Enterococci are Gram-positive commensals of the mammalian intestinal tract and harbor intrinsic resistance to broad-spectrum cephalosporins. Disruption of colonization resistance in humans by antibiotics allows enterococci to proliferate in the gut and cause disseminated infections.
KEYWORDS: Enterococcus, Lactobacillus, antibiotic resistance, bacterial dissemination, ceftriaxone, intestinal colonization, intestinal homeostasis
ABSTRACT
Enterococci are Gram-positive commensals of the mammalian intestinal tract and harbor intrinsic resistance to broad-spectrum cephalosporins. Disruption of colonization resistance in humans by antibiotics allows enterococci to proliferate in the gut and cause disseminated infections. In this study, we used Enterococcus faecalis (EF)-colonized mice to study the dynamics of enterococci, commensal microbiota, and the host in response to systemic ceftriaxone administration. We found that the mouse model recapitulates intestinal proliferation and dissemination of enterococci seen in humans. Employing a ceftriaxone-sensitive strain of enterococci (E. faecalis JL308), we showed that increased intestinal abundance is critical for the systemic dissemination of enterococci. Investigation of the impact of ceftriaxone on the mucosal barrier defenses and integrity suggested that translocation of enterococci across the intestinal mucosa was not associated with intestinal pathology or increased permeability. Ceftriaxone-induced alteration of intestinal microbial composition was associated with transient increase in the abundance of multiple bacterial operational taxonomic units (OTUs) in addition to enterococci, for example, lactobacilli, which also disseminated to the extraintestinal organs. Collectively, these results emphasize that ceftriaxone-induced disruption of colonization resistance and alteration of mucosal homeostasis facilitate increased intestinal abundance of a limited number of commensals along with enterococci, allowing their translocation and systemic dissemination in a healthy host.
INTRODUCTION
The mammalian gastrointestinal (GI) tract is colonized by a diverse community of microorganisms that play critical roles in host physiology, including metabolic regulation and epithelial and immune development (1–5). This microbial ecosystem, the microbiota, includes commensals, symbionts, and opportunists (6). Enterococci, which inhabit the GI tracts of vertebrates and many insects, are known opportunistic pathogens (7, 8). They do not cause disease in healthy hosts but can cause life-threatening infections in critically ill and immunocompromised individuals (9–13).
Enterococci harbor intrinsic resistance to cephalosporins (14) and have acquired resistance to several other broad-spectrum antibiotics (15–18). Extensive use of antibiotics, including aminoglycosides, linezolids, and vancomycin, along with the ability of enterococci to rapidly respond and adapt to selection pressure, has resulted in the emergence of multidrug-resistant (MDR) strains (15, 19). Lack of therapeutic resources to treat nosocomial infections, especially those caused by MDR strains of enterococci, has posed concerns in the community and health care environments (20, 21).
Treatment with broad-spectrum antibiotics depletes commensal inhabitants in the GI tract and disrupts colonization resistance, an important protective attribute provided primarily by the microbiota to limit pathogenic access in the host (22, 23). This facilitates enterococcal proliferation in the intestinal tract (10) and subsequent systemic spread. Therefore, understanding host-enterococcus interactions in the context of antibiotic exposure is critical in the development of strategies to prevent enterococcal infections.
Host invasion by enteric pathogens results in diverse pathogen-specific outcomes, ranging from local intestinal infection to systemic inflammation and tissue damage (24–28). Conversely, commensal translocation is highly compartmentalized to establish and maintain homeostasis and prevent chronic systemic immune responses in the host (4, 29–31). However, systemic dissemination of commensals, including enterococci, can occur during intestinal damage or debilitating conditions, such as stroke, injury, antibiotic treatment, and diseases like inflammatory bowel disease (32–34). The mechanism of transmission of enterococci from the intestinal lumen to the bloodstream is not clearly understood. It is also unclear whether intestinal proliferation and systemic dissemination are enterococcus-specific phenomena or generic mechanisms among commensals that occur in healthy hosts when colonization resistance is disrupted by treatment with broad-spectrum antibiotics.
In this study, we focused on understanding the effects of cephalosporin treatment on commensal and enterococcal dynamics in the host. To determine the specificity of enterococcal proliferation and dissemination in response to cephalosporins, we established a ceftriaxone (broad-spectrum cephalosporin) treatment model in mice stably colonized with laboratory strains of Enterococcus faecalis (35). This mouse model recapitulates several aspects of cephalosporin treatment in humans, which allows the investigation of ceftriaxone-mediated alterations in the host as well as commensal dynamics during intestinal enterococcal proliferation and systemic spread from the GI tract.
RESULTS
Systemic ceftriaxone administration results in intestinal expansion and systemic dissemination of enterococcal populations.
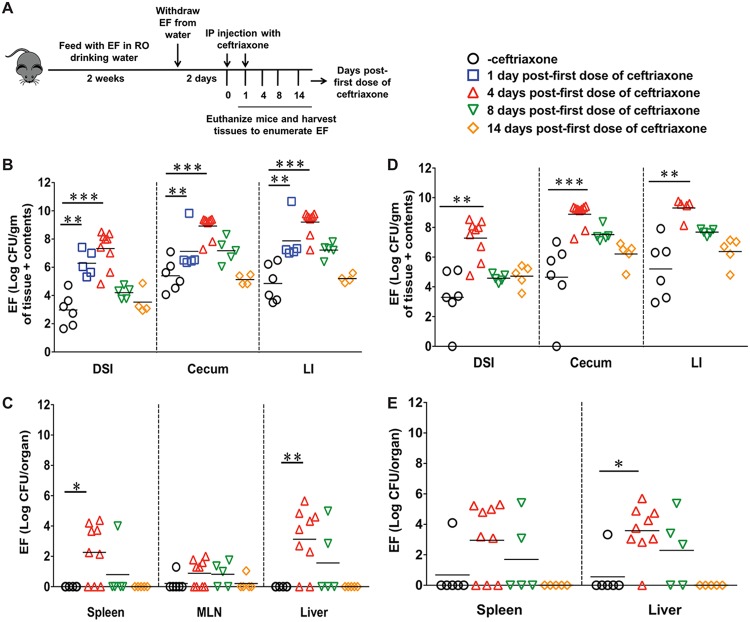
To study the dynamics of intestinal enterococcal populations in response to ceftriaxone administration, C57BL/6J mice were colonized with a rifampin-resistant strain of Enterococcus faecalis, CK135 (termed EFCK135) (36), and injected with ceftriaxone (Fig. 1A). We observed increased numbers of EFCK135 throughout the intestinal tract in response to ceftriaxone treatment (Fig. 1B). At day 1 after the first dose of ceftriaxone, the number of CFU of EFCK135 in the intestinal tract was 2 to 3 logs higher in the ceftriaxone-treated group than in the saline-treated controls (Fig. 1B). At day 4, we noticed a 4-log average increase of EFCK135 CFU in the intestines of ceftriaxone-treated mice that recovered to the baseline by 14 days (Fig. 1B). Although EFCK135 was detected at low levels in liver, spleen, and mesenteric lymph node (MLN) at 3 days after the first dose of ceftriaxone (data not shown), we found increased accumulation in the spleen and liver at day 4 that was 2 to 3 logs higher in the ceftriaxone-treated mice than in the controls (Fig. 1C). The number of CFU of EFCK135 recovered from the MLN at day 4 was comparatively lower than that obtained from the spleen and the liver (Fig. 1C). We also determined CFU of total intestinal enterococci to assess the effect of ceftriaxone on the native enterococcal populations. Like EFCK135, we observed an increased abundance of total enterococci, suggesting ceftriaxone-induced expansion of the indigenous enterococcal populations in the intestinal tract at day 4 (Fig. 1D). These CFU declined at day 8 and returned to the baseline by day 14 after the first dose of ceftriaxone (Fig. 1D). Dissemination of total enterococci to the liver and spleen was also comparable to that of EFCK135 at day 4, with a gradual decline in CFU over time (Fig. 1E). Both EFCK135 and total enterococci were cleared systemically by 14 days after the first dose of ceftriaxone administration (Fig. 1C and E).
FIG 1.
Ceftriaxone-mediated expansion and dissemination of enterococci. (A) Schematic representation of ceftriaxone treatment in C57BL/6J mice. EFCK135-colonized mice were intraperitoneally injected with ceftriaxone or physiological-grade saline for 2 consecutive days and then euthanized at days 1, 4, 8, and 14 after the first dose of ceftriaxone. (B and C) Determination of EFCK135 CFU in the distal small intestine (DSI), cecum, and large intestine (LI) of saline- and ceftriaxone-treated mice (B) and in the liver, spleen, and MLN (C) at 4, 8, and 14 days after the first dose of ceftriaxone. (D and E) CFU of total enterococci were determined in the intestinal tissues (D) and in the liver and spleen (E) at days 4, 8, and 14 after the first dose of ceftriaxone. n ≥ 4 mice per group. Statistics were done using unpaired Mann-Whitney U test (B and D) and Fisher’s exact test (C and E). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Ceftriaxone-induced systemic dissemination of enterococci is not associated with intestinal pathology, GI inflammation, or increased intestinal permeability.
