Abstract
Background
Obesity is one of the main causes of morbidity and mortality worldwide. More than 120 genes have been shown to be associated with obesity related phenotypes. The aim of this study was to determine the effect of selected genetic polymorphisms in Uncoupling protein 1 (UCP1) and Niemann-Pick C1 (NPC1) genes in an obese population in Saudi Arabia.
Methods
The genotypes of rs1800592, rs10011540 and rs3811791 (UCP1 gene) and rs1805081 and rs1805082 (NPC1 gene) were determined in a total of 492 subjects using TaqMan chemistry by Real-time PCR. In addition, capillary sequencing assay was performed to identify two specific polymorphisms viz., rs45539933 (exon 2) and rs2270565 (exon 5) of UCP1 gene.
Results
A significant association of UCP1 polymorphisms rs1800592 [OR, 1.52 (1.10–2.08); p = 0.009] was observed in the obese cohort after adjusting with age, sex and type 2 diabetes. Further BMI based stratification revealed that this association was inconsistent with both moderate and extreme obese cohort. A significant association of UCP1 polymorphisms rs3811791 was observed only in the moderate-obese cohort [OR = 2.89 (1.33–6.25); p = 0.007] but not in the extreme-obese cohort indicating an overlying genetic complexity between moderate-obesity and extreme-obesity. The risk allele frequencies, which were higher in moderate-obese cohort, had abnormal HDL, LDL and triglyceride levels.
Conclusion
The rs1800592 and rs3811791 of UCP1 gene are associated with obesity in general and in the moderate-obese group in particular. The associated UCP1 polymorphisms in the moderate-obese group may regulate the impaired energy metabolism which plays a significant role in the initial stages of obesity.
Electronic supplementary material
The online version of this article (10.1186/s12881-018-0715-5) contains supplementary material, which is available to authorized users.
Keywords: UCP1, NCP1, Moderate-obese, Extreme-obese, Diabetes, HDL, LDL, Cholesterol, Metabolic disorder
Background
Obesity represent a serious public health problem worldwide and is associated with co-existing diseases, including cardiovascular diseases, type 2 diabetes mellitus (T2DM), musculoskeletal conditions and various cancers [1–3]. The prevalence of obesity in a population is an indicator of its health status and in recent years obesity has reached epidemic proportions. In the last few decades, Saudi Arabia has witnessed an increased prevalence of obesity [4]. A recent survey revealed that overall, 28.7% of 10,735 Saudi nationals recruited for a Saudi Health Information Survey in 2013 were obese, with a higher prevalence in females than males (33.5% vs 24.1%) [5].
Obesity and weight gain have been reported to be associated with several genes in addition to known factors, such as diet and lack of exercise [6]. It has also been reported that obesity is influenced by genetic variations and ethnicity [7, 8]. The polymorphisms which are involved in obesity were shown to be discordant in their association in various e ethnic populations. Therefore, genetic variations in ethnic populations need to be determined to validate the genetic significance of the polymorphisms. The majority of the genetic variations in obesity are related to the genes associated with energy metabolism. Uncoupling proteins are associated with the pathogenesis of obesity and T2DM by deregulation of energy expenditure, thermogenesis and reduction in oxidative stress [9]. Several studies have reported that polymorphisms of the UCP1 gene such as, g.-3826A > G (rs1800592), g.-1766A > G (rs10011540) and g.-112A > C (rs3811791) in the promoter region, and p.Ala64Thr (rs45539933) and p.Met299Leu (rs2270565) in the codon region are associated with obesity and T2DM [10–15]. The Niemann-Pick C1 gene (NPC1), another reported genetic determinant of obesity, is a gene for transmembrane glycoprotein located in the limiting membrane of late endosome/lysosome (LE/LY) and mediates intracellular trafficking of sterols [16–18]. It has been reported that rs1805081 (p.His215Arg) and rs1805082 (p.Ile858Val) polymorphisms of the NPC1 gene are associated with early-onset and morbid adult obesity in a European population and Chinese children [19–21]. A Genome Wide Association Study conducted on Mexican children found a significant association of risk allele NPC1 rs1805081 with increased fasting glucose levels and decreased fasting serum insulin levels [22].
The UCP1 and NPC1 genes are known to be involved in the regulation of energy metabolism and the role of the polymorphisms in these genes with respect to obesity is arguable due to the diverse results of studies performed in different ethnicities. The aim of this study is to determine the association of the polymorphisms of the UCP1 [rs1800592, rs10011540, rs3811791, rs45539933 and rs2270565] and NPC1 [rs1805081 and rs1805082] genes in a Saudi population.
Materials and methods
Subjects eligibility and recruitment
A total of 337 obese patients and 155 non-obese control subjects attending King Fahd Hospital of the University were included in the study. All patients and controls were Saudi origin. The inclusion criteria for obese patients included BMI ≥30 kg/m2 and age between 18 and 60 years. The control group comprised healthy subjects with a BMI < 30 kg/m2. The patient cohort was grouped as moderate-obese and extreme-obese based on the heterogeneity of variations in suspected etiology, prevalence, mortality rate and anthropometric measures, mainly BMI. The moderate-obese cohort comprised patients with a BMI ≥30–39.9 kg/m2 and the extreme-obese cohort with a BMI ≥40 kg/m2. These cohorts were further subdivided based on age, gender, abnormal biochemical parameters and co-existing conditions. Written informed consent was obtained from all participants. The study was approved by Institutional Review Board of Imam Abdulrahman Bin Faisal University of Dammam (IRB-2013-01-008).
Sample collection and biochemical parameters estimation
Five mL of whole blood was collected in EDTA anti-coagulated vacutainers from patient and control subjects after an overnight fast. Biochemical parameters, including total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), triglycerides levels, fasting blood glucose (FBG) level and insulin levels were determined at King Fahd Hospital of the University using Siemens Dimension RxL chemistry system (Siemens, Erlangen, Germany) and other details of co-existing medical conditions were collected from the hospital medical records.
Mutation detection by TaqMan SNP genotyping assay
Genomic DNA was isolated using Promega DNA isolation kit (Promega, Madison, USA) according to the manufacturer’s instructions. Concentration and purity of isolated DNA were determined using Nanodrop spectrophotometer and then stored at − 20 °C until the day of mutation analysis. TaqMan chemistry based Real-Time PCR method was used to detect the SNPs of UCP1 (rs1800592, rs10011540 and rs3811791) and NPC1 (rs1805081 and rs1805082). TaqMan probes were synthesized by Applied Bio systems, (Thermo Scientific, CA, USA) which detect both wild and mutant alleles. The assay was conducted as per the manufacturer’s instructions. ABI 7500 fast real-time PCR system proprietary software (Thermo Scientific, CA, USA) was used for analysis and interpretation of the results.
