Abstract
Background
Colorectal sessile serrated adenoma/polyps (SSA/Ps) are considered early precursor lesions in the serrated neoplasia pathway. Recent studies have shown associations of SSA/Ps with lost MLH1 expression, a CpG island methylator phenotype, and BRAF mutations. However, the molecular biological features of SSA/Ps with early neoplastic progression have not yet been fully elucidated, owing to the rarity of cases of SSA/P with advanced histology such as cytologic dysplasia or invasive carcinoma. In this study, we aimed to elucidate the molecular biological features of SSA/Ps with dysplasia/carcinoma, representing relatively early stages of the serrated neoplasia pathway.
Methods
We performed immunostaining for β-catenin, MLH1, and mucins (e.g., MUC2, MUC5AC, MUC6, and CD10); targeted next-generation sequencing; and microsatellite instability (MSI) testing in 8 SSA/P lesions comprised of 4 SSA/Ps with high-grade dysplasia and 4 SSA/Ps with submucosal carcinoma.
Results
Lost MLH1 expression was found in 5 cases. All lesions studied were positive for nuclear β-catenin expression. Regarding phenotypic mucin expression, all lesions were positive for MUC2, but negative for CD10. MUC5AC and MUC6 positivity was observed in 7 cases. Genetically, the most frequently mutated gene was BRAF (7 cases), and other mutations were detected in FBXW7 (3 cases); TP53 (2 cases), and KIT, PTEN, SMAD4, and SMARCB1 (1 case each). Furthermore, 4 of 8 lesions were MSI-high and the remaining 4 lesions were microsatellite-stable (MSS). Interestingly, all 4 MSI-high lesions displayed MLH1 loss, 3 of which harbored a FBXW7 mutation, but not a TP53 mutation. However, 2 MSS lesions harbored a TP53 mutation, although none harbored a FBXW7 mutation.
Conclusions
SSA/Ps with dysplasia/carcinoma frequently harbored BRAF mutations. Activation of the WNT/β-catenin signaling pathway may facilitate the development of dysplasia in SSA/Ps and progression to carcinoma. Furthermore, our results suggested that these lesions might be associated with both MSI-high and MSS colorectal cancer, which might be distinguished by distinct molecular biological features such as lost MLH1 expression, FBXW7 mutations, and TP53 mutations.
Keywords: Sessile serrated adenoma/polyp, Colorectal carcinoma, MLH1, FBXW7, TP53, Microsatellite instability
Background
In 2003, Torlakovic et al. [1] reported evidence of abnormal proliferation in colorectal serrated polyps that superficially resembled hyperplastic polyps (but that could be distinguished histologically based on their abnormal architectural features) and introduced the terms “sessile serrated polyp” and “sessile serrated adenoma” to describe their observations. Currently, this category is designated as “sessile serrated adenoma/polyp (SSA/P),” as recommended by the World Health Organization [2]. SSA/P is considered as an early precursor lesion in the serrated neoplasia pathway, which largely results in colorectal carcinomas with high levels of microsatellite instability (MSI-high) [3–5]. Recent studies have shown associations of SSA/Ps and those with dysplasia/carcinoma with DNA methylation or lost protein expression of DNA-repair genes (i.e., MLH1) [1, 4, 6–9], a CpG island methylator phenotype [3, 4, 6, 7], and BRAF mutations [3, 4, 6–13]. This pathway is thought to be distinct from the conventional adenoma-carcinoma pathway, where adenomas progress to invasive colorectal carcinomas through the influence of several genetic alterations including adenomatous polyposis coli (APC) and KRAS mutations [4, 6, 10, 11, 14, 15].
Recently, targeted next-generation sequencing (NGS) has shown unprecedented potential for detecting underlying changes in the genetic architecture of cancer in a comprehensive and economically feasible manner, and the development of its platforms has enabled comprehensive analysis of genetic alterations in tumors [16, 17]. A comprehensive understanding of the genetic alterations associated with cancer could improve the molecular biological classifications of tumors and identify effective molecularly targeted therapies.
At present, the molecular biological features underlying the development of colorectal serrated neoplasia remain unclear. Hence, the aim of this study was to employ NGS to elucidate the molecular biological features of SSA/Ps with dysplasia/carcinoma, representing relatively early stages of the serrated neoplasia pathway.