To determine whether ceftriaxone-induced translocation and dissemination of enterococci are associated with intestinal mucosal pathology, we first performed histopathological examination of intestinal tissue sections collected from saline- and ceftriaxone-treated mice. Hematoxylin and eosin (H&E) staining of these tissues showed no evidence of mucosal architecture disruption, intestinal damage, or immune cell infiltration in response to ceftriaxone treatment (Fig. 2A). To analyze the inflammatory milieu of the GI tract in response to ceftriaxone, we measured transcriptional expression of proinflammatory cytokines interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α) in the distal small intestine (DSI) at day 4 after the first dose of ceftriaxone administration. IL-6 was not detected in either saline- or ceftriaxone-treated mice (data not shown), and TNF-α levels were found unchanged between these groups (Fig. 2B).
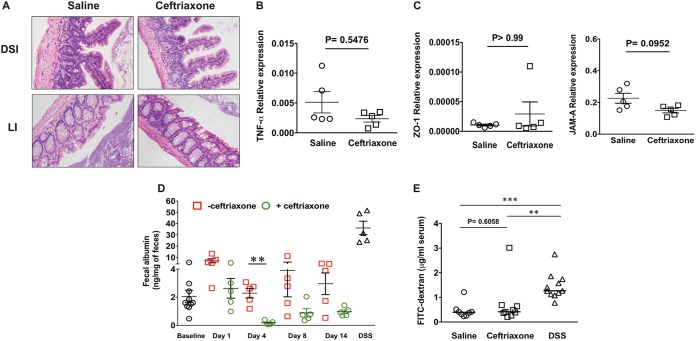
FIG 2.
Systemic ceftriaxone administration does not result in intestinal pathology or increased gut leakiness. (A) H&E staining of distal small intestine (DSI) and large intestine (LI) tissue sections collected from mice treated with saline or ceftriaxone and euthanized at day 4 after the first dose of ceftriaxone; original magnification, ×20. Whole-tissue sections were analyzed, and at least 2 different fields were captured per tissue. (B and C) Relative expression of TNF-α (B) and tight junction proteins ZO-1 and JAM-A (C) in the DSI of mice treated with saline or ceftriaxone by qRT-PCR. (D) Measurement of fecal albumin by ELISA as a determinant of intestinal permeability in saline- and ceftriaxone-treated mice at different time points after the first dose of ceftriaxone. (E) Measurement of FITC-dextran in the serum of saline- and ceftriaxone-treated mice at day 4 after the first dose of ceftriaxone. DSS-treated mice were used as a positive-control group in panels D and E. n ≥ 5 mice per group. **, P < 0.01; ***, P < 0.001 (by Mann-Whitney U test).
To determine whether ceftriaxone-induced translocation of enterococci across the intestinal epithelium occurs due to increased permeability of the gut, we next measured relative mRNA expression of representative tight-junction proteins ZO-1 and JAM-A by quantitative reverse transcription-PCR (qRT-PCR). We did not observe alterations in the transcriptional expression of these tight junction proteins in response to ceftriaxone challenge (Fig. 2C), suggesting no disruption of the intestinal tight junctions. To further explore alteration in the intestinal permeability after ceftriaxone treatment, we quantitatively measured albumin levels in the feces of mice that received either saline (control) or ceftriaxone (37, 38), demonstrating no increase in albumin levels in response to ceftriaxone treatment at day 4 or at an earlier time compared to the level for saline-treated mice (Fig. 2D). In addition, we performed an oral challenge with fluorescein isothiocyanate (FITC)-dextran in mice treated with either saline or ceftriaxone at day 4 after ceftriaxone treatment and measured FITC-dextran concentrations in their serum. We did not observe significant differences in the concentrations of FITC-dextran between the treatment groups (Fig. 2E). Mice treated with dextran sodium sulfate (DSS) were used as positive controls in both experiments (Fig. 2D and E). These results demonstrate that translocation and dissemination of EFCK135 during ceftriaxone exposure were not associated with increased intestinal permeability or mucosal damage.
Ceftriaxone treatment results in alteration of small intestinal innate immune defenses.
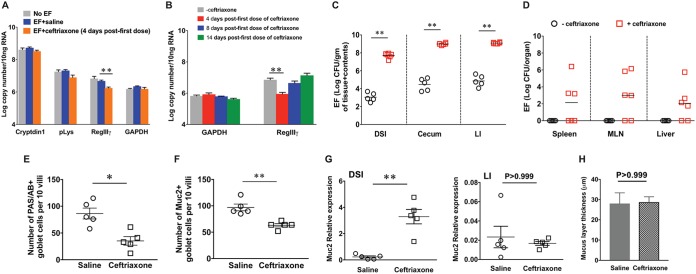
The intestinal epithelial barrier consists of a single layer of epithelial cells connected by junctional proteins. This barrier is fortified by mucus and antimicrobial peptides (AMPs) secreted by goblet cells and small intestinal Paneth cells, respectively (39). An intact mucus layer allows segregation of the intestinal microbiota from the epithelial surface and contributes to the maintenance of homeostasis (40–42). Previous reports suggested that antibiotic-induced downregulation of regenerating islet-derived protein IIIγ (RegIIIγ), an intestinal antimicrobial lectin, can cause decreased luminal killing of enterococci and subsequently allow dissemination (43, 44). To determine the effect of ceftriaxone treatment on these intestinal AMPs in our animal model, we assessed mRNA expression of representative Paneth cell-secreted AMPs. Transcriptional expression of constitutively expressed AMPs such as Cryptdin1 and Paneth cell lysozyme (pLys) was found to be unaltered, but that of RegIIIγ, which is induced by bacterial colonization (45, 46), was significantly reduced at day 4 after the first dose of ceftriaxone administration (Fig. 3A). RegIIIγ expression was partially restored by 8 days and recovered to baseline levels by 14 days after the first dose of ceftriaxone (Fig. 3B). To determine whether RegIIIγ was essential for the intestinal containment of enterococci in our animal model, we colonized RegIIIγ−/− mice (47) with EFCK135 and then treated them with either saline or ceftriaxone. The baseline colonization level of EFCK135 in the intestinal tracts of RegIIIγ−/− mice was comparable to that in wild-type (WT) mice (data not shown), and there was no evidence of systemic EFCK135 transmission (Fig. 3D). Upon ceftriaxone administration, RegIIIγ−/− mice exhibited similar levels of luminal proliferation and extraintestinal dissemination of EFCK135 (Fig. 3C and D), as observed in WT mice. These results suggest that RegIIIγ is not solely responsible for restricting the abundance of EFCK135 in the intestinal tract and is not essential for preventing their translocation across the mucosa. However, in the absence of RegIIIγ, it is possible that other compensatory mechanisms contribute to maintaining the barrier. For example, IgA, which coats intestinal commensals and restricts their mucosal penetration, was shown to be induced in RegIIIγ−/− mice (47). This suggests that IgA serves as a compensatory effector molecule in these mice; however, in our animals, luminal IgA levels were not measured after ceftriaxone administration.
FIG 3.
Ceftriaxone treatment results in alterations of intestinal innate barrier defenses. (A and B) qRT-PCR results of Paneth cell antimicrobials from DSI of saline- and ceftriaxone-treated mice colonized with EFCK135 and euthanized at day 4 after the first dose of ceftriaxone administration (A) and RegIIIγ at different time points (B). GAPDH was used as an internal control. CFU of EFCK135 in the intestinal sections (C) and in the liver, spleen, and MLN (D) of RegIIIγ−/− mice treated with saline or ceftriaxone and euthanized at day 4 after the first dose of ceftriaxone. (E) Enumeration of PAS/AB+ goblet cells per 10 villi in the distal small intestinal tissue sections collected from saline- and ceftriaxone-treated mice. (F) Enumeration of Muc2+ cells per 10 villi in the distal small intestinal tissue sections collected from saline- and ceftriaxone-treated mice. (G) Relative expression of Muc2 by qRT-PCR in the DSI and large intestinal sections from saline- and ceftriaxone-treated mice at day 4 after ceftriaxone administration. (H) Measurement of mucus layer thickness in the large intestines of saline- and ceftriaxone-treated mice at day 4 after ceftriaxone treatment. n = 5 mice per group. *, P values were <0.05 (*) and <0.01 (**) by unpaired Mann-Whitney U test.