Mutation detection by capillary sequencing assay
Distribution of polymorphisms and novel mutations on exon 2 and exon 5 of UCP1 gene was carried out using capillary sequencing by ABI 3500 genetic analyzer (Thermo Scientific, CA, USA) as previously reported [23]. The targeted gene sequence was amplified using polymerase chain reaction with specific primers for Exon 2 (Fwd-5’TCTGCACCTTTCTTATTTC3’ Rev-5’TCTCGCCAATTTGTTATGAA3’) and Exon 5 (Fwd-5’CAAAAGTCTGATGTTGAC3’ Rev-5’GAAATCTGTGGCAAGGAAAAGT3’) on a thermal cycler S1000 (Bio Rad, Hercules, California, USA). BigDye Direct sequencing master mix was used to perform the cycle sequencing reaction. The Sequencing Install Standard and BigDye® Terminator v3.1 Kit. POP7 polymer and 50 cm capillary (Thermo Scientific, CA, USA) were used in this procedure. 10 μl of purified product was loaded in 96 well plates and analyzed using ABI genetic analyzer 3500 (Thermo Scientific, CA, USA) for sequence detection.
The DNA sequence was then viewed on sequence analysis software. Sequence alignment was performed using the NCBI alignment and codon code analyzer software with reference sequence of UCP1 gene (NG_012139.1).
Statistical analysis
Collected data were summarized as mean ± SD. The patient and control demographic parameters, including biochemical and clinical data, were tested for statistical difference using students’ “t” test for continuous variables and Chi-square test for discrete variables with one degree of freedom. Risk allele frequencies (RAF) were estimated by direct counting of the test allele divided by the total number of alleles. Multiple variable logistic regression model using age, sex, and absence/presence of T2D as covariates was performed to assess the association of these SNPs with obesity. The p value < 0.0125 has been considered as significant for regression analysis as per Bonferroni-correction. All statistical analyses were performed using SPSS software (version19) and GraphPad Prism 7.03.
Results
Clinical, biochemical and genotypic characteristics of the Unstratified case and control subjects
A total of 337 obese patients (Male = 138, Female = 199) with a mean BMI of 39.59 ± 10.32 kg/m2 and a mean age of 47.41 ± 12.79 years were included in this study. The control population included 155 healthy volunteers (Male = 76, Female = 79) with a mean BMI of 24.09 ± 2.6 and a mean age of 43.86 ± 14.54 years. Of the 337 obese patients, 235 were T2DM patients and 85 had hypertension (HTN). The levels of FBG, triglycerides, and HDL were significantly different (p < 0.05) between the patient and control groups. The clinical and biochemical parameters of the patients and controls are presented in Table 1.
Table 1.
Clinical and biochemical parameters of the study cohort
| Clinical and biochemical parameters | Control (n = 155) (mean ± SD) | Patient (n = 337) (mean ± SD) | p-value |
|---|---|---|---|
| Age (years) | 43.86 ± 14.54 | 47.41 ± 12.79 | 0.006 |
| Male / Female, n (%) | 76 (49) / 79 (51) | 138 (41) / 199 (59) | 0.097 |
| BMI (kg/m2) | 24.09 ± 2.6 | 39.59 ± 10.32 | < 0.005 |
| FBG (mg/dL) | 120.58 ± 56.75 | 152.08 ± 71.66 | < 0.005 |
| Triglycerides (mg/dL) | 100.00 ± 62.45 | 136.85 ± 78.48 | < 0.005 |
| LDL (mg/dL) | 115.25 ± 42.90 | 111.59 ± 36.56 | 0.331 |
| Cholesterol (mg/dL) | 189.34 ± 134.62 | 179.58 ± 40.89 | 0.225 |
| HDL (mg/dL) | 48.52 ± 14.13 | 45.18 ± 12.71 | 0.009 |
| T2DM, n (%) | 56 (36.12) | 235 (69.73) | < 0.005 |
| HTN, n (%) | 0 | 85 (25.22) | < 0.005 |
| CVD, n (%) | 0 | 47 (13.94) | < 0.005 |
Data with significant p-value (< 0.05) are shown in bold
FBG fasting blood glucose, LDL low density lipoprotein, HDL high density lipoprotein, T2DM type-2 diabetes mellitus, HTN hypertension, CVD cardiovascular disease
The allele frequencies of the SNPs rs1800592, rs10011540, rs3811791 (UCP1) and rs1805081 and rs1805082 (NPC1) are listed in Table 2 and genotype frequencies are mentioned in Additional file 1: Table S1. The mutant allele G of SNP rs1800592 on UCP1 showed a significant association with obesity [OR, 1.52 (1.10–2.08); p = 0.009]. The distribution of all other SNPs in the patient and control population did not reveal any statistical significance. All genotype frequencies of the control group were consistent with Hardy-Weinberg equilibrium. Sequencing for Exon 2 and Exon 5 of UCP1 revealed three genetic variants, Leu59Gln (1.51%), Ala64Thr (11.36%) and Met229Leu (14.39%) in the studied population and all were genotypically heterozygous. Therefore, these variants were excluded from the association analysis.
Table 2.
