Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by deficits in social interaction and communication, along with repetitive and restrictive patterns of behaviors or interests. Normal brain development is crucial to behavior and cognition in adulthood. Abnormal brain development, such as synaptic and myelin dysfunction, is involved in the pathogenesis of ASD. Microtubules and microtubule-associated proteins (MAPs) are important in regulating the processes of brain development, including neuron production and synaptic formation, as well as myelination. Increasing evidence suggests that the level of MAPs are changed in autistic patients and mouse models of ASD. Here, we discuss the roles of MAPs.
Keywords: Autism spectrum disorder, Microtubule-associated proteins, Synapse, Myelin
Introduction
Autism spectrum disorder (ASD) is a severe neurodevelopmental disorder with a high prevalence. It is characterized by difficulties in reciprocal social interaction and communication, and repetitive and restrictive patterns of behavior or interests. In consideration of the poor social adaptability, autistic patients need lifelong care which imposes huge economic and mental burdens on their families. The number of patients who have been diagnosed with ASD has risen dramatically around the world over the last decades [1]. The most recent prevalence of ASD among children aged 8 years is ~ 1 in 68 children, as estimated by Developmental Disabilities Monitoring Network in 11 sites, USA, 2012 [2].
It has been demonstrated that a complicated interplay of both genetic and environment factors is involved in ASD [3]. However, to date, the precise pathogenesis of ASD remains undefined. A few pharmacological treatments may release some of the associated symptoms, but there are no therapeutic options that target the core symptoms of autism. In this review, we describe the current status of the pathogenesis of ASD and highlight the current research on microtubule-associated proteins (MAPs).
Genetic, Environmental, and Neuroimmune Factors in ASD
Genetic and Epigenetic Factors in ASD
Emerging evidence suggests that genetic factors play important roles in ASD. For instance, a child who has an affected sibling is more likely to be diagnosed with ASD [4], and parents of such children show subtle cognitive or behavioral deficits compared to controls [5]. ASD studies based on both human genetics and animal models have demonstrated that the interruption of processes such as synapse formation, neuron production, and spine stability are important features of ASD etiology. Therefore, changes in common molecular pathways involved in those factors may contribute to the pathogenesis of ASD [6, 7]. Individual base-pairs (single-nucleotide variants), millions of base-pairs [copy-number variants (CNVs)] and de novo genetic variants have been found in hundreds of genes, which are mostly rare and cover the whole spectrum of mutations [8]. For instance, Ziats et al. [9] concluded that CNVs associated with synaptogenesis, axon guidance, and neuronal motility are involved in the pathogenesis of ASD. Duplication and deletion of the mutations involved in CNVs can interrupt gene structure and function and often result in developmental delays [10]. And some risk genes for ASD have been identified, such as NEUROLIGIN4 (NLGN4), NLG1, SHANK3, CNTN4, SYNGAP1, and CNTNAP2 [10]. Studies focusing on these genes have identified cellular pathways that are changed in ASD during the process of neuronal development (Fig. 1) [11].
Fig. 1.
Diagram of ASD risk genes involved in brain development. Image adapted from Lin YC et al. Front Cell Neurosci 2016 [11].
Despite the fact that many ASD-associated genes have been identified, these coding variants only account for a tiny segment of ASD cases; a large proportion of heritability still cannot be explained by genetic findings. So far, many studies have demonstrated that epigenetic modifications caused by non-coding genetic variation play an important role in neuronal differentiation and communication, and abnormal epigenetics is involved in the development of the nervous system and can lead to neurobehavioral deficiencies and psychiatric disorders [12, 13]. Varadinova et al. [10] found that prenatal and early postnatal exposure to alcohol, stress, toxins, or drugs can easily damage brain development by changing the epigenetic status of genes in important periods of neuronal and synaptic organization and epigenetic modification in brain development. The specific mechanisms of epigenetic modification, including DNA methylation, non-coding RNA, and post-translational histone changes [14], can have impact on the quantity and quality of gene products at different levels [15, 16]. A hypothesis has been put forward that epigenetics can bridge the genetic and environmental factors that can result in DNA hypomethylation or demethylation in specific genomic regions in certain tissues [13, 17, 18]. Sun et al. reported that > 5000 enhancer or promoter loci changed up or down in the ASD cerebral cortex, demonstrating that aberrations in histone acetylation are widely distributed in the ASD cerebral cortex [19].
Environmental Factors in ASD
Some environmental factors, including maternal viral infection, bacterial infection, influenza, and febrile episodes during pregnancy, are involved in the etiology of ASD via activation of the maternal immune system, and can result in altered fetal brain development. Epidemiological research has demonstrated that gestational infections are tied to the core features of autism by interrupting fetal brain development [20]. Lee et al. [21] found a positive correlation between severe maternal infection and an increased risk of ASD, supporting the hypothesis that immune-mediated mechanisms contribute to the phenotypic deficits. Furthermore, in mouse models, injecting either influenza, double-stranded RNA poly (I:C), or bacterial lipopolysaccharide during pregnancy induced an active immune response, and the offspring of these mice showed behavioral deficits in adulthood. Besides, changed cytokine levels were found in the serum, placenta, and amniotic fluid, as well as in the fetal brain [22].