Goblet cells and their secreted mucus contribute to the physical separation of the microbiota from the host epithelium and limit bacterial translocation across the mucosal surface (48). To investigate the effect of ceftriaxone on goblet cells and mucus, we performed periodic acid-Schiff/alcian blue (PAS/AB) staining of terminal ileal sections from conventional mice treated with saline or ceftriaxone and assessed the number of goblet cells across the length of the villi. The number of PAS/AB+ goblet cells was significantly reduced and was similar to the number of Muc2+ goblet cells in the ceftriaxone-treated mice compared with the controls (Fig. 3E and F). However, mRNA expression of Muc2 was significantly increased in the DSI of ceftriaxone-treated mice compared with the controls, suggesting increased biosynthesis and possibly secretion of mucus in response to ceftriaxone in the small intestines of conventional mice (Fig. 3G). Measurement of mucus layer thickness in the large intestines of saline- and ceftriaxone-treated mice showed no significant differences between the treatment groups, which also supported unaltered Muc2 relative expression in the large intestine at day 4 (Fig. 3G and H). These results suggest a compensatory role of mucus in the luminal containment of enterococci in the small intestine and inhibition of their increased translocation across the intestinal mucosal surface.
Intestinal proliferation of enterococci is associated with altered composition of the indigenous microbiota.
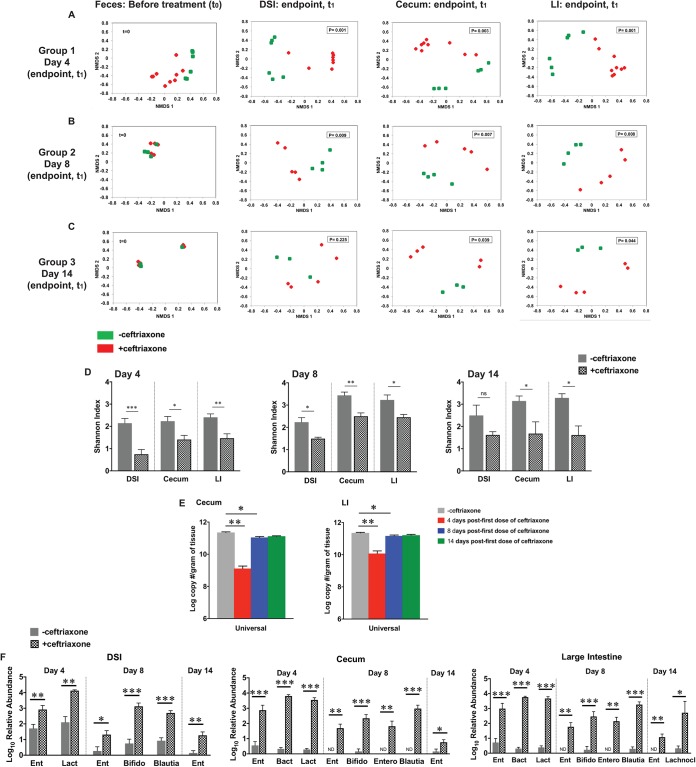
To further explore the effect of ceftriaxone on the intestinal barrier, we focused on the microbiota, which acts as a barrier against pathogenic and opportunistic infections in the host (49). We performed 16S rRNA gene sequencing with DNA extracted from the feces, DSI, cecum, and large intestines of EFCK135-colonized conventional mice treated with either saline or ceftriaxone to determine the effect of ceftriaxone on the composition of the intestinal microbiota at three time points. Complete litters of mice were used for each time point. Group 1 (day 4 time point) included 2 litters, whereas groups 2 (day 8) and 3 (day 14) each corresponded to a single litter. Comparison of baseline microbiota composition between littermates showed no statistical significance at day 0 (t0) (Fig. 4A to C). Each litter of mice was subsequently divided into 2 treatment groups; one group received saline and the other received ceftriaxone for 2 consecutive days before euthanization at days 4, 8, and 14, respectively. Microbiota composition was significantly altered throughout the intestinal tract at day 4 after the first dose of ceftriaxone treatment, as shown by beta (Fig. 4A to C)- and alpha (Fig. 4D)-diversity analyses. Additionally, these results were supported by absolute quantification of bacterial 16S rRNA gene copies by qRT-PCR, which revealed significant reduction in the total microbial load at day 4 in the cecum and large intestine of ceftriaxone-treated mice compared with that of the controls (Fig. 4E). OTU analysis revealed that the relative abundance of 6 bacterial taxa, including Bacteroidales and Lachnospiraceae, was significantly reduced throughout the intestinal tract at day 4 in response to ceftriaxone exposure (Table S3), but that of Enterococcus (GBKMun50) was found to be significantly elevated (Fig. 4F). Interestingly, we also noticed a significant increase in the relative abundance of a limited number of bacterial OTUs in response to ceftriaxone, including those of Lactobacillus (Unc00cd4) and Bacteroides (GG7Spe54) (Fig. 4F and Table S2 in the supplemental material). Determination of relative abundance of bacterial families supported our OTU analyses depicted in Fig. 4F and showed increased percentages of Lactobacillaceae in the DSI and Lactobacillaceae and Bacteroidaceae in the cecum and LI in addition to Enterococcaceae at day 4 after ceftriaxone administration (Fig. S1). At day 8, we noticed dynamic alterations in the relative abundance of multiple bacterial OTUs (Tables S2 and S3), including Enterobacter (GWTAmn51), Blautia (Unc01277), and Bifidobacterium (GWMPse11), which increased significantly in the intestinal tract (Fig. 4F). By day 14, differences in microbiota composition between saline- and ceftriaxone-treated mice were considerably reduced although still significant in the cecum and the large intestine (Fig. 4C and F). Relative abundance of most transiently increased bacterial taxa recovered to the baseline by day 14, except Enterococcus (GBKMun50), which remained persistently elevated in the ceftriaxone-treated group (Fig. 4F). We also noticed that ceftriaxone-induced depletion in the relative abundance of several native bacterial OTUs continued until day 14 (Table S3). These results were consistent with our alpha-diversity analysis, which showed significant alterations in the microbial diversity at day 8 and even at day 14 after ceftriaxone challenge (Fig. 4D), suggesting persistent disruption of the intestinal microbial community after antibiotic treatment.
FIG 4.
Ceftriaxone treatment alters the composition of the intestinal microbiota. (A to C) Baseline microbiota composition was determined by 16S rRNA gene sequencing using fecal DNA collected from EFCK135-colonized mice before treatment (t = 0). Mice represented in groups 1, 2, and 3 were subsequently divided into 2 treatment groups that received either saline (green) or ceftriaxone (red) and then were euthanized at the indicated time points (t1). 16S rRNA gene sequencing was performed with DNA extracted from the DSI, cecum, and large intestine posteuthanization. Nonparametric ordination of Bray-Curtis distance showed spatial distribution of the fecal samples before treatment and segregation of samples into saline- and ceftriaxone-treated groups at 4, 8, and 14 days after the first dose of ceftriaxone (P values were calculated using Adonis). Each ordination figure shows the relative pairwise Bray-Curtis distance (or dissimilarity in beta biodiversity) between the samples and the two treatment groups. Even though the axes in each figure are shown with a harmonized range of −0.8 to 0.8, it is not meaningful to compare across the figures. (D) Shannon’s diversity index to determine ceftriaxone-induced alterations in the diversity of native microbial populations in the intestinal tract. (E) Absolute quantification of total intestinal bacteria (universal) in the cecum and LI of EFCK135-colonized mice treated with saline or ceftriaxone by qRT-PCR. (F) Relative abundance of bacterial OTUs in the DSI, cecum, and large intestine of EFCK135-colonized mice treated with saline or ceftriaxone and euthanized at 4, 8, and 14 days after the first dose of ceftriaxone. OTU IDs are mentioned in parentheses. Ent, Enterococcus species (GBKMun50); Lact, Lactobacillus species (Unc00cd4); Bact, Bacteroides species (GG7Spe54); Bifido, Bifidobacterium species (GWMPse11); Blau, Blautia species (Unc01277); Entero, Enterobacter species (GWTAmn51); Lachnocl, Lachnoclostridium species (Unc06361). n ≥ 4 mice per group. P values were <0.05 (*), <0.01 (**), and <0.001 (***) by Student’s t test in (A to D and F) and Mann-Whitney U test in panel E.
Increased intestinal abundance of enterococci in response to ceftriaxone is important for their systemic dissemination.