Allelic distribution among patient and control cohort
| Gene | SNP | Allele | Control (n) | Cases (n) | Model 1a | Model 2b | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 98.75%CI | p-value | OR | 98.75%CI | p value | ||||||
| UCP1 | rs1800592 | A | 227 | 443 | Ref | ||||||
| G | 83 | 231 | 1.42 | 0.97–2.08 | 0.019 | 1.52 | 1.01–2.27 | 0.009 | 0.23 | ||
| rs10011540 | T | 283 | 627 | Ref | |||||||
| G | 27 | 47 | 0.78 | 0.42–1.47 | 0.338 | 0.78 | 0.40–1.52 | 0.361 | 0.85 | ||
| rs3811791 | T | 298 | 624 | Ref | |||||||
| C | 12 | 50 | 1.98 | 0.87–4.5 | 0.036 | 2.06 | 0.87–4.90 | 0.036 | 0.09 | ||
| NPC1 | rs1805081 | T | 275 | 605 | Ref | ||||||
| C | 35 | 69 | 0.89 | 0.51–1.55 | 0.617 | 0.84 | 0.47–1.52 | 0.487 | 0.43 | ||
| rs1805082 | T | 225 | 491 | Ref | |||||||
| C | 85 | 183 | 0.98 | 0.67–1.44 | 0.930 | 0.99 | 0.66–1.49 | 0.963 | 0.17 | ||
a Unadjusted and bAdjusted for Age, Gender and T2DM
Data with significant p-value (< 0.0125) are shown in bold
Clinical, biochemical and genotypic characteristics of BMI stratified cohort
The association of rs1800592 and rs3811791 SNPs with obesity is arguable as results in different populations [10] and also in stratified obesity groups are controversial [24, 25]. To shed some light on the association, the patient group was further classified according to BMI namely moderate-obese and extreme-obese. The risk status of these cohorts was analyzed. The clinical and biochemical parameters of these cohorts are shown in Table 3. There were no significant differences among the biochemical parameters after stratification except for HDL which showed a significant difference in the extreme-obese cohort (p = 0.007). The allelic frequency distribution for UCP1 and NPC1 polymorphism was analyzed for the stratified cohorts, moderate-obese and extreme-obese (Table 4), similarly the frequencies of genotype in stratified cohort are given in Additional file 1: Table S2. The significant SNPs rs1800592 was not significantly associated with both stratified cohort, whereas another UCP1 SNP rs3811791 was strongly associated with the moderate-obese (BMI 30–39.9 kg/m2) patients [OR = 2.89 (1.33–6.25); p = 0.007] but not with the extreme obese after stratification (Table 4).
Table 3.
Clinical and biochemical parameters of study cohort after stratification into two groups based on their BMI
| Clinical and biochemical parameters | Control (n = 155) (mean ± SD) | Moderate obese (n = 207) (mean ± SD) | p-value | Extreme obese (n = 130) (mean ± SD) | p-value |
|---|---|---|---|---|---|
| Age (years) | 43.86 ± 14.54 | 50.45 ± 11.17 | < 0.005 | 42.57 ± 13.72 | 0.248 |
| Male / Female, n (%) | 76 (49) / 79 (51) | 91 (43.96) / 116 (56.03) | 0.394 | 47 (36.15) / 83 (63.84) | 0.031 |
| BMI kg/m2 | 24.09 ± 2.60 | 34.15 ± 2.69 | < 0.005 | 48.26 ± 11.94 | < 0.005 |
| FBG (mg/dL) | 120.58 ± 56.75 | 161.33 ± 69.77 | < 0.005 | 137.33 ± 72.41 | 0.029 |
| Triglycerides (mg/dL) | 100.00 ± 62.45 | 133.91 ± 66.23 | < 0.005 | 141.54 ± 94.87 | < 0.005 |
| LDL (mg/dL) | 115.25 ± 42.90 | 107.81 ± 37.11 | 0.078 | 117.62 ± 34.97 | 0.614 |
| Cholesterol (mg/dL) | 189.34 ± 134.62 | 176.86 ± 40.23 | 0.208 | 183.92 ± 41.72 | 0.659 |
| HDL (mg/dL) | 48.52 ± 14.13 | 45.78 ± 13.13 | 0.058 | 44.23 ± 12.00 | 0.007 |
| T2DM, n (%) | 56 (36.12) | 175 (84.54) | < 0.005 | 60 (46.15) | 0.092 |
| HTN, n (%) | 0 | 60 (28.98) | < 0.005 | 25 (19.23) | < 0.005 |
| CVD, n (%) | 0 | 34 (16.42) | < 0.005 | 13 (10) | < 0.005 |
Data with significant p-value (< 0.05) are shown in bold
BMI body mass index, FBG fasting blood glucose, LDL low density lipoprotein, HDL high density lipoprotein, T2DM type-2 diabetes mellitus, HTN hypertension, CVD cardiovascular disease
Table 4.
Allelic distribution of patient population after stratifying to moderate-obese and extreme-obese groups based on their BMI
| Gene | SNP | Allele | Control | Moderate obese | Model 1a | Model 2b | Extreme obese | Model 1a | Model 2b | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 98.75%CI | P value | OR | 98.75%CI | p value | OR | 98.75%CI | P value | OR | 98.75%CI | p value | ||||||
| UCP1 | rs1800592 | A | 227 | 271 | Ref | 172 | Ref | ||||||||||
| G | 83 | 143 | 1.44 | 1.96–2.18 | 0.026 | 1.52 | 0.95–2.46 | 0.027 | 88 | 1.39 | 0.88–2.21 | 0.067 | 1.39 | 0.88–2.23 | 0.072 | ||
| rs10011540 | T | 283 | 382 | Ref | 245 | Ref | |||||||||||
| G | 27 | 32 | 0.87 | 0.44–1.73 | 0.633 | 1.01 | 0.46–2.23 | 0.967 | 15 | 0.64 | 0.28–1.48 | 0.183 | 0.58 | 0.25–1.37 | 0.113 | ||
| rs3811791 | T | 298 | 378 | Ref | 246 | Ref | |||||||||||
| C | 12 | 36 | 2.36 | 1.0–5.56 | 0.011 | 2.89 | 1.08–7.73 | 0.007 | 14 | 1.41 | 0.52–3.86 | 0.390 | 1.32 | 0.47–3.71 | 0.494 | ||
| NPC1 | rs1805081 | T | 275 | 371 | Ref | 234 | Ref | ||||||||||
| C | 35 | 43 | 0.91 | 0.50–1.66 | 0.698 | 0.78 | 0.39–1.56 | 0.376 | 26 | 0.87 | 0.44–1.73 | 0.619 | 0.86 | 0.43–1.75 | 0.612 | ||
| rs1805082 | T | 225 | 306 | Ref | 185 | Ref | |||||||||||
| C | 85 | 108 | 0.93 | 0.61–1.42 | 0.688 | 0.91 | 0.56–1.49 | 0.637 | 75 | 1.07 | 0.67–1.71 | 0.705 | 1.04 | 0.65–1.68 | 0.813 | ||
Data with significant p-value (< 0.0125) are shown in bold
aUnadjusted and bAdjusted for Age, Gender and T2DM
Associated UCP1 SNPs (rs1800592 and rs3811791) vs biochemical parameters and co-existing diseases
To verify the association of UCP1 risk alleles (rs1800592 and rs3811791) with the abnormal biochemical parameters and co-existing diseases such as T2DM and HTN in the control, moderate-obese and extreme-obese cohorts, the risk allele frequencies (RAF) of both SNPs were calculated and plotted as shown in Fig. 1. The RAF of SNP rs1800592 was higher in the moderate-obese cohort with abnormal HDL and LDL levels compared to the control and extreme-obese cohorts. In rs3811791, the RAF was equally distributed to each cohort for low HDL level but for high LDL level, it was higher in the moderate-obese cohort (Fig. 1a). For hypercholesterolemia, the RAF of rs1800592 was higher in the extreme-obese cohort whereas, rs3811791 was higher in the moderate-obese. The RAF of rs1800592 was higher in the moderate-obese cohort for hypertriglyceridemia whereas, that of rs3811791 did not show any difference between these cohorts (Fig. 1b) (Additional file 1: Table S3).