Above all, brain development is associated with the action of neuroimmune factors which are critical for neuronal migration, axonal growth, neuronal positioning, and cortical lamination, as well as dendrite and synapse formation. Therefore, altered cytokine levels are involved in neurodevelopmental disorders such as ASD [23]. A set of increased cytokines has been identified in the brain of autistic patients; these include interleukin (IL)6, IL-10, tumor necrosis factor-α, transforming growth factor-ß1, and macrophage chemoattractant protein-1 [24, 25]. IL-6, an important neuroimmune factor, signals via the heterodimerization of gp130 and the IL-6 receptor on the cell surface [22]. It has been reported that increased IL-6 mediates neuroanatomical abnormalities by interrupting the balance of excitatory and inhibitory synaptic formations and synaptic transmission as well as prolonging the length of dendritic spines which is linked to synaptic function [26–28]. By activating IL-6 in the fetal brain, gestational infections can activate a maternal immune response that is associated with abnormal fetal brain development, and can lead to offspring with the core features of ASD [22]. Smith et al. [22] found that maternal immune activation (MIA) does not result in behavioral deficits in IL-6 knockout mice, unlike those seen in the offspring of wild-type mice after MIA. Wei et al. [22, 29] demonstrated that inhibition of IL-6 trans-signaling increases sociability in an autistic mouse model. Therefore, increased IL-6 is implicated in the pathogenesis of ASD.
Moreover, some studies have suggested that ASD patients are more likely to have at least one gastrointestinal symptom [30], and this may contribute to the behavioral impairments. A gut-brain axis hypothesis has been proposed, in which gut microbiota are believed to affect brain excitability via endocrine, metabolic, or immunological mechanisms that are likely to provoke the pathogenesis of ASD [31–33].
Synapse and Myelin Dysfunction in ASD
Abnormal brain development, such as synapse and myelin dysfunction, is involved in the pathogenesis of ASD. To better illustrate the pathogenesis of ASD, genetic studies have focused on the processes of brain development. For instance, some risk genes that are key regulators of synaptic plasticity have been found in genetic studies and many animal studies of ASD [34]. These genes encode various proteins including synaptic scaffolding proteins, receptors, and cell adhesion molecules, as well as proteins involved in chromatin remodeling, transcription, protein synthesis or degradation, or actin cytoskeletal dynamics. Some of these risk genes are governed by neuronal activity and can influence neuronal connectivity and synaptic plasticity. In addition, it is certain that sensory input and intrinsic brain activity are influenced by synaptic efficacy and are increased or decreased by neuronal connectivity. Studies have revealed that an imbalance between excitatory and inhibitory synapses induces dysfunction of the brain, and is involved in the pathology of ASD [34, 35]. Magnetic resonance spectroscopic studies have found that the glutamate signal varies depending on the diverse phenotypes of ASD [36]. Glutamate is the most important excitatory neurotransmitter, giving rise to excitatory synaptic transmission by binding to glutamate receptors [35]. There is no notable change in the number of glutamatergic synapses and the levels of excitatory synaptic proteins, as well as in the density and postsynaptic density thickness of dendritic spines in the BTBR mouse, a model of autism [37], but the evoked release of glutamate is reduced [35]. An imbalance between excitation and inhibition could be a fundamental component of the pathogenesis of autism.
In ASD, neuronal communication is compromised, and the axon is an important conduit of information communication between neurons [38]. The myelin produced by oligodendrocytes is essential for achieving and maintaining optimal brain function [39]. The myelination process may underlie many cognitive and behavioral abilities. Furthermore, the myelination process may be involved in many neurodevelopmental disorders, such as schizophrenia, Down syndrome, and bipolar disorders [40]. Myelination-related proteins such as myelin basic protein, myelin-associated glycoprotein, and proteolipid protein 1 are important in the formation of myelin and the maintenance of its stability. It has been reported that abnormal myelin development occurs in BTBR mice compared to control mice. For example, there are fewer myelinated axons and less myelin in the cortex of BTBR mice [41]. Similarly, Jones-Davis et al. suggested that many abnormalities, such as a complete loss of the corpus callosum, a reduction in the length and area of the hippocampal commissure, and a slightly enlarged anterior commissure, occur in the formation of commissural structures [42]. In the same way, abnormalities in both white matter microstructure and the corpus callosum have also been found in a study using MRI technology [43].
Microtubule-Associated Protein in ASD
Physiological and Pathological Roles of Microtubule-Associated Protein in the Central Neural System
The complicated architectures of neuronal networks and synaptic connections are responsible for the maintenance of suitable brain function [44]. Many neurodevelopmental disorders present abnormal physical and functional connections, and these abnormalities can lead to the disturbances in corticogenesis and changes in cortical laminar architecture [44, 45]. Some steps in brain development are associated with neurodevelopmental disorders linked with altered cortical development. These steps include cellular proliferation, migration, and differentiation [45]. Furthermore, abnormal neuron production and migration and altered synaptogenesis can result in aberrant microcircuit assembly in many regions [46]. Cytoskeletal elements, including the microtubular network, are needed for suitable brain connectivity and neuronal morphogenesis and differentiation, and sufficient evidence suggests that microtubules play a fundamental role in adjusting all the processes of neuron and brain development [44, 45].
