High-risk hematological malignancies are a privileged setting for infection by opportunistic microbes, with invasive mycosis being one of the most serious complications. Recently, genetic background has emerged as an unanticipated risk factor.
KEYWORDS: fungal infection, PTX3, SNP, Dectin-1, oncohematology
ABSTRACT
High-risk hematological malignancies are a privileged setting for infection by opportunistic microbes, with invasive mycosis being one of the most serious complications. Recently, genetic background has emerged as an unanticipated risk factor. For this reason, polymorphisms for genes encoding archetypal receptors involved in the opsonic and nonopsonic clearance of microbes, pentraxin-3 (PTX3) and Dectin-1, respectively, were studied and correlated with the risk of infection. Fungal, bacterial, and viral infections were registered for a group of 198 patients with high-risk hematological malignancies. Polymorphisms for the pentraxin-3 gene (PTX3) showed a significant association with the risk of fungal infection by Candida spp. and, especially, by Aspergillus spp. This link remained even for patients undergoing antifungal prophylaxis, thus demonstrating the clinical relevance of PTX3 in the defense against fungi. CLEC7A polymorphisms did not show any definite correlation with the risk of invasive mycosis, nor did they influence the expression of Dectin-1 isoforms generated by alternative splicing. The PTX3 mRNA expression level was significantly lower in samples from healthy volunteers who showed these polymorphisms, although no differences were observed in the extents of induction elicited by bacterial lipopolysaccharide and heat-killed Candida albicans, thus suggesting that the expression of PTX3 at the start of infection may influence the clinical outcome. PTX3 mRNA expression can be a good biomarker to establish proper antifungal prophylaxis in immunodepressed patients.
INTRODUCTION
Management of opportunistic infections in patients with hematological malignancies is a difficult task. Immunosuppression due to the progress of the disease and the risks associated with chemotherapy and hematopoietic stem cell transplantation (HSCT) pave the way for microbial invasion. Despite the use of antifungal therapy, infection by Candida spp. and Aspergillus spp. remains an important cause of mortality (1, 2). In addition, several studies have confirmed the existence of risk factors associated with genetic polymorphisms in the immune system (3–7). In this regard, polymorphisms in genes coding for components of innate immunity, such as the pattern recognition receptors (PRRs) Dectin-1 and long pentraxin 3 (PTX3), have been identified as risk factors in oncohematology patients (3, 8–11).
Dectin-1 is a C-type lectin transmembrane receptor encoded by the CLEC7A gene that, through alternative splicing, generates various receptor isoforms, with the A and B isoforms being the only functional ones (12). Both isoforms recognize the most abundant fungal cell wall polysaccharides, i.e., the β(1→3)- and/or β(1→6)-linked glucans, trigger nonopsonic phagocytosis, and initiate a proinflammatory response (13). Among the several CLEC7A polymorphisms studied in oncohematology patients, rs16910526 has been associated with risks of both candidiasis and aspergillosis. This single nucleotide polymorphism (SNP) entails a loss of function of the protein by generating an early stop codon leading to the loss of amino acids in the C-terminal domain, which affects β-glucan recognition (14). The presence of β-glucans on some bacteria entails their recognition by Dectin-1. Nonetheless, mycobacteria, which do not express β-glucans, may also be recognized (15, 16). In addition, rs2078178 and rs16910631 polymorphisms have been associated with ulcerative colitis (17) and Asperger disease (18), i.e., diseases associated with immune dysfunction in the gastrointestinal tract.
PTX3 is a soluble receptor encoded as a single transcript by the PTX3 gene, which participates in opsonic phagocytosis by interacting with both complement components and Fcγ receptors (FcγRs). Both in vitro and in vivo studies have shown the direct binding of PTX3 to the surface of fungi, bacteria, and viruses (19, 20). Genetic variants of PTX3 in oncohematology patients have been studied previously (10, 11), and PTX3 SNPs rs2305619, rs3816527, and rs1840680 have been associated with an increased risk of invasive mycosis (particularly aspergillosis). However, this association was not observed in either non-HSCT patients (10) or oncohematology patients with preexistent neutropenia (11). These polymorphisms have also shown a strong association with the risks of tuberculosis and colonization by Pseudomonas spp. (21, 22), which highlights an overall role of PTX3 in the innate immune response.
Given that both opsonic and nonopsonic phagocytoses are involved in host defense, we have selected model receptors for each mechanism of phagocytosis to disclose the role of their polymorphisms in the risk of infection in a series of patients with hematological malignancies. We selected polymorphisms rs16910526, rs2078178, and rs16910631 for CLEC7A and rs3816527, rs2305619, and rs1840680 for PTX3. Genotyping studies showed a marked association between PTX3 SNPs and the risk of fungal infections. Irrespective of the PTX3 genotype, the extent of PTX3 mRNA expression correlated with susceptibility to infection, thus suggesting that a high level of PTX3 expression is a key factor for an efficient innate immune response to fungal infection.
RESULTS
Patients.
A retrospective study was carried out with 198 oncohematology patients, whose clinical characteristics are shown in Table 1. The most frequent clinical diagnoses were acute myeloblastic leukemia (33.8%), lymphoma (31.3%), and acute lymphoblastic leukemia (10.6%). Approximately 41% of patients underwent HSCT: 58% underwent autologous and 42% underwent allogeneic HSCT. Table 2 shows the percentages of patients affected by different microorganisms (78.8%): bacteria were most frequently detected (66.7%), followed by fungal infections (46.5%) and, to a lesser extent, viral infections (17.7%). Diagnosis of fungal infection was carried out according to revised EORTC/MSG criteria (39). Candida species infections were the most common (38.4% of the total number of patients), 14.5% of which were considered invasive because of positive cultures in samples from at least two different tissues. The remainder showed positive cultures for Candida in mucosae, urine, and stool, which were considered evidence of colonization. Aspergillus species infections were detected in 9.6% of the patients, 63.2% of which were considered invasive according to EORTC/MSG criteria. Yeasts such as Saccharomyces cerevisiae, Geotrichum candidum, and Rhodotorula mucilaginosa were identified in a few patients.
TABLE 1.
Demographic and clinical characteristics
| Parametera | Value |
|---|---|
| Demographic variables | |
| No. of male patients/no. of female patients (% of male patients/% of female patients) | 117/81 (59.1/40.9) |
| Median age (yr) (range) | 62 (4–92) |
| No. (%) of patients with hematological malignancy | |
| AML | 67 (33.8) |
| Lymphoma | 62 (31.3) |
| ALL | 21 (10.6) |
| MM | 20 (10.1) |
| MDS | 17 (8.6) |
| CLL | 8 (4) |
| Other | 3 (1.5) |
| No. (%) of patients with HSCT | 81 (40.9) |
| Allogenic | 34 (42) |
| Autologous | 47 (58) |
Abbreviations: AML, acute myeloblastic leukemia; ALL, acute lymphoblastic leukemia; MM, multiple myeloma; MDS, myelodysplastic syndrome; CLL, chronic lymphocytic leukemia; HSCT, hematopoietic stem cell transplantation.
TABLE 2.
Percentages of patients affected by pathogens, the recognition of which has been associated with Dectin-1 and PTX3
| Pathogen | No (%) of patients affecteda |
|---|---|
| Bacteria | 132 (66.7) |
| Genus Pseudomonas | 29 (14.6) |
| Klebsiella pneumoniae | 15 (7.6) |
| Genus Staphylococcus | 61 (30.8) |
| Staphylococcus aureus | 16 (8.1) |
| Mycobacterium tuberculosis | 2 (1) |
| Fungi | 92 (46.5) |
| Genus Candida | 76 (38.4) |
| Colonization | 65 (32.8; 85.5*) |
| Invasive disease | 11 (5.6; 14.5*) |
| Genus Aspergillus | 19 (9.6) |
| Colonization | 7 (3.54; 36.8*) |
| Invasive disease | 12 (6.1; 63.2*) |
| Other yeasts | 10 (5.1) |
| Viruses | 35 (17.7) |
| Cytomegalovirus | 12 (6.1) |
Percentages were calculated based on the total number of patients, except for the values marked with an asterisk, which refer to the number of patients infected with the respective pathogen.
SNPs and risk of infection.
