The MarR-like protein PapX represses the transcription of the flagellar master regulator genes flhDC in uropathogenic Escherichia coli (UPEC), the primary cause of uncomplicated urinary tract infections (UTIs). PapX is encoded by the pap operon, which also encodes the adherence factors termed P fimbriae.
KEYWORDS: UPEC, adherence, fimbriae, flagellar motility, gene regulation
ABSTRACT
The MarR-like protein PapX represses the transcription of the flagellar master regulator genes flhDC in uropathogenic Escherichia coli (UPEC), the primary cause of uncomplicated urinary tract infections (UTIs). PapX is encoded by the pap operon, which also encodes the adherence factors termed P fimbriae. Both adherence and motility are critical for productive colonization of the urinary tract. However, the mechanisms involved in coordinating the transition between adherence and motility are not well characterized. UPEC strain CFT073 carries both papX and a homolog, focX, located in the foc operon encoding F1C fimbriae. In this study, we characterized the dose effects of “X” genes on flagellar gene expression and cross talk between focX and papX. We found that both FocX and PapX repress flhD transcription. However, we determined that the ΔpapX mutant was hypermotile, while the loss of focX did not affect motility. We further investigated this phenotype and found that FocX functions as a repressor of papX. Additionally, we identified a proximal independent promoter upstream of both focX and papX and assessed the expression of focX and papX during culture in human urine and on LB agar plates compared to LB medium. Finally, we characterized the contributions of PapX and FocX to fitness in the ascending murine model of UTI and observed a subtle, but not statistically significant, fitness defect in colonization of the kidneys. Altogether, these results expand our understanding of the impact of carrying multiple X genes on the coordinated regulation of motility and adherence in UPEC.
INTRODUCTION
Escherichia coli, a common, and typically commensal, bacterial species, regularly colonizes the human gastrointestinal tract (1, 2). Some strains of E. coli, broadly referred to as extraintestinal pathogenic E. coli (ExPEC), are equipped with virulence and accessory genes that are lacking from their commensal counterparts and promote infections outside the intestine, including the urinary tract (3). Uropathogenic E. coli (UPEC) strains are the primary cause of approximately 80% of all community-acquired urinary tract infections (UTIs), which are a pervasive and costly public health burden afflicting approximately one-half of all women and one-fifth of men at least once in their lifetime (4, 5). Most UTIs are established when bacteria contaminate the periurethral area and migrate to the bladder via the urethra. While many UTIs are self-limiting and resolve within a few days, in some cases, UPEC may further ascend via the ureters and cause a more severe secondary infection in the kidneys, called pyelonephritis, which increases the risk of renal scarring, sepsis, and death (6).
Extracellular polymeric filaments called flagella promote swimming motility in bacteria and facilitate UPEC ascension of the urethra and ureters during UTIs (7, 8). While the ability to produce flagella is not required for colonization of the urinary tract, strains capable of flagellum-mediated motility persist longer within the bladder and kidneys and colonize to higher levels (9, 10). Moreover, expression of fliC, the subunit that polymerizes to form the flagellum, coincides with ascension of the urinary tract and colonization of the kidneys in the murine model of UTI (11). However, fliC is poorly expressed in UPEC strains isolated from the urine of women experiencing acute cystitis as well as during in vitro culture in human urine (12, 13). Additionally, flagella are energetically costly to produce, and monomeric FliC can be detected by Toll-like receptor 5 (TLR5) found on the uroepithelium (14, 15). Thus, expression of flagella during UTI may be detrimental for UPEC survival and contribute to the transient expression of flagellar genes observed during infection (11, 16).
During a UTI, bacteria transition between motile and adherent states through coordinated cross talk between genes encoding flagella and adherence factors, termed fimbriae (11, 17, 18). Adherence of bacteria to host cells is critical for colonization of the urinary tract, and UPEC isolates are more likely to encode fimbriae that bind to cells found within the urinary tract (19–21). UPEC strain CFT073 encodes 12 fimbriae, including F1C and two separate P fimbriae (22). F1C fimbriae, encoded by the foc operon, bind glycolipids found on the kidney epithelium and endothelium, and cystitis UPEC isolates were more likely than fecal E. coli isolates to encode F1C fimbriae (23–25). Similarly, pyelonephritis-associated pili (Pap), or P fimbriae, bind the P blood group antigen enriched on human kidney epithelial cells and erythrocytes, and UPEC strains harboring the pap operon are more likely to cause pyelonephritis (26, 27).
Differing from most fimbrial operons, only one of the two pap operons and the single foc operon carry a 3′-terminal gene encoding a MarR-like transcription factor, PapX or FocX, respectively. MarR-like proteins share a winged helix-turn-helix structure (wHTH), bind as dimers to palindromic DNA sequences, and have been shown to mediate the regulation of numerous genes, including those involved in resistance to antibiotics, oxidative stress, and low pH as well as motility (28–31). We have previously shown that PapX binds to a 29-bp palindromic DNA sequence centered 410 bp upstream of the flhDC translational start site, encoding the master transcriptional regulator FlhD4C2 of flagellar gene expression. Overproduction of PapX represses the transcription of flhD and subsequently reduces swimming motility (17, 32, 33). Consistent with our findings in CFT073, papX was also identified in a transposon directed insertion sequencing (TraDIS) screen for genes affecting motility in E. coli strain EC958 (34). Thus, PapX is directly involved in regulatory cross talk between genes associated with adherence and motility.
FocX shares 96.7% amino acid sequence identity with PapX and therefore is predicted to also function as a repressor of motility. However, the function of FocX in UPEC has not been well characterized, and the impact of encoding multiple homologous “X” proteins on motility in UPEC is poorly understood. In this study, we found that both PapX and FocX repress flhD expression as well as swimming motility when overproduced. However, we have discovered that FocX can also repress the expression of papX and that cross talk between “X” genes affects motility. Additionally, we characterized a proximal promoter for focX and papX, suggesting that focX and papX can be expressed independently from their respective fimbrial operons. However, we found that the expression of papX positively correlated with pap expression during in vitro culture on LB agar plates. Furthermore, we assessed the relative fitness of either the single ΔpapX or the double ΔfocX ΔpapX mutant compared to the wild type by in vivo competitive cochallenge in CBA/J mice and observed a slight, but not statistically significant, decrease in kidney colonization. Since UPEC isolates are more likely than commensal strains to carry at least two X genes, investigating the interactions between PapX and FocX is imperative to understand how UPEC regulates motility and responds to environmental signals within the urinary tract.
RESULTS
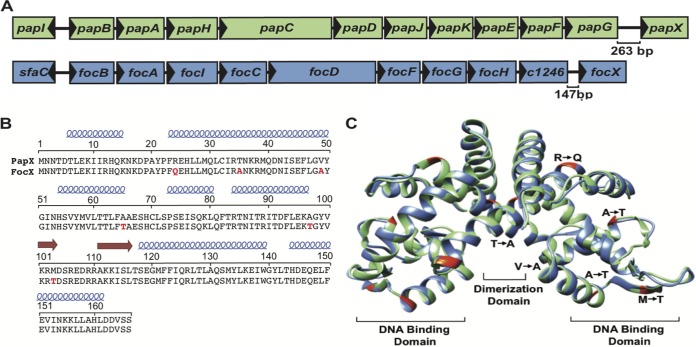
PapX and FocX share high sequence and structural similarities.
The E. coli CFT073 genome harbors both the pap and foc operons, which encode the homologous MarR-like proteins PapX and FocX, respectively (Fig. 1A) (22). The predicted structures of PapX and FocX were created by using I-TASSER and were modeled as dimers based on the solved dimer structure of the MarR-like protein HucR (35). While MarR-like proteins contain a conserved wHTH DNA binding motif, the majority of these proteins share limited amino acid sequence identity (∼25 to 35%) (36). Despite being encoded by different fimbrial operons, PapX and FocX share high amino acid sequence identity (96.7%) (Fig. 1B) and predicted structural homology (Fig. 1C) (32). In MarR-like proteins, key structural motifs include the dimerization domain between subunits, the DNA recognition helices that bind palindromic DNA sequences within the major groove, and the wing domain that interacts with residues within the DNA minor groove (37, 38). Therefore, amino acid changes within these structural domains are more likely to alter DNA binding-site recognition and protein function. There are three amino acid differences within key structural areas between PapX and FocX: T35A within the dimerization domain, A97T within the DNA binding helix, and M103T within the wing domain. Therefore, while we predict that PapX and FocX share the same function based on their high structural and amino acid sequence similarities, it is not known if these substitutions affect protein function.