Methods
Patients and materials
Eight colorectal lesions (from 8 different patients) were resected endoscopically or surgically at Juntendo University Hospital between 2014 and 2016, and used in this study. These lesions comprised 4 SSA/Ps with high-grade dysplasia and 4 SSA/Ps with submucosal carcinoma. The diagnosis of SSA/P was based on the following criteria described by Torlakovic et al. [1]: the presence of serration at the base of crypts, irregularly dilated crypts, irregularly branching crypts, and horizontally and/or laterally arranged basal crypts. The histologic features of the high-grade dysplasia were assessed as described previously [2, 12], as follows: a tubular, tubulovillous, or fused glandular pattern (mimicking conventional adenomatous high-grade dysplasia) or a serrated glandular pattern (preserving the serrated or saw-toothed structure with infolding of the crypt epithelium), which consisted of cuboidal and eosinophilic dysplastic cells with substantially larger nuclei and irregular thickening of the nuclear membrane (i.e. ‘serrated-type’ high-grade dysplasia). Submucosal carcinoma was defined as an obvious epithelial neoplasm histologically atypical enough to be diagnosed as at least high-grade dysplasia that was invading the muscularis mucosa into the submucosa. The inclusion criteria for SSA/P with high-grade dysplasia or with submucosal carcinoma included a component of ordinary SSA/P visible at the lesion edge that comprised at least 3 crypts, with an SSA/P-type histology required in 1 crypt. All samples were reviewed independently by 2 authors (TY and TM).
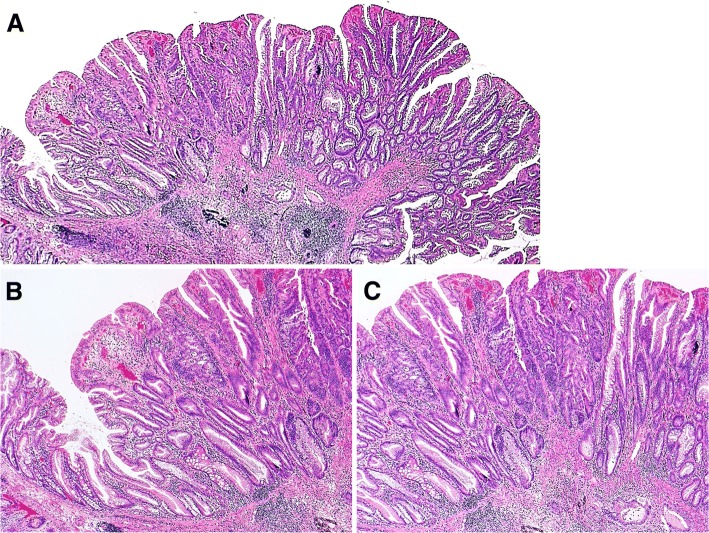
Typical morphologies in representative cases of SSA/P with submucosal carcinoma are shown in Fig. 1.
Fig. 1.
Typical morphologies in a representative case of sessile serrated adenoma/polyp (SSA/P) with submucosal carcinoma (Case #6). a Dilated crypts with deep serration was seen on both sides of the lesion, and high-grade dysplasia with submucosal invasion was seen in the middle. b High-power field of (a) (the left side). Crypts with a serrated architecture included those that were irregularly dilated, irregularly branched, and horizontally arranged (basal), corresponding to SSA/P. c High-power field of (a) (the middle). Well-differentiated tubular adenocarcinoma was observed invading the submucosa. Adjacent SSA/P areas were observed towards both sides of the panel. An abrupt transition was evident between the 2 adjacent regions
Clinicopathological analysis
For each patient, we recorded the age, sex, tumor location (proximal colon defined as proximal to the splenic flexure, remaining colon defined as distal), macroscopic type, whole tumor size (including both high-grade dysplasia or invasive carcinoma and in situ SSA/P), histological type (based on the World Health Organization’s histological classification of tumors), depth of submucosal invasion, lymphovascular invasion, and lymph node metastasis.
Immunohistochemical analysis
Four-micrometer-thick serial tissue sections prepared from formalin-fixed and paraffin-embedded tissues were subjected to immunohistochemistry. Staining was performed using a Dako EnVision Kit with antibodies against MLH1 (ab14206; 1:50 dilution; Abcam, CA, USA) and β-catenin (clone 14, 1:200 dilution; BD Bioscience, San Diego, CA, USA). Tumor phenotypes were evaluated by immunostaining with antibodies from Novocastra (Newcastle upon Tyne, UK; 1:100 dilution) against MUC2 (NCL-MUC-2), MUC5AC (NCL-MUC-5 AC), MUC6 (NCL-MUC-6), and CD10 (NCL-CD10–270). Appropriate positive and negative controls were used for each antibody.
Loss of MLH1 expression was noted when one or more clusters of tumor cells (minimal, focal, or multifocal) or all malignant cells showed no nuclear staining, compared with positive nuclear staining in normal epithelial cells and lymphocytes [1]. Expression of β-catenin, which generally showed an inverse relationship between membranous and nuclear (with cytoplasmic) reactivity, was only evaluated in terms of nuclear localization in this study. Nuclear β-catenin expression was considered as positive when distinct and strong nuclear staining was observed in over 5% of the tumor cells [9]. Membrane staining for CD10, and cytoplasmic staining for MUC2, MUC5AC, and MUC6 were judged as positive, when over 5% of tumor cells showed a positive reaction for each marker. The immunohistochemical staining results were evaluated by 2 authors (TY and TM).