The association between the different SNPs and the risk of both colonization and infection was adjusted for sex, age, and hematological malignancy. After confirming that all SNPs were in Hardy-Weinberg equilibrium (HWE), at a P value of >0.05, within each group studied, none of the studied SNPs displayed any association with the risk of either bacterial or viral infections. In contrast, there was a significant association between PTX3 SNP rs3816527 and the presence of Candida and Aspergillus spp. (Table 3). The calculation of odds ratio (OR) values, according to genotype distribution, revealed that the C allele of rs3816527 (on the basis of the best model fit [recessive model]) behaved as a risk factor (ORCC+CA = 2.29 [95% confidence interval {CI}, 1.20 to 4.36] [P value of 0.01]). However, when the incidences of fungal colonization and invasive disease (36.34% and 11.1%, respectively) were evaluated separately, the analysis exhibited a significant association with invasive mycosis only. As shown in Table 4, the AA genotype for intronic SNPs (rs2305619 and rs1840680) displayed a significant risk (ORrs2305619AA = 3.28 [95% CI, 1.24 to 8.69] [P value of 0.02]; ORrs1840680AA = 3.98 [95% CI, 1.52 to 10.4] [P value of 0.0058]). Although the analysis was not significant for rs3816527, the CC genotype exhibited a certain degree of association (ORrs3816527CC = 2.55 [95% CI, 0.9 to 7.16] [P value of 0.086]) (Table 4). Moreover, the risk of invasive aspergillosis showed a robust association for three PTX3 SNPs (ORrs2305619AA = 4.2 [95% CI, 1.22 to 14.57] [P value of 0.026]; ORrs3816527CC = 5.1 [95% CI, 1.45 to 17.97] [P value of 0.013]; ORrs1840680AA = 4.15 [95% CI, 1.21 to 14.3] [P value of 0.027]) (Table 5). PTX3 also exhibited a significant correlation with the risk of developing invasive candidiasis but only for SNP rs1840680 (OR = 3.8 [95% CI, 1.05 to 13.57] [P value of 0.048]) (Table 6). Antifungal prophylaxis (AP) is crucial for high-risk patients and is currently mandatory under these conditions (23). Nonetheless, despite AP with fluconazole, posaconazole, or micafungin, 11.43% of patients experienced invasive mycosis, whereas colonization was detected in 37.86% of patients (Table 7). A new analysis adjusted for the above-mentioned covariates plus AP showed similar results. New association analyses were performed to evaluate the risks of fungal colonization and systemic infection in patients who received AP. PTX3 SNPs displayed no association with the risk of fungal colonization. However, it is worth noting that there was a significant association of systemic aspergillosis for rs3816527 and a clear trend for intronic PTX3 SNPs (ORrs3816527CC = 5.64 [95% CI, 1.18 to 26.88] [P value of 0.034]) (Table 8).
TABLE 3.
Genotype distribution of PTX3 SNPs in patients with and without detection of Candida spp. and/or Aspergillus spp. and association thereofa
| SNP | Model | Genotype | No. (%) of patients with Candida or Aspergillus species detection |
No. (%) of patients without Candida or Aspergillus species detection |
OR (95% CI) | P value | BIC |
|---|---|---|---|---|---|---|---|
| rs2305619 | Codominant | AA | 23 (25.8) | 18 (16.5) | 1.00 | ||
| AG | 46 (51.7) | 58 (53.2) | 1.65 (0.77–3.52) | 0.18 | 318.6 | ||
| GG | 20 (22.5) | 33 (30.3) | 2.22 (0.94–5.24) | ||||
| Dominant | AA | 23 (25.8) | 18 (16.5) | 1.00 | |||
| AG+GG | 66 (74.2) | 91 (83.5) | 1.83 (0.89–3.76) | 0.099 | 314.1 | ||
| Recessive | AA+AG | 69 (77.5) | 76 (69.7) | 1.00 | |||
| GG | 20 (22.5) | 33 (30.3) | 1.55 (0.8–3.01) | 0.19 | 315 | ||
| rs3816527 | Codominant | CC | 19 (21.4) | 15 (13.8) | 1.00 | ||
| CA | 49 (55.1) | 52 (47.7) | 1.37 (0.6–3.1) | 0.069 | 316.7 | ||
| AA | 21 (23.6) | 42 (38.5) | 2.56 (1.05–6.2) | ||||
| Dominant | CC | 19 (21.4) | 15 (13.8) | 1.00 | |||
| CA+AA | 70 (78.7) | 94 (86.2) | 1.74 (0.8–3.78) | 0.16 | 314.8 | ||
| Recessive | CC+CA | 68 (76.4) | 67 (61.5) | 1.00 | |||
| AA | 21 (23.6) | 42 (38.5) | 2.02 (1.07–3.84) | 0.029 | 312 | ||
| rs1840680 | Codominant | AA | 24 (27) | 18 (16.5) | 1.00 | ||
| AG | 45 (50.6) | 55 (50.5) | 1.68 (0.79–3.57) | 0.092 | 317.3 | ||
| GG | 20 (22.5) | 36 (33) | 2.54 (1.09–5.94) | ||||
| Dominant | AA | 24 (27) | 18 (16.5) | 1.00 | |||
| AG+GG | 65 (73) | 91 (83.5) | 1.94 (0.95–3.98) | 0.066 | 313.4 | ||
| Recessive | AA+AG | 69 (77.5) | 73 (67) | 1.00 | |||
| GG | 20 (22.5) | 36 (33) | 1.76 (0.92–1.76) | 0.086 | 313.8 | ||
Model adjusted for sex, age, and hematological malignancy. For 198 patients, the SNPStats program established the most frequent genotype as the default reference and different inheritance models. The best-fit model is indicated in boldface type, as are the significant associations at a P value of <0.05. Abbreviations: OR, odds ratio; CI, confidence interval; BIC, Bayesian information criterion.
TABLE 4.
Genetic association of PTX3 SNPs with invasive fungal infectiona
| SNP | Model | Genotype | No. (%) of patients with IFI | No. (%) of patients without IFI | OR (95% CI) | P value | BIC |
|---|---|---|---|---|---|---|---|
| rs2305619 | Codominant | AA | 9 (40.9) | 32 (18.2) | 1.00 | ||
| AG | 9 (40.9) | 95 (54) | 2.97 (1.04–8.4) | 0.059 | 184.5 | ||
| GG | 4 (18.2) | 49 (27.8) | 4.04 (1.1–14.96) | ||||
| Dominant | AA | 9 (40.9) | 32 (18.2) | 1.00 | |||
| AG+GG | 13 (59.1) | 144 (81.8) | 3.28 (1.24–8.69) | 0.02 | 179.5 | ||
| Recessive | AA+AG | 18 (81.8) | 127 (72.2) | 1.00 | |||
| GG | 4 (18.2) | 49 (27.8) | 2.03 (0.63–6.50) | 0.21 | 183.3 | ||
| rs3816527 | Codominant | CC | 7 (31.8) | 27 (15.3) | 1.00 | ||
| CA | 10 (45.5) | 91 (51.7) | 2.17 (0.72–6.55) | 0.17 | 186.7 | ||
| AA | 5 (22.7) | 58 (33) | 3.35 (0.9–12.16) | ||||
| Dominant | CC | 7 (31.8) | 27 (15.3) | 1.00 | |||
| CA+AA | 15 (68.2) | 149 (84.7) | 2.55 (0.9–7.16) | 0.086 | 182 | ||
| Recessive | CC+CA | 17 (77.3) | 118 (67) | 1.00 | |||
| AA | 5 (22.7) | 58 (33) | 1.97 (0.67–5.8) | 0.2 | 183.3 | ||
| rs1840680 | Codominant | AA | 10 (45.5) | 32 (18.2) | 1.00 | ||
| AG | 8 (36.4) | 92 (52.3) | 3.58 (1.26–10.2) | 0.02 | 182.4 | ||
| GG | 4 (18.2) | 52 (29.6) | 4.87 (1.33–17.8) | ||||
| Dominant | AA | 10 (45.5) | 32 (18.2) | 1.00 | |||
| AG+GG | 12 (54.5) | 144 (81.8) | 3.98 (1.52–10.4) | 0.0058 | 177.3 | ||
| Recessive | AA+AG | 18 (81.8) | 124 (70.5) | 1.00 | |||
| GG | 4 (18.2) | 52 (29.6) | 2.24 (0.7–7.2) | 0.15 | 182.8 | ||
Association analysis for 198 patients was adjusted for sex, age, and hematological malignancy. The SNPStats program established the dominant model as the best-fit model, which is indicated in boldface type, as are its significant associations at a P value of <0.05. IFI, invasive fungal infection.
TABLE 5.