Microtubules are cylindrical polymers which are assembled from α-β tubulin heterodimers [47]. Microtubules, actin microfilaments, and intermediate filaments constitute the cytoskeleton, which is a dynamic structure giving the cell its shape and resistance to deformation [45]. Microtubules spread throughout the cell body, not only providing rigidity, but running dynamic processes such as mitotic spindle formation and intracellular cargo transport [45]. During neuronal development, microtubules are homogeneously oriented in the axon, have various orientations and acquire different post-translational modifications and cytoskeletal density in the dendrites. These structures establish synaptic contacts and contribute to the creation of effective functional connectivity network [45, 48]. And some studies have suggested that microtubules play a vital role in cognitive functions and behaviors, as they are essential in the growth and maintenance of the axon, the development and plasticity of the dendritic spines, and the migration of developing neurons to their destinations.
Microtubule dynamics are adjusted by microtubule-interacting proteins and various pathways, such as the Wnt-dishevelled, NF-κB, Rho-associated coiled-coil kinase, Reelin, and Notch pathways [45, 49, 50]. The Wnt-dishevelled pathway reinforces microtubule stability in the axon and plays a vital role in the formation of looped microtubules at enlarged growth cones [45]. Wnt signaling not only adjusts multiple developmental steps, such as cell proliferation, specification, migration, and differentiation, but also influences the neuronal differentiation of cortical intermediate progenitors [51]. In addition, tau, MAPs, and microtubule-stabilizing proteins regulate Wnt/Ca2+ signaling [45]. The NF-κB pathway regulates axon initiation, extension, guidance, and branching, adjusts dendritic arbor size and complexity, and influences dendritic spine density in mature neurons [45, 52]. Although the mechanisms by which NF-κB plays its role in cytoskeletal remodeling are unclear, fortunately, it is clear that MAP2 and MAP1B are targets of NF-κB transcription [53].
To realize microtubules, precise modulation of the cytoskeleton that involves MAPs is required [54, 55]. MAPs, binding along the full length of microtubules, are helpful for axon outgrowth and pathfinding [55]. For instance, MAPs mediate microtubule dynamics by changing the rates of microtubule growth and shrinkage, and modifying the frequency of disintegration or recovery [56]. MAPs have been divided into various categories, such as microtubule motors, the microtubule-severing protein katanin, and structural MAPs that include MAP1, MAP2, tau and MAP6 (also known as STOP for Stable Tubule Only Polypeptide) [44, 57]. MAP1 family proteins, which bind along the microtubule lattice, are classical MAPs [44]. MAP1A and MAP1B are found in neurons, where they are regarded as dominant proteins in the formation and development of axons and dendrites [44]. Binding to microtubules and actin filaments, MAP1B is detectable in axons, dendrites, and growth cones throughout the central nervous system and throughout development [58]. A specific developmental brain defect characterized by agenesis of the corpus callosum, the aberrant formation of Probst bundles, and the abnormal growth of fibers in thick bundles in the lower layers of the cortex, occurs in MAP1B-deficient mice [58]. In addition, MAP1B deficiency also affects the peripheral nervous system; the axons of sciatic nerve in MAP1B-deficient mice have smaller diameters and thinner myelin sheaths of the remaining axons [58]. The MAP2/tau family includes MAP2, MAP4, Tau, and other isoforms in other animals. Members of this family not only stabilize microtubules and regulate microtubule networks in axons and dendrites, but also interact with numerous proteins. For example, by binding to microtubules, they regulate microtubule stability by inhibiting depolymerization and increasing the microtubule density and rigidity [59]. For signal transduction and integration, MAP2 binds to F-actin to modulate neurite initiation, interacts with tyrosine kinase Src, adapter protein Grb 2, and tyrosine kinase Fyn, and interacts with the neurofilaments of the cross-bridges between microtubules and neurofilaments. Furthermore, tau binds to Fyn, Src, presenilin 1, and calmodulin, each interaction having a different function [59].
Expression and Possible Mechanism of Action of Microtubule-Associated Protein in ASD
A strictly-controlled process of cytoskeletal structuring is involved in the complicated and plastic morphology of neurons. Hence, there are tight relationships between deficiencies in cytoskeletal protein-encoding genes and many neurodevelopmental disorders. Many autism-linked brain abnormalities including variations in mini-columnar and laminar cortical organization, synaptic abnormalities, and faulty links in neuronal circuits, are linked with microtubule-related gene mutations and changes. MAPs are important in regulating cytoskeletal organization and dendritic arborization, and thus maintaining normal neuronal function and network formation [45].