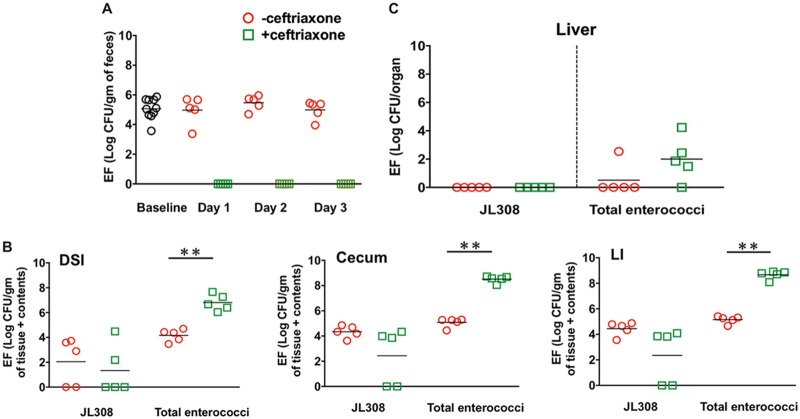
The association between enterococcal expansion and dissemination in response to ceftriaxone administration (Fig.1) suggested that increased luminal abundance is required for enterococcal translocation. To test this hypothesis, we colonized mice with EFJL308, a ceftriaxone-sensitive strain of enterococci that does not expand in response to ceftriaxone, and then challenged mice systemically with ceftriaxone. Up to 3 days after the first dose of ceftriaxone administration, we were unable to recover any EFJL308 clones from the feces of ceftriaxone-treated mice (Fig. 5A). However, at day 4, EFJL308 clones were detected in the intestines of 3 out of 5 mice, with the CFU being almost comparable to their baseline colonization levels (Fig. 5B). This was likely due to decreased concentrations of ceftriaxone in the local intestinal environment, as recovered clones retained their sensitivity to ceftriaxone (data not shown). CFU determination of total enterococci, which in this experiment was used as an internal control, showed significant expansion of indigenous enterococcal populations throughout the intestinal tracts of ceftriaxone-treated mice compared with the controls (Fig. 5B). Importantly, while we did not observe EFJL308 dissemination to the liver, we did detect indigenous enterococci (Fig. 5C), suggesting that intestinal proliferation of enterococci in response to ceftriaxone treatment is important for their systemic dissemination in the host.
FIG 5.
Intestinal abundance of enterococci is important for their systemic dissemination in the host. Mice colonized with EFJL308 were injected with saline or ceftriaxone and euthanized at day 4 after the first dose of ceftriaxone. (A) CFU of EFJL308 were determined in the feces of mice at day 0 (baseline) and at different time points after ceftriaxone administration. (B and C) Enumeration of EFJL308 CFU in the intestinal sections and liver, respectively, of mice at euthanization. n = 5 mice per group. **, P = 0.0079 by unpaired Mann-Whitney U test.
Systemic ceftriaxone challenge mediates increased intestinal abundance and extraintestinal dissemination of lactobacilli in the absence of enterococci.
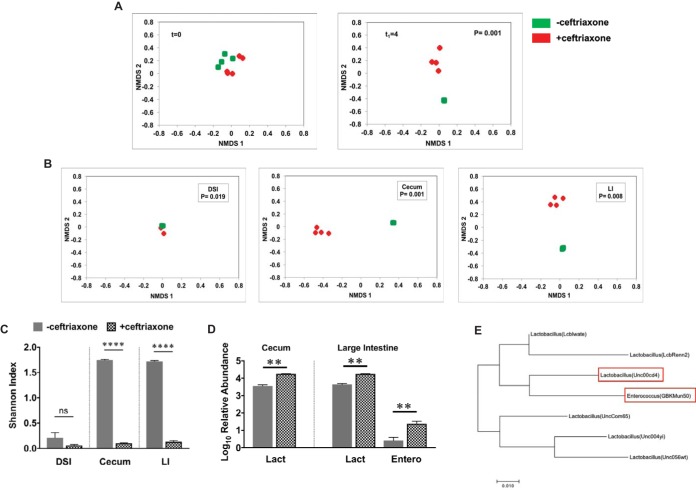
To investigate whether increased abundance and systemic dissemination in response to ceftriaxone challenge are enterococcus-specific phenomena or common to other GI commensals, we used defined-flora mice (devoid of cocci, including enterococci) and treated them with either saline or ceftriaxone. We performed 16S rRNA gene sequencing with DNA extracted from the feces, intestinal tissues, and extraintestinal organs, such as liver, spleen, and MLN, to identify bacterial taxa that have expanded and disseminated in the absence of enterococci at day 4 after the first dose of ceftriaxone. Fecal DNA was extracted from these mice before ceftriaxone challenge and at day 4 after the first dose of ceftriaxone to determine alterations in their microbiota composition. At day 0, the fecal microbiota compositions were closely aligned, but following ceftriaxone treatment, at day 4, they became significantly separated (Fig. 6A). We observed significant shifts in the microbial composition throughout the intestinal tract in response to ceftriaxone, which was indicated by highly segregated saline- and ceftriaxone-treated samples in the NMDS plots (Fig. 6B). Determination of alpha diversity by Shannon index analysis further showed that the overall richness of the indigenous microbial populations was significantly reduced in response to ceftriaxone administration, most notably in the cecum and the large intestine; however, that in the DSI was relatively unaltered (Fig. 6C). Analysis of bacterial families and OTUs revealed increased relative abundance of Lactobacillaceae, specifically the OTU Unc00cd4 in both cecum and large intestine (Fig. 6D and Fig. S2), as we had observed in the conventional mice. Relative abundance of Enterobacteriaceae (UncG3786) was also elevated in the large intestines of ceftriaxone-treated mice (Fig. 6D). However, we could not detect any bacterial OTUs that were relatively increased in the DSI, and Lactobacillaceae were detected as the dominant taxa irrespective of ceftriaxone treatment (Fig. S2). Since a Lactobacillus species (Unc00cd4) was the only bacterial OTU that was found to be consistently increased in the conventional as well as in the defined-flora mice upon ceftriaxone exposure and lactobacilli and enterococci are taxonomically related, we compared the evolutionary distance between several Lactobacillus OTUs and an Enterococcus species (GBKMun50) from the 16S analysis data. Interestingly, phylogenetic analysis revealed that the Lactobacillus species (Unc00cd4) shares closer evolutionary association with Enterococcus species (GBKMun50) than with other Lactobacillus OTUs included in this analysis (Fig. 6E). To further explore links between increased intestinal abundance and dissemination, we analyzed microbial populations in the extraintestinal tissues by 16S rRNA gene sequencing. However, low bacterial biomass from the extraintestinal tissues prevented reliable detection of any translocating bacterial strains. We then used standard culturing techniques to detect lactobacilli in the extraintestinal organs of saline- and ceftriaxone-treated mice. We anaerobically cultured extraintestinal tissue homogenates on MRS and LAMVAB, selective media for lactobacilli (50, 51), and recovered colonies from the liver tissues of 50% of the ceftriaxone-treated mice (data not shown). Full-length 16S rRNA gene sequencing from at least 50 single colonies from anaerobically cultured liver and spleen homogenates detected Lactobacillus murinus and Lactobacillus animalis as the disseminating organisms. These species shared 99% sequence similarity with the Lactobacillus OTU Unc00cd4, whose relative abundance in the intestinal tracts of conventional as well as defined-flora mice was significantly increased in response to ceftriaxone exposure. Collectively, these data suggest that extraintestinal dissemination in response to systemic ceftriaxone challenge is not an enterococcus-specific phenomenon, as we observed dissemination of selected lactobacillus species in the liver of defined-flora mice after ceftriaxone treatment.
FIG 6.
Ceftriaxone treatment results in increased intestinal abundance of other commensals in the absence of enterococci. (A) Fecal DNA was extracted from defined-flora mice (lacking cocci including enterococci) at day 0 (t = 0; before treatment) and day 4 (t1 = 4; endpoint) after the first dose of ceftriaxone for 16S rRNA gene sequencing. NMDS plots showing even distribution of the samples at day 0 (left) and distantly separated saline (green)- and ceftriaxone (red)-treated samples at day 4 (right). (B) NMDS plots showing separation of saline- and ceftriaxone-treated samples by Bray-Curtis beta diversity analysis at day 4 after the first dose of ceftriaxone administration in the DSI, cecum, and large intestine of defined-flora mice. (C) Shannon’s diversity index to determine ceftriaxone-induced alterations in the diversity of native microbial populations in the intestinal tracts of defined-flora mice at day 4 after first dose of ceftriaxone administration. (D) Relative abundance of bacterial OTUs in the intestinal tracts of defined-flora mice treated with saline or ceftriaxone at day 4 after ceftriaxone challenge (Lact, Lactobacillus species [Unc00cd4]; Entero, Enterobacteriaceae species [UncG3786]). (E) Molecular phylogenetic analysis by the maximum likelihood method showing evolutionary distance comparison between several OTUs of Lactobacillus species and an Enterococcus species (GBKMun50). The tree is drawn to scale, and the highest log likelihood (−702.23) is shown. n = 4 to 5 mice per group. ns, not significant; **, P < 0.01; ****, P < 0.0001 (by Student’s t test).