Genotype association of PTX3 SNPs with the risk of developing invasive aspergillosisa
| SNP | Model | Genotype | No. (%) of patients with IA | No. (%) of patients without IA | OR (95% CI) | P value | BIC |
|---|---|---|---|---|---|---|---|
| rs2305619 | Codominant | AA | 6 (50) | 35 (18.8) | 1.00 | ||
| AG | 3 (25) | 101 (54.3) | 5.3 (1.21–23.26) | 0.069 | 134 | ||
| GG | 3 (25) | 50 (26.9) | 3.1 (0.68–14.14) | ||||
| Dominant | AA | 6 (50) | 35 (18.8) | 1.00 | |||
| AG+GG | 6 (50) | 151 (81.2) | 4.2 (1.22–14.57) | 0.026 | 129.1 | ||
| Recessive | AA+AG | 9 (75) | 136 (73.1) | 1.00 | |||
| GG | 3 (25) | 50 (26.9) | 1.27 (0.32–5.13) | 0.73 | 133.9 | ||
| rs3816527 | Codominant | CC | 6 (50) | 28 (15.1) | 1.00 | ||
| CA | 3 (25) | 98 (52.7) | 5.88 (1.3–26.27) | 0.044 | 133.1 | ||
| AA | 3 (25) | 60 (32.3) | 4.3 (0.94–19.78) | ||||
| Dominant | CC | 6 (50) | 28 (15.1) | 1.00 | |||
| CA+AA | 6 (50) | 158 (85) | 5.1 (1.45–17.97) | 0.013 | 127.9 | ||
| Recessive | CC+CA | 9 (75) | 126 (67.7) | 1.00 | |||
| AA | 3 (25) | 60 (32.3) | 1.63 (0.41–6.52) | 0.48 | 133.5 | ||
| rs1840680 | Codominant | AA | 6 (50) | 36 (19.4) | 1.00 | ||
| AG | 3 (25) | 97 (52.1) | 5.06 (1.15–22.2) | 0.076 | 134.2 | ||
| GG | 3 (25) | 53 (28.5) | 3.2 (0.7–14.62) | ||||
| Dominant | AA | 6 (50) | 36 (19.4) | 1.00 | |||
| AG+GG | 6 (50) | 150 (80.7) | 4.15 (1.21–14.3) | 0.027 | 129.2 | ||
| Recessive | AA+AG | 9 (75) | 133 (71.5) | 1.00 | |||
| GG | 3 (25) | 53 (28.5) | 1.36 (0.34–5.51) | 0.66 | 133.9 | ||
Model adjusted for sex, age, and hematological malignancy for the 198 high-risk hematology patients. The dominant model was established as the best-fit model and is indicated in boldface type, as are its significant associations at a P value of <0.05. IA, invasive aspergillosis.
TABLE 6.
Influence of PTX3 SNPs on the risk of invasive candidiasis in high-risk oncohematology patientsa
| SNP | Model | Genotype | No. (%) of patients with candidiasis |
No. (%) of patients without candidiasis |
OR (95% CI) | P value | BIC |
|---|---|---|---|---|---|---|---|
| rs2305619 | Codominant | AA | 4 (36.4) | 37 (19.8) | 1.00 | ||
| AG | 6 (54.5) | 98 (52.4) | 1.94 (0.5–7.58) | 0.18 | 137 | ||
| GG | 1 (9.1) | 52 (27.8) | 6.66 (0.69–64.0) | ||||
| Dominant | AA | 4 (36.4) | 37 (19.8) | 1.00 | |||
| AG+GG | 7 (63.6) | 150 (80.2) | 2.62 (0.7–9.8) | 0.17 | 133.3 | ||
| Recessive | AA+AG | 10 (90.9) | 135 (72.2) | 1.00 | |||
| GG | 1 (9.1) | 52 (27.8) | 4.26 (0.5–34.66) | 0.11 | 132.6 | ||
| rs3816527 | Codominant | CC | 2 (18.2) | 32 (17.1) | 1.00 | ||
| CA | 7 (63.6) | 94 (50.3) | 0.87 (0.16–4.65) | 0.45 | 138.9 | ||
| AA | 2 (18.2) | 61 (32.6) | 2.3 (0.3–17.85) | ||||
| Dominant | CC | 2 (18.2) | 32 (17.1) | 1.00 | |||
| CA+AA | 9 (81.8) | 155 (82.9) | 1.19 (0.24–6.04) | 0.83 | 135.1 | ||
| Recessive | CC+CA | 9 (81.8) | 126 (67.4) | 1.00 | |||
| AA | 2 (18.2) | 61 (32.6) | 2.55 (0.52–12.5) | 0.21 | 133.6 | ||
| rs1840680 | Codominant | AA | 5 (45.5) | 37 (19.8) | 1.00 | ||
| AG | 5 (45.5) | 95 (50.8) | 2.75 (0.73–10.4) | 0.07 | 135.1 | ||
| GG | 1 (9.1) | 55 (29.4) | 9.12 (0.99–84.3) | ||||
| Dominant | AA | 5 (45.5) | 37 (19.8) | 1.00 | |||
| AG+GG | 6 (54.5) | 150 (80.2) | 3.8 (1.05–13.57) | 0.048 | 131.3 | ||
| Recessive | AA+AG | 10 (90.9) | 132 (70.6) | 1.00 | |||
| GG | 1 (9.1) | 55 (29.4) | 4.8 (0.59–39.22) | 0.076 | 132 | ||
Association analysis for the risk of invasive candidiasis in 198 patients. The study, adjusted for sex, age, and hematological malignancy, showed a significant association for SNP rs1840680. The significant associations at a P value of <0.05 are indicated in boldface type.
TABLE 7.
Influence of antifungal prophylaxis on the incidence of fungal colonization and systemic infectiona
| Parameter | No. of patients | % of patients |
|---|---|---|
| With antifungal prophylaxis (fluconazole, posaconazole, and/or micafungin) | 140 | 70.7 |
| Fungal colonization | 53 | 37.86 |
| Genus Candida | 47 | 88.7 |
| Genus Aspergillus | 7 | 13.2 |
| Invasive fungal infection | 16 | 11.43 |
| Genus Candida | 9 | 56.25 |
| Genus Aspergillus | 8 | 50 |
| Without antifungal prophylaxis | 58 | 29.3 |
| Fungal colonization | 18 | 31.03 |
| Genus Candida | 18 | 100 |
| Genus Aspergillus | 0 | 0 |
| Invasive fungal infection | 6 | 10.34 |
| Genus Candida | 2 | 33.33 |
| Genus Aspergillus | 4 | 66.66 |
The numbers of patients with and without antifungal prophylaxis and the respective percentages are represented, as are the incidences of acquired fungal colonization and invasive infection.
TABLE 8.
Genetic susceptibility of PTX3 SNPs to invasive aspergillosis in patients with antifungal prophylaxisa
| SNP | Model | Genotype | No. (%) of patients with IA | No. (%) of patients without IA | OR (95% CI) | P value | BIC |
|---|---|---|---|---|---|---|---|
| rs2305619 | Codominant | AA | 4 (50) | 28 (21.2) | 1.00 | ||
| AG | 3 (37.5) | 67 (50.8) | 3.25 (0.65–16.4) | 0.19 | 101.1 | ||
| GG | 1 (12.5) | 37 (28) | 6.13 (0.6–62.33) | ||||
| Dominant | AA | 4 (50) | 28 (21.2) | 1.00 | |||
| AG+GG | 4 (50) | 104 (78.8) | 3.94 (0.87–17.8) | 0.08 | 96.4 | ||
| Recessive | AA+AG | 7 (87.5) | 95 (72) | 1.00 | |||
| GG | 1 (12.5) | 37 (28) | 3.1 (0.35–27.36) | 0.26 | 98.2 | ||
| rs3816527 | Codominant | CC | 4 (50) | 22 (16.7) | 1.00 | ||
| CA | 3 (37.5) | 65 (49.2) | 4.35 (0.81–23.3) | 0.083 | 99.5 | ||
| AA | 1 (12.5) | 45 (34.1) | 9.55 (0.91–99.6) | ||||
| Dominant | CC | 4 (50) | 22 (16.7) | 1.00 | |||
| CA+AA | 4 (50) | 110 (83.3) | 5.64 (1.18–26.9) | 0.034 | 95 | ||
| Recessive | CC+CA | 7 (87.5) | 87 (65.9) | 1.00 | |||
| AA | 1 (12.5) | 45 (34.1) | 3.97 (0.44–35.6) | 0.16 | 97.5 | ||
| rs1840680 | Codominant | AA | 4 (50) | 29 (22) | 1.00 | ||
| AG | 3 (37.5) | 63 (47.7) | 3.14 (0.62–15.8) | 0.18 | 101 | ||
| GG | 1 (12.5) | 40 (30.3) | 6.5 (0.64–67.25) | ||||
| Dominant | AA | 4 (50) | 29 (22) | 1.00 | |||
| AG+GG | 4 (50) | 103 (78) | 3.94 (0.87–17.8) | 0.08 | 96.4 | ||
| Recessive | AA+AG | 7 (87.5) | 92 (69.7) | 1.00 | |||
| GG | 1 (12.5) | 40 (30.3) | 3.4 (0.38–30.63) | 0.22 | 98 | ||
For 140 patients undergoing antifungal prophylaxis, the analysis model was adjusted for sex, age, and hematological malignancy. The SNPStats program recognized the most frequent genotype as the default reference genotype and established the corresponding inheritance models. The best-fit model is indicated in boldface type, as are the significant associations at a P value of <0.05. IA, invasive aspergillosis.