The FMR1 knock-out mouse, a model of ASD, shows numerous neuroanatomical changes including dysregulated dendritic spine morphology, abnormal axonal growth, and altered synaptic development [60, 61]. In another ASD model, the valproic acid rat model, numerous genes are differentially expressed [62]. These differentially-expressed genes are involved in nervous system development including neurogenesis, neuron maturation and differentiation, synapse formation, and maturation [62, 63]. Unfortunately, studies of MAPs in these rodent models are still lacking. In schizophrenia, structural alterations, including synaptic pruning defects and spine and dendrite atrophy, have been detected in the cortex. And changes in brain structure have been associated with changes in plasticity including decreased pruning and defects in spinogenesis [64]. Tangled lamination, delayed cortical development, and altered dendrite development, are correlated with Down syndrome. In terms of dendrite development, the abnormalities can be represented as reduced number of spines, changed morphology, and deficient cortical layering [45, 65]. There is sufficient evidence to suggest that many neurodevelopment disorders are characterized by cytoskeletal abnormalities [38, 39, 45, 64, 65]. As a result, MAPs are important in these disorders. Systematic investigation of the MAPs involved in the mediation of autism-like behaviors is as yet lacking [66].
Here, several MAPs associated with some neurological disorders are described. By interacting with MAP1B, KIRREL3 (a synaptic molecule of the immunoglobulin superfamily) is involved in ASD pathogenesis. MAP1B plays a vital role in controlling neuronal morphogenesis, and its defect results in altered actin microfilament polymerization and changed activity of GTPases [67]. Autism susceptibility candidate 2, a gene associated with many neurodevelopmental disorders including ASD, interacts with several MAPs and influences neuronal migration [68]. By phosphorylating tau protein and related MAPs, microtubule affinity-regulating kinases adjust microtubule dynamics in neurons and thus participate in the pathology of ASD [45, 69]. Low MAP1B and MAP2 immunoreactivity has been found in the brains of individuals with schizophrenia, and MAP6-null mice display several features associated with schizophrenia such as cognitive deficits, anxiety, and hyperactivity [70, 71]. The increased expression of DYRK1A (dual-specificity tyrosine-phosphorylation-regulated kinase 1A), a gene associated with the neuropathological features of Down syndrome, has been detected in individuals with this syndrome, and its overexpression leads to decreased dendritic spine formation in cultured hippocampal neurons. Also, the dysregulation of cytoskeletal proteins such as tubulin, actin and MAPs are associated with these anatomical abnormalities [72, 73].
STOP/MAP-6 are calmodulin-regulated proteins responsible for the high degree of stabilization shown by neuronal microtubules and the establishment of neuronal architecture and synaptic plasticity [41]. Changes in microtubule dynamics and stability have an impact on brain functions, and such changes are related to the physiopathology of neurodegenerative and psychiatric diseases, such as cognition, memory, attention, and executive function [74]. Wei et al. [41, 75] found that STOP/MAP-6 is significantly reduced in the plasma of autistic patients and in the cerebral cortex of BTBR mice compared with controls. STOP/MAP6-null mice exhibit several synaptic abnormalities and behavioral changes, including disorganized activity, social interaction, and maternal behavior, and the deletion of STOP/MAP-6 in mice results in changes in mood and cognitive performance [66, 75, 76].
Overall, STOP/MAP-6 is reckoned to be involved in ASD, and a possible mechanism of action of STOP/MAP-6 protein in the pathogenesis of ASD has been proposed. STOP/MAP-6 has different isoforms: N- and E-isoforms expressed by neurons, and A- and O-isoforms expressed by astrocytes and oligodendrocytes [77]. Oligodendrocytes are involved in the maintenance of the myelin sheets wrapping neurons, astrocytes maintain homeostasis in the CNS, and neuronal STOP/MAP-6 isoforms participate in the maintenance of microtubule stability [71, 77]. Sufficient evidence suggests that STOP/MAP-6 protects microtubules against depolymerization when exposed to cold or depolymerizing drugs, and helps microtubules to remain stable. Deletion of STOP/MAP-6 in rats disrupts the synaptic plasticity and neurotransmission associated with severe behavioral disorders [57, 71]. These different isoforms of STOP/MAP-6 have various microtubule-stabilizing actions. All of these activities are accomplished by binding two sets of microtubule-stabilizing motifs, the Mn and Mc modules, and these activities are regulated by Ca2+-calmodulin [77, 78]. Therefore, one possible hypothesis is that STOP/MAP-6 damages myelin development in oligodendrocytes, impairs microtubule dynamics and stability, and induces a chain of abnormalities in synaptic function and myelination [75]. Wei et al. [35] have reported a lower level of evoked glutamate release in the cerebral cortex of BTBR mice than in B6 mice. Similarly, it has been reported that STOP knockout mice have a lower density of synaptic vesicles and less glutamate is released into the synaptic cleft [79]. As a result, a glutamatergic hypothesis has been proposed [35, 37, 79]