FIG 1.
PapX and FocX share high sequence and structural homology. (A) Schematic of the pap1 and foc operons in CFT073. (B) The amino acid sequences of PapX and FocX were manually aligned, and residues in red signify differences in FocX compared to PapX. The predicted location of α-helices (blue) and β-sheets (red arrows) are shown above the amino acid sequence. (C) The predicted dimer structures of PapX (green) and FocX (blue) were created by using I-TASSER and aligned to the known structure of the dimer HucR by using Chimera (not shown). Amino acid differences between FocX and PapX are highlighted in red and labeled on one monomer.
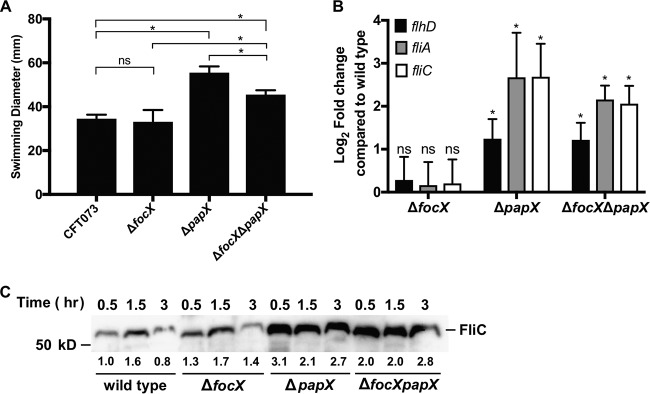
Loss of papX, but not focX, increases swimming motility.
To determine the effects of PapX and FocX on motility, we measured the motility of the CFT073 wild-type, ΔfocX mutant, ΔpapX mutant, and ΔfocX ΔpapX double mutant strains using a swimming motility assay. We observed that the loss of focX did not affect swimming motility (Fig. 2A). In contrast, the loss of papX resulted in a hypermotile phenotype, and this result was consistent with previous studies showing that PapX represses motility (17, 32, 33, 39). Interestingly, the loss of both focX and papX resulted in an intermediate level of swimming that was significantly greater than that of the wild type but not as robust as that of the ΔpapX mutant, suggesting that there may be cross talk between focX and papX.
FIG 2.
Effects of focX and papX expression on swimming motility. (A) Swimming diameter (millimeters) was measured for CFT073 and isogenic mutants after 16 to 18 h of incubation at 30°C. Data represent results for five biological replicates, with the error bars showing the standard deviations. Tukey’s multiple-comparison test following ANOVA was used for statistical analysis. *, P < 0.05; ns, not significant. (B) qPCR of flagellar genes flhD, fliA, and fliC using cDNA collected from wild-type E. coli CFT073 and ΔfocX, ΔpapX, and ΔfocX ΔpapX constructs cultured in tryptone medium until the OD600 reached 0.3. Data represent the averages of results from three experiments. Standard deviations are shown, and Student’s t test was used for statistical analysis. *, P < 0.05. No statistical difference was found between the ΔpapX and ΔpapX ΔfocX strains using a Mann-Whitney test. (C) Immunoblotting to detect FliC levels from whole-cell lysates of wild-type CFT073 and the ΔfocX, ΔpapX, and ΔpapX ΔfocX constructs cultured in tryptone broth. Relative quantification of FliC (shown below the protein band) was obtained by using Image Lab 5.2.1 and represents the average fold change from two independent experiments compared to FliC levels of the wild type at 0.5 h and normalized to a conserved nonspecific band.
E. coli strains differ in their capacities for motility due in part to heterogeneity in the presence of insertion sequence (IS) elements upstream of flhDC as well as variability in the encoded transcriptional regulators of flagellar genes (40–42). Therefore, to compare the functions of PapX in additional UPEC strains, we compared motility between the wild type and a ΔpapX mutant in UPEC cystitis isolates F11 and HM69 (see Fig. S1 in the supplemental material). Both strains F11 and HM69 carry the pap operon, including papX, but not the foc operon. We observed that the deletion of papX resulted in a significant increase in motility compared to the wild type in F11 (121%) and HM69 (117%); however, the hypermotile phenotypes were not as robust as what was observed in CFT073 (161%) (Fig. 2A). Therefore, these data support that PapX functions through a conserved mechanism in UPEC but that the impact of PapX on motility is dependent on the strain background.
We have previously shown that PapX inhibits swimming motility by repressing the expression of flhDC, resulting in the downregulation of additional flagellar genes (32, 33, 39). To verify that the observed motility phenotypes correspond to changes in the expression of flagellar genes, we performed quantitative PCR (qPCR) to quantify the changes in the mRNA abundances of flhD, fliA, and fliC between CFT073 and the ΔfocX, ΔpapX, and ΔfocX ΔpapX constructs. RNA was collected from bacteria cultured in tryptone medium to an optical density at 600 nm (OD600) of 0.3. Growth conditions were chosen based on previous work demonstrating that the production of flagella in CFT073 is elevated during early-logarithmic-phase growth in tryptone medium (18). Consistent with our swimming results, the loss of focX did not alter the expression of flhD, fliA, or fliC (Fig. 2B). However, we observed a significant increase in the expression of flhD, fliA, and fliC in both the ΔpapX (log2 fold change [FC], 1.2 to 2.7) and the double ΔfocX ΔpapX (log2 FC, 1.2 to 2.1) mutants compared to the wild type.
To determine if the qPCR results correlated with an increase in flagellum production, we performed immunoblotting to compare FliC (flagellin) levels between wild-type CFT073 and the ΔfocX, ΔpapX, and ΔfocX ΔpapX constructs. Bacteria were cultured in tryptone medium, and the levels of FliC were assessed from normalized whole-cell lysates collected at 0.5, 1.5, and 3 h, representing early-, mid-, and late-logarithmic-phase growth (Fig. S2). In wild-type CFT073, we observed a peak in FliC production at 1.5 h, and this result was consistent with data from previous studies characterizing the temporal production of FliC (18) (Fig. 2C). Additionally, we observed increases in FliC production in the ΔpapX and ΔfocX ΔpapX constructs at all observed time points, which were quantified by densitometry based on the averages of data from two replicates. We did not detect an increase in FliC production in the ΔfocX construct compared to the wild type. Therefore, these results show that the deletion of papX, compared to focX, has a great impact on swimming motility, flagellar gene expression, and FliC production in CFT073.
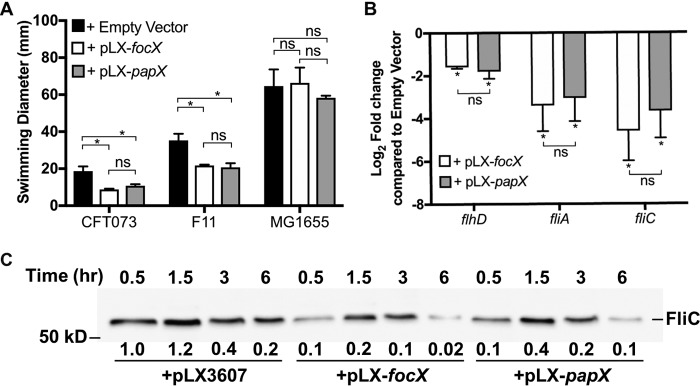
Both PapX and FocX function as repressors of motility.
Based on the amino acid sequence and structural similarities between PapX and FocX, we predicted them to share the same function as transcriptional repressors of motility. However, compared to the wild type, the deletion of focX did not significantly increase swimming motility. Therefore, to assess PapX and FocX function independent of their native expression, we overexpressed papX and focX in CFT073 and performed a swimming motility assay (Fig. 3A). Compared to CFT073 carrying the empty vector pLX3607, expression of either papX (pLX-papX) or focX (pLX-focX) resulted in approximately a 50% reduction in swimming motility. In addition, complementation of focX and papX in the ΔfocX, ΔpapX, and ΔfocX ΔpapX mutants resulted in comparable levels of inhibition of motility (see Fig. S3 in the supplemental material). To assess whether this phenotype was specific to CFT073, we repeated this assay in the cystitis isolate F11 and observed a similar decrease in motility following the overproduction of either FocX or PapX. We have previously shown that the commensal E. coli K-12 MG1655 strain lacks the PapX binding site upstream of flhDC, and therefore, the overproduction of PapX does not reduce motility (32). To determine if the function of FocX also depends on the presence of the PapX binding site upstream of flhDC, we assessed the swimming motility of K-12 MG1655 carrying pLX3607, pLX-papX, or pLX-focX. We observed that the expression of either papX or focX did not decrease motility compared to the empty vector. Therefore, these data support that PapX and FocX share the same function as repressors of motility and that this mechanism is dependent on the presence of the PapX binding site upstream of flhDC.