DISCUSSION
Translocation of intestinal commensals is compartmentalized to prevent deleterious chronic systemic immune responses in the host. This compartmentalization is achieved by regulated mechanisms that include commensal transcytosis through M cells into the underlying Peyer’s patches, where they are engulfed by dendritic cells (DCs) or directly sampled by the lamina propria resident DCs via dendrites to generate protective adaptive immune responses (52, 53). These immune responses, in turn, strengthen the intestinal mucosal barrier, help establish homeostasis, and contain the microbiota within the intestinal lumen. Colonization resistance, which is established by complex microbiota-host interactions, protects the host from exogenous pathogens (22, 49). Consequently, disruption of colonization resistance predisposes the host to infections by pathogens and opportunists. As native GI residents, enterococci do not cause complications in the healthy host. However, immunosuppression, underlying diseases, or prolonged antibiotic exposure allows these opportunists to proliferate in the intestinal tract and cause disseminated infections. In this study, we investigated the effects of ceftriaxone on the host mucosal barrier and the dynamics of commensals, including enterococci. We developed a mouse model that recapitulates ceftriaxone-mediated enterococcal dynamics described in humans. Using this model, we showed that proliferation of enterococci in the intestinal lumen preceded their dissemination to liver, spleen, and MLN, suggesting an association between expansion of the enterococcal populations and dissemination. Our experiments with a ceftriaxone-sensitive strain of enterococci (EFJL308) uncoupled these two phenomena and emphasized the requirement of intestinal expansion of enterococcal populations for dissemination during ceftriaxone exposure.
Recent studies have shown that antibiotic treatment can alter the regulation of the intestinal mucosal barrier (54–56) and downregulate the expression of RegIIIγ (43), which in turn could lead to translocation of commensals across the intestinal epithelium. We have found that systemic ceftriaxone administration was associated with significant reduction in the transcriptional expression of RegIIIγ; however, our studies in RegIIIγ knockout mice suggest that it is not essential in the intestinal containment or extraintestinal dynamics of enterococci. Lack of intestinal pathology, inflammation, or increased intestinal permeability in response to ceftriaxone challenge suggests that translocation of enterococci across the mucosal surface does not occur due to altered barrier function. Instead this phenomenon could represent a regulated course of commensal translocation, which is now saturated by enterococci due to ceftriaxone-induced luminal domination by these bacteria. In support of this notion, we also observed increased intestinal abundance and extraintestinal dissemination of selected species of lactobacilli in response to ceftriaxone exposure. These results suggest that ceftriaxone-induced extraintestinal dissemination is not an enterococcus-specific phenomenon. Luminal enterococci could be directly captured by the interepithelial DCs via dendrites or phagocytosed by the lamina propria macrophages. The ability of these mononuclear phagocytes to migrate from the periphery into the bloodstream or to the lymphatic system could contribute to enterococcal dissemination. Additionally, goblet cell-associated passages (GAPs) (57) or endocytosis by epithelial cells (58–61) could serve as portals for enterococcal entry into the underlying lamina propria. Nevertheless, further studies in this area will be needed to achieve a better understanding of these mechanisms.
Previous studies in RegIIIγ−/− mice highlight the significance of microbiota-dependent compensatory adaptive immune responses (47) that could be beneficial in preventing bacterial passage across the intestinal mucosa. Mucus also actively participates in the physical separation of the microbiota from the host (48). We leveraged these insights to investigate the relationship between ceftriaxone-mediated altered innate barrier defenses and enterococcal dynamics in the gut and mucus in conventional mice. Increased transcriptional expression of Muc2 during ceftriaxone exposure suggests increased production and most likely secretion of mucus. Since mucin production could be modulated by microbial cues (62), our observation with Muc2 could be interpreted as a compensatory mechanism critical for the luminal entrapment of the rapidly proliferating pool of enterococci to reduce their contact with the epithelium. Decreased numbers of PAS/AB+ goblet cells indicate reduced storage or increased secretion of mucus in response to ceftriaxone challenge, which is consistent with our findings with Muc2 expression.
Prolonged alteration of the indigenous microbial composition even at day 14 after the first dose of ceftriaxone suggested persistent disruption of the intestinal microbial communities (63, 64). Several bacterial OTUs were either increased or decreased at day 8, which could be considered a transitional phase with alterations in bacterial abundance during recovery. The elimination half-life of intravenously introduced ceftriaxone in humans after a single dose is ∼6 h (65). Therefore, it seems reasonable to speculate that in our study, several bacterial OTUs or taxa were not able to recolonize their native ecological niches to the original threshold levels even long after ceftriaxone was eliminated from the system.
Interestingly, we identified a Lactobacillus species (Unc00cd4) that was phylogenetically closer to an Enterococcus species (GBKMun50) than other lactobacilli. This Lactobacillus species also exhibited a transient increase in its relative abundance in the intestinal tracts of conventional as well as defined-flora mice in response to ceftriaxone treatment. Culturing on selective media for lactobacilli and full-length 16S rRNA gene sequencing from single bacterial isolates revealed that Lactobacillus murinus and Lactobacillus animalis disseminated to the extraintestinal organs in response to ceftriaxone. These species shared 99% sequence similarity with the Lactobacillus OTU Unc00cd4, whose relative abundance in the intestinal tracts of conventional as well as defined-flora mice was significantly increased in response to ceftriaxone exposure. These observations pose questions as to whether these species of lactobacilli are resistant to ceftriaxone and therefore behaved like enterococci during ceftriaxone treatment. Alternatively, ceftriaxone-induced modulations in the intestinal microbiota composition could allow these species of lactobacilli to respond to certain metabolites or microbial attributes in their immediate environment, thereby resulting in their expansion and likely dissemination. Interestingly, we did not observe increased numbers of lactobacilli in the intestinal lumen of the ceftriaxone-treated mice by our culturing analyses. A possible explanation is that a relatively unaltered background of lactobacillus population obscured the increased abundance of specific lactobacillus species in response to ceftriaxone. However, this abundance was apparently sufficient to saturate the microenvironment and dominate the commensal translocation pathway, thereby allowing lactobacilli to disseminate. Nonetheless, further work is needed to achieve an in-depth understanding of the molecular mechanisms behind these observations.
Previous studies have shown that translocation of lactobacilli is generally compartmentalized within the MLNs of conventional as well as gnotobiotic mice (66, 67). Here, we show for the first time that certain species of lactobacilli possibly pursue different translocation routes and eventually disseminate to the liver and spleen in response to systemic ceftriaxone challenge. Alternatively, it is also possible that lactobacillus translocation to the liver and spleen is a part of their conventional compartmentalization mechanism that, although inconspicuous under steady-state conditions, may become prominent during antibiotic exposure. Altogether, our study emphasizes that disruption of colonization resistance by broad-spectrum antibiotics could enhance a general pathway of commensal dissemination, allowing transiently dominant species of selected intestinal commensals to saturate this pathway in a healthy host.
MATERIALS AND METHODS
Animals.
All experiments were performed using protocols approved by the committee for animal care and use at the Medical College of Wisconsin. Five-week-old male C57BL/6J mice were obtained from the Jackson Laboratory (room RB08, JAXWEST facility, Sacramento, CA). Upon arrival at our facility, mice were acclimatized for 1 week before being used for any experiments. All mice were fed a standard chow diet (PicoLab laboratory rodent diet) ad libitum and reverse osmosis water and maintained under specific-pathogen-free conditions throughout the course of the experiments. Where mentioned, facility-bred C57BL/6J mice were used and treated the same way as the vendor-obtained mice. C57BL/6N defined-flora mice (Taconic Biosciences), which were devoid of cocci and non-spore-forming rod bacteria, were housed in biocontainment (BCU) cages in a HEPA filter-provisioned rack or in the gnotobiotic facility. Mice were fed with autoclaved chow (LabDiet) and autoclaved water until completion of the experiments.
Antibiotic treatment.
Mice were injected with ceftriaxone (ceftriaxone for injection USP; Apotex Corp.) intraperitoneally once daily for 2 consecutive days. Each mouse received a daily dose of 2.5 mg ceftriaxone in a volume of 100 μl normal or physiological-grade saline. Control mice received equal volumes of normal saline.
Bacterial strains and colonization of mice with enterococci.
Overnight cultures of EFCK135 (spontaneous rifampin-resistant derivative of OG1 [rpoB H486Y]) or EFJL308 (pbp5 mutant [Δpbp5 strain containing an in-frame deletion of the pbp5 locus], a ceftriaxone-sensitive derivative of OG1 [rpoB H486Y]) (36), were grown in brain heart infusion (BHI) broth (BD) or Mueller-Hinton (MH) broth (Difco), respectively, supplemented with rifampin (200 μg/ml) (BHI-RIF or MH-RIF), and pelleted by centrifugation at 5,000 rpm for 10 min. After decanting the supernatant, the pellets were washed with autoclaved water 2 to 3 times to remove excess media and antibiotics. Final pellets were resuspended in autoclaved water, and bacterial cell concentration was measured according to the optical density at 600 nm (OD600) with a NanoDrop device (NanoDrop 2000 spectrophotometer; Thermo Scientific). For colonization, mice were fed 5 × 108 cells/ml of bacteria in 250 ml of drinking water as described before (35).