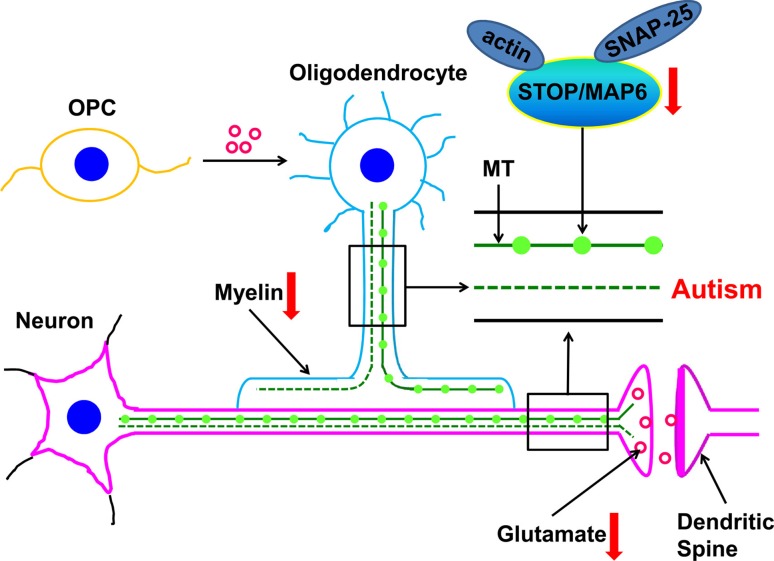
Above all, on the basis of the evidence from the literature and results in animal and human studies, the mechanism of STOP/MAP-6 can be proposed. On the one hand, reduced STOP/MAP6 protein impacts the vesicle density and glutamate release in excitatory synapses. On the other hand, it may affect the myelin development in oligodendrocytes, and the consequent abnormalities in synaptic function and myelination could mediate the behavioral phenotypes of ASD. The mechanisms by which STOP/MAP6 is involved in ASD are illustrated in Fig. 2 [41, 75]. On the basis of the literature, we can conclude that decreased STOP expression in the brain is involved in the mediation of autism-like behaviors by impairing myelination in oligodendrocytes and synaptic function in neurons.
Fig. 2.
Possible pathological mechanisms by which STOP/MAP6 protein is involved in ASD. MT, microtubule; OPC, oligodendrocyte precursor cell; SNAP-25, synaptosomal-associated protein 25; STOP/MAP6, stable tubule only polypeptide/microtubule-associated protein 6. Image adapted from Wei et al. Psychiatry Res 2016 [75].
Acknowledgements
This review was supported by National Natural Science Foundation of China (81671364 and 81201061), a China Postdoctoral Science Foundation Funded Project (2017M611195), and the Outstanding Youth Talents Program of Shanxi Province, China (2015009).
Compliance with Ethical Standards
Conflicts of interest
The authors declare that there are no conflicts of interest.
Contributor Information
Hongen Wei, Email: hongen.wei@sxmu.edu.cn.
Fengyun Hu, Email: fengyun71@163.com.
References
- 1.Davidovitch M, Hemo B, Manning-Courtney P, Fombonne E. Prevalence and incidence of autism spectrum disorder in an Israeli population. J Autism Dev Disord. 2013;43:785–793. doi: 10.1007/s10803-012-1611-z. [DOI] [PubMed] [Google Scholar]
- 2.Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years–Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahams BS, Geschwind DH. Connecting genes to brain in the autism spectrum disorders. Arch Neurol. 2010;67:395–399. doi: 10.1001/archneurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jorde LB, Hasstedt SJ, Ritvo ER, Mason-Brothers A, Freeman BJ, Pingree C, et al. Complex segregation analysis of autism. Am J Hum Genet. 1991;49:932–938. [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop DV, Maybery M, Maley A, Wong D, Hill W, Hallmayer J. Using self-report to identify the broad phenotype in parents of children with autistic spectrum disorders: a study using the Autism-Spectrum Quotient. J Child Psychol Psychiatry. 2004;45:1431–1436. doi: 10.1111/j.1469-7610.2004.00325.x. [DOI] [PubMed] [Google Scholar]
- 6.Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vorstman JAS, Parr JR, Moreno-De-Luca D, Anney RJL, Nurnberger JI, Jr, Hallmayer JF. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet. 2017;18:362–376. doi: 10.1038/nrg.2017.4. [DOI] [PubMed] [Google Scholar]
- 9.Ziats MN, Grosvenor LP, Rennert OM. Functional genomics of human brain development and implications for autism spectrum disorders. Transl Psychiatry. 2015;5:e665. doi: 10.1038/tp.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varadinova M, Boyadjieva N. Epigenetic mechanisms: A possible link between autism spectrum disorders and fetal alcohol spectrum disorders. Pharmacol Res. 2015;102:71–80. doi: 10.1016/j.phrs.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Lin YC, Frei JA, Kilander MB, Shen W, Blatt GJ. A subset of autism-associated genes regulate the structural stability of neurons. Front Cell Neurosci. 2016;10:263. doi: 10.3389/fncel.2016.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomalski P, Johnson MH. The effects of early adversity on the adult and developing brain. Curr Opin Psychiatry. 2010;23:233–238. doi: 10.1097/YCO.0b013e3283387a8c. [DOI] [PubMed] [Google Scholar]