FIG 3.
Expression of either focX or papX represses flagellar gene expression and motility. (A) Bars representing the average diameters (millimeters) of swimming motility of bacteria following 16 to 18 h of incubation at 30°C. Data represent results of five biological replicates, with the error bars showing the standard deviations. Tukey’s multiple-comparison test following ANOVA was used for statistical analysis. *, P < 0.05; ns, not significant. (B) qPCR of the flagellar genes flhD, fliA, and fliC using cDNA synthesized from mRNA collected from E. coli CFT073 ΔpapX ΔfocX carrying pLX3607, pLX-focX, or pLX-papX cultured in tryptone medium until the OD600 reached 0.3. Data represent the averages of results from three experiments with standard deviations. Student’s t test was used for statistical analysis. (C) Immunoblotting to detect FliC levels from whole-cell lysates of wild-type CFT073 carrying pLX3607, pLX-focX, or pLX-papX. Relative quantification of FliC was obtained by using Image Lab 5.2.1 and represents the average fold change from two independent experiments compared to FliC levels of the ΔpapX ΔfocX/pLX3607 strain at 0.5 h and normalized to a conserved nonspecific band.
To determine if the reduction in motility following the overexpression of focX or papX was a result of decreased expression of flagellar genes, we used qPCR to assess the gene expression levels of flhD, fliA, and fliC (class I, class II, and class III, respectively) in the ΔfocX ΔpapX double mutant carrying pLX3607, pLX-papX, or pLX-focX. We assessed flagellar gene expression in the double mutant background, versus the wild type, to eliminate any interference due to native levels of PapX and FocX. RNA was collected from bacteria cultured to an OD600 of 0.3 in tryptone medium. We found that the expression of either papX or focX, compared to the empty vector, resulted in a comparable decrease in the transcription of flhD, fliA, and fliC (Fig. 3B). To verify that our qPCR data correlated with a decrease in flagellin production, we performed immunoblotting for FliC using the ΔfocX ΔpapX mutant carrying pLX3607, pLX-focX, or pLX-papX. Bacteria were cultured in tryptone medium, and whole-cell lysates were collected at 0.5, 1.5, 3, and 6 h and normalized by the OD600. We observed that the overexpression of either focX or papX resulted in a decrease in FliC production at all observed time points compared to the empty vector (Fig. 3C). These data support that both FocX and PapX, when ectopically expressed, can repress flhD, resulting in decreased flagellar production and motility.
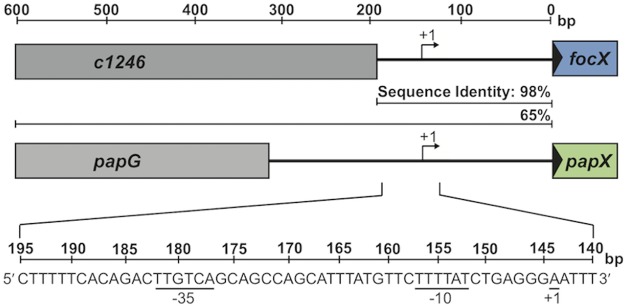
focX and papX are transcribed from a proximal promoter.
We have previously shown that papX is transcribed as part of the pap operon and have confirmed that focX is also transcribed as part of the foc operon (see Fig. S4 in the supplemental material) (33). However, the papX homolog sfaX is transcribed as part of the sfa operon as well as from an independent proximal promoter (43). Since the upstream DNA sequences of papX, focX, and sfaX share high sequence identity, papX and focX likely share a similar proximal promoter. We used 5′ rapid amplification of cDNA ends (RACE) to map the transcriptional start sites of both papX and focX to an adenosine residue located 144 bp upstream of their predicted ATG start codon (Fig. 4). We manually identified putative −10 and −35 regions separated by a 19-bp spacer that resemble the consensus sequence of a bacterial σ70 promoter. This site was shared between papX and focX but was located 44 bp upstream of the transcriptional start site identified for sfaX (43). The presence of an additional proximal promoter upstream of focX and papX suggests that both these genes can be transcribed independently and from their associated fimbrial operons.
FIG 4.
focX and papX are transcribed from a proximal promoter. Shown is a schematic of the transcriptional start sites of focX and papX identified by using 5′ RACE, followed by DNA sequencing. cDNA was synthesized from mRNA collected from either the ΔpapX or ΔfocX strain. A reference horizontal line measuring the DNA sequence length (base pairs) is included at the top. The transcriptional start site (+1) of either focX or papX is located 144 bp upstream of the initial ATG site of each gene. Putative −10 and −35 sites with similarity to the bacterial σ70 promoter are underlined. The DNA sequence identities (percent) of the upstream DNA sequences between papX and focX in CFT073 are shown for two DNA regions (200 bp and 600 bp) upstream of the ATG start site of focX or papX.
Expression of papX or focX trends with the expression of the preceding fimbrial operon.
Since focX and papX are also transcribed from an independent proximal promoter, we investigated the expression of focX and papX compared to the foc and pap operons, respectively, under different environmental conditions. Previous work by Hancock et al. determined that the pap and foc genes are upregulated in CFT073 during planktonic growth in human urine compared to morpholinepropanesulfonic acid (MOPS), but the expression of focX or papX was not investigated (44). Therefore, we used qPCR to quantify the expression of papA, papX, focA, focX, and fliC in wild-type CFT073 during culture to the mid-logarithmic growth phase in pooled human urine compared to LB medium. We included fliC to assess correlations of focX and papX with motility under these growth conditions. We found that culture in human urine compared to LB medium led to significant increases in the transcription of focA (log2 FC, 2.8) and papX (log2 FC, 1.3), but we did not detect a statistically significant change in focX or papA (Fig. 5A). Additionally, we observed a decrease in fliC expression (log2 FC, −5.48), and this finding was consistent with previous work showing decreased motility in UPEC strains cultured in human urine (13).
FIG 5.
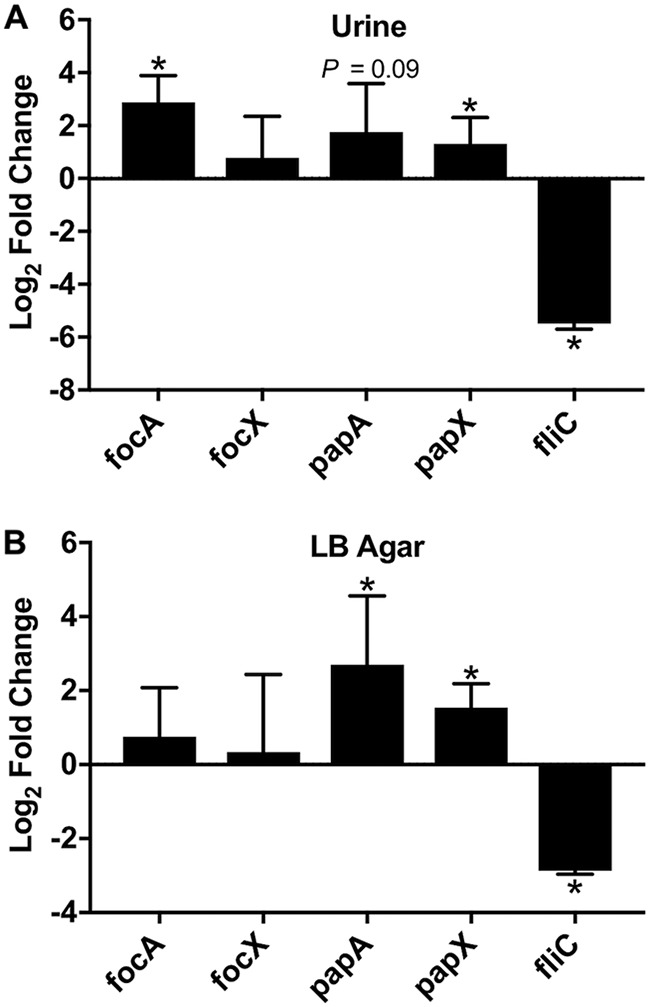
Expression of foc, pap, and fliC in strains grown in human urine or on LB agar. Shown are data from qPCR of focA, focX, papA, papX, and fliC using cDNA collected from wild-type E. coli CFT073 cultured in human urine until an OD600 of 0.3 was reached (A) or on LB agar for 24 h compared to cDNA collected from bacteria cultured to the mid-logarithmic growth phase in LB medium (B). Data represent the averages of results from three experiments, with error bars showing standard deviations. Student’s t test was used to calculate statistical significance. *, P < 0.05.
