudson River fish to toxicants Eds. Waldman JR, Limburg KE, and Strayer D Hudson River Fishes and their Environment. AFS Symposium; 51:331–364. [Google Scholar]
- Wirgin I, Grunwald C, Stabile J, Waldman JR 2010. A genetic contribution to the delineation of distinct population segments of shortnose sturgeon (Acipenser brevirostrum). Conserv. Genet. 11:689–708. [Google Scholar]
- Wirgin I, Kreamer G-L, Grunwald C, Squibb K, Garte SJ 1992. Effects of prior exposure history on cytochrome P4501A induction by PCB congener 77 in Atlantic tomcod. Mar. Environ. Res 34:103–108. [Google Scholar]
- Wirgin I, Roy NK, Loftus M, Chambers RC, Franks DG, Hahn ME 2011. Mechanistic basis of resistance to PCBs in Atlantic tomcod from the Hudson River, USA. Science 331:1322–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Maceda L, Grunwald C, King T 2015. Population origin of Atlantic sturgeon bycaught in U.S. Atlantic coast fisheries. J. Fish Biol 85:1251–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Weis JS, McElroy AE 2006. Physiological and genetic aspects of toxicity in Hudson River species, pp 441–464 In: Levinton J, Waldman JR. Eds. The Hudson River Ecosystem, Cambridge University Press. [Google Scholar]
- Yuan Z, Courtenay SC, Chambers RC, Wirgin I 2006. Evidence of spatially extensive resistance to PCBs in an anadromous fish of the Hudson River. Environ. Health Perspect. 114:77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Courtenay S, Wirgin I 2006. Comparison of hepatic and extra hepatic induction of cytochrome P4501A by graded doses of aryl hydrocarbon receptor agonists in Atlantic tomcod from two populations. Aquat. Toxicol 76:306–320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.