DNA extraction
Genomic DNA was extracted from 5 formalin-fixed paraffin-embedded sections (10-mm-thick) using a QIAamp DNA FFPE Tissue Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s instructions. Sections were stained lightly with hematoxylin, and only the dysplastic or carcinomatous areas in the lesions were microdissected with direct observation of the tissue under a light microscope. The DNA quality and integrity were checked spectrophotometrically.
Targeted NGS
The 50-gene Ion AmpliSeq Cancer Hotspot Panel v2 (Life Technologies) was used with the Ion-Torrent ™ Personal Genome Machine platform (Life Technologies, Foster city, CA, USA) in all experiments. This panel is designed to amplify 207 amplicons covering approximately 2800 mutations deposited in the COSMIC database from 50 oncogenes and tumor-suppressor genes commonly mutated in human cancers (ABL1, AKT1, ALK, APC, ATM, BRAF, CDH1, CDKN2A, CSF1R, CTNNB1, EGFR, ERBB2, ERBB4, EZH2, FBXW7, FGFR1, FGFR2, FGFR3, FLT3, GNA11, GNAS, GNAQ, HNF1A, HRAS, IDH1, IDH2, JAK2, JAK3, KDR, KIT, KRAS, MET, MLH1, MPL, NOTCH1, NPM1, NRAS, PDGFRA, PIK3CA, PTEN, PTPN11, RB1, RET, SMAD4, SMARCB1, SMO, SRC, STK11, TP53, and VHL). The Ion AmpliSeq Library Kit, version 2.0 (Life Technologies) was used to amplify 10 ng of DNA according to the manufacturer’s instructions. Sequencing beads were templated and enriched using the Hi-Q Template OT2 200 Kit, and sequencing was performed on 318v2 chips using the Hi-Q Sequencing Kit (Life Technologies) according to the manufacturer’s protocols. Signal processing, mapping, and quality control were performed with Torrent Suite, v.5.0 (Life Technologies). Sequence variants were called using Ion Reporter, v5.2 using the AmpliSeq CHPv2 single-sample workflow and default settings. Variants were categorized according to whether they comprised a nonsynonymous or frameshift mutation, or stop codon in the exonic region. The limit of detection was a 5% mutational allelic frequency at 500 × coverage or a 3% mutational allelic frequency at 1000 × coverage for each tested region. The minimum coverage depth was 500 × .
MSI testing
DNA extracted from microdissected paraffin-embedded tumor sections and non-neoplastic tissues was analyzed by a polymerase chain reaction–based method, followed by capillary electrophoretic detection. MSI detection was performed using a panel of 5 mononucleotide microsatellite markers (BAT-25, BAT-26, NR-21, NR-24, and MONO-27). In accordance with National Cancer Institute guidelines, MSI at 2 loci or more was defined as MSI-high, instability at a single locus was defined as low levels of MSI (MSI-low), and no instability at any of the loci tested was defined as microsatellite-stable (MSS).
Results
Clinicopathological analysis
The detailed clinicopathological findings of the 8 SSA/P lesions are shown in Table 1. This study included 2 male and 6 female SSA/P patients, with ages ranging from 61 to 79 years (mean 69 years). Six lesions were within the proximal colon, and the remaining 2 lesions were in the distal colon. Macroscopically, sessile morphology was frequently observed among the studied SSA/P lesions. The mean tumor size was 18 mm (range: 10 to 31 mm).
Table 1.
Summary of clinicopathological features in each case of sessile serrated adenoma/polyp (SSA/P) with dysplasia and invasive adenocarcinoma
| Case No. | Age | Sex | Location | Macroscopic type | Size (mm) | Histological type | Depth of invasion (μm) | Mucinous component | Lymphatic invasion | Vascular invasion | Lymph node metastasis | Removal method |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 67 | F | A | Sessile | 10 | HGD | Mucosa | – | – | – | – | EMR |
| 2 | 69 | M | S | Semipedunculated | 10 | HGD | Mucosa | – | – | – | – | EMR |
| 3 | 79 | F | C | Sessile | 14 | HGD | Mucosa | – | – | – | – | EMR |
| 4 | 61 | F | A | Sessile | 18 | HGD | Mucosa | – | – | – | – | ESD |
| 5 | 61 | M | D | Sessile | 31 | Well-ACA | Submucosa (400) | – | – | – | – | ESD + OPE |
| 6 | 75 | F | A | Sessile | 15 | Well-ACA | Submucosa (1100) | – | – | – | – | EMR + OPE |
| 7 | 73 | F | A | Sessile | 25 | Mod-ACA | Submucosa (2000) | – | + | – | + | OPE |
| 8 | 65 | F | T | Sessile | 19 | Mod-ACA | Submucosa (4000) | + | – | – | – | OPE |
M Male, F Female, C Cecum, A Ascending colon, T Transverse colon, D Descending colon, S Sigmoid colon, HGD High-grade dysplasia, Well-ACA Well-differentiated adenocarcinoma, Mod-ACA Moderately-differentiated adenocarcinoma, EMR Endoscopic mucosal resection, ESD Endoscopic submucosal dissection, OPE Operation, + present; − absent
Histologically, 4 lesions were high-grade dysplasias, and the other 4 lesions were well-to-moderately differentiated tubular adenocarcinomas invading into the submucosa, of which 1 lesion was accompanied with a mucinous component in the submucosa. Lymphatic invasion and lymph node metastasis were found in 1 case of SSA/P with carcinoma.