Previous studies have shown that the production of P fimbriae is elevated in the E. coli CFT073 strain cultured on LB agar compared to planktonic growth (45). Therefore, we assessed the expression of papA, papX, focA, focX, and fliC by qPCR following a 24-h incubation on LB agar plates at 37°C compared to culture in LB medium to the mid-logarithmic growth phase. Consistent with previous findings, we observed that the expression levels of papA (log2 FC, 2.7) and papX (log2 FC, 1.5) were significantly increased, but we did not observe a change in the expression of focA and focX (Fig. 5B). Additionally, fliC expression was decreased (log2 FC, −2.8) following incubation on LB agar plates. Overall, the relative expression levels of focX and papX were lower than that of the reference gene gapA under all three tested growth conditions (see Fig. S5 in the supplemental material).
FocX functions as a repressor of papX.
We have previously shown that the deletion of papX in CFT073 does not affect the production of P fimbriae; however, the effect of FocX on the expression of genes within the foc operon has not been characterized (33). Therefore, we performed qPCR to assess the changes in the gene expression levels of focA, focX, papA, and papX following the deletion of either focX or papX. cDNA was synthesized from mRNA collected from wild-type CFT073 and the ΔfocX and ΔpapX constructs cultured to the mid-logarithmic growth phase in tryptone medium. We found that the deletion of focX did not affect focA or papA expression but resulted in higher expression levels of papX (log2 FC, 2.06) (Fig. 6A). In contrast, the deletion of papX did not result in any significant changes in the gene expression of focA, focX, or papA. These data suggest that FocX functions as a repressor of papX and that the regulatory mechanism is not a result of decreased expression of the pap operon. Therefore, the regulation of papX may be occurring at the proximal papX promoter. However, we did not identify a binding site upstream of either focX or papX that matched the characterized PapX binding site upstream of flhDC, suggesting that regulation of papX may be indirect or occurs through a degenerate DNA binding site (32).
FIG 6.
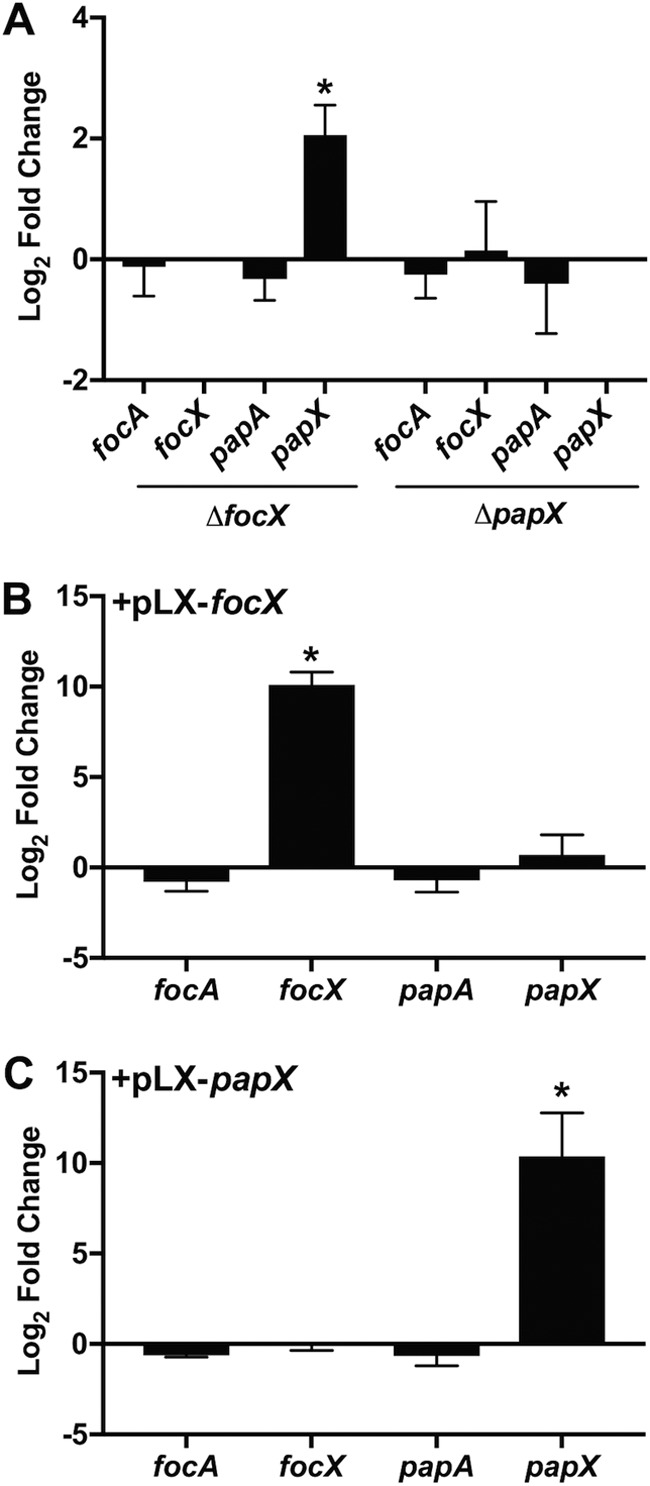
Loss of focX results in elevated papX expression. qPCR was performed to calculate the average (n = 3) log2 fold changes of mRNA abundances of focA, focX, papA, and papX using cDNA collected from the E. coli CFT073 wild-type, ΔfocX, or ΔpapX strain (A) or the wild type carrying pLX3607, pLX-focX (B), or pLX-papX (C) cultured in tryptone medium until an OD600 of 0.3 was reached. Errors are shown as standard deviations, and statistical significance was calculated using Student’s t test. *, P < 0.05.
We also assessed the changes in the gene expression levels of focA, focX, papA, and papX in CFT073 when either focX or papX was ectopically expressed. cDNA was synthesized from mRNA collected from CFT073 harboring pLX3607, pLX-focX, or pLX-papX cultured to the mid-logarithmic growth phase in tryptone medium. We found that the production of FocX did not affect the expression of focA, papA, or papX (Fig. 6B). Similarly, the production of PapX did not impact the expression of focA, focX, or papA. Therefore, the function of FocX as a repressor of papX may be dependent on the protein concentration, as we did not observe a decrease in papX expression in response to high levels of FocX.
papX provides a subtle fitness advantage in colonization of the kidneys during murine UTI.