- 13.Kubota T, Miyake K, Hirasawa T. Epigenetic understanding of gene-environment interactions in psychiatric disorders: a new concept of clinical genetics. Clin Epigenetics. 2012;4:1. doi: 10.1186/1868-7083-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Homs A, Codina-Sola M, Rodriguez-Santiago B, Villanueva CM, Monk D, Cusco I, et al. Genetic and epigenetic methylation defects and implication of the ERMN gene in autism spectrum disorders. Transl Psychiatry. 2016;6:e855. doi: 10.1038/tp.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–632. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaSalle JM, Powell WT, Yasui DH. Epigenetic layers and players underlying neurodevelopment. Trends Neurosci. 2013;36:460–470. doi: 10.1016/j.tins.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135:1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 18.Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W, Poschmann J, Cruz-Herrera Del Rosario R, Parikshak NN, Hajan HS, Kumar V, et al. Histone acetylome-wide association study of autism spectrum disorder. Cell. 2016;167(1385–1397):e1311. doi: 10.1016/j.cell.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 20.Mazina V, Gerdts J, Trinh S, Ankenman K, Ward T, Dennis MY, et al. Epigenetics of autism-related impairment: copy number variation and maternal infection. J Dev Behav Pediatr. 2015;36:61–67. doi: 10.1097/DBP.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee BK, Magnusson C, Gardner RM, Blomstrom A, Newschaffer CJ, Burstyn I, et al. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain Behav Immun. 2015;44:100–105. doi: 10.1016/j.bbi.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007;27:10695–10702. doi: 10.1523/JNEUROSCI.2178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei H, Alberts I, Li X. Brain IL-6 and autism. Neuroscience. 2013;252:320–325. doi: 10.1016/j.neuroscience.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 24.Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 25.Chez MG, Dowling T, Patel PB, Khanna P, Kominsky M. Elevation of tumor necrosis factor-alpha in cerebrospinal fluid of autistic children. Pediatr Neurol. 2007;36:361–365. doi: 10.1016/j.pediatrneurol.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Wei H, Chadman KK, McCloskey DP, Sheikh AM, Malik M, Brown WT, et al. Brain IL-6 elevation causes neuronal circuitry imbalances and mediates autism-like behaviors. Biochim Biophys Acta. 2012;1822:831–842. doi: 10.1016/j.bbadis.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Wei H, Mori S, Hua K, Li X. Alteration of brain volume in IL-6 overexpressing mice related to autism. Int J Dev Neurosci. 2012;30:554–559. doi: 10.1016/j.ijdevneu.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Wei H, Zou H, Sheikh AM, Malik M, Dobkin C, Brown WT, et al. IL-6 is increased in the cerebellum of autistic brain and alters neural cell adhesion, migration and synaptic formation. J Neuroinflammation. 2011;8:52. doi: 10.1186/1742-2094-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei H, Ma Y, Liu J, Ding C, Jin G, Wang Y, et al. Inhibition of IL-6 trans-signaling in the brain increases sociability in the BTBR mouse model of autism. Biochim Biophys Acta. 2016;1862:1918–1925. doi: 10.1016/j.bbadis.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Dev Disord. 2014;44:1117–1127. doi: 10.1007/s10803-013-1973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams BL, Hornig M, Buie T, Bauman ML, Cho Paik M, Wick I, et al. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6:e24585. doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coury DL, Ashwood P, Fasano A, Fuchs G, Geraghty M, Kaul A, et al. Gastrointestinal conditions in children with autism spectrum disorder: developing a research agenda. Pediatrics. 2012;130(Suppl 2):S160–S168. doi: 10.1542/peds.2012-0900N. [DOI] [PubMed] [Google Scholar]
- 34.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 35.Wei H, Ma Y, Ding C, Jin G, Liu J, Chang Q, et al. Reduced glutamate release in adult btbr mouse model of autism spectrum disorder. Neurochem Res. 2016;41:3129–3137. doi: 10.1007/s11064-016-2035-5. [DOI] [PubMed] [Google Scholar]
- 36.Tebartz van Elst L, Maier S, Fangmeier T, Endres D, Mueller GT, Nickel K, et al. Disturbed cingulate glutamate metabolism in adults with high-functioning autism spectrum disorder: evidence in support of the excitatory/inhibitory imbalance hypothesis. Mol Psychiatry. 2014;19:1314–1325. doi: 10.1038/mp.2014.62. [DOI] [PubMed] [Google Scholar]
- 37.Wei H, Ding C, Jin G, Yin H, Liu J, Hu F. Abnormal glutamate release in aged BTBR mouse model of autism. Int J Clin Exp Pathol. 2015;8:10689–10697. [PMC free article] [PubMed] [Google Scholar]