Immunohistochemical analysis
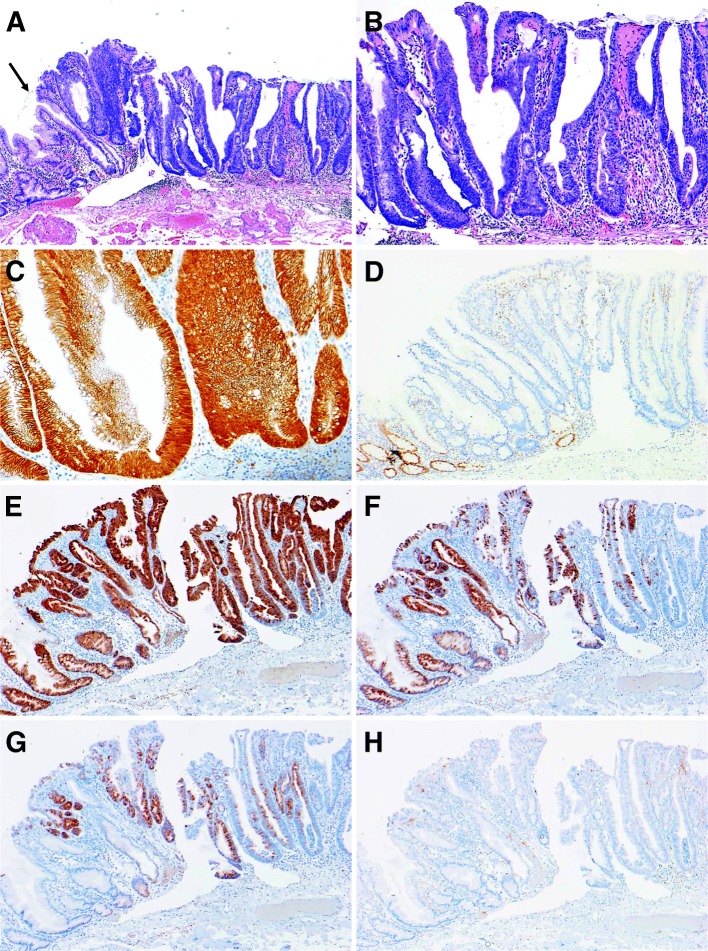
Loss of MLH1 expression was found in 5 cases. Without exception, all lesions were positive for nuclear β-catenin expression. Regarding phenotypic mucin expression, all lesions were positive for MUC2, but negative for CD10. MUC5AC and MUC6 were positive in 7 of 8 cases (88%). Immunohistochemical staining of these proteins in a representative case is illustrated in Fig. 2.
Fig. 2.
Immunohistochemical staining in a representative case of sessile serrated adenoma/polyp (SSA/P) with dysplasia (Case #2). a Dilated crypts and deep serration, corresponding to SSA/P, were seen on the left side, and high-grade dysplasia without submucosal invasion was seen on the right side. An abrupt transition was seen between the 2 adjacent regions (arrow). b High-power field of (a) (the right side). A view shows high-grade dysplasia without submucosal invasion, which was pathologically consistent with SSA/P with cytologic dysplasia. The lesion showed nuclear β-catenin expression (c) and a loss of MLH1 expression (d). Regarding phenotypic mucin expression, the lesion was positive for MUC2 (e), MUC5AC (f), and MUC6 (g) expression, and negative for CD10 (H) expression
Mutation analysis
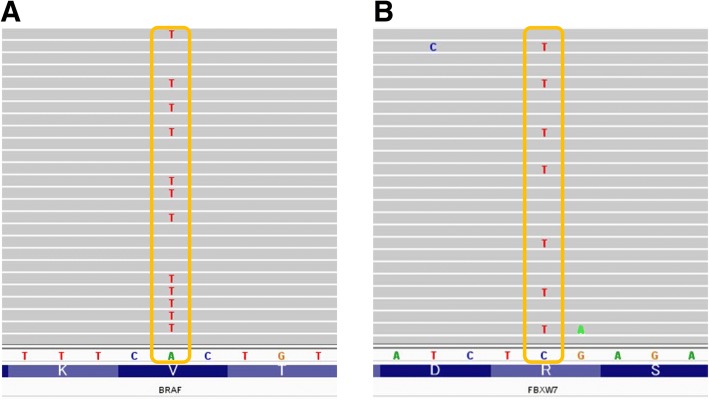
All SSA/P lesions had acceptable DNA integrity (the average RQ value was 0.18) and were subjected to NGS. The genetic alterations identified by NGS in 8 SSA/P lesions are summarized in Table 2. The most frequently mutated gene was BRAF (7 of 8; 88%), and the other detected mutations were in FBXW7 (3 of 8; 38%), TP53 (2 of 8; 25%), KIT (1 of 8; 13%), PTEN (1 of 8; 13%), SMAD4 (1 of 8; 13%), and SMARCB1 (1 of 8; 13%). The only BRAF mutation found was V600E (c.1799 T > A). The FBXW7 mutations found were R278X (c.832 C > T), R479Q (c.1436 G > A), and Q508Q (c.1524 A > G). Examples of data analysis using the Integrative Genomics Viewer ™ software are shown in Fig. 3.