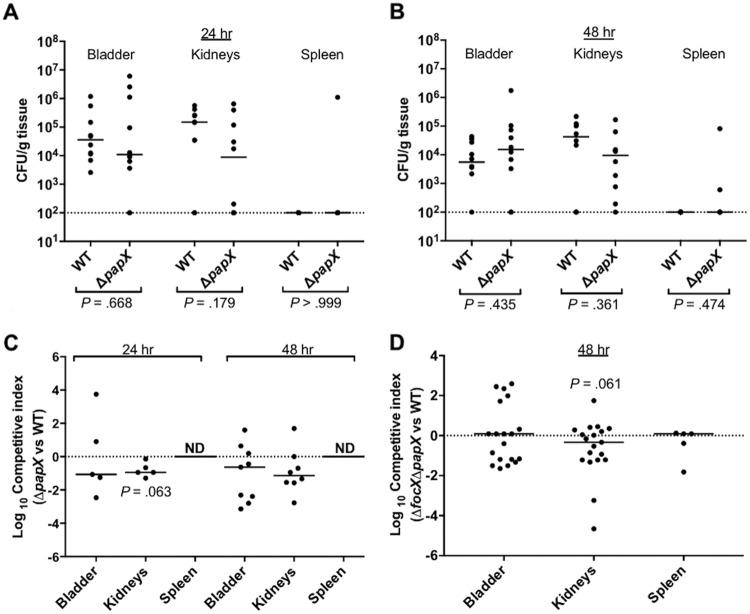
While P fimbriae have been shown to promote colonization of the kidneys during murine UTI, papX has not been confirmed as a fitness factor during in vivo infection (46, 47). Instead, PapX has been hypothesized to negatively impact colonization during UTI, as a ΔpapX mutant showed a slight increase in kidney colonization compared to wild-type CFT073 during an experimental cochallenge in transurethrally inoculated CBA/J mice (33). However, the contribution of PapX during an ascending UTI may be more observable following intraurethral inoculation, since transurethral inoculation places a high number of bacteria directly into the bladder and increases the occurrence of vesicoureteral reflux into the kidneys in the murine model (48). Also, previous work by Lane et al. demonstrated that flagella are a fitness factor for colonization of the bladder and kidneys through independent infections by wild-type CFT073 and a ΔfliC mutant following intraurethral inoculation in CBA/J mice (9, 11). Therefore, to assess the contribution of papX to in vivo colonization, we performed an independent infection of CBA/J mice intraurethrally inoculated with either wild-type CFT073 or the ΔpapX mutant. Mice were sacrificed at 24 or 48 h, and the bladder, kidneys, and spleen were homogenized and plated to enumerate the bacterial load. We did not observe any significant differences between the wild type and the ΔpapX mutant in the colonization of the bladder, kidneys, or spleen after either 24 or 48 h (Fig. 7A and B). While we observed high levels of colonization of three spleens infected with the ΔpapX mutant, we did not have sufficient statistical power to make any conclusions about the role of papX in dissemination.
FIG 7.
The contribution of PapX to in vivo colonization is subtle and observable only following intraurethral inoculation. (A and B) Independent challenges of CBA/J mice (n = 10) inoculated with a 10-μl suspension of 107 CFU/ml of either wild-type (WT) CFT073 or the ΔpapX mutant directly into the urethra. Mice were sacrificed at either 24 h (A) or 48 h (B) postinoculation, and the bladders, kidneys, and spleens were homogenized and plated to enumerate the bacterial load and are presented as CFU per gram of tissue. Statistical differences in colonization were determined by using a Mann-Whitney test. (C) Cochallenge infection of mice (n = 10) using a 1:1 mixture of the wild type and the ΔpapX mutant. CBA/J mice were intraurethrally inoculated with a 10-μl suspension of a total of 107 CFU/ml. Mice were sacrificed at either 24 h (n = 5) or 48 h (n = 10) postinoculation, and the bladders, kidneys, and spleens were homogenized and plated to enumerate the bacterial load. Relative fitness is presented as a competitive index. (D) Cochallenge infection of mice (n = 20) using a 1:1 mixture of the wild type and the ΔfocX ΔpapX mutant. Mice were transurethrally inoculated with a 50-μl suspension of a total of 108 CFU/ml. Relative fitness after 48 h postinoculation is presented as a competitive index. Horizontal bars represent the median values for the populations, and the dotted line represents the limit of detection (2 × 102 CFU/g). Statistical differences for cochallenge infections were determined by using a Wilcoxon signed-rank test. ND, not determined.
In some cases, a cochallenge model has been shown to be more sensitive than independent challenge for the detection of subtle fitness defects during UTI (49). Thus, to measure relative fitness, CBA/J mice were intraurethrally inoculated with a 1:1 ratio of wild-type CFT073 and the ΔpapX mutant. We did not observe any statistically significant differences between the wild type and the ΔpapX mutant in colonization of the bladder, kidneys, or spleen after either 24 or 48 h (Fig. 7C). However, the ΔpapX mutant tended to have a decrease in colonization of the kidneys after 24 h (P = 0.063).
It is possible that FocX can compensate for PapX during UTI and thereby mask fitness defects of the ΔpapX mutant during infection. Therefore, we performed a cochallenge infection in CBA/J mice transurethrally inoculated with a 1:1 ratio of wild-type CFT073 and the ΔfocX ΔpapX double mutant. Mice were sacrificed at 48 h, and the bladder, kidneys, and spleen were homogenized and plated to enumerate the bacterial load. We did not observe any fitness defect in the colonization of the bladder or spleen, but we observed a subtle, but not statistically significant, decrease in colonization of the kidneys (P = 0.061) (Fig. 7D).
DISCUSSION
UPEC strains rely on flagella and fimbriae to successfully ascend and colonize the diverse niches within the urinary tract (7, 10, 50). However, it is not practical for a bacterium to be both motile and adherent simultaneously. Thus, UPEC isolates likely possess mechanisms that mediate a rapid transition between adherent and motile states in response to environmental signals during a UTI. One such mechanism is through the MarR-like protein PapX, which is encoded as part of the pap operon and functions as a transcriptional repressor of the flagellar master regulator genes flhDC (17, 32, 33). PapX is one member of a subset of 17-kDa MarR-like proteins encoded within fimbrial operons that have been identified in E. coli, and the presence of UPEC strains harboring multiple MarR-like “X” proteins raises the notion that these proteins may function to cooperatively regulate motility (32). However, another “X” gene, vatX, has also been identified in strain CFT073 (51). While VatX shares 44% amino acid identity with PapX, neither the deletion nor the overexpression of vatX impacted the motility of CFT073 (52). Hence, the study of PapX and its homologs is central to our understanding of UPEC ascension and colonization of the urinary tract.
In this study, we investigated the regulation of focX and papX gene expression and characterized the effect of encoding multiple PapX homologs on motility. We found that ectopic expression of focX or papX resulted in comparable inhibition of motility that corresponded with decreased flagellar gene expression (flhD, fliA, and fliC) and reduced flagellin (FliC) production. Therefore, FocX and PapX share the same function as repressors of motility. However, in E. coli, a single bacterium typically expresses one dominant fimbrial type at one time, and since focX and papX are located within different fimbrial operons, they are likely expressed in an individual bacterium at different times (53, 54). Therefore, during infection, UPEC strains encoding multiple X proteins may be better suited to rapidly repress motility in response to a range of niche-specific cues. Since both P and F1C fimbriae mediate adherence to renal cells, the function of PapX and FocX may be particularly important during kidney colonization (25, 27). We predicted that the production of PapX would improve the success of kidney colonization since the loss of motility improves bacterial attachment. Also, reducing flagellum production likely allows bacteria to better evade host defenses, as the monomeric flagellin FliC is recognized by TLR5 found on host epithelial cells. The activation of TLR5 can lead to the rapid recruitment of neutrophils, inflammation, and, subsequently, bacterial clearance (14, 55). However, we observed during cochallenge in the murine model of ascending UTI that an isogenic ΔpapX mutant had only a slight decrease in colonization of the kidneys (P = 0.063). While we predict that FocX shares the same function as PapX, deletion of both X genes did not substantially affect kidney colonization (P = 0.061). Evaluation of the relative fitness of the ΔfocX ΔpapX double mutant instead by intraurethral inoculation may demonstrate a greater effect on colonization. Since there are few studies in the literature exploring the effects of hypermotility on kidney colonization during UTI, further investigation of the function of X genes during infection would broaden our understanding of the role of motility in the development of pyelonephritis.
In this study, we also identified a shared transcriptional start site located 144 bp upstream of the ATG start codon of both focX and papX, suggesting that focX and papX can also be regulated independently of their fimbrial operon. A similar transcriptional schema has also been observed for the papX homolog sfaX, which is transcribed from an independent promoter in addition to being expressed by the sfa operon, encoding S fimbriae (43). Thus, dual transcription from two promoters appears to be a conserved regulatory feature of MarR-like proteins encoded within fimbrial operons. In the case of papX and focX, we found that, in general, the expression of focX and papX mimicked the expression of their preceding fimbrial operons during in vitro culture on LB agar plates, as elevated expression of papA was paralleled by an increase in papX expression. Overall, the regulation of X genes from an independent promoter may allow for control over flagellar gene expression while independently preserving the appropriate regulation of fimbria production, allowing for fine-tuned transitions in the regulation of adherence and motility factors during ascension from the bladder to the kidneys.