Tissue collection.
Control and ceftriaxone-treated mice were euthanized at days 1, 4, 8, and 14 after the first dose of ceftriaxone by CO2 asphyxiation followed by cervical dislocation. Tissues were harvested in the order of liver, spleen, and mesenteric lymph node (MLN), followed by intestinal tract for further analysis. Liver, spleen, and MLN were each collected in 1 ml sterile phosphate-buffered saline (PBS) (pH 7.4), whereas the intestinal tract was divided into small intestine, cecum, and large intestine, and each was collected in 2 ml sterile PBS. For RNA extraction, 1.5- to 3-cm section of the distal small intestine (DSI), 1 cm of the distal end of the large intestine and cecum tip were excised, intestinal contents were removed, and the tissues were collected immediately in RNAlater (Qiagen). Tissues were kept at 4°C with rocking overnight before storing at −80°C until use.
Determination of CFU.
All tissues were weighed before homogenization. Entire homogenized samples of liver, spleen, and MLN were plated on BHI-RIF or MH-RIF plates to enumerate EFCK135 or EFJL308, respectively. Intestinal samples were serially diluted in autoclaved MilliQ water and cultured on the above-mentioned plates with appropriate antibiotic selection. EFCK135 or EFJL308 was enumerated in the samples after overnight incubation at 37°C and normalized to tissue weight (per gram for intestinal samples and per organ for liver, spleen, and MLN). Total enterococci were assessed in the tissues by culturing tissue homogenates on m-Enterococcus agar (Difco) plates, and CFU were determined after 24 to 36 h of incubation at 37°C. To enumerate and isolate lactobacilli, liver, spleen, and MLN were cultured on LBS (BD), Lactobacilli MRS (Difco), and LAMVAB (50) agars and incubated anaerobically (Coy anaerobic chamber) for 48 to 72 h. LAMVAB agar medium was manually prepared by following the protocol mentioned in reference 50, with minor modifications. MRS broth (Difco) was used as the base for the medium, and 15 g/liter of agar was used instead of 40 g/liter.
RNA extraction from tissues and cDNA synthesis.
RNAlater-preserved tissues were homogenized manually using a pestle tissue grinder assembly (Thomas Scientific), and RNA was extracted from the DSI, cecum, and large intestine using an RNeasy minikit (Qiagen) by following the manufacturer’s instructions. RNA concentrations were determined using a NanoDrop device. First-strand cDNA synthesis was performed with 500 ng of RNA using an iScript cDNA synthesis kit (Bio-Rad Laboratories) by following kit instructions.
Single-colony PCR and quantitative real-time PCR.
For a single colony, a 200-μl reaction mixture was prepared that contained 40 μl of 5× HF buffer, 16 μl of deoxynucleoside triphosphate mixture (10 mM; Promega), 1 μl of Phusion polymerase (Thermo Scientific), 500 nM (each) forward and reverse primers (100 μM) (universal forward primer [UNI8F] 5′-AGAGTTTGATCMTGGCTCAG-3′ and universal reverse primer [1492R] 5′-TACGGYTACCTTGTTACGACTT-3′) (68), and 141 μl of ultrapure water (Life Technologies). PCR conditions included an initial cell lysis step at 95°C for 5 min, followed by 25 cycles of sequential denaturation for 30 s at 95°C, 30 s of annealing at 50°C, 3.30 min of extension at 72°C, and a final extension of 10 min at 72°C. Amplified PCR products were purified using a PCR purification kit (Qiagen) and sent to Retrogen for full-length 16S rRNA gene sequencing.
Quantitative real-time PCR (qRT-PCR) was performed using iTaq universal SYBR green supermix (Bio-Rad), prepared DNA or cDNA, and specific primers listed in Table S1 in the supplemental material. qRT-PCR was run on either a StepOnePlus real-time PCR system (Applied Biosystems) or a CFXConnect real-time system (Bio-Rad). Standard curves were used for the absolute quantification of bacterial 16S rRNA gene copies (69) and Paneth cell AMPs Cryptdin1, Paneth cell lysozyme (pLys), and RegIIIγ (70, 71). The quantitative PCR (qPCR) program consisted of two steps: a holding stage, where initial denaturation of the DNA template occurs at 95°C for 3 min, followed by an amplification stage which has 40 cycles, with each cycle consisting of denaturation at 95°C for 10 s and primer annealing and extension for 45 s at temperatures listed in Table S1. Relative quantification of mRNA levels was determined for Muc2 (72), ZO-1 (73), JAM-A (74), and TNF-α (75) using the ΔCT method. The qPCR conditions included an initial denaturation at 95°C for 10 min, followed by 40 cycles of amplification with each cycle, consisting of denaturation at 95°C for 15 s and annealing and extension for 1 min at the listed temperatures (Table S1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (76) or β-actin (55) was used for normalization, and relative mRNA expression was expressed as 2−ΔCT (77).
Histology and mucus staining.
Sections of DSI (∼1.5 cm), cecum tip, and distal large intestine (∼1 cm) were excised at euthanization and immediately placed in zinc formalin (ThermoFisher Scientific) for histology. Fixed tissues were processed by the Core Histology facility at the Childrens’ Research Institute (CRI), Children’s Hospital of Wisconsin, and then stained with hematoxylin and eosin (H&E) for characterization of tissue architecture and histopathology. For mucus staining, ∼1.5-cm sections of the terminal ileum were excised, immediately placed in Carnoy’s fixative, and incubated on ice for 2 h before transferring the tissues into 100% ethanol (72). Fixed tissues were cut into 4-μm sections and stained with PAS/AB. PAS/AB+ goblet cells were then enumerated in 10 villi per sample. The thickness of the mucus layer was measured in the large intestinal sections stained with PAS/AB. Stained tissue sections were scanned, and the width of the mucus layer was determined by at least 10 measurements per tissue section using NDP.view2 software. For immunohistochemical analyses, longitudinal cross-sections of DSI were deparaffinized and rehydrated. Staining was performed by the Core Histology facility at the CRI using Dako Autostainer Plus by following a standard protocol. Briefly, antigen retrieval was performed by placing the slides in citrate buffer (Dako) (pH 6.0) at 90°C for 20 min, followed by cooling to room temperature and blocking. Tissues were then stained with murine Muc2 (H-300; Santa Cruz Biotechnology) antibody at 4°C for 90 min, followed by incubation with a goat anti-rabbit secondary antibody, staining with DAB for 2 min, and counterstaining with hematoxylin.
Fecal albumin measurement.
Fecal albumin concentrations were determined in saline- and ceftriaxone-treated mice using a mouse albumin enzyme-linked immunosorbent assay (ELISA) kit (Bethyl Laboratories). A group of 5 mice were fed with 4% DSS (Affymetrix) in autoclaved drinking water for 7 days to induce colitis and included in this experiment as a positive control. Fecal pellets were collected from mice at days 1, 4, 8, and 14 after the first dose of ceftriaxone and at day 7 from DSS-treated mice and processed using a standard protocol (37), with some modifications. Briefly, 100 mg of feces was collected in 1 ml cold dilution buffer (provided in the kit) and incubated on ice for 15 min. Samples were then vortexed for 30 s and centrifuged at 13,000 rpm for 5 min at 4°C to pellet bacteria and debris. Supernatants were then diluted at a 1:10 ratio in dilution buffer for saline- and ceftriaxone-treated mice and 1:100 for DSS-treated mice and used to calculate fecal albumin levels by following the manufacturer’s instructions.
FITC-dextran assay.
Mice were treated with saline or ceftriaxone and euthanized at day 4 after the first dose of ceftriaxone to measure intestinal permeability. As previously described, mice fed with 4% DSS (Affymetrix) were used as a positive-control group and euthanized after 7 days. On the day of euthanization, mice were deprived of food and water for 4 h before 150 mg of freshly prepared FITC-dextran (Sigma) in PBS was administered by oral gavage. After 4 h, mice were anesthetized by isoflurane (Phoenix Pharmaceutical) inhalation and blood was collected by cardiac puncture. Blood was immediately transferred into Microtainer serum separator tubes (BD), and serum was isolated by centrifugation at 1,500 × g for 10 min. Serum was diluted with an equal volume of 1× PBS, and 100 μl of the diluted serum was pipetted into a 96-well plate in duplicates. The concentration of FITC was measured in the diluted serum by a fluorometer (Synergy H1), with excitation and emission wavelengths of 485 nm and 528 nm, respectively, using a standard curve prepared with serially diluted concentrations of FITC-dextran in 1× PBS.