Fig. 1.
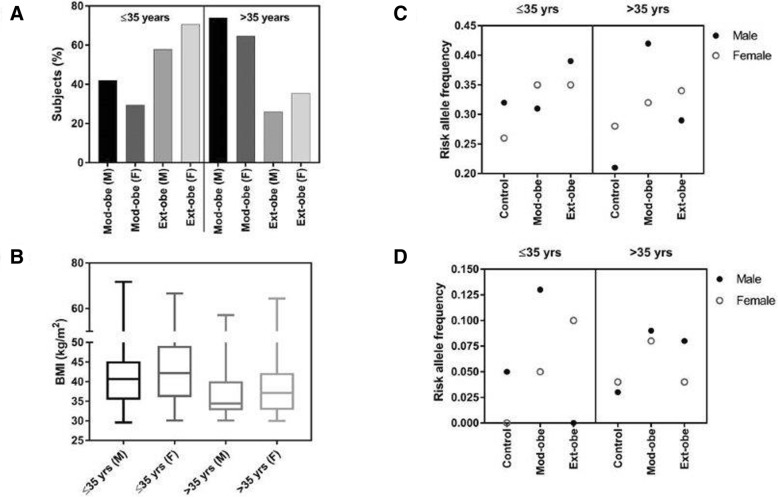
Frequency of the risk alleles of rs1800592 and rs3811791 in patients with abnormal HDL and LDL (a) and hypercholesterolemia and hypertriglyceridemia (b). Number of T2DM and HTN patients in the whole patient group and after the stratification (c); and the frequency of the risk alleles in these stratified cohort (d). Mod-obe: moderate-obese; Ext-obe: extreme-obese; T2DM: type-2 diabetes mellitus and HTN: hypertension
T2DM (70%) and HTN (25.2%) were co-existing diseases in the total patient cohort. In 207 moderate-obese subjects, 84% had T2DM and 25% had HTN, whereas in the 130 extreme-obese subjects, 47.3% had T2DM and 19.2% had HTN (Fig. 1c). Allele frequency analysis showed that the RAF of rs1800592 was higher in the moderate-obese cohort in both T2DM and HTN patients, whereas the other risk allele of rs3811791 was not remarkably changed after stratification (Fig. 1d).
Associated UCP1 SNPs (rs1800592 and rs3811791) vs age and gender with BMI
It has been reported that the prevalence of obesity is higher in young females in the Saudi population than males. In our study, it was found that the majority of the subjects were female in both stratified groups. To confirm if there was any association of age and gender with BMI, the patient population was subdivided based on age (≤35 years and > 35 years) and gender. We observed a higher number of young females (≤35 years) in the extreme-obese cohort whereas, the males > 35 years of age dominated the moderate-obese cohort (Fig. 2a). To determine the association of age and gender with BMI, a box plot with BMI on Y-axis and subdivided patient groups on X-axis was plotted as shown in Fig. 2b. In both age groups, the increased BMI was associated with female subjects (≤35 years: mean BMI = 43.26 ± 8.58 kg/m2; and > 35 years: mean BMI = 38.64 ± 7.27 kg/m2).
Fig. 2.
Percentage of subjects after subdividing the total patient cohort based on their age, sex and BMI (a); Box plot representing the association of BMI with these subdivided groups (b); Frequency of risk alleles in these subdivided groups: risk allele frequency of rs1800592 (c) and risk allele frequency of rs3811791 (d). Mod-obe (M): moderate-obese male, Mod-obe (F): moderate-obese female, Ext-obe (M): extreme-obese male and Ext-obe (F): extreme-obese female
To verify whether these two risk alleles of UCP1 had any association with age and gender, the RAF was studied after subdividing the patient group based on age and gender. The RAF of rs1800592 was higher in the extreme-obese males in ≤35 years sub group whereas, in the > 35 subgroup, the RAF in the moderate-obese males was high (Fig. 2c). For rs3811791, the RAF was higher in females aged ≤35 years with extreme-obesity and in males aged ≤35 years with moderate-obesity. In the > 35 years group, the RAF was high in both male and female in the moderate-obese cohort (Fig. 2d).
Discussion
This study reports the association of common UCP1 polymorphisms in an obese population in Saudi Arabia. The UCP1 gene is considered to be a candidate gene for obesity and T2DM as the polymorphism of this gene reduces the mitochondrial membrane potential and mediates proton leak [26]. Mutations in these genes reduce the availability of functional proteins, which in turn, could reduce energy expenditure by increasing coupling of oxidative phosphorylation, thereby contributing to the development of obesity.
We selected the most studied polymorphisms of the UCP1 gene with respect to obesity. Many studies have reported that polymorphisms (rs1800592, rs10011540, rs3811791) of the UCP1 promoter region and rs45539933 and rs2270565 of the UCP1 coding region are associated with obesity and T2DM. Among these polymorphisms, rs1800592 was the most studied polymorphism and the results were highly controversial in different populations [10]. To the best of our knowledge, no previous studies have reported an association of rs1800592 polymorphism from our geographic region. The importance of rs1800592 polymorphism in regulating the expression of the UCP1 gene has previously been reported in obese subjects [27]. The presence of rs1800592 polymorphism in the UCP1 gene was first identified in 1994 in a pilot study conducted on 261 Canadian patients and was associated with obesity and weight gain [28, 29]. Subsequently, several studies reported the status of this polymorphism with obesity and other associated parameters in different populations, but it still remains arguable as it exhibits different allele frequencies in various ethnic populations.