Regarding CLEC7A SNPs, a significant association with the different risk groups could not be observed. However, rs16910526 exhibited a certain degree of protective association in those patients presenting with Candida spp. and/or Aspergillus spp. (ORrs16910526TT = 2.28 [95% CI, 0.95 to 5.51] [P value of 0.058]) (see Table S1 in the supplemental material), particularly in those with fungal colonization (Table S2). Moreover, when the analysis was restricted to patients who received AP, the G allele for rs16910526 showed a significant protective effect versus the risk of fungal colonization (Table S3), particularly by Candida (Table S4). Although the relevance of this finding may be tempered by the low overall frequency of this polymorphism in patients with AP (GG, 1%; GT, 13%), it is noteworthy that neither of the two patients homozygous for rs16910526 had colonization. However, the only carrier of the GG genotype, who did not receive AP, experienced fungal colonization by Candida spp. (Table S5). Within the group of patients with AP, rs2078178 also showed a significant association with the risk of fungal colonization, although it did not exhibit any trend in other analysis groups (Table S6). In the case of rs16910631, the TT genotype was not observed, which explains why this polymorphism was not included for further analysis.
Association of PTX3 SNP haplotypes with mycosis risk.
After confirming the strong association of PTX3 SNPs with the risk of invasive aspergillosis, their haplotype distribution was assessed, discarding possible confusion covariates. As shown in Table 9, the SNPStats program established that haplotype A-C-A (SNP positions rs2305619-rs3816527-rs1840680) was significantly associated with the risk of invasive aspergillosis, where 62.5% of carriers of this haplotype developed infection, versus 41.1% of patients who did not develop it (Table 9). Even in patients who received AP, this haplotype showed a strong trend toward association (Table 9).
TABLE 9.
Risk assessment of invasive aspergillosis for the haplotypes corresponding to rs2305619, rs3816527, and rs1840680a
| Haplotype at rs2305619-rs3816527-rs1840680 | Association risk for main haplotypes formed by PTX3 SNPs (frequency) |
|||||
|---|---|---|---|---|---|---|
| 198 patients |
140 patients with APc |
|||||
| Total | IAb | No IAb | Total | IA | No IA | |
| A-C-A | 0.424 | 0.625 | 0.411 | 0.429 | 0.687 | 0.413 |
| G-A-G | 0.523 | 0.375 | 0.532 | 0.514 | 0.312 | 0.526 |
Haplotype analyses were performed both on the overall population of 198 patients and on the 140 patients with AP. The assessment was adjusted for sex, age, hematological malignancy, and AP using the SNPStats program. Data represent the distributions of the two most frequent haplotypes and the association of the A-C-A haplotype with distinct infection groups. Associations at a P value of <0.05 are significant.
OR, 2.95 (95% CI, 1.1 to 7.91) (P value of 0.032).
OR, 3.12 (95% CI, 0.98 to 9.99) (P value of 0.057).
Meta-analysis for PTX3 SNPs.
In order to confirm the risk genotype, a meta-analysis was performed with the inclusion criteria described in Materials and Methods. Data from studies by Cunha et al. (10) and Brunel et al. (11) were selected for combined analysis with our data. As shown in Fig. S1 in the supplemental material, only the AA genotype for rs3816527 and the GG genotype for intronic SNPs were checked as risk factors for invasive aspergillosis. Meta-analysis displayed these genotypes as risk factors (ORrs2305619GG = 2.37 [95% CI, 1.23 to 4.57] [P value of 0.01]; ORrs3816527AA = 2.08 [95% CI, 1.01 to 4.28] [P value of 0.049]; ORrs1840680GG = 2.30 [95% CI, 1.14 to 4.61] [P value of 0.019]). In contrast, our data reflected a protector effect for these genotypes but with great uncertainty (ORrs2305619GG = 0.91 [95% CI, 0.24 to 3.48]; ORrs3816527AA = 0.70 [95% CI, 0.18 to 2.68]; ORrs1840680GG = 0.84 [95% CI, 0.22 to 3.21]). Although meta-analysis reflected a significant publication bias for SNP rs3816527 (Egger’s P value of 0.0438), there was no clear evidence of publication bias for the other SNPs (Egger’s P value of >0.05) (Fig. S2). Nevertheless, these genotypes did not show any significant effect in our study, as previously shown. On the other hand, our risk genotypes for this invasive mycosis could not be assessed by meta-analysis due to the lack of information in the studies by Cunha et al. and Brunel et al. regarding the number of individuals for the remainder of the genotypes.
Prediction of SNP pathogenicity.
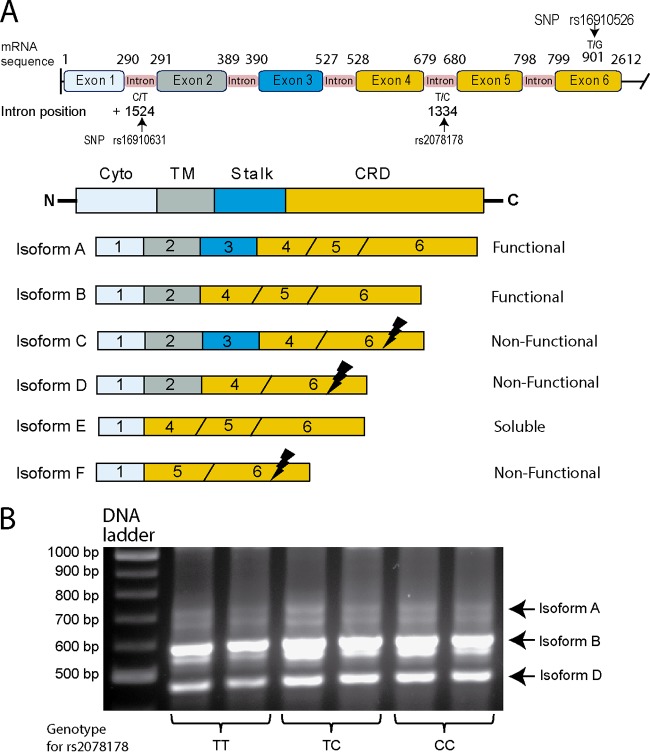
Given the significant association between SNP rs2078178 in CLEC7A and the risk of fungal colonization in the patients who received AP, the possible influence of this SNP on the alternative splicing of CLEC7A was assessed (Fig. 1A). In silico analysis did not show any impact on splicing by using the Human Splicing Finder program. Nevertheless, we sought the presence of the different transcripts of Dectin-1 in monocytes from healthy volunteer donors according to their genotypes. As shown in Fig. 1B, the two functional isoforms and the nonfunctional D isoform were expressed in monocytes independently of their genotype.
FIG 1.
Genomic structure of Dectin-1, isoforms resulting from alternative splicing, and influence of the rs2078178 genotype. (A) The structures of CLEC7A and the different isoforms resulting from alternative splicing are shown together with the positions of the polymorphisms. The black lightning bolts indicate the positions of the stop codons introduced through frameshifts generated by alternative splicing. The diagram has been constructed based on the sequence under GenBank accession number NG_016291.1 and data reported previously by Willment et al. (12). (B) Monocytes from buffy coats from healthy donors of the different rs2078178 genotypes were isolated and cultured for mRNA collection. The different isoforms of Dectin-1 were amplified by PCR and separated by electrophoresis on an agarose gel. Abbreviations: Cyto, cytoplasmic tail; TM, transmembrane region; CRD, carbohydrate recognition domain.
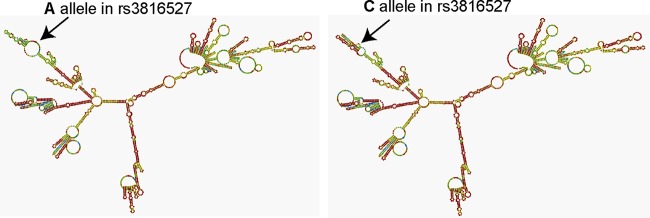
Regarding PTX3 SNPs, the change of nucleotide C for A in exonic SNP rs3816527 is responsible for the substitution of aspartic acid for alanine in the 48th position, the consequences of which were checked by the SIFT program. This program determined that both amino acids are tolerated at the 48th position (see Table S7 in the supplemental material). However, the RNAfold program showed that the exchange of both nucleotides could affect mRNA folding (Fig. 2). Unexpectedly, the folding pattern observed was somewhat different from the results reported by Cunha et al. (see Fig. S6 in reference 10). On this basis, the potential splice-site effects of SNP rs3816527 showed that allele C might create a new cryptic splicing site, which could compete against the canonical splicing site (Table 10). Consequently, the C allele would modify exon 2 expression and generate a smaller transcript (Fig. 3A). According to the primers used for amplifying PTX3 mRNA in healthy donors, a normal transcript of 170 bp must be present in samples with the A allele, a 147-bp transcript must be present in samples with the C allele, and both transcripts must be present in the case of the CA genotype. However, independently of their genotype, all samples showed a band of 170 bp and some bands with higher mobility, even after stimulation (Fig. 3B). In the case of intronic PTX3 SNPs (rs2305619 and rs1840680), the Human Splicing Finder program did not detect any type of impact on splicing (data not shown).