DNA extraction.
Bacterial genomic DNA was extracted from murine feces and tissues using the MO BIO Powerlyzer Powersoil DNA isolation kit (MO BIO Laboratories, Carlsbad, California), with minor modifications (35). Approximately 250 mg of feces was suspended in the bead buffer provided with the MO BIO DNA extraction kit, and we proceeded with the DNA isolation protocol. One ml of homogenized DSI and large intestine and 500 µl of cecal homogenate were pelleted by centrifugation at 13,000 rpm for 10 min, supernatants were discarded, and the pellets were used for genomic DNA isolation. For DNA extraction from extraintestinal tissues, 400 μl of liver homogenate was centrifuged as mentioned above, and the pellets were used for the isolation. Spleen and MLN were collected in 750 μl of bead buffer and homogenized, and the entire homogenates were used for DNA extraction.
16S rRNA gene sequencing and data processing.
Extracted DNA samples from murine feces and tissues were sent to Diversigen (Baylor College of Medicine, Waco, TX) for 16S rRNA gene sequencing. Details of sample handling, 16S rRNA gene amplification, sequencing, and data processing were provided by Diversigen. Samples with low bacterial genomic DNA content were further processed in Diversigen by following an adapted version of the MO BIO Powersoil DNA isolation protocol before subjecting them to 16S rRNA gene sequencing. Briefly, following addition of C1 solution, 50 μl of C2 buffer and 50 μl of C3 buffer were added to each sample tube, incubated for 5 min at 4°C, and then proceeded to the next step. Finally, DNA was eluted in 50 μl of C6 solution.
16S rRNA gene sequencing methods were adapted from the methods developed for the NIH Human Microbiome Project (78, 79). The V4 region of the 16S rRNA gene was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2× 250-bp paired-end protocol, yielding paired-end reads that overlap almost completely. The primers used for amplification contain adapters for MiSeq sequencing and single-end barcodes allowing pooling and direct sequencing of PCR products (80). The 16S rRNA gene pipeline data incorporate phylogenetic and alignment-based approaches to maximize data resolution. The read pairs were demultiplexed based on the unique molecular barcodes, and reads were merged using USEARCH v7.0.1090 (81), allowing zero mismatches and a minimum overlap of 50 bases. Merged reads were trimmed at the first base with Q5. In addition, a quality filter was applied to the resulting merged reads, and reads containing above 0.05 expected errors were discarded. 16S rRNA gene sequences were demultiplexed using QIIME (82) and clustered into OTUs at a similarity cutoff value of 97% using the UPARSE algorithm. OTUs were mapped to an optimized version of the SILVA Database containing only the 16S V4 region to determine taxonomies (83, 84). Abundances were recovered by mapping the demultiplexed reads to the UPARSE OTUs. A custom script constructed a rarefied OTU table from the output files generated in the previous two steps for downstream analyses of alpha-diversity, beta-diversity (85), and phylogenetic trends.
Bioinformatics and phylogenetic analysis.
16S rRNA gene sequencing data were analyzed using the Vegan (86) and Ecodist (87) packages in R, 3.0.2 (88). Sequence counts of each sample were normalized to the average sequencing depth, and the Bray-Curtis metric was used to assess intersample (beta) diversity. Microbial species richness or diversity within a sample (alpha diversity) was measured by Shannon’s diversity index. Statistical significance for differences in microbiome diversity between groups was determined using Adonis (in Vegan). NMDS ordination (in Ecodist) was used to visualize group clustering and diversity distance between samples. Log-transformed abundance data (log10 of sequence count + 1) was used to normalize the data, and statistically significant differences in OTU abundance between groups were determined by heteroscedastic, two-sided Student’s t tests. The false-positive rate due to multiple testing was corrected by estimating the false discovery rate, or q value (89).
Evolutionary distance comparison was performed using MEGA7 software (90).
Statistical analysis.
Statistical significance was determined using two-tailed Student’s t test (nonparametric Mann-Whitney U test), and geometric means or means with standard errors of means (SEM) were calculated for the results using GraphPad Prism software, version 7.0b (GraphPad Software Inc., CA), unless otherwise stated. To compare CFU of E. faecalis in the extraintestinal tissues, Fisher’s exact test (GraphPad QuickCalcs; GraphPad Software) was performed. Statistical significance is represented as P values of <0.001 (***), <0.01 (**), and <0.05 (*).
Accession number(s).
16S rRNA gene sequences used for microbiome analyses were submitted to the NCBI database under Bioproject accession no. PRJNA489674 and study accession no. SRP161800.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants R01 GM099526 from the National Institutes of Health (NIH) to N.H.S., R01 AI081692 and OD006447 to C.J.K., and from the Children’s Research Institute of Children’s Hospital of Wisconsin to N.H.S. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Pippa M. Simpson and Amy Y. Pan for providing advice regarding statistical analyses of the data presented here. We acknowledge the contribution of the Children’s Hospital of Wisconsin (CHW) Children’s Research Institute (CRI) Histology Core for slide preparation, staining, and scanning of stained slides. We also thank Michele A. Battle for kindly providing DSS and valuable advice to perform intestinal permeability experiments.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00674-18.
REFERENCES
- 1.Brown EM, Sadarangani M, Finlay BB. 2013. The role of the immune system in governing host-microbe interactions in the intestine. Nat Immunol 14:660–667. doi: 10.1038/ni.2611. [DOI] [PubMed] [Google Scholar]
- 2.Charbonneau MR, Blanton LV, DiGiulio DB, Relman DA, Lebrilla CB, Mills DA, Gordon JI. 2016. A microbial perspective of human developmental biology. Nature 535:48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenburg JL, Backhed F. 2016. Diet-microbiota interactions as moderators of human metabolism. Nature 535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda K, Littman DR. 2016. The microbiota in adaptive immune homeostasis and disease. Nature 535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 5.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tannock GW, Cook G. 2002. Enterococci as members of the intestinal microflora of humans, p 101–132. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 8.Aarestrup FM, Butaye P, Witte W. 2002. Nonhuman reservoirs of enterococci, p 55–99. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, DC. [Google Scholar]
- 9.Orloff SL, Busch AM, Olyaei AJ, Corless CL, Benner KG, Flora KD, Rosen HR, Rabkin JM. 1999. Vancomycin-resistant Enterococcus in liver transplant patients. Am J Surg 177:418–422. doi: 10.1016/S0002-9610(99)00083-5. [DOI] [PubMed] [Google Scholar]
- 10.Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montecalvo MA, Horowitz H, Gedris C, Carbonaro C, Tenover FC, Issah A, Cook P, Wormser GP. 1994. Outbreak of vancomycin-, ampicillin-, and aminoglycoside-resistant Enterococcus faecium bacteremia in an adult oncology unit. Antimicrob Agents Chemother 38:1363–1367. doi: 10.1128/AAC.38.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Handwerger S, Raucher B, Altarac D, Monka J, Marchione S, Singh KV, Murray BE, Wolff J, Walters B. 1993. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin Infect Dis 16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 13.Lautenbach E, Bilker WB, Brennan PJ. 1999. Enterococcal bacteremia: risk factors for vancomycin resistance and predictors of mortality. Infect Control Hosp Epidemiol 20:318–323. doi: 10.1086/501624. [DOI] [PubMed] [Google Scholar]
- 14.Murray BE. 1990. The life and times of the Enterococcus. Clin Microbiol Rev 3:46–65. doi: 10.1128/CMR.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollenbeck BL, Rice LB. 2012. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 3:421–433. doi: 10.4161/viru.21282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bush LM, Calmon J, Cherney CL, Wendeler M, Pitsakis P, Poupard J, Levison ME, Johnson CC. 1989. High-level penicillin resistance among isolates of enterococci. Implications for treatment of enterococcal infections. Ann Intern Med 110:515–520. doi: 10.7326/0003-4819-110-7-515. [DOI] [PubMed] [Google Scholar]
- 17.Sapico FL, Canawati HN, Ginunas VJ, Gilmore DS, Montgomerie JZ, Tuddenham WJ, Facklam RR. 1989. Enterococci highly resistant to penicillin and ampicillin: an emerging clinical problem? J Clin Microbiol 27:2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horodniceanu T, Bougueleret L, El-Solh N, Bieth G, Delbos F. 1979. High-level, plasmid-borne resistance to gentamicin in Streptococcus faecalis subsp. zymogenes. Antimicrob Agents Chemother 16:686–689. doi: 10.1128/AAC.16.5.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huycke MM, Sahm DF, Gilmore MS. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis 4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arias CA, Murray BE. 2012. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10:266–278. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller WR, Munita JM, Arias CA. 2014. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 12:1221–1236. doi: 10.1586/14787210.2014.956092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buffie CG, Pamer EG. 2013. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Covington A, Pamer EG. 2017. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ribet D, Cossart P. 2015. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect 17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Romero S, Grompone G, Carayol N, Mounier J, Guadagnini S, Prevost MC, Sansonetti PJ, Van Nhieu GT. 2011. ATP-mediated Erk1/2 activation stimulates bacterial capture by filopodia, which precedes Shigella invasion of epithelial cells. Cell Host Microbe 9:508–519. doi: 10.1016/j.chom.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vazquez-Torres A, Jones-Carson J, Baumler AJ, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks WT, Fang FC. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 27.Drevets DA. 1999. Dissemination of Listeria monocytogenes by infected phagocytes. Infect Immun 67:3512–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson EJ, Harris JB, Morris JG Jr, Calderwood SB, Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol 7:693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper LV. 2009. Do symbiotic bacteria subvert host immunity? Nat Rev Microbiol 7:367–374. doi: 10.1038/nrmicro2114. [DOI] [PubMed] [Google Scholar]