In this study, we found that UCP1 gene polymorphism rs1800592 is significantly associated with increased BMI. When the patient cohort was stratified based on their BMI, other UCP1 SNP, rs3811791, was associated with moderate-obese patients. Several independent studies conducted in different ethnicities supported the association between the G-allele of rs1800592 and obesity, BMI or other obesity-related parameters [30–32]. On the other hand, a number of studies have reported a lack of association of rs1800592 with an obese population with different ethnic background [33–37]. Previously, the association of another significant UCP1 SNP rs3811791, was reported in Japanese and Indian diabetic patients [13, 38]. For the first time, this study reports the association of rs3811791 SNP with obesity, specifically in moderate-obese patients. We could not determine any association of other UCP1 polymorphisms, namely rs10011540, rs45539933 and rs2270565 in this population. The association of NPC1 polymorphisms rs1805081 and rs1805082 has been reported in European subjects [19]. However, in a study conducted on obese Chinese children, rs1805081 was not significantly associated [20]. Obesity in the population included in the current study was not associated with the reported NPC1 polymorphisms, which is in accordance with an earlier study conducted in Saudi Arabia, indicating that this SNP is neither associated with obesity nor BMI [39].
Several studies have reported that the G-allele of rs1800592 is associated with a low level of HDL [40], and high level of triglyceride [41] and LDL [42] in obese subjects in different populations. Similarly, in this study, we observed an increased RAF (G-allele) with lower HDL and higher LDL and hypertriglyceridemia in the moderate-obese cohort than in the extreme-obese and control cohorts. These observations reflect the effective involvement of UCP1-mediated pathways in the regulation of obesity-related metabolic parameters in moderate-obese subjects. However, in extreme-obese cases, other functional pathways are effectively involved which may increase BMI, thereby increasing the risk of metabolic complications.
UCP1 is predominantly expressed in brown adipose tissue and eminently participates in the process of thermogenesis [43, 44]. Recent studies conducted in animal models using targeted chemical uncouplers and adipose tissue- and skeletal muscle-targeted overexpression of UCP1 resulted in decreased hypertriglyceridemia, glucose homeostasis by increased insulin sensitivity and glucose uptake and as well as a decreased level of diet- and genetic-induced obesity [44–48]. There was a significant number of diabetic patients in both moderate-obese (four-fold increase) and extreme-obese cohorts (two-fold increase) compared to the control cohort. The increased number of T2DM patients and increased RAF of rs1800592 in the moderate-obese cohort sheds light on the association of this SNP with obesity associated with T2DM. The association of rs1800592 with T2DM is controversial as the studies conducted in different ethnicities exhibited varying results [31, 33, 35, 49]. UCP1 may play a major role in inducing insulin-resistance and diabetes in moderate-obese cases. This observation may also help to accelerate the investigation on how these two complicated conditions, obesity and T2DM, are inter-related with each other.
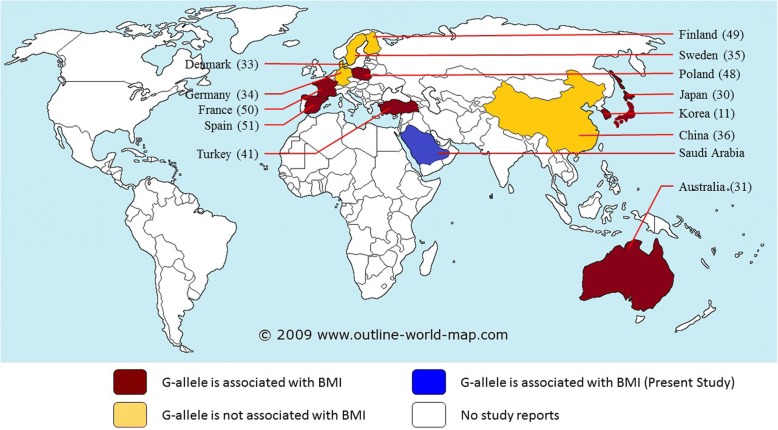
A schematic representation of the available data which specifically reported how this rs1800592 risk allele (allele G) is associated with obeisty worldwide is shown in Fig. 3 [11, 30, 31, 33–36, 41, 48–51]. A recent population-based study reported that BMI levels were increasing in the Saudi population, with a more rapid increase in females than males [4]. Similarly, Memish et al. (2014) reported that the level of obesity in Saudi females was higher than that in males (33.5% vs 24.1%) [5]. The present study revealed a higher ratio of young obese female patients within the extreme-obese cohort compared to the moderate-obese cohort.
Fig. 3.
Schematic diagram represents how the risk allele G of rs1800592 is distributed in different population worldwide
Conclusion
The present study reveals a significant association of rs1800592 and rs3811791 polymorphisms in the promoter region of the UCP1 gene with obese population in Saudi Arabia. The associated UCP1 polymorphisms in the moderate-obese group may regulate impaired energy metabolism which plays a significant role in the initial stages of obesity. NPC1 polymorphisms were not found to be an important risk factor for obesity in Saudi Arabia.
Additional file
Table S1. Genotypic distribution among patient and control cohort. Genotypic odds ratio for all cases and controls, unadjusted and adjusted for Age, Sex and T2D. Table S2. Genotypic distribution among stratified cohort. Genotypic odds ratio among patient population stratified for BMI; moderate-obese and extreme-obese groups. Table S3. Distribution of risk alleles in normal and abnormal levels of biochemical parameters. Association of the risk alleles frs1800592 and rs3811791 with HDL, LDL, Triglycerides and total cholesterol. (DOCX 42 kb)
Acknowledgements
The authors acknowledge the Deanship of Scientific Research, Imam Abdulrahman Bin Faisal University for funding the project (#2013178). We thank Professor Sulaiman Bah for his contribution in the statistical analysis And Mr. Geoffrey James Tam Moro for technical assistance in conducting experiments.
Funding
Deanship of Scientific Research, Imam Abdulrahman Bin Faisal University (#2013178).
Availability of data and materials
All the study materials and analysis data are available with the corresponding author.