FIG 2.
Impact of SNP rs3816527 on the secondary structure of PTX3 mRNA. The effect of the exchange of nucleotides (A allele or C allele) is shown by the position of the arrows.
TABLE 10.
Possible effects of SNP rs3816527 on alternative splicing of PTX3 pre-mRNAa
| 3′ reference motif | Reference score | Mutated motif | Mutation score | Variation (%) |
|---|---|---|---|---|
| gtgtgtatcccgtactctagCCA | 4.47 | gtgtgtatcccgtactctagCCA | 4.47 | +0 |
| CGCCGTGCGACTGCGGTCAGGAG | 1.69 | cgccgtgcgcctgcggtcagGAG | 4.1 | +342.6 |
The Human Splicing Finder system allowed the identification and prediction of a new possible cryptic site on PTX3 pre-mRNA with a high score variation. The C/A change (boldface) generates a cryptic site (italics). As a result, 23 nucleotides of exon 2 (uppercase) become part of the intron sequence (lowercase).
FIG 3.
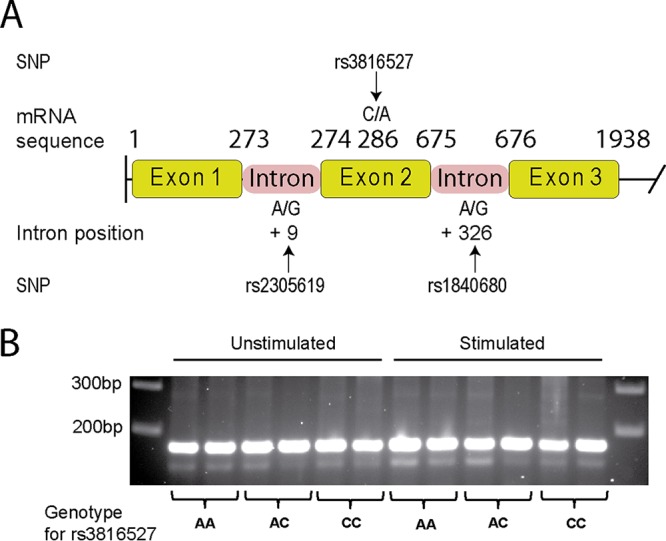
Structure of the PTX3 gene, positions of the polymorphisms, and expression of PTX3 transcripts of the different PTX3rs3816527 genotypes. (A) Structure of the PTX3 gene and positions of the polymorphisms. The diagram has been constructed based on the sequence reported under GenBank accession number NG_051000.1. (B) Samples of buffy coats were used for rs3816527 genotyping and processed to obtain mRNA from monocytes cultured in the presence and absence of heat-killed C. albicans. PTX3 transcripts from each sample were generated by RT-PCR and separated by electrophoresis on an agarose gel.
PTX3 expression in monocytes.
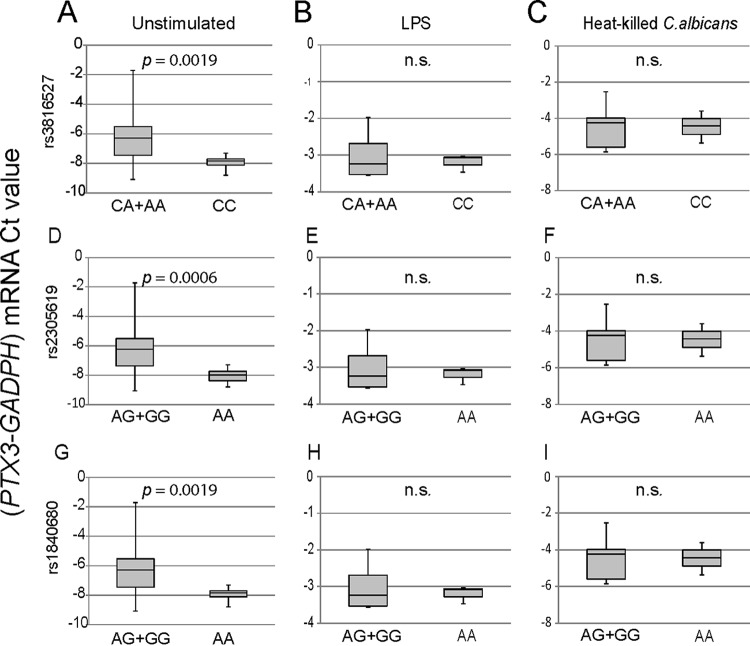
To address possible functional consequences associated with the different polymorphisms, the expression of PTX3 mRNA was assayed in monocytes under resting conditions and after stimulation with lipopolysaccharide (LPS) and heat-killed Candida conidia. As shown in Fig. 4A, the basal PTX3 mRNA expression level was significantly lower in carriers of the rs3816527 CC genotype than in the CA+AA group. In the case of both intronic SNPs, the PTX3 mRNA expression level was significantly lower in the carriers of AA genotype than in the AG+GG groups (Fig. 4A). In contrast, although PTX3 mRNA expression was increased by the stimuli, no differences in expression were observed between genotypic groups of each SNP (Fig. 4B and C).
FIG 4.
Impact of PTX3 SNPs on PTX3 mRNA expression. After genotyping and processing of buffy coats, monocytes were cultured in the presence and absence of a stimulus (LPS or heat-killed C. albicans) for 6 h. PTX3 transcripts were amplified by quantitative reverse transcription-PCR in duplicate, and the expression level from each sample was calculated as the difference between the mean CT values of the targeted gene (PTX3) and the mean CT values of the housekeeping gene (GAPDH) (y axis, PTX3-GAPDH mRNA CT value). For each SNP, two comparisons of genetic groups were performed: CC and CA+AA for rs3816527 and AA and AG+GG for both intronic SNPs (rs2305619 and rs1840680). The groups passed the Shapiro-Wilks normality test. PTX3 mRNA expression values in monocytes for the indicated genotypes in the absence (A) and presence (B and C) of stimuli are presented in box plot graphs. Results represent data from at least three independent experiments. P values determined by Welch’s t test are shown. n.s., not significant.
DISCUSSION
Natural immunity is the first line of defense against infection. However, individual susceptibility to specific microbes is variable and difficult to assess. This makes the identification of host biomarkers to predict the risk of infection by opportunistic microbes an imperative issue. On this basis, the assessment of risk factors associated with polymorphisms in receptors involved in the early recognition of microbes by the innate immune system is of clinical relevance (24).
rs16910526 is the Dectin-1 polymorphism for which loss of function is best explained (14); however, it did not exhibit a correlation with invasive mycosis (Candida spp. and Aspergillus spp.). Other studies have shown that rs16910526 has a limited influence on susceptibility to invasive aspergillosis, although it might be important in susceptible patients not submitted to HSCT (25, 26). In the present study, a significant protective effect of rs16910526 on fungal colonization was observed in patients receiving AP. Although the relevance of this finding may be tempered by the low frequency of this polymorphism, it is worth noting that G allele carriers displayed a lower rate of fungal colonization (7.5% with fungal colonization versus 18.4% without fungal colonization) and that the rate of colonization was lower in patients with AP than in those who did not receive AP (7.5% and 16.7%, respectively) (see Table S5 in the supplemental material). An interpretation of this finding may be that the benefit obtained from AP can be associated with the prevention of colonization, particularly by Candida. However, the redundancy of mechanisms involved in pathogen recognition and the possibility that it may be part of a larger haplotype involving other genetic variants in neighboring genes should be taken into account. In fact, a study using monocytes showed that Dectin-1-defective human macrophages produce proinflammatory cytokines in response to Aspergillus (25). This ability to retain the capacity to respond to infection in the absence of functional Dectin-1 highlights the relevance of alternative routes and PRRs in antifungal defense (13, 27, 28). Both the low frequency and the presence of alternative mechanisms of defense could also explain the lack of an association of the rs16910631 and rs2078178 Dectin-1 SNPs with fungal infection as well as the surprising correlation between rs2078178 and fungal colonization in patients with AP. These polymorphisms have been associated with ulcerative colitis and Asperger disease. As both pathologies are characterized by the presence of dysbiosis (29, 30), the involvement of these SNPs in pathogenesis would agree with the inability of Clec7a−/− mice to lead an effective immune response against specific intestinal fungi that might contribute to dysbiosis. However, evidence disclosing whether these polymorphisms influence CLEC7A expression has not been reported, and the present study did not show any difference in the expression of CLEC7A isoforms that could be associated with these genotypes in human monocytes.