- 38.Zikopoulos B, Barbas H. Changes in prefrontal axons may disrupt the network in autism. J Neurosci. 2010;30:14595–14609. doi: 10.1523/JNEUROSCI.2257-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu H, Bi W, Liu C, Zhao Y, Zhang D, Yue W. A hypothesis-driven pathway analysis reveals myelin-related pathways that contribute to the risk of schizophrenia and bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2014;51:140–145. doi: 10.1016/j.pnpbp.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Haroutunian V, Katsel P, Roussos P, Davis KL, Altshuler LL, Bartzokis G. Myelination, oligodendrocytes, and serious mental illness. Glia. 2014;62:1856–1877. doi: 10.1002/glia.22716. [DOI] [PubMed] [Google Scholar]
- 41.Wei H, Ma Y, Liu J, Ding C, Hu F, Yu L. Proteomic analysis of cortical brain tissue from the BTBR mouse model of autism: Evidence for changes in STOP and myelin-related proteins. Neuroscience. 2016;312:26–34. doi: 10.1016/j.neuroscience.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 42.Jones-Davis DM, Yang M, Rider E, Osbun NC, da Gente GJ, Li J, et al. Quantitative trait loci for interhemispheric commissure development and social behaviors in the BTBR T(+) tf/J mouse model of autism. PLoS One. 2013;8:e61829. doi: 10.1371/journal.pone.0061829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolff JJ, Gerig G, Lewis JD, Soda T, Styner MA, Vachet C, et al. Altered corpus callosum morphology associated with autism over the first 2 years of life. Brain. 2015;138:2046–2058. doi: 10.1093/brain/awv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halpain S, Dehmelt L. The MAP1 family of microtubule-associated proteins. Genome Biol. 2006;7:224. doi: 10.1186/gb-2006-7-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonini SA, Mastinu A, Ferrari-Toninelli G, Memo M. Potential role of microtubule stabilizing agents in neurodevelopmental disorders. Int J Mol Sci 2017, 18. pii: E1627. [DOI] [PMC free article] [PubMed]
- 46.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Venoux M, Delmouly K, Milhavet O, Vidal-Eychenie S, Giorgi D, Rouquier S. Gene organization, evolution and expression of the microtubule-associated protein ASAP (MAP9) BMC Genomics. 2008;9:406. doi: 10.1186/1471-2164-9-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barnes AP, Polleux F. Establishment of axon-dendrite polarity in developing neurons. Annu Rev Neurosci. 2009;32:347–381. doi: 10.1146/annurev.neuro.31.060407.125536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonini SA, Ferrari-Toninelli G, Montinaro M, Memo M. Notch signalling in adult neurons: a potential target for microtubule stabilization. Ther Adv Neurol Disord. 2013;6:375–385. doi: 10.1177/1756285613490051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Q, Liu J, Fang A, Li R, Bai Y, Kriegstein AR, et al. The dynamics of neuronal migration. Adv Exp Med Biol. 2014;800:25–36. doi: 10.1007/978-94-007-7687-6_2. [DOI] [PubMed] [Google Scholar]
- 51.Munji RN, Choe Y, Li G, Siegenthaler JA, Pleasure SJ. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J Neurosci. 2011;31:1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutierrez H, Davies AM. Regulation of neural process growth, elaboration and structural plasticity by NF-κB. Trends Neurosci. 2011;34:316–325. doi: 10.1016/j.tins.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Gu X, Ma Y, Calicchio ML, Kong D, Teng YD, et al. Nna1 mediates Purkinje cell dendritic development via lysyl oxidase propeptide and NF-kappaB signaling. Neuron. 2010;68:45–60. doi: 10.1016/j.neuron.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baas PW, Ahmad FJ. Beyond taxol: microtubule-based treatment of disease and injury of the nervous system. Brain. 2013;136:2937–2951. doi: 10.1093/brain/awt153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu G, Dwyer T. Microtubule dynamics in axon guidance. Neurosci Bull. 2014;30:569–583. doi: 10.1007/s12264-014-1444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowne-Anderson H, Hibbel A, Howard J. Regulation of Microtubule Growth and Catastrophe: Unifying Theory and Experiment. Trends Cell Biol. 2015;25:769–779. doi: 10.1016/j.tcb.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deloulme JC, Gory-Faure S, Mauconduit F, Chauvet S, Jonckheere J, Boulan B, et al. Microtubule-associated protein 6 mediates neuronal connectivity through Semaphorin 3E-dependent signalling for axonal growth. Nat Commun. 2015;6:7246. doi: 10.1038/ncomms8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meixner A, Haverkamp S, Wassle H, Fuhrer S, Thalhammer J, Kropf N, et al. MAP1B is required for axon guidance and Is involved in the development of the central and peripheral nervous system. J Cell Biol. 2000;151:1169–1178. doi: 10.1083/jcb.151.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biol. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu B, Zhang Y, Zhan S, Wang X, Zhang H, Meng X, et al. Proteomic Profiling of Brain and Testis Reveals the Diverse Changes in Ribosomal Proteins in fmr1 Knockout Mice. Neuroscience. 