Table 2.
Summary of molecular biological features in each case of sessile serrated adenoma/polyp (SSA/P) with dysplasia and invasive adenocarcinoma
| Case No. | Group | Immunohistochemistry | Next generation sequencing analysis (Gene mutations) | MSI analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLH1 loss | β-catenin | MUC2 | MUC5AC | MUC6 | CD10 | BRAF | FBXW7 | KIT | PTEN | SMAD4 | SMARCB1 | TP53 | |||
| 1 | Dysplasia | – | + | + | + | + | – | + | – | – | – | – | – | + | MSS |
| 2 | Dysplasia | + | + | + | + | + | – | + | – | – | – | – | – | – | MSI-high |
| 3 | Dysplasia | + | + | + | + | + | – | + | + | + | – | + | – | – | MSI-high |
| 4 | Dysplasia | – | + | + | + | + | – | + | – | – | + | – | + | – | MSS |
| 5 | Carcinoma | + | + | + | + | + | – | + | – | – | – | – | – | – | MSS |
| 6 | Carcinoma | – | + | + | – | – | – | + | – | – | – | – | – | + | MSS |
| 7 | Carcinoma | + | + | + | + | + | – | – | + | – | – | – | – | – | MSI-high |
| 8 | Carcinoma | + | + | + | + | + | – | + | + | – | – | – | – | – | MSI-high |
Dysplasia, SSA/P with high-grade dysplasia; Carcinoma, SSA/P with submucosal carcinoma; MSI-high, Microsatellite instability-high; MSS, Microsatellite-stable; +, present; −, absent
Fig. 3.
Representative examples of mutations detected by next-generation sequencing. a Case #1, BRAF mutation, V600E (c.1799 T > A). b Case #3, FBXW7 mutation, R479Q (c.1436 G > A)
MSI testing
Of the 8 SSA/P lesions, 4 lesions (including 2 SSA/Ps with dysplasia and 2 SSA/Ps with carcinoma) were MSI-high and the remaining 4 lesions were MSS (Table 2).
Associations between immunohistochemistry, mutation analysis, and MSI
All 4 MSI-high SSA/P lesions showed a loss of MLH1 expression, and 3 of those lesions harbored an FBXW7 mutation, but not a TP53 mutation. In contrast, among the SSA/P lesion with MSS, only 1 showed loss of MLH1 expression, 2 cases harbored a TP53 mutation, and no cases harbored an FBXW7 mutation.
Discussion
Rare occurrences of BRAF mutations have been documented for conventional colorectal carcinomas, although they are frequent in dysplasia/carcinoma arising from SSA/Ps (50–90%) [3, 4, 9, 10, 12, 13]. In this study, BRAF mutations were detected in 7 of 8 SSA/P lesions, in accordance with previous reports [3, 4, 9, 10, 12, 13]. BRAF mutations result in activation of the RAS-RAF-MAPK pathway, and therefore our findings and previous reports [3, 4, 9, 10, 12, 13] suggest that those signaling pathways might be activated in serrated neoplasia.
We investigated the core protein-expression levels of MUC2, MUC5AC, MUC6, and CD10 because altered expressions of these proteins may be significantly correlated with the biological behavior of colorectal carcinoma and, possibly its prognosis. In this study, MUC2, MUC5AC, and MUC6 expression was frequently positive in the serrated lesions. MUC2 is a goblet cell-type mucin predominantly expressed in the colon. In contrast, MUC5AC and MUC6 are two gastric type mucins that are expressed in the surface foveolar epithelium and deep antral/pyloric glands, respectively, but not normally in colonic mucosa. MUC5AC and MUC6 expression have been strongly associated with tumorigenesis via the serrated neoplasia pathway [18–20], in agreement with our current findings.