PTX3 SNPs have been associated with both fungal and bacterial infections, including those caused by Pseudomonas aeruginosa and Mycobacterium tuberculosis (21, 22), due to the ability of PTX3 to opsonize microbial surfaces. Regarding fungal infections, invasive mycoses represent a great risk for morbidity and mortality of patients with high-risk hematological malignancies. Although the number of patients with invasive mycosis was relatively low (11.1%), a significant association with PTX3 polymorphisms was observed, particularly with systemic aspergillosis. Strikingly, this association was maintained in those patients who had received antifungal prophylaxis. Regarding the risk of developing candidiasis, only polymorphism rs1840680 displayed a significant correlation. This highlights the role of PTX3 in antifungal defense through the opsonization of Candida spp. either directly (31) or in combination with mannose-binding lectin (MBL) and complement components (32). In keeping with data from previous reports (10, 11, 21, 22, 33, 34), these results highlight the importance of these polymorphisms in protection against both fungal and bacterial infections, suggesting that PTX3 expression is a significant risk biomarker. In accordance with our risk alleles, the C allele for rs3816527 and the A allele for both rs2305619 and rs1840680 were associated with pulmonary tuberculosis and Pseudomonas species colonization (21, 22). Conversely, risk alleles for invasive mycosis and, particularly, for aspergillosis were the A allele for rs3816527 and the G allele for intronic SNPs, both in solid-organ transplant recipients (33, 34) and in nonneutropenic hematology patients (10, 11). Similar results were observed with risk haplotype A-C-A (rs2305619-rs3816527-rs1840680) in the present study, which agrees with the risk haplotypes obtained in the analyses by Olesen et al. and Chiarini et al. (21, 22) but not with those in other studies of fungal infection (10, 11, 33, 34). Although combined analysis of data from our studies and those from the studies by Cunha et al. (10) and Brunel et al. (11) allowed the establishment of the AA genotype for rs3816527 and the GG genotype for intronic SNPs as risk genotypes, the lack of data did not allow verification of our risk genotype. In addition, despite considering data from the studies by Cunha et al. and Brunel et al. as best fitting with our inclusion criteria, it has to be taken into account that in both studies, the risk association was established in patients without neutropenia, while in the present study, all patients had severe neutropenia, a condition that restricts phagocytic defense to mononuclear cells. These cells exhibit opsonic phagocytosis via CR3 and CR4 integrins and FcγRs, but they also express C-type lectin receptors for nonopsonic phagocytosis. In contrast, the unopsonized killing of Candida by human polymorphonuclear leukocytes depends not on Dectin-1 but on CR3 (35). The distinct array of receptors involved in fungal recognition in polymorphonuclear leukocytes and mononuclear phagocytes may explain this discrepancy. Thus, as reflected by Egger’s P value and the trim-and-fill method, further studies of the association between these SNPs and hematology patients, distinguishing those with and those without neutropenia, are needed.
In order to resolve these divergences, the molecular consequences of these SNPs were assessed. In silico studies showed that the exchange of amino acids induced by missense variant rs3816527 did not influence either protein folding or structural stability, in keeping with the report by Cunha et al. showing that the ability to bind fungi was guaranteed with both amino acids (10). However, the RNAfold program showed that mRNA stability could be affected by this exchange of nucleotides, and the Human Splicing Finder tool showed that the presence of the C allele for rs3816527 could create a new cryptic splicing site, which would encompass the loss of 23 bp in exon 2. However, no differences in the sizes of transcripts were found. In contrast, the rs3816527 CC genotype showed lower levels of PTX3 mRNA expression in resting monocytes than in the CA+AA group. Similarly, in both intronic SNPs, the PTX3 mRNA expression level was significantly lower in carriers of the AA genotype than in carriers of the AG or GG genotype. These results were consistent with the predicted alterations in the PTX3 mRNA structure and agreed with the observed clinical associations in our study. In contrast, Cunha et al. (10) observed that the A allele for rs3816527 and the G allele for rs2305619 showed lower levels of PTX3 mRNA in neutrophil precursors. In keeping with our data, these samples belonged to healthy individuals who showed no infections. To relate the mRNA expression data with the response to pathogen-associated molecular patterns (PAMPs), monocytes were cultured in the presence of both heat-killed C. albicans and LPS. It could be posited that the difference between genotypes would remain after stimulation, according to the different rates of infection incidence. However, no significant differences in the extents of PTX3 mRNA induction were found. An overall interpretation could be that a low expression level of PTX3 in mononuclear phagocytes at the time of initial contact with the fungal burden may be critical to cope successfully with the cargo and to control the spread of invasion, given the limited supply of polymorphonuclear leukocytes in these patients. It is also possible that PTX3 deficiency adversely affects antifungal mechanisms mediated by other innate immune receptors (36) or the mobilization of phagocytes (37). Given the various PAMPs expressed in fungi and the formation of a phagocytic synapse in mononuclear phagocytes, critical for fungal recognition (38), it seems likely that the presence of PTX3 in the initial phase of infection might be a crucial factor for a proper response. Even though no modifications in the PTX3 structure were recognized as a result of the D48A substitution in rs3816527, damage to its electrostatic potential or interactions with other proteins cannot be ruled out. The fact that the 48th position is fringed by two cysteine residues involved in the formation of PTX3 protein complex octamers supports this possibility. To establish a better correlation between PTX3 RNA expression and infection, patient samples should have been collected during an infection period. However, the clinical protocol was designed to use a small amount of peripheral blood at the time of high-risk hematological malignancy diagnosis. Despite this limitation in this retrospective study, the clinical results confirm the significant role of PTX3 in defense against fungi, particularly against Aspergillus spp., and highlight the presence, in a homozygous state, of the C allele for rs3816527 and of the A allele for rs2305619 and rs1840680 as hazard factors in high-risk hematology patients (both transplanted and nontransplanted). However, given the discrepancies with some of the previously reported results, further investigations are warranted to establish the real risk genotype in PTX3 SNPs under definite criteria for patient enrollment. This would facilitate early prophylaxis and appropriate treatment in patients with high-risk hematological malignancies. A corollary to this study is that genetic associations with a particular disease should be established with caution, given the difficulty in replicating data from clinical studies due to the involvement of populations that differ in terms of baseline characteristics and/or immunosuppressive regimens (5).
MATERIALS AND METHODS
Study populations.
This is a retrospective study of a group of 198 patients with high-risk hematological malignancies enrolled between 2013 and 2017 from both the Hospital Universitario Río-Hortega (HURH) and the Hospital Clínico Universitario of Valladolid (HCUV). Clinical diagnoses included acute leukemias, high-risk myelodysplastic syndromes, highly aggressive lymphomas, and chronic lymphoblastic leukemia. Patients who underwent HSCT were also included in the study. The inclusion criteria were intensive chemotherapy and/or high doses of corticoids and episodes of severe neutropenia (<500 cells/mm3) for at least 7 days. The observational period was defined as the interval between the date of the initial high-risk diagnosis and the end of clinical monitoring. Bacterial, viral, and fungal infections were monitored, as were the prophylactic and therapeutic strategies employed. Fungal infections were detected mainly by microscopy and culture and diagnosed based on updated criteria reported by the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group (EORTC/IFICG) (39). Clinical and laboratory data were obtained through detailed review of hospital records.
For genetic studies, the time of extraction of the samples was decided by clinical criteria at the time of establishment of a high-risk diagnosis. For transplant patients, only samples collected prior to HSCT were studied. For complementary functional studies, buffy coat samples from healthy volunteer donors were provided by the Centro de Hemoterapia y Hemodonación de Castilla y León. All determinations and genetic analyses in hematological patients as well as the use of buffy coat samples were performed after obtaining written informed consent in accordance with the Declaration of Helsinki, and anonymity of the data was guaranteed. The study protocol for patients was approved by the ethical review committees of both hospitals.
Genotyping.
Genomic DNA was extracted from 400-μl samples of peripheral blood or buffy coats from healthy donors, using MagNA Pure compact nucleic acid isolation kit I (Roche, Mannheim, Germany). Genotyping of CLEC7A and PTX3 polymorphisms studied here was performed by using the Kompetitive allele-specific PCR (KASP) assay. Primers, designed by using Primer-BLAST software, are shown in Table S8 in the supplemental material. Samples of 4 ng genomic DNA were amplified by using a Roche LightCycler 480 instrument (Roche Austria, Vienna, Austria). PCR fluorescent endpoint readings were performed by using the LightCycler 480 real-time PCR system (Roche, Germany).
Isolation of human monocytes.