Additionally, an independent proximal promoter may also serve as a site for cross talk between X genes or other transcription factors. Genes encoding MarR-like proteins are commonly autoregulated, where increased protein production creates a negative-feedback loop that inhibits transcription (56, 57). Since FocX and PapX share the same function, we predict that PapX is capable of autoregulation; however, future studies are needed to confirm this regulatory mechanism. Also, we observed that FocX functions as a repressor of papX, and this regulation likely occurs at the proximal independent promoter of papX, as we did not observe any changes in the expression of the rest of the pap operon. However, the overexpression of focX had no observable effect on papX expression. It is possible that the FocX protein levels present in CFT073/pLX3607 are sufficient for complete repression of papX. Thus, increasing the protein concentration of FocX (CFT073/pLX-focX) may not translate into a more robust decrease in papX transcription. Also, FocX binding may compete with a transcriptional activator of papX. For example, the MarR-like protein SlyA promotes gene expression by antagonizing the binding of the nucleoid protein H-NS (58, 59). While there is limited information on the regulators of papX, sfaX is downregulated by the nucleoid protein H-NS (43). The role of H-NS as a regulator of papX has not been well characterized. Interestingly, the transcriptional start site of sfaX was located 44 bp downstream from the start site identified for focX and papX, which may allow H-NS to serve different roles in regulating X genes. For example, H-NS, or another transcription factor, may instead act as a transcriptional activator of papX. Therefore, the absence of focX may allow for the recruitment of a transcriptional activator of papX, while the overproduction of FocX does not directly induce the repression of papX.
In contrast, we did not observe that PapX repressed the expression of focX, and it is not clear if a required regulatory element is absent from the focX promoter or if expression would be better observed under a different condition. In general, cross talk between focX and papX may serve as a mechanism to limit the negative consequences of extended repression of motility due to excessive X protein production. Also, maintenance of X protein levels appears to be critical for the mechanism behind the repression of motility by PapX and FocX, as we observed in our swimming motility assays that the deletion of focX had no effect on motility, while the ΔfocX ΔpapX double mutant had an intermediate motility phenotype compared to the hypermotile ΔpapX mutant. We propose that increased levels of PapX in the focX mutant may compensate for the loss of focX and thus result in a degree of motility similar to that of the wild type. However, in the double mutant, it may be that the loss of both focX and papX affects the recruitment of a transcriptional activator of flhD or allows another regulator of flhDC expression to partially compensate for the absence of X proteins, resulting in an intermediate motility phenotype. The mechanism of PapX and FocX regulation of flhDC is not clearly defined. The PapX binding site upstream of flhDC does not overlap binding sites of other characterized transcription factors, and the impact of PapX binding within the flhDC promoter on downstream access to DNA binding sites for other regulators of flhDC is not well characterized (32, 60, 61).
In relation to the flhDC promoter, the PapX binding site is centered 220 bp upstream of the flhDC transcriptional start site (32, 40). There are numerous transcription factors that have been linked either directly or indirectly to the regulation of flhDC expression. For example, the nucleoid-associated protein H-NS promotes flhDC expression in part through binding to sites within the flhDC promoter as well as repressing the expression of hdfR, encoding a transcriptional repressor of flhDC (62, 63). Additionally, multiple insertion sequence (IS) elements have been found to integrate into the flhDC promoter and affect transcription (42). Therefore, a possible molecular mechanism is that the binding of PapX or FocX affects the accessibility of DNA binding sites within the flhDC promoter for other regulatory elements, including other transcription factors and IS elements. Indeed, the IS5 element in the E. coli K-12 MG1655 strain disrupts the PapX binding site and likely explains why ectopic expression of papX in this strain does not impact motility. Thus, preservation of the PapX binding site in the flhDC promoter may be more strongly conserved in E. coli strains carrying P, S, or F1C fimbriae than in commensal E. coli isolates. The complex interplay of regulators of flhDC expression emphasizes the lengths that bacteria employ to control motility, and in the context of UTIs, MarR-like X proteins may provide a mechanism to mediate fine-tuned coordinated transitions between motility and adherence allowing for better evasion of the host immune system and improved colonization of the upper urinary tract.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in this study are listed in Table S1 in the supplemental material. Unless otherwise noted, bacteria were cultured in LB medium (10 g tryptone, 5 g yeast extract, 0.5 g NaCl/liter) at 37°C with aeration. UPEC strain CFT073 was isolated from the blood and urine of a patient hospitalized with acute pyelonephritis, and strains F11 and HM69 were isolated from the urine of women experiencing cystitis (64–66). Human urine was collected from at least three women, pooled, filter sterilized, and stored at −20°C in accordance with the recommendations of the University of Michigan Institutional Review Board (approval HUM00004949). When necessary, the following antibiotics were added: ampicillin (100 μg/ml), kanamycin (25 μg/ml), and chloramphenicol (20 μg/ml).
Mutant and plasmid construction.
The primers used in our study are listed in Table S2 in the supplemental material. The ΔpapX mutant was constructed as described previously (39). Lambda red-mediated recombineering was used to construct the ΔfocX and ΔfocX ΔpapX mutants. In brief, the chloramphenicol resistance gene (cat) was amplified from pKD4 by PCR using EasyA polymerase (Agilent) and primers KO-F and KO-R and then transformed into competent CFT073 cells carrying pKD46 harboring the lambda red operon. Transformed bacteria were cultured at 30°C for 2.5 h, plated on LB agar containing chloramphenicol, and cultured overnight at 37°C (67). The resulting colonies were screened by PCR using Taq polymerase (New England BioLabs) and primers scrnF and scrnR for the deletion of focX. The same overall strategy was used to construct the ΔfocX mutants in E. coli F11 and HM69, but the kanamycin resistance gene (kan) was instead amplified from pKD3 for transformation.
To construct the ΔfocX ΔpapX double mutant, primers KO-F and KO-R, which have homology to both papX and focX, were used with EasyA polymerase to PCR amplify the cat gene from pKD3. The resulting PCR product was transformed into competent CFT073 cells carrying pKD46. Colonies were screened by PCR using Taq polymerase and primers scrnF and scrnR to verify the deletion of either focX or papX. To remove the second X gene, the kanamycin resistance gene was PCR amplified from pKD4 using primers KO-F2 and KO-R2, which flank the first set of primers, and transformed into the competent single mutant strain carrying pKD46. Primers scrnF2 and scrnR2 were used with Taq polymerase to screen for the deletion of the second X gene, and the deletion of both focX and papX was verified by DNA sequencing.
To induce the expression of papX, we previously cloned papX into the vector pLX3607 under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter, referred to as pLX-papX (68). A similar strategy was used to control the expression of focX. In brief, focX was amplified from CFT073 ΔpapX by PCR using EasyA polymerase and primers pLXfocX-F and pLXfocX-R. Both the resulting PCR product and pLX3607 were digested by NcoI and HindIII, ligated together using T4 DNA ligase (New England BioLabs) to generate pLX-focX, and transformed into E. coli Top10 cells (Invitrogen). Transformants were plated on LB medium with ampicillin, and the resulting colonies were screened by PCR using primers scrn_pLX-F and scrn_pLX-R to verify plasmid construction. pLX-focX was obtained by using a miniprep kit (Qiagen) from bacterial cultures grown overnight and transformed into CFT073. We utilized “leaky” expression from the Plac promoter in the absence of the IPTG inducer to assess the ectopic expression of papX or focX.
Swimming motility assay.
Stationary cultures grown overnight were diluted to an OD600 of 1.0 and stab inoculated into 0.025% agar motility plates (10 g tryptone, 5 g NaCl, 1.25 g agar/liter). Plates were incubated for 16 to 18 h at 30°C, and the diameter of bacterial spread was averaged as a quantification of swimming motility. Tukey’s multiple-comparison test following analysis of variance (ANOVA) was used for statistical analysis, with the error bars representing the standard deviations.
qPCR.