2018;371:469–483. doi: 10.1016/j.neuroscience.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 61.Klemmer P, Meredith RM, Holmgren CD, Klychnikov OI, Stahl-Zeng J, Loos M, et al. Proteomics, ultrastructure, and physiology of hippocampal synapses in a fragile X syndrome mouse model reveal presynaptic phenotype. J Biol Chem. 2011;286:25495–25504. doi: 10.1074/jbc.M110.210260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang R, Zhou J, Ren J, Sun S, Di Y, Wang H, et al. Transcriptional and splicing dysregulation in the prefrontal cortex in valproic acid rat model of autism. Reprod Toxicol. 2018;77:53–61. doi: 10.1016/j.reprotox.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 63.Barrett CE, Hennessey TM, Gordon KM, Ryan SJ, McNair ML, Ressler KJ, et al. Developmental disruption of amygdala transcriptome and socioemotional behavior in rats exposed to valproic acid prenatally. Mol Autism. 2017;8:42. doi: 10.1186/s13229-017-0160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson JS, Nielsen JA, Ferguson MA, Burback MC, Cox ET, Dai L, et al. Abnormal brain synchrony in Down Syndrome. Neuroimage Clin. 2013;2:703–715. doi: 10.1016/j.nicl.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Broek JA, Guest PC, Rahmoune H, Bahn S. Proteomic analysis of post mortem brain tissue from autism patients: evidence for opposite changes in prefrontal cortex and cerebellum in synaptic connectivity-related proteins. Mol Autism. 2014;5:41. doi: 10.1186/2040-2392-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guerin A, Stavropoulos DJ, Diab Y, Chenier S, Christensen H, Kahr WH, et al. Interstitial deletion of 11q-implicating the KIRREL3 gene in the neurocognitive delay associated with Jacobsen syndrome. Am J Med Genet A. 2012;158A:2551–2556. doi: 10.1002/ajmg.a.35621. [DOI] [PubMed] [Google Scholar]
- 68.Sultana R, Yu CE, Yu J, Munson J, Chen D, Hua W, et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 69.Bernard LP, Zhang H. MARK/Par1 Kinase Is Activated Downstream of NMDA Receptors through a PKA-Dependent Mechanism. PLoS One. 2015;10:e0124816. doi: 10.1371/journal.pone.0124816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Javitt DC, Sweet RA. Auditory dysfunction in schizophrenia: integrating clinical and basic features. Nat Rev Neurosci. 2015;16:535–550. doi: 10.1038/nrn4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lefevre J, Savarin P, Gans P, Hamon L, Clement MJ, David MO, et al. Structural basis for the association of MAP6 protein with microtubules and its regulation by calmodulin. J Biol Chem. 2013;288:24910–24922. doi: 10.1074/jbc.M113.457267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dowjat K, Adayev T, Kaczmarski W, Wegiel J, Hwang YW. Gene dosage-dependent association of DYRK1A with the cytoskeleton in the brain and lymphocytes of down syndrome patients. J Neuropathol Exp Neurol. 2012;71:1100–1112. doi: 10.1097/NEN.0b013e31827733c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tejedor FJ, Hammerle B. MNB/DYRK1A as a multiple regulator of neuronal development. FEBS J. 2011;278:223–235. doi: 10.1111/j.1742-4658.2010.07954.x. [DOI] [PubMed] [Google Scholar]
- 74.Volle J, Brocard J, Saoud M, Gory-Faure S, Brunelin J, Andrieux A, et al. Reduced expression of STOP/MAP6 in mice leads to cognitive deficits. Schizophr Bull. 2013;39:969–978. doi: 10.1093/schbul/sbs113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei H, Sun S, Li Y, Yu S. Reduced plasma levels of microtubule-associated STOP/MAP6 protein in autistic patients. Psychiatry Res. 2016;245:116–118. doi: 10.1016/j.psychres.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 76.Eastwood SL, Lyon L, George L, Andrieux A, Job D, Harrison PJ. Altered expression of synaptic protein mRNAs in STOP (MAP6) mutant mice. J Psychopharmacol. 2007;21:635–644. doi: 10.1177/0269881106068825. [DOI] [PubMed] [Google Scholar]
- 77.Galiano MR, Bosc C, Schweitzer A, Andrieux A, Job D, Hallak ME. Astrocytes and oligodendrocytes express different STOP protein isoforms. J Neurosci Res. 2004;78:329–337. doi: 10.1002/jnr.20260. [DOI] [PubMed] [Google Scholar]
- 78.Bosc C, Frank R, Denarier E, Ronjat M, Schweitzer A, Wehland J, et al. Identification of novel bifunctional calmodulin-binding and microtubule-stabilizing motifs in STOP proteins. J Biol Chem. 2001;276:30904–30913. doi: 10.1074/jbc.M011614200. [DOI] [PubMed] [Google Scholar]
- 79.Kajitani K, Thorne M, Samson M, Robertson GS. Nitric oxide synthase mediates the ability of darbepoetin alpha to improve the cognitive performance of STOP null mice. Neuropsychopharmacology. 2010;35:1718–1728. doi: 10.1038/npp.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]