Abbreviations
- BMI
Body mass index
- EDTA
Ethylenediamine tetra acetic acid
- ELISA
Enzyme-linked immunosorbent assay
- HDL
High-density lipoprotein
- LDL
Low-density lipoprotein
- LE/LY
Late endosome/lysosome
- NPC1
Niemann-Pick C1
- PCR
Polymerase Chain Reaction
- SNP
Single Nucleotide Polymorphism
- T2DM
Type 2 diabetes mellitus
- UCP1
Uncoupling protein 1
Authors’ contributions
SC, CV and CC designed the study, performed the assay, and drafted the manuscript. MI, AAB, ZAA and KAA was involved in the sample collection, collection of data and follow-up of the study subjects. CV and SA performed statistical analysis. AAA provided critical review of the manuscript. All authors have read and reviewed the manuscript and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of Imam Abdulrahman Bin Faisal University of Dammam (IRB-2013-01-008). Written informed consent was obtained from all participants.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shahanas Chathoth, Phone: +966 506770540, Email: schathoth@iau.edu.sa, Email: shahanasc@gmail.com.
Mona H. Ismail, Email: moismail@iau.edu.sa
Chittibabu Vatte, Email: cbvatte@iau.edu.sa.
Cyril Cyrus, Email: ccyrus@iau.edu.sa.
Zhara Al Ali, Email: Zhara.a.alali@hotmail.com.
Khandaker Ahtesham Ahmed, Email: pavelmb@gmail.com.
Sadananda Acharya, Email: sabaikady@iau.edu.sa.
Aisha Mohammed Al Barqi, Email: abarqi@iau.edu.sa.
Amein Al Ali, Email: aalali@iau.edu.sa.
References
- 1.Haslam DW, James WP. Obesity. Lancet. 2005;366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Yach D, Stuckler D, Brownell KD. Epidemiological and economic consequences of the global epidemics of obesity and diabetes. Nat Med. 2006;12:62–66. doi: 10.1038/nm0106-62. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M, Leitzmann M, Freisling H, Bray F, Romieu I, Renehan A, Soerjomataram I. Obesity and cancer: an update of the global impact. Cancer Epidemiol. 2016;41:8–15. doi: 10.1016/j.canep.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hazzaa HM, Abahussain NA, Al-Sobayel HI, Qahwaji DM, Musaiger AO. Lifestyle factors associated with overweight and obesity among Saudi adolescents. BMC Public Health. 2012;12:354. doi: 10.1186/1471-2458-12-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memish ZA, El Bcheraoui C, Tuffaha M, Robinson M, Daoud F, Jaber S, Mikhitarian S, Al Saeedi M, AlMazroa MA, Mokdad AH, Al Rabeeah AA. Obesity and associated factors-Kingdom of Saudi Arabia, 2013. Prev Chronic Dis. 2014;11:E174. doi: 10.5888/pcd11.140236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papoutsakis C, Dedoussis GV. Gene-diet interactions in childhood obesity: paucity of evidence as the epidemic of childhood obesity continues to rise. Pers Med. 2007;4:133–146. doi: 10.2217/17410541.4.2.133. [DOI] [PubMed] [Google Scholar]
- 7.Qian L, Xu K, Xu X, Gu R, Liu X, Shan S, Yang T. UCP2 -866G/a, Ala55Val and UCP3 -55C/T polymorphisms in association with obesity susceptibility - a meta-analysis study. PLoS One. 2013;8(4):e58939. doi: 10.1371/journal.pone.0058939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brondani LA, de Souza BM, Assmann TS, Bouças AP, Bauer AC, Canani LH, Crispim D. Association of the UCP polymorphisms with susceptibility to obesity: case-control study and meta-analysis. Mol Biol Rep. 2014;41(8):5053–5067. doi: 10.1007/s11033-014-3371-7. [DOI] [PubMed] [Google Scholar]
- 9.Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 10.Brondani LA, Assmann TS, Duarte GC, Gross JL, Canani LH, Crispim D. The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus. Arq Bras Endocrinol Metabol. 2012;56:215–225. doi: 10.1590/S0004-27302012000400001. [DOI] [PubMed] [Google Scholar]
- 11.Kim KS, Cho DY, Kim YJ, Choi SM, Kim JY, Shin S, Yoon YS. The finding of new genetic polymorphism of UCP-1 A-1766G and its effects on body fat accumulation. Biochim Biophys Acta. 2005;1741:149–155. doi: 10.1016/j.bbadis.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 12.Shin HD, Kim KS, Cha MH, Yoon Y. The effects of UCP-1 polymorphisms on obesity phenotypes among Korean female subjects. Biochem Biophys Res Commun. 2005;335:624–630. doi: 10.1016/j.bbrc.2005.07.096. [DOI] [PubMed] [Google Scholar]
- 13.Mori H, Okazawa K, Iwamoto E, Maeda M, Hashiramoto M, Kasuga M. A polymorphism in the 5’untranslated region and a Met229→Leu variant in exon 5 of the human UCP1 gene are associated with susceptibility to type II diabetes mellitus. Diabetologia. 2001;44:373–376. doi: 10.1007/s001250051629. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama K, Miyashita H, Yanagisawa Y, Iwamoto S. Seasonal effects of UCP1 gene polymorphism on visceral fat accumulation in Japanese adults. PLoS One. 2013;8:e74720. doi: 10.1371/journal.pone.0074720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicoletti CF, de Oliveira AP, Brochado MJ, de Oliveira BP, Pinhel MA, Marchini JS, dos Santos JE, Salgado Junior W, Silva Junior WA, Nonino CB. UCP1 3826 a>G polymorphism affects weight, fat mass, and risk of type 2 diabetes mellitus in grade III obese patients. Nutrition. 2016;32:83–87. doi: 10.1016/j.nut.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 16.Catalán V, Gómez-Ambrosi J, Rodríguez A, Silva C, Rotellar F, Gil MJ, Cienfuegos JA, Salvador J, Frühbeck G. Expression of caveolin-1 in human adipose tissue is upregulated in obesity and obesity-associated type 2 diabetes mellitus and related to inflammation. Clin Endocrinol. 2008;68:213–219. doi: 10.1111/j.1365-2265.2007.03021.x. [DOI] [PubMed] [Google Scholar]