Stationary cultures were diluted 1:100 into 25 ml of either LB medium, tryptone medium (10 g tryptone, 5 g NaCl/liter), or sterilized pooled human urine with ampicillin when needed for plasmid maintenance. Strains were cultured at 37°C with aeration until the mid-logarithmic growth phase was reached, and an aliquot was then collected for RNA extraction and treated with a stop solution (95% ethanol [EtOH], 5% phenol) to preserve RNA stability. RNA was also collected from bacteria that were plated on LB agar and incubated at 37°C for 24 h. Plated bacteria were resuspended in 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4/liter [pH 7.4]) and treated with the stop solution. For RNA extraction, bacterial cells were lysed by using 1 mg/ml of lysozyme, and RNA was purified by using the RNeasy kit (Qiagen) according to the manufacturer’s guidelines. RNA samples were treated with DNase I (Ambion), and the removal of genomic DNA was confirmed by PCR using Taq polymerase and primers gapA-F and gapA-R. RNA was converted to cDNA by using Superscript III (Invitrogen) and purified by using the PCR cleanup kit (Epoch Life Science). qPCR was conducted by using PowerUP SYBR green (Invitrogen), 4 ng of total cDNA as the template, and the primers listed in Table S2 in the supplemental material. To improve specificity in detecting papX and focX, qPCR was performed by using a higher annealing temperature and the primers labeled qPCR2 in Table S2. Threshold cycle (CT) values were normalized to the values for gapA, a reference gene, and analyzed by using the ΔΔCT method (68). Data are shown as the log2 fold changes (FC) of data from three biological replicates. Student’s t test was used for statistical analysis.
Immunoblotting for FliC detection.
Production of FliC was determined by immunoblotting using standardized whole-cell lysates using a previously described method (39). Stationary bacterial cultures were diluted 1:100 in 20 ml of tryptone medium and cultured at 37°C with aeration. Samples were taken at 0.5, 1.5, 3, and 6 h; centrifuged at 1,500 rpm for 10 min to limit the shearing of flagella; suspended in 2× SDS loading buffer (100 mM Tris-Cl [pH 6.8], 4% SDS, 0.2% bromophenol blue, 20% glycerol, 200 mM dithiothreitol [DTT]); and boiled for 10 min at 95°C. Cultures were normalized by the OD600 to load individual lanes of a 10% SDS-polyacrylamide gel with similar total protein content. Samples were subjected to SDS-PAGE, followed by transfer to a polyvinylidene difluoride membrane (Immobilon-P; Millipore Corp.). The blot was incubated with a 1:10,000 dilution of rabbit polyclonal antiserum to H1 flagella (Statens Serum Institute, Copenhagen, Denmark), followed by secondary incubation with a 1:20,000 dilution of peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma). The Clarity Western ECL substrate kit (Bio-Rad) was used to develop the blot. Data were normalized to the value for a nonspecific band, which served as an additional standard for a loading control.
5′ rapid amplification of cDNA ends.
Cultures of ΔpapX and ΔfocX strains grown overnight were diluted 1:100 in LB medium and cultured at 37°C to the mid-logarithmic growth phase. An aliquot was collected for RNA extraction and treated with a stop solution (95% EtOH, 5% phenol) to preserve RNA stability. To determine the transcriptional start site of papX and focX, we used a 5′ rapid amplification of cDNA ends (RACE) kit (Invitrogen) according to the manufacturer’s guidelines, and the primers are listed in Table S2 in the supplemental material. cDNA was inserted into pCR2.1-TOPO via the TOPO TA cloning kit (Invitrogen) and transformed into E. coli Top10 cells (Invitrogen). Transformants were plated on LB with ampicillin, and plasmids were isolated by miniprep and sequenced to determine the papX and focX transcription initiation sites.
Cochallenge and independent infections in the mouse model of UTI.
Six- to eight-week-old female CBA/J mice (Jackson Laboratories) were infected as previously described (11, 69, 70). Briefly, bacteria were grown to stationary phase in LB medium and, to induce motility were then diluted 1:50 in fresh LB medium and cultured with aeration at 37°C until an OD600 of ∼0.3 was reached. Bacteria were harvested by centrifugation (1,500 rpm), resuspended in sterile PBS, and adjusted by the OD to a final total concentration of 107 CFU/ml. For independent intraurethral infections by E. coli CFT073 and the ΔpapX construct, mice were inoculated over a 6-s period with a low-dose inoculum (10 µl of 107 CFU/ml) of each strain into the proximal end of the urethra. For cochallenge infections by E. coli CFT073 and the ΔpapX construct, mice were inoculated intraurethrally by using the same technique but were instead inoculated with a 1:1 mixture of the wild type and the ΔpapX mutant (10 µl of a total of 107 CFU/ml).
A transurethral infection model was used for cochallenge infections by E. coli CFT073 and the ΔfocX ΔpapX construct. Bacteria were grown to stationary phase in LB medium and then harvested by centrifugation (4,000 rpm), resuspended in sterile PBS, and adjusted by the OD to a final total concentration of 108 CFU/ml. Mice were transurethrally inoculated (50 µl of 108 CFU/ml) with a 1:1 mixture of the wild type and the ΔfocX ΔpapX mutant. Dilutions of the initial inoculums were plated to verify the input CFU per milliliter.
For all infections, after either 24 or 48 h postinoculation, mice were sacrificed, and the bladder, kidneys, and spleen were removed; homogenized in PBS (GLH homogenizer; Omni International); and plated onto LB agar by using an Autoplate 400 spiral plater (Spiral Biotech). Bacterial counts were enumerated by using a QCount automated plate counter (Spiral Biotech) to determine the output CFU per gram of tissue. Statistically significant differences were determined for independent infections by using a Mann-Whitney test. For cochallenge infections, competitive indices (CI) were calculated as the ratio of mutant to wild-type bacteria in the output over the ratio of mutant to wild-type bacteria in the input inoculum. Statistically significant differences were determined by using a Wilcoxon signed-rank test. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Michigan (approval PRO00007111).
Supplementary Material
ACKNOWLEDGMENTS
Our work is supported by Public Health Service grants AI43363 and AI59722 from the National Institutes of Health.
We thank Sara Smith for her assistance in the mouse experiments.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00338-18.
REFERENCES
- 1.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettelheim KA, Bushrod FM, Chandler ME, Cooke EM, O’Farrell S, Shooter RA. 1974. Escherichia coli serotype distribution in man and animals. J Hyg (Lond) 73:467–471. [PMC free article] [PubMed] [Google Scholar]
- 3.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J Infect Dis 181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 4.Foxman B, Brown P. 2003. Epidemiology of urinary tract infections: transmission and risk factors, incidence, and costs. Infect Dis Clin North Am 17:227–241. doi: 10.1016/S0891-5520(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 5.Ronald A. 2003. The etiology of urinary tract infection: traditional and emerging pathogens. Dis Mon 49:71–82. doi: 10.1067/mda.2003.8. [DOI] [PubMed] [Google Scholar]
- 6.Foxman B. 2014. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Schwan WR. 2008. Flagella allow uropathogenic Escherichia coli ascension into murine kidneys. Int J Med Microbiol 298:441–447. doi: 10.1016/j.ijmm.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 9.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HL. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect Immun 73:7644–7656. doi: 10.1128/IAI.73.11.7644-7656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci U S A 104:16669–16674. doi: 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HL. 2014. Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc Natl Acad Sci U S A 111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog 6:e1001187. doi: 10.1371/journal.ppat.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol 4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 15.Smith DR, Chapman MR. 2010. Economical evolution: microbes reduce the synthetic cost of extracellular proteins. mBio 1:e00131-10. doi: 10.1128/mBio.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Rasko DA, Lockatell CV, Johnson DE, Mobley HL. 2001. Repression of bacterial motility by a novel fimbrial gene product. EMBO J 20:4854–4862. doi: 10.1093/emboj/20.17.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane MC, Simms AN, Mobley HL. 2007. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J Bacteriol 189:5523–5533. doi: 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco M, Blanco JE, Alonso MP, Mora A, Balsalobre C, Munoa F, Juarez A, Blanco J. 1997. Detection of pap, sfa and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res Microbiol 148:745–755. doi: 10.1016/S0923-2508(97)82450-3. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1995. Distribution of virulence factors in Escherichia coli isolated from urine of cystitis patients. Microbiol Immunol 39:401–404. doi: 10.1111/j.1348-0421.1995.tb02219.x. [DOI] [PubMed] [Google Scholar]