The WNT signaling pathway involves β-catenin and plays a crucial role in the development of colorectal carcinomas through a conventional adenoma-carcinoma progression [21]. Several previous reports including our own [8, 9, 12, 13, 22] have also demonstrated that nuclear β-catenin accumulation is common in SSA/Ps with dysplasia and invasive carcinoma, but not in those without dysplasia. This observation implies that activation of the WNT/β-catenin signaling pathway is involved in the development of dysplasia in SSA/Ps and progression to carcinoma. Although APC and CTNNB1 mutations are major causes of WNT/β-catenin signaling activation in conventional-type adenomas, these genetic alterations are absent in SSA/Ps [13]. In this study, all serrated lesions examined also displayed nuclear β-catenin immunoreactivity, whereas no mutations in WNT/β-catenin signaling-associated genes (such as APC and CTNNB1) were found in the studied lesions. Previously, we reported that activation of the WNT/β-catenin signaling pathway might be associated with methylation of the associated genes, including SFRP4, MCC, and AXIN2 [9]. This association could explain at least some of the discrepancies between β-catenin immunoreactivity and the lack of associated gene mutations.
The MSI phenotype has been regarded as a main subtype of colorectal cancers. Sporadic colorectal cancers with the MSI-high phenotype account for approximately 3–15% of all colorectal cancers [23]. MSI is a unique molecular alteration induced by deficiencies in the DNA-mismatch repair system and is characterized by unstable (variable length) microsatellites, a type of simple DNA sequence repeat. Some previous studies have shown that serrated neoplasia is associated with MSI-high colorectal carcinomas, which is accompanied with DNA methylation or loss of protein expression of DNA-repair genes such as MLH1 [3–5]. In our study, all 4 MSI-high SSA/P lesions showed a loss of MLH1 expression, in accordance with previous reports [3–5].
A recent report by the Cancer Genome Atlas Project elucidated the molecular landscape of colorectal cancers and revealed that the hypermutated phenotype mainly overlaps with MSI status in colorectal cancers [24]. Many types of genetic mutations can occur in MSI-high colorectal cancers. Indeed, a previous report showed that mutations in various genes, including the tumor-suppressor gene PTEN and the oncogene PIK3CA, were caused by the instability of microsatellites in MSI-high colorectal cancers [25]. In this study, no MSI-high lesions harbored PTEN or PIK3CA mutations. However, it is interesting that 3 of 4 MSI-high lesions harbored an FBXW7 mutation, whereas no MSS lesions harbored an FBXW7 mutation. FBXW7 is a tumor-suppressor gene located on human chromosome 4q that encodes a substrate-recognition component of SKP1–Cullin1–F-box protein-ubiquitin E3 ligase complexes [26]. These specific E3 ligase complexes negatively regulate the intracellular abundance of an expanding list of key oncogenic proteins such as cyclin E [27], c-JUN [28, 29], c-MYC [30, 31], MCL1 (myeloid cell leukemia 1) [32, 33], NOTCH [34–36], AURKA (aurora kinase A) [37, 38], KLF5 (Krüppel-like factor 5) [39], mTOR [40], and TGIF1 [41]. Therefore, the loss of FBXW7 function results in accumulation of its substrates, which leads to oncogenesis and progression of multiple cancers including colorectal cancers [42, 43]. A study of over 500 primary tumors of diverse tissue origins suggested that FBXW7 mutations occurred in approximately 6% of all evaluated tumors. Of these, the most commonly affected tumors were cholangiocarcinoma (35%), T-cell acute lymphocytic leukemia (31%), endometrial cancer (9%), and gastric cancer (6%) [43]. FBXW7 has also consistently been identified as one of the most commonly mutated genes in colorectal cancer, being observed in 6 to 10% of all cases [43, 44]. Furthermore Chang et al. reported that FBXW7-mutant colorectal cancers had significant associations with MSI-high tumors in a large-scale study of 1519 cases [45]. Our findings and those previous reports indicated that FBXW7 mutations might potentially be involved in the progression of MSI-high serrated lesions.