Human monocytes were isolated from buffy coats of healthy donors by two different density gradient centrifugations: a first centrifugation using a Ficoll-Paque solution (GE Healthcare Bio-Sciences, Uppsala, Sweden) to obtain mononuclear cells and a second centrifugation based on a discontinuous density gradient constituted by an OptiPrep solution (Axis-Shield PoC AC, Oslo, Sweden), a Ficoll-Paque solution, and a mixture composed of HEPES-buffered saline containing OptiPrep, bovine serum albumin (BSA), and 1 mM EDTA, in order to separate lymphocytes from monocytes. Monocytes were collected and adhered to 100-mm plastic dishes for 2 h at 37°C to discard nonadherent cells and guarantee a high purity of monocytes. Monocytes were cultured overnight at a density of 1 × 107 cells/plate in RPMI 1640 medium (Lonza, Vervier, Belgium) supplemented with 2 mM l-glutamine, 100 U/ml penicillin-streptomycin, and 5% heat-inactivated human serum. To address the changes in the expression of PTX3 upon stimulation by pathogen-associated molecular patterns (PAMPs), adhered monocytes were stimulated for 6 h with 10 μg/ml lipopolysaccharide (LPS) and heat-killed Candida albicans at a concentration of 5 conidia per cell. At the end of this period, RNA was isolated and used for the assay of PTX3 mRNA expression by reverse transcription and real-time PCR (RT-PCR).
Pathogenicity prediction for PTX3 SNPs.
The influence of the D48A substitution associated with the exonic SNP rs3816527 on PTX3 folding and structural stability was studied with the SIFT program (http://sift.jcvi.org/). The secondary structure of single-stranded RNA was predicted by using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi), while the potential splice-site effects of SNPs were predicted by using the Human Splicing Finder tool (http://www.umd.be/HSF3/HSF.shtml).
Analysis of mRNA expression.
Total RNA from healthy control monocytes was obtained by using TRIzol-chloroform extraction according to the manufacturer’s instructions and then used for reverse transcription. RT-PCR was performed in duplicate by using a Kapa SYBR fast quantitative PCR (qPCR) kit optimized for the LightCycler 480 system (Kapa Biosystems, Boston, MA, USA) on a Roche LightCycler 480 instrument. Amplification efficiencies were validated and normalized against the value for the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. Relative expression levels of both PTX3 and CLEC7A mRNAs were calculated as the difference between the mean threshold cycle (CT) values of targeted genes and the mean CT values of the housekeeping gene (GAPDH). The primer sequences, designed by using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/), are listed in Table S9 in the supplemental material. In addition, PCR products were separated by electrophoresis on agarose gels stained with gel red nucleic acid gel stain.
Statistical analysis.
Descriptive analysis of patients was performed by using IBM SPSS Statistics 20 (IBM, Chicago, IL, USA). The clinical variables were assessed, and their absolute values, frequencies, and percentages are presented. The microorganisms so far associated with the response orchestrated by Dectin-1 or PTX3 were considered appropriate groups to assess the possible influence of SNPs. These microorganisms included cytomegalovirus, the genus Pseudomonas, the genus Staphylococcus, Klebsiella pneumoniae, Mycobacterium tuberculosis, the genus Candida, and the genus Aspergillus. Genotypic analysis of the different SNPs was carried out to assess HWE, allele and genotype distributions and association tests. This was performed by using the SNPStats program (http://bioinfo.iconcologia.net/SNPstats). This Web application establishes the most frequent genotype as the default reference and generates odds ratios (OR), 95% confidence intervals (CI), and P values for the three main inheritance models: codominant, dominant, and recessive. Model fit was evaluated based on the Bayesian information criterion (BIC), with a lower BIC value indicating a better fit. All analyses were adjusted for sex, age, hematological malignancy, and antifungal prophylaxis in order to discard these possible confusion covariates. Finally, haplotype frequencies of PTX3 SNPs and their association risk with different infection groups were also tested by using this Web tool.
Systematic review with meta-analysis for PTX3 SNPs.
A literature search of published studies in the PubMed database was performed by using the keywords “PTX3,” “polymorphisms,” and “infections.” Of the 11 studies obtained, only 2 reports were selected under the following inclusion criteria: (i) study SNPs (rs2305619, rs3816527, and rs1840680), (ii) type of study (analysis of the association of PTX3 SNPs with fungal infection), and (iii) patients (patients with hematological disease). A flow chart showing the stages of database searching and study selection is provided in Fig. S3 in the supplemental material. The OR from different studies were used to assess the influence of the reported SNPs on invasive aspergillosis in hematology patients. The heterogeneity of studies was checked by using Cochran’s chi-square test (Q-test), considering a P value of <0.05 to be statistically significant heterogeneity, and the I2 statistic, where an I2 value of >75% was considered to indicate high heterogeneity. A random-effects-model meta-analysis was performed for each SNP, as recommended previously (40). We used the Der Simmonian-Laird approach to estimate the variance between studies. Finally, publication bias was assessed by using a funnel plot, with filled studies estimated from the trim-and-fill method, and the Egger regression asymmetry test. All meta-analyses were conducted with the metafor R package, version 2.0-0.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Centro de Hemoterapia y Hemodonación de Castilla y León (CHEMCYL) (Valladolid, Spain) for providing buffy coats and coworkers at the hematologic services of both the Hospital Universitario Río Hortega and the Hospital Clínico de Valladolid for their kind assistance.
M. Carmen Herrero-Sánchez is a recipient of a postdoctoral grant of the Sociedad Española de Hematología y Hemoterapia. This work was supported by the Plan Nacional de Salud y Farmacia under grants SAF2013-44521-R and SAF2017-83079-R, the Gerencia Regional de Salud de Castilla y León under grant GRS 726/A13, the Junta de Castilla y León/Fondo Social Europeo under grant CSI035P17, and the Fundación Domingo Martínez.
We declare no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00709-18.
REFERENCES
- 1.Ruiz-Camps I, Aguado JM, Almirante B, Bouza E, Ferrer-Barbera CF, Len O, Lopez-Cerero L, Rodríguez-Tudela JL, Ruiz M, Solé A, Vallejo C, Vazquez L, Zaragoza R, Cuenca-Estrella M. 2011. Guidelines for the prevention of invasive mould diseases caused by filamentous fungi by the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC). Clin Microbiol Infect 17:1–24. doi: 10.1111/j.1469-0691.2011.03477.x. [DOI] [PubMed] [Google Scholar]
- 2.Ruping MJ, Vehreschild JJ, Cornely OA. 2008. Patients at high risk of invasive fungal infections: when and how to treat. Drugs 68:1941–1962. doi: 10.2165/00003495-200868140-00002. [DOI] [PubMed] [Google Scholar]
- 3.Fisher CE, Hohl TM, Fan W, Storer BE, Levine DM, Zhao LP, Martin PJ, Warren EH, Boeckh M, Hansen JA. 2017. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood 129:2693–2701. doi: 10.1182/blood-2016-10-743294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pothlichet J, Quintana-Murci L. 2013. The genetics of innate immunity sensors and human disease. Int Rev Immunol 32:157–208. doi: 10.3109/08830185.2013.777064. [DOI] [PubMed] [Google Scholar]
- 5.Wójtowicz A, Bochud PY. 2015. Host genetics of invasive Aspergillus and Candida infections. Semin Immunopathol 37:173–186. doi: 10.1007/s00281-014-0468-y. [DOI] [PubMed] [Google Scholar]
- 6.Smeekens SP, van de Veerdonk FL, Kullberg BJ, Netea MG. 2013. Genetic susceptibility to Candida infections. EMBO Mol Med 5:805–813. doi: 10.1002/emmm.201201678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunha C, Aversa F, Romani L, Carvalho A. 2013. Human genetic susceptibility to invasive aspergillosis. PLoS Pathog 9:e1003434. doi: 10.1371/journal.ppat.1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plantinga TS, van der Velden WJ, Ferwerda B, van Spriel AB, Adema G, Feuth T, Donnelly JP, Brown GD, Kullberg BJ, Blijlevens NM, Netea MG. 2009. Early stop polymorphism in human DECTIN-1 is associated with increased candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis 49:724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 9.Cunha C, Di Ianni M, Bozza S, Giovannini G, Zagarella S, Zelante T, D’Angelo C, Pierini A, Pitzurra L, Falzetti F, Carotti A, Perruccio K, Latgé JP, Rodrigues F, Velardi A, Aversa F, Romani L, Carvalho A. 2010. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood 116:5394–5402. doi: 10.1182/blood-2010-04-279307. [DOI] [PubMed] [Google Scholar]