- 22.Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, Buckles EL, Liou S-R, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HLT, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan AS, Kniep B, Oelschlaeger TA, Van Die I, Korhonen T, Hacker J. 2000. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect Immun 68:3541–3547. doi: 10.1128/IAI.68.6.3541-3547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsumori K, Terai A, Yamamoto S, Yoshida O. 1998. Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol Med Microbiol 21:261–268. doi: 10.1111/j.1574-695X.1998.tb01173.x. [DOI] [PubMed] [Google Scholar]
- 25.Bäckhed F, Alsén B, Roche N, Angström J, von Euler A, Breimer ME, Westerlund-Wikström B, Teneberg S, Richter-Dahlfors A. 2002. Identification of target tissue glycosphingolipid receptors for uropathogenic, F1C-fimbriated Escherichia coli and its role in mucosal inflammation. J Biol Chem 277:18198–18205. doi: 10.1074/jbc.M111640200. [DOI] [PubMed] [Google Scholar]
- 26.Mobley HL, Jarvis KG, Elwood JP, Whittle DI, Lockatell CV, Russell RG, Johnson DE, Donnenberg MS, Warren JW. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol Microbiol 10:143–155. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 27.Korhonen TK, Virkola R, Holthofer H. 1986. Localization of binding sites for purified Escherichia coli P fimbriae in the human kidney. Infect Immun 54:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulavik MC, Gambino LF, Miller PF. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med 1:436–446. doi: 10.1007/BF03401581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson SP, Grove A. 2004. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J Biol Chem 279:51442–51450. doi: 10.1074/jbc.M405586200. [DOI] [PubMed] [Google Scholar]
- 30.Deochand DK, Grove A. 2017. MarR family transcription factors: dynamic variations on a common scaffold. Crit Rev Biochem Mol Biol 52:595–613. doi: 10.1080/10409238.2017.1344612. [DOI] [PubMed] [Google Scholar]
- 31.Deochand DK, Meariman JK, Grove A. 2016. pH-dependent DNA distortion and repression of gene expression by Pectobacterium atrosepticum PecS. ACS Chem Biol 11:2049–2056. doi: 10.1021/acschembio.6b00168. [DOI] [PubMed] [Google Scholar]
- 32.Reiss DJ, Mobley HL. 2011. Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J Biol Chem 286:44726–44738. doi: 10.1074/jbc.M111.290684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simms AN, Mobley HL. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect Immun 76:4833–4841. doi: 10.1128/IAI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kakkanat A, Phan MD, Lo AW, Beatson SA, Schembri MA. 2017. Novel genes associated with enhanced motility of Escherichia coli ST131. PLoS One 12:e0176290. doi: 10.1371/journal.pone.0176290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deochand DK, Perera IC, Crochet RB, Gilbert NC, Newcomer ME, Grove A. 2016. Histidine switch controlling pH-dependent protein folding and DNA binding in a transcription factor at the core of synthetic network devices. Mol Biosyst 12:2417–2426. doi: 10.1039/C6MB00304D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duval V, McMurry LM, Foster K, Head JF, Levy SB. 2013. Mutational analysis of the multiple-antibiotic resistance regulator MarR reveals a ligand binding pocket at the interface between the dimerization and DNA binding domains. J Bacteriol 195:3341–3351. doi: 10.1128/JB.02224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 A resolution. Nat Struct Biol 8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 38.Bordelon T, Wilkinson SP, Grove A, Newcomer ME. 2006. The crystal structure of the transcriptional regulator HucR from Deinococcus radiodurans reveals a repressor preconfigured for DNA binding. J Mol Biol 360:168–177. doi: 10.1016/j.jmb.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Simms AN, Mobley HL. 2008. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J Bacteriol 190:3747–3756. doi: 10.1128/JB.01870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker CS, Pruss BM, Matsumura P. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol 186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet 3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahrner KA, Berg HC. 2015. Mutations that stimulate flhDC expression in Escherichia coli K-12. J Bacteriol 197:3087–3096. doi: 10.1128/JB.00455-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjostrom AE, Sonden B, Muller C, Rydstrom A, Dobrindt U, Wai SN, Uhlin BE. 2009. Analysis of the sfaX(II) locus in the Escherichia coli meningitis isolate IHE3034 reveals two novel regulatory genes within the promoter-distal region of the main S fimbrial operon. Microb Pathog 46:150–158. doi: 10.1016/j.micpath.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Hancock V, Vejborg RM, Klemm P. 2010. Functional genomics of probiotic Escherichia coli Nissle 1917 and 83972, and UPEC strain CFT073: comparison of transcriptomes, growth and biofilm formation. Mol Genet Genomics 284:437–454. doi: 10.1007/s00438-010-0578-8. [DOI] [PubMed] [Google Scholar]
- 45.Blyn LB, Braaten BA, White-Ziegler CA, Rolfson DH, Low DA. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J 8:613–620. doi: 10.1002/j.1460-2075.1989.tb03416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts JA, Hardaway K, Kaack B, Fussell EN, Baskin G. 1984. Prevention of pyelonephritis by immunization with P-fimbriae. J Urol 131:602–607. doi: 10.1016/S0022-5347(17)50513-3. [DOI] [PubMed] [Google Scholar]
- 47.Buckles EL, Luterbach CL, Wang X, Lockatell CV, Johnson DE, Mobley HL, Donnenberg MS. 2015. Signature-tagged mutagenesis and co-infection studies demonstrate the importance of P fimbriae in a murine model of urinary tract infection. Pathog Dis 73:ftv014. doi: 10.1093/femspd/ftv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopkins WJ, Hall JA, Conway BP, Uehling DT. 1995. Induction of urinary tract infection by intraurethral inoculation with Escherichia coli: refining the murine model. J Infect Dis 171:462–465. doi: 10.1093/infdis/171.2.462. [DOI] [PubMed] [Google Scholar]
- 49.Beuzon CR, Holden DW. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3:1345–1352. doi: 10.1016/S1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 50.Lane MC, Mobley HL. 2007. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72:19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 51.Parreira VR, Gyles CL. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect Immun 71:5087–5096. doi: 10.1128/IAI.71.9.5087-5096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nichols KB, Totsika M, Moriel DG, Lo AW, Yang J, Wurpel DJ, Rossiter AE, Strugnell RA, Henderson IR, Ulett GC, Beatson SA, Schembri MA. 2016. Molecular characterization of the vacuolating autotransporter toxin in uropathogenic Escherichia coli. J Bacteriol 198:1487–1498. doi: 10.1128/JB.00791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marklund BI, Tennent JM, Garcia E, Hamers A, Baga M, Lindberg F, Gaastra W, Normark S. 1992. Horizontal gene transfer of the Escherichia coli pap and prs pili operons as a mechanism for the development of tissue-specific adhesive properties. Mol Microbiol 6:2225–2242. doi: 10.1111/j.1365-2958.1992.tb01399.x. [DOI] [PubMed] [Google Scholar]
- 54.Snyder JA, Haugen BJ, Lockatell CV, Maroncle N, Hagan EC, Johnson DE, Welch RA, Mobley HL. 2005. Coordinate expression of fimbriae in uropathogenic Escherichia coli. Infect Immun 73:7588–7596. doi: 10.1128/IAI.73.11.7588-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. 2012. Structural basis of TLR5-flagellin recognition and signaling. Science 335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grove A. 2013. MarR family transcription factors. Curr Biol 23:R142–R143. doi: 10.1016/j.cub.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 58.Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, Hokamp K, Hinton JC. 2016. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet 12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Curran TD, Abacha F, Hibberd SP, Rolfe MD, Lacey MM, Green J. 2017. Identification of new members of the Escherichia coli K-12 MG1655 SlyA regulon. Microbiology 163:400–409. doi: 10.1099/mic.0.000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 61.Wang X, Wood TK. 2011. IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J 5:1517–1525. doi: 10.1038/ismej.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ko M, Park C. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Bacteriol 182:4670–4672. doi: 10.1128/JB.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol 181:7500–7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subashchandrabose S, Hazen TH, Rasko DA, Mobley HL. 2013. Draft genome sequences of five recent human uropathogenic Escherichia coli isolates. Pathog Dis 69:66–70. doi: 10.1111/2049-632X.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stapleton A, Moseley S, Stamm WE. 1991. Urovirulence determinants in Escherichia coli isolates causing first-episode and recurrent cystitis in women. J Infect Dis 163:773–779. doi: 10.1093/infdis/163.4.773. [DOI] [PubMed] [Google Scholar]
- 67.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg EC. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson DE, Lockatell CV, Russell RG, Hebel JR, Island MD, Stapleton A, Stamm WE, Warren JW. 1998. Comparison of Escherichia coli strains recovered from human cystitis and pyelonephritis infections in transurethrally challenged mice. Infect Immun 66:3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.