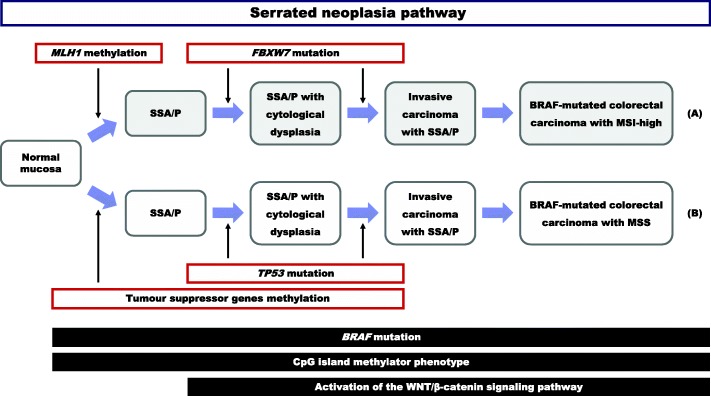
Activation of the second arm of the serrated neoplasia pathway, also driven by CpG island methylation of unspecified tumor-suppressor genes but not DNA-repair genes, may indicate progression to BRAF-mutant MSS carcinoma [46, 47]. Furthermore, as reported in earlier works [48–50], a strong inverse correlation was found between TP53 alterations and the MSI phenotype. In this study, 2 of 4 lesions with MSS harbored a TP53 mutation, whereas no MSI-high lesions harbored a TP53 mutation, similar to previous observations [48–50]. The TP53 gene encodes a tumor-suppressor protein containing transcriptional-activation, DNA-binding, and oligomerization domains. The encoded protein responds to diverse cellular stresses to regulate the expression of target genes, thereby inducing cell cycle arrest, apoptosis, senescence, DNA repair, or changes in metabolism [51]. Mutations in the TP53 gene are reportedly associated with a variety of human cancers, including colon, breast, lung, and brain cancers [51]. Our findings and those previous reports indicated that TP53 mutations are potentially involved in the progression of MSS serrated lesions. A schematic depiction of differences in the expression of key proteins and genetic alterations in the serrated neoplasia pathway is shown in Fig. 4.
Fig. 4.
Schematic representation of differences in the molecular biological expressions and genetic alterations in the serrated neoplasia pathway. Sessile serrated adenoma/polyp (SSA/P) is an early precursor lesion in the serrated neoplasia pathway that progresses to cytological dysplasia and results in BRAF-mutated colorectal carcinomas that are commonly high levels of microsatellite instability (MSI-high) (diagram A) or microsatellite-stable (MSS) (diagram B). Both pathways are associated with a CpG island methylator phenotype and WNT/β-catenin signaling activation. a The upper arm, driven by BRAF mutation and MLH1 methylation, indicates progression to BRAF-mutated MSI-high carcinoma. FBXW7 mutations are potentially involved in progression of this pathway. b The lower arm, driven by BRAF mutation and methylation of unspecified tumor-suppressor genes, involves progression to BRAF-mutated MSS carcinoma. TP53 mutations are potentially involved in progression of this pathway
Our study had a major limitation; the sample size was very small. A major impediment to investigating dysplasia/carcinoma arising in SSA/P is the rarity of this lesion. Additionally, a sufficient amount of specimen is necessary for NGS analysis, which limited the lesions that could be studied to those of 10 mm in diameter or more. These limitations lowered the statistical power of this study. Particularly, although the high rate of FBXW7 or TP53 mutations in the studied lesions is important, the possibility that this result was a chance occurrence cannot be denied, considering that only 2 or 3 cases of the studied lesions harbored TP53 or FBXW7 mutations, respectively.
To the best of our knowledge, few comprehensive genetic studies have been conducted in SSA/Ps with dysplasia and invasive carcinoma. Our results revealed mutations in BRAF, FBXW7, TP53, KIT, PTEN, SMAD4, and SMARCB1. Furthermore, it is very interesting that all MSI-high lesions might be associated with MLH1 expression loss and mutation of FBXW7, but not TP53. Our results may help clarify the detailed mechanism of serrated neoplasia development. A comprehensive understanding of genetic alterations associated with the serrated neoplasia pathway could help identify effective molecularly targeted therapies.
Conclusions
In conclusion, colorectal SSA/Ps with dysplasia and invasive carcinoma frequently harbored BRAF mutations and showed nuclear β-catenin expression. Furthermore, these lesions might not only be associated with MSI-high colorectal cancer, but also MSS, and MSI-high and MSS serrated lesions might have distinct genetic features (such as FBXW7 and TP53 mutations). BRAF-mutant MSS colon carcinomas are particularly important because they have a dismal prognosis and an aggressive clinical course with adverse histologic features, such as lymphatic and perineural invasion and high tumor budding [52, 53]. Further investigations are required to elucidate molecular biological characteristics in the serrated neoplasia pathway in greater detail.
Acknowledgements
The work was supported in part by a Grant-in-Aid from the Japan Society for the Promotion of Science (#18 K15796 to T. Murakami).
Competing interests
The authors declare that they have no competing interests.
Funding
No funds have been received for this study.
Availability of data and materials
The data supporting the conclusions are included in the article. Raw data are available upon request.
Authors’ contributions
TM, YA, and TS designed the study; TM, YA, NY, TH, NS, TS, and NS performed technical procedures and acquired pathological and clinical data; TM, YA, AN, and TY analyzed and interpreted the data; TM drafted the manuscript; all authors reviewed and approved the final version of the manuscript. All individuals listed as co-authors of the manuscript have significantly contributed to this study.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Ethical Committee of Juntendo University School of Medicine (reference number 2017166). The committee determined that additional informed consent was not required due to the retrospective nature of our analysis.