- 17.Jelinek D, Heidenreich RA, Erickson RP, Garver WS. Decreased Npc1 gene dosage in mice is associated with weight gain. Obesity. 2009;18:1457–1459. doi: 10.1038/oby.2009.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garver WS, Horwood JL, Erickson RP, Heidenreich RA, Jelinek D, Meaney JF. The Niemann-pick C1 gene interacts with a high-fat diet to promote weight gain in young mice. Obesity. 2010;18:S54. doi: 10.1038/oby.2009.432. [DOI] [PubMed] [Google Scholar]
- 19.Meyre D, Delplanque J, Chèvre JC, Lecoeur C, Lobbens S, Gallina S, Durand E, Vatin V, Degraeve F, Proença C, Gaget S, Körner A, Kovacs P, Kiess W, Tichet J, Marre M, Hartikainen AL, Horber F, Potoczna N, Hercberg S, Levy-Marchal C, Pattou F, Heude B, Tauber M, McCarthy MI, Blakemore AI, Montpetit A, Polychronakos C, Weill J, Coin LJ, Asher J, Elliott P, Järvelin MR, Visvikis-Siest S, Balkau B, Sladek R, Balding D, Walley A, Dina C, Froguel P. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 20.Wu L, Xi B, Zhang M, Shen Y, Zhao X, Cheng H, Hou D, Sun D, Ott J, Wang X, Mi J. Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children. Diabetes. 2010;59:3085–3089. doi: 10.2337/db10-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xi B, Wang C, Wu L, Zhang M, Shen Y, Zhao X, Wang X, Mi J. Influence of physical inactivity on associations between single nucleotide polymorphisms and genetic predisposition to childhood obesity. Am J Epidemiol. 2011;173:1256–1262. doi: 10.1093/aje/kwr008. [DOI] [PubMed] [Google Scholar]
- 22.Mejía-Benítez A, Klünder-Klünder M, Yengo L, Meyre D, Aradillas C, Cruz E, Pérez-Luque E, Malacara JM, Garay ME, Peralta-Romero J, Flores-Huerta S, García-Mena J, Froguel P, Cruz M, Bonnefond A. Analysis of the contribution of FTO, NPC1, ENPP1, NEGR1, GNPDA2 and MC4R genes to obesity in Mexican children. BMC Med Genet. 2013;14:21. doi: 10.1186/1471-2350-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Amri AM, Vatte C, Cyrus C, Chathoth S, Hashim TM, Mohamed YS, Al Ali R, Alsaid A, Al Ali A. Novel mutations of PIK3CA gene in head and neck squamous cell carcinoma. Cancer Biomark. 2016;16(3):377–383. doi: 10.3233/CBM-160576. [DOI] [PubMed] [Google Scholar]
- 24.Ramos AV, Bastos-Rodrigues L, Resende BA, Friedman E, Campanha-Versiani L, Miranda DM, Sarquis M, De Marco L. The contribution of FTO and UCP-1 SNPs to extreme obesity, diabetes and cardiovascular risk in Brazilian individuals. BMC Med Genet. 2012;13:101. doi: 10.1186/1471-2350-13-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brondani LA, Assmann TS, de Souza BM, Bouças AP, Canani LH, Crispim D. Meta-analysis reveals the association of common variants in the uncoupling protein (UCP) 1-3 genes with body mass index variability. PLoS One. 2014;9(5):e96411. doi: 10.1371/journal.pone.0096411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalgaard LT, Pedersen O. Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and type II diabetes. Diabetologia. 2001;44:946–965. doi: 10.1007/s001250100596. [DOI] [PubMed] [Google Scholar]
- 27.Esterbauer H, Oberkofler H, Liu YM, Breban D, Hell E, Krempler F, Patsch W. Uncoupling protein-1 mRNA expression in obese human subjects: the role of sequence variations at the uncoupling protein-1 gene locus. J Lipid Res. 1998;39(4):834–844. [PubMed] [Google Scholar]
- 28.Oppert JM, Vohl MC, Chagnon M, Dionne FT, Cassard-Doulcier AM, Ricquier D, Pérusse L, Bouchard C. DNA polymorphism in the uncoupling protein (UCP) gene and human body fat. Int J Obes Relat Metab Disord. 1994;18(8):526–531. [PubMed] [Google Scholar]
- 29.Cassard-Doulcier AM, Bouillaud F, Chagnon M, Gelly C, Dionne FT, Oppert JM, Bouchard C, Chagnon Y, Ricquier D. The Bcl I polymorphism of the human uncoupling protein (ucp) gene is due to a point mutation in the 5′-flanking region. Int J Obes Relat Metab Disord. 1996;20(3):278–279. [PubMed] [Google Scholar]
- 30.Hayakawa T, Nagai Y, Taniguchi M, Yamashita H, Takamura T, Abe T, Nomura G, Kobayashi K. Phenotypic characterization of the beta3-adrenergic receptor mutation and the uncoupling protein 1 polymorphism in Japanese men. Metabolism. 1999;48(5):636–640. doi: 10.1016/S0026-0495(99)90063-X. [DOI] [PubMed] [Google Scholar]
- 31.Heilbronn LK, Kind KL, Pancewicz E, Morris AM, Noakes M, Clifton PM. Association of −3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia. 2000;43(2):242–244. doi: 10.1007/s001250050036. [DOI] [PubMed] [Google Scholar]
- 32.Ramis JM, González-Sánchez JL, Proenza AM, Martínez-Larrad MT, Fernández-Pérez C, Palou A, Serrano-Ríos M. The Arg64 allele of the beta 3-adrenoceptor gene but not the −3826G allele of the uncoupling protein 1 gene is associated with increased leptin levels in the Spanish population. Metabolism. 2004;53(11):1411–1416. doi: 10.1016/j.metabol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Urhammer SA, Hansen T, Borch-Johnsen K, Pedersen O. Studies of the synergistic effect of the Trp/Arg64 polymorphism of the beta3-adrenergic receptor gene and the −3826 A-->G variant of the uncoupling protein-1 gene on features of obesity and insulin resistance in a population-based sample of 379 young Danish subjects. J Clin Endocrinol Metab. 2000;85(9):3151–3154. doi: 10.1210/jcem.85.9.6790. [DOI] [PubMed] [Google Scholar]
- 34.Nieters A, Becker N, Linseisen J. Polymorphisms in candidate obesity genes and their interaction with dietary intake of n-6 polyunsaturated fatty acids affect obesity risk in a sub-sample of the EPIC-Heidelberg cohort. Eur J Nutr. 2002;41(5):210–221. doi: 10.1007/s00394-002-0378-y. [DOI] [PubMed] [Google Scholar]