- 30.Macpherson AJ, McCoy KD, Johansen FE, Brandtzaeg P. 2008. The immune geography of IgA induction and function. Mucosal Immunol 1:11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 31.Macpherson AJ, Uhr T. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 32.Stanley D, Mason LJ, Mackin KE, Srikhanta YN, Lyras D, Prakash MD, Nurgali K, Venegas A, Hill MD, Moore RJ, Wong CH. 2016. Translocation and dissemination of commensal bacteria in post-stroke infection. Nat Med 22:1277–1284. doi: 10.1038/nm.4194. [DOI] [PubMed] [Google Scholar]
- 33.Kirkland D, Benson A, Mirpuri J, Pifer R, Hou B, DeFranco AL, Yarovinsky F. 2012. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity 36:228–238. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fung TC, Artis D, Sonnenberg GF. 2014. Anatomical localization of commensal bacteria in immune cell homeostasis and disease. Immunol Rev 260:35–49. doi: 10.1111/imr.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristich CJ, Little JL. 2012. Mutations in the beta subunit of RNA polymerase alter intrinsic cephalosporin resistance in enterococci. Antimicrob Agents Chemother 56:2022–2027. doi: 10.1128/AAC.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartmann P, Chen P, Wang HJ, Wang L, McCole DF, Brandl K, Starkel P, Belzer C, Hellerbrand C, Tsukamoto H, Ho SB, Schnabl B. 2013. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology 58:108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Fouts DE, Starkel P, Hartmann P, Chen P, Llorente C, DePew J, Moncera K, Ho SB, Brenner DA, Hooper LV, Schnabl B. 2016. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rescigno M. 2011. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol 32:256–264. doi: 10.1016/j.it.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Bevins CL, Salzman NH. 2011. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol 9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 41.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 42.Hansson GC. 2012. Role of mucus layers in gut infection and inflammation. Curr Opin Microbiol 15:57–62. doi: 10.1016/j.mib.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. 2008. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. 2010. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis 201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. 2007. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med 204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A 105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamada N, Chen GY, Inohara N, Nunez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartemink R, Domenech VR, Rombouts FM. 1997. LAMVAB—a new selective medium for the isolation of lactobacilli from faeces. J Microbiol Methods 29:77–84. doi: 10.1016/S0167-7012(97)00025-0. [DOI] [Google Scholar]
- 51.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Frätzer C, Krannich A, Gollasch M, Grohme DA, Côrte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, Müller DN. 2017. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551:585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. 2013. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol 6:666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coombes JL, Powrie F. 2008. Dendritic cells in intestinal immune regulation. Nat Rev Immunol 8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tulstrup MV, Christensen EG, Carvalho V, Linninge C, Ahrne S, Hojberg O, Licht TR, Bahl MI. 2015. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS One 10:e0144854. doi: 10.1371/journal.pone.0144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nevado R, Forcen R, Layunta E, Murillo MD, Grasa L. 2015. Neomycin and bacitracin reduce the intestinal permeability in mice and increase the expression of some tight-junction proteins. Rev Esp Enferm Dig 107:672–676. doi: 10.17235/reed.2015.3868/2015. [DOI] [PubMed] [Google Scholar]
- 56.Hendrickx AP, Top J, Bayjanov JR, Kemperman H, Rogers MR, Paganelli FL, Bonten MJ, Willems RJ. 2015. Antibiotic-driven dysbiosis mediates intraluminal agglutination and alternative segregation of Enterococcus faecium from the intestinal epithelium. mBio 6:e01346-15. doi: 10.1128/mBio.01346-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. 2015. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol 8:198–210. doi: 10.1038/mi.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu LL, Peng WH, Kuo WT, Huang CY, Ni YH, Lu KS, Turner JR, Yu LC. 2014. Commensal bacterial endocytosis in epithelial cells is dependent on myosin light chain kinase-activated brush border fanning by interferon-gamma. Am J Pathol 184:2260–2274. doi: 10.1016/j.ajpath.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clark E, Hoare C, Tanianis-Hughes J, Carlson GL, Warhurst G. 2005. Interferon gamma induces translocation of commensal Escherichia coli across gut epithelial cells via a lipid raft-mediated process. Gastroenterology 128:1258–1267. doi: 10.1053/j.gastro.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 60.Danielsen EM, Hansen GH. 2003. Lipid rafts in epithelial brush borders: atypical membrane microdomains with specialized functions. Biochim Biophys Acta 1617:1–9. doi: 10.1016/j.bbamem.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 61.Kalischuk LD, Inglis GD, Buret AG. 2009. Campylobacter jejuni induces transcellular translocation of commensal bacteria via lipid rafts. Gut Pathog 1:2. doi: 10.1186/1757-4749-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGuckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 63.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, Ubeda C, Xavier J, Pamer EG. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun 80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jernberg C, Lofmark S, Edlund C, Jansson JK. 2007. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 65.Patel IH, Chen S, Parsonnet M, Hackman MR, Brooks MA, Konikoff J, Kaplan SA. 1981. Pharmacokinetics of ceftriaxone in humans. Antimicrob Agents Chemother 20:634–641. doi: 10.1128/AAC.20.5.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berg RD, Garlington AW. 1979. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun 23:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma L, Deitch E, Specian R, Steffen E, Berg R. 1990. Translocation of Lactobacillus murinus from the gastrointestinal tract. Curr Microbiol 20:177–184. doi: 10.1007/BF02091994. [DOI] [Google Scholar]
- 68.Kong Y, Xia Y, Nielsen JL, Nielsen PH. 2007. Structure and function of the microbial community in a full-scale enhanced biological phosphorus removal plant. Microbiology 153:4061–4073. doi: 10.1099/mic.0.2007/007245-0. [DOI] [PubMed] [Google Scholar]
- 69.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. 2008. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun 76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karlsson J, Putsep K, Chu H, Kays RJ, Bevins CL, Andersson M. 2008. Regional variations in Paneth cell antimicrobial peptide expression along the mouse intestinal tract. BMC Immunol 9:37. doi: 10.1186/1471-2172-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wehkamp J, Chu H, Shen B, Feathers RW, Kays RJ, Lee SK, Bevins CL. 2006. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett 580:5344–5350. doi: 10.1016/j.febslet.2006.08.083. [DOI] [PubMed] [Google Scholar]
- 72.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. 2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun 79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu T, Shi Y, Du J, Ge X, Teng X, Liu L, Wang E, Zhao Q. 2016. Vitamin D treatment attenuates 2,4,6-trinitrobenzene sulphonic acid (TNBS)-induced colitis but not oxazolone-induced colitis. Sci Rep 6:32889. doi: 10.1038/srep32889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hwang I, An BS, Yang H, Kang HS, Jung EM, Jeung EB. 2013. Tissue-specific expression of occludin, zona occludens-1, and junction adhesion molecule A in the duodenum, ileum, colon, kidney, liver, lung, brain, and skeletal muscle of C57BL mice. J Physiol Pharmacol 64:11–18. [PubMed] [Google Scholar]
- 75.Lim MX, Png CW, Tay CY, Teo JD, Jiao H, Lehming N, Tan KS, Zhang Y. 2014. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect Immun 82:4789–4801. doi: 10.1128/IAI.02279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alex P, Zachos NC, Nguyen T, Gonzales L, Chen TE, Conklin LS, Centola M, Li X. 2009. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis 15:341–352. doi: 10.1002/ibd.20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 78.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 82.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 84.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oksanen J. 2013. Vegan: community ecology package. R package, version 2.0-10. https://cran.r-project.org/web/packages/vegan/index.html.
- 87.Goslee SC, Urban DL. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J Stat Soft 22:1–19. doi: 10.18637/jss.v022.i07. [DOI] [Google Scholar]
- 88.R Foundation for Statistical Computing. 2014. R: a language and environment for statistical computing. http://www.r-project.org/.
- 89.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.