- 10.Cunha C, Aversa F, Lacerda JF, Busca A, Kurzai O, Grube M, Löffler J, Maertens JA, Bell AS, Inforzato A, Barbati E, Almeida B, Santos e Sousa P, Barbui A, Potenza L, Caira M, Rodrigues F, Salvatori G, Pagano L, Luppi M, Mantovani A, Velardi A, Romani L, Carvalho A. 2014. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med 370:421–432. doi: 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- 11.Brunel A-S, Wójtowicz A, Lamoth F, Spertini O, Neofytos D, Calandra T, Marchetti O, Bochud P-Y. 7 June 2018. Pentraxin-3 polymorphisms and invasive mold infections in acute leukemia patients with intensive chemotherapy. Haematologica doi: 10.3324/haematol.2018.195453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willment JA, Gordon S, Brown GD. 2001. Characterization of the human beta-glucan receptor and its alternatively spliced isoforms. J Biol Chem 276:43818–43823. doi: 10.1074/jbc.M107715200. [DOI] [PubMed] [Google Scholar]
- 13.Brown GD. 2006. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol 6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 14.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, Jacobs L, Jansen T, Verheijen K, Masthoff L, Morré SA, Vriend G, Williams DL, Perfect JR, Joosten LA, Wijmenga C, van der Meer JW, Adema GJ, Kullberg BJ, Brown GD, Netea MG. 2009. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med 361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yadav M, Schorey JS. 2006. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood 108:3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothfuchs AG, Bafica A, Feng CG, Egen JG, Williams DL, Brown GD, Sher A. 2007. Dectin-1 interaction with Mycobacterium tuberculosis leads to enhanced IL-12p40 production by splenic dendritic cells. J Immunol 179:3463–3471. doi: 10.4049/jimmunol.179.6.3463. [DOI] [PubMed] [Google Scholar]
- 17.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. 2012. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennabi M, Delorme R, Oliveira J, Fortier C, Lajnef M, Boukouaci W, Feugeas JP, Marzais F, Gaman A, Charron D, Ghaleh B, Krishnamoorthy R, Leboyer M, Tamouza R. 2015. Dectin-1 polymorphism: a genetic disease specifier in autism spectrum disorders? PLoS One 10:e0137339. doi: 10.1371/journal.pone.0137339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inforzato A, Reading PC, Barbati E, Bottazzi B, Garlanda C, Mantovani A. 2012. The “sweet” side of a long pentraxin: how glycosylation affects PTX3 functions in innate immunity and inflammation. Front Immunol 3:407. doi: 10.3389/fimmu.2012.00407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunes P, Holubcova Z, Kolackova M, Krejsek J. 2012. Pentraxin 3 (PTX 3): an endogenous modulator of the inflammatory response. Mediators Inflamm 2012:920517. doi: 10.1155/2012/920517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olesen R, Wejse C, Velez DR, Bisseye C, Sodemann M, Aaby P, Rabna P, Worwui A, Chapman H, Diatta M, Adegbola RA, Hill PC, Østergaard L, Williams SM, Sirugo G. 2007. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun 8:456–467. doi: 10.1038/sj.gene.6364410. [DOI] [PubMed] [Google Scholar]
- 22.Chiarini M, Sabelli C, Melotti P, Garlanda C, Savoldi G, Mazza C, Padoan R, Plebani A, Mantovani A, Notarangelo LD, Assael BM, Badolato R. 2010. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun 11:665–670. doi: 10.1038/gene.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vazquez L. 2016. Antifungal prophylaxis in immunocompromised patients. Mediterr J Hematol Infect Dis 8:e2016040. doi: 10.4084/mjhid.2016.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochud PY, Bochud M, Telenti A, Calandra T. 2007. Innate immunogenetics: a tool for exploring new frontiers of host defence. Lancet Infect Dis 7:531–542. doi: 10.1016/S1473-3099(07)70185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai LY, de Boer MG, van der Velden WJ, Plantinga TS, van Spriel AB, Jacobs C, Halkes CJ, Vonk AG, Blijlevens NM, van Dissel JT, Donnelly PJ, Kullberg BJ, Maertens J, Netea MG. 2011. The Y238X stop codon polymorphism in the human beta-glucan receptor dectin-1 and susceptibility to invasive aspergillosis. J Infect Dis 203:736–743. doi: 10.1093/infdis/jiq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sainz J, Lupianez CB, Segura-Catena J, Vazquez L, Rios R, Oyonarte S, Hemminki K, Försti A, Jurado M. 2012. Dectin-1 and DC-SIGN polymorphisms associated with invasive pulmonary aspergillosis infection. PLoS One 7:e32273. doi: 10.1371/journal.pone.0032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang J, Kim BM, Chang CH. 2014. Co-stimulation of TLR4 and Dectin-1 induces the production of inflammatory cytokines but not TGF-beta for Th17 cell differentiation. Immune Netw 14:30–37. doi: 10.4110/in.2014.14.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dennehy KM, Ferwerda G, Faro-Trindade I, Pyz E, Willment JA, Taylor PR, Kerrigan A, Tsoni SV, Gordon S, Meyer-Wentrup F, Adema GJ, Kullberg BJ, Schweighoffer E, Tybulewicz V, Mora-Montes HM, Gow NA, Williams DL, Netea MG, Brown GD. 2008. Syk kinase is required for collaborative cytokine production induced through Dectin-1 and Toll-like receptors. Eur J Immunol 38:500–506. doi: 10.1002/eji.200737741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagao-Kitamoto H, Kamada N. 2017. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw 17:1–12. doi: 10.4110/in.2017.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buie T, Campbell DB, Fuchs GJ, Furuta GT, Levy J, Vandewater J, Whitaker AH, Atkins D, Bauman ML, Beaudet AL, Carr EG, Gershon MD, Hyman SL, Jirapinyo P, Jyonouchi H, Kooros K, Kushak R, Levitt P, Levy SE, Lewis JD, Murray KF, Natowicz MR, Sabra A, Wershil BK, Weston SC, Zeltzer L, Winter H. 2010. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics 125(Suppl 1):S1–S18. doi: 10.1542/peds.2009-1878C. [DOI] [PubMed] [Google Scholar]
- 31.Tierney L, Linde J, Müller S, Brunke S, Molina JC, Hube B, Schöck U, Guthke R, Kuchler K. 2012. An interspecies regulatory network inferred from simultaneous RNA-seq of Candida albicans invading innate immune cells. Front Microbiol 3:85. doi: 10.3389/fmicb.2012.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma YJ, Doni A, Skjoedt MO, Honore C, Arendrup M, Mantovani A, Garred P. 2011. Heterocomplexes of mannose-binding lectin and the pentraxins PTX3 or serum amyloid P component trigger cross-activation of the complement system. J Biol Chem 286:3405–3417. doi: 10.1074/jbc.M110.190637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wójtowicz A, Lecompte TD, Bibert S, Manuel O, Rueger S, Berger C, Boggian K, Cusini A, Garzoni C, Hirsch H, Khanna N, Mueller NJ, Meylan PR, Pascual M, van Delden C, Bochud PY. 2015. PTX3 polymorphisms and invasive mold infections after solid organ transplant. Clin Infect Dis 61:619–622. doi: 10.1093/cid/civ386. [DOI] [PubMed] [Google Scholar]
- 34.Cunha C, Monteiro AA, Oliveira-Coelho A, Kühne J, Rodrigues F, Sasaki SD, Schio SM, Camargo JJ, Mantovani A, Carvalho A, Pasqualotto AC. 2015. PTX3-based genetic testing for risk of aspergillosis after lung transplant. Clin Infect Dis 61:1893–1894. doi: 10.1093/cid/civ679. [DOI] [PubMed] [Google Scholar]
- 35.Gazendam RP, van Hamme JL, Tool AT, van Houdt M, Verkuijlen PJ, Herbst M, Liese JG, van de Veerdonk FL, Roos D, van den Berg TK, Kuijpers TW. 2014. Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood 124:590–597. doi: 10.1182/blood-2014-01-551473. [DOI] [PubMed] [Google Scholar]
- 36.Bozza S, Bistoni F, Gaziano R, Pitzurra L, Zelante T, Bonifazi P, Perruccio K, Bellocchio S, Neri M, Iorio AM, Salvatori G, De Santis R, Calvitti M, Doni A, Garlanda C, Mantovani A, Romani L. 2006. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood 108:3387–3396. doi: 10.1182/blood-2006-03-009266. [DOI] [PubMed] [Google Scholar]
- 37.Deban L, Russo RC, Sironi M, Moalli F, Scanziani M, Zambelli V, Cuccovillo I, Bastone A, Gobbi M, Valentino S, Doni A, Garlanda C, Danese S, Salvatori G, Sassano M, Evangelista V, Rossi B, Zenaro E, Constantin G, Laudanna C, Bottazzi B, Mantovani A. 2010. Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat Immunol 11:328–334. doi: 10.1038/ni.1854. [DOI] [PubMed] [Google Scholar]
- 38.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, Underhill DM. 2011. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Muñoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group Consensus Group. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46:1813–1821. doi: 10.1086/588660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borenstein M, Hedges LV, Higgins JP, Rothstein HR. 2011. Introduction to meta-analysis. Wiley, Hoboken, NJ. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.