- 35.Mottagui-Tabar S, Hoffstedt J, Brookes AJ, Jiao H, Arner P, Dahlman I. Association of ADRB1 and UCP3 gene polymorphisms with insulin sensitivity but not obesity. Horm Res. 2008;69(1):31–36. doi: 10.1159/000111793. [DOI] [PubMed] [Google Scholar]
- 36.Shen ZN, Wang XS, Bai H, Fan P, Liu R, Liu Y, Liu BW. Analysis of -3826A/G polymorphism in the promoter of the uncoupling protein-1 gene in Chinese non-obese and obese populations. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26(5):555–561. doi: 10.3760/cma.j.issn.1003-9406.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 37.Lee KH, Chai VY, Kanachamy SS, Say YH. Association of UCP1 -3826A/G and UCP3 -55C/T gene polymorphisms with obesity and its related traits among multi-ethnic Malaysians. Ethn Dis. 2015;25:65–71. [PubMed] [Google Scholar]
- 38.Vimaleswaran KS, Radha V, Ghosh S, Majumder PP, Rao MR, Mohan V. A haplotype at the UCP1 gene locus contributes to genetic risk for type 2 diabetes in Asian Indians (CURES-72) Metab Syndr Relat Disord. 2010;8(1):63–68. doi: 10.1089/met.2009.0039. [DOI] [PubMed] [Google Scholar]
- 39.Al-Daghri NM, Cagliani R, Forni D, Alokail MS, Pozzoli U, Alkharfy KM, Sabico S, Clerici M, Sironi M. Mammalian NPC1 genes may undergo positive selection and human polymorphisms associate with type 2 diabetes. BMC Med. 2012;10:140. doi: 10.1186/1741-7015-10-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamada T, Kotani K, Nagai N, Tsuzaki K, Matsuoka Y, Sano Y, Fujibayashi M, Kiyohara N, Tanaka S, Yoshimura M, Egawa K, Kitagawa Y, Kiso Y, Moritani T, Sakane N. Low-calorie diet-induced reduction in serum HDL cholesterol is ameliorated in obese women with the -3826 G allele in the uncoupling protein-1 gene. Tohoku J Exp Med. 2009;219(4):337–342. doi: 10.1620/tjem.219.337. [DOI] [PubMed] [Google Scholar]
- 41.Proenza AM, Poissonnet CM, Ozata M, Ozen S, Guran S, Palou A, Strosberg AD. Association of sets of alleles of genes encoding beta3-adrenoreceptor, uncoupling protein 1 and lipoprotein lipase with increased risk of metabolic complications in obesity. Int J Obes Relat Metab Disord. 2000;24(1):93–100. doi: 10.1038/sj.ijo.0801091. [DOI] [PubMed] [Google Scholar]
- 42.Nicholls DG, Locke RM. Thermogenic mechanisms in brown fat. Physiol Rev. 1984;64:1e64. doi: 10.1152/physrev.1984.64.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Ost M, Keipert S, Klaus S. Targeted mitochondrial uncoupling beyond UCP1 - the fine line between death and metabolic health. Biochimie. 2016;134:77–85. doi: 10.1016/j.biochi.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Perry RJ, Zhang D, Zhang XM, Boyer JL, Shulman GI. Controlled-release mitochondrial protonophore reverses diabetes and steatohepatitis in rats. Science. 2015;347(6227):1253–1256. doi: 10.1126/science.aaa0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossmeis M, Syrový I, Baumruk F, Flachs P, Janovská P, Kopecký J. Decreased fatty acid synthesis due to mitochondrial uncoupling in adipose tissue. FASEB J. 2000;14(12):1793–1800. doi: 10.1096/fj.99-0965com. [DOI] [PubMed] [Google Scholar]
- 46.Yamada T, Katagiri H, Ishigaki Y, Ogihara T, Imai J, Uno K, Hasegawa Y, Gao J, Ishihara H, Niijima A, Mano H, Aburatani H, Asano T, Oka Y. Signals from intra-abdominal fat modulate insulin and leptin sensitivity through different mechanisms: neuronal involvement in food-intake regulation. Cell Metab. 2006;3(3):223–229. doi: 10.1016/j.cmet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Kopecky J, Clarke G, Enerbäck S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J Clin Invest. 1995;96(6):2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kieć-Wilk B, Wybrańska I, Malczewska-Malec M, Leszczyńska-Gołabek L, Partyka L, Niedbał S, Jabrocka A, Dembińska-Kieć A. Correlation of the -3826A>G polymorphism in the promoter of the uncoupling protein 1 gene with obesity and metabolic disorders in obese families from southern Poland. J Physiol Pharmacol. 2002;53(3):477–490. [PubMed] [Google Scholar]
- 49.Fogelholm M, Valve R, Kukkonen-Harjula K, Nenonen A, Hakkarainen V, Laakso M, Uusitupa M. Additive effects of the mutations in the beta3-adrenergic receptor and uncoupling protein-1 genes on weight loss and weight maintenance in Finnish women. J Clin Endocrinol Metab. 1998;83(12):4246–4250. doi: 10.1210/jcem.83.12.5339. [DOI] [PubMed] [Google Scholar]
- 50.Clément K, Ruiz J, Cassard-Doulcier AM, Bouillaud F, Ricquier D, Basdevant A, Guy-Grand B, Froguel P. Additive effect of A-->G (−3826) variant of the uncoupling protein gene and the Trp64Arg mutation of the beta 3-adrenergic receptor gene on weight gain in morbid obesity. Int J Obes Relat Metab Disord. 1996;20(12):1062–1066. [PubMed] [Google Scholar]
- 51.Forga LI, Corbalán M, Marti A, Fuentes C, Martínez-González MA, Martínez A. Influence of the polymorphism 03826 a --> G in the UCP1 gene on the components of metabolic syndrome. An Sist Sanit Navar. 2003;26(2):231–236. doi: 10.4321/S1137-66272003000300005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genotypic distribution among patient and control cohort. Genotypic odds ratio for all cases and controls, unadjusted and adjusted for Age, Sex and T2D. Table S2. Genotypic distribution among stratified cohort. Genotypic odds ratio among patient population stratified for BMI; moderate-obese and extreme-obese groups. Table S3. Distribution of risk alleles in normal and abnormal levels of biochemical parameters. Association of the risk alleles frs1800592 and rs3811791 with HDL, LDL, Triglycerides and total cholesterol. (DOCX 42 kb)
Data Availability Statement
All the study materials and analysis data are available with the corresponding author.