Abbreviations
- APC
Adenomatous polyposis coli
- MSI
Microsatellite instability
- MSI-high
High levels of microsatellite instability
- MSI-low
Low levels of MSI
- MSS
Microsatellite-stable
- NGS
Next-generation sequencing
- SSA/P
Sessile serrated adenoma/polyp
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Takashi Murakami, Phone: 81-3-3813-3111, Email: t-murakm@juntendo.ac.jp.
Yoichi Akazawa, Email: yakazawa@juntendo.ac.jp.
Noboru Yatagai, Email: nyataga@juntendo.ac.jp.
Takafumi Hiromoto, Email: thiromo@juntendo.ac.jp.
Noriko Sasahara, Email: n-sasa@juntendo.ac.jp.
Tsuyoshi Saito, Email: tysaitou@juntendo.ac.jp.
Naoto Sakamoto, Email: sakamoto@juntendo.ac.jp.
Akihito Nagahara, Email: nagahara@juntendo.ac.jp.
Takashi Yao, Email: tyao@juntendo.ac.jp.
References
- 1.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65–81. doi: 10.1097/00000478-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Snover DC, Ahnen DJ, Burt RW, Odze RD. Serrated polyps of the colon and rectum and serrated polyposis. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO classification of tumours of the digestive system. Lyon: IARC Press; 2010. pp. 160–165. [Google Scholar]
- 3.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien MJ, Yang S, Mack C, Xu H, Huang CS, Mulcahy E, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol. 2006;30:1491–1501. doi: 10.1097/01.pas.0000213313.36306.85. [DOI] [PubMed] [Google Scholar]
- 5.Patil DT, Shadrach BL, Rybicki LA, Leach BH, Pai RK. Proximal colon cancers and the serrated pathway: a systematic analysis of precursor histology and BRAF mutation status. Mod Pathol. 2012;25:1423–1431. doi: 10.1038/modpathol.2012.98. [DOI] [PubMed] [Google Scholar]
- 6.Kim YH, Kakar S, Cun L, Deng G, Kim YS. Distinct CpG island methylation profiles and BRAF mutation status in serrated and adenomatous colorectal polyps. Int J Cancer. 2008;123:2587–2593. doi: 10.1002/ijc.23840. [DOI] [PubMed] [Google Scholar]
- 7.Kim KM, Lee EJ, Ha S, Kang SY, Jang KT, Park CK, et al. Molecular features of colorectal hyperplastic polyps and sessile serrated adenoma/polyps from Korea. Am J Surg Pathol. 2011;35:1274–1286. doi: 10.1097/PAS.0b013e318224cd2e. [DOI] [PubMed] [Google Scholar]
- 8.Dhir M, Yachida S, Van Neste L, Glöckner SC, Jeschke J, Pappou EP, et al. Sessile serrated adenomas and classical adenomas: an epigenetic perspective on premalignant neoplastic lesions of the gastrointestinal tract. Int J Cancer. 2011;129:1889–1898. doi: 10.1002/ijc.25847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami T, Mitomi H, Saito T, Takahashi M, Sakamoto N, Fukui N, et al. Distinct WNT/β-catenin signaling activation in the serrated neoplasia pathway and the adenoma-carcinoma sequence of the colorectum. Mod Pathol. 2015;28:146–158. doi: 10.1038/modpathol.2014.41. [DOI] [PubMed] [Google Scholar]
- 10.Jass JR, Baker K, Zlobec I, Higuchi T, Barker M, Buchanan D, et al. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a ‘fusion’ pathway to colorectal cancer. Histopathology. 2006;49:121–131. doi: 10.1111/j.1365-2559.2006.02466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spring KJ, Zhao ZZ, Karamatic R, Walsh MD, Whitehall VL, Pike T, et al. High prevalence of sessile serrated adenomas with BRAF mutations: a prospective study of patients undergoing colonoscopy. Gastroenterology. 2006;131:1400–1407. doi: 10.1053/j.gastro.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 12.Fujita K, Yamamoto H, Matsumoto T, Hirahashi M, Gushima M, Kishimoto J, et al. Sessile serrated adenoma with early neoplastic progression: a clinicopathologic and molecular study. Am J Surg Pathol. 2011;35:295–304. doi: 10.1097/PAS.0b013e318205df36. [DOI] [PubMed] [Google Scholar]
- 13.Yachida S, Mudali S, Martin SA, Montgomery EA, Iacobuzio-Donahue CA. Beta-catenin nuclear labeling is a common feature of sessile serrated adenomas and correlates with early neoplastic progression following BRAF activation. Am J Surg Pathol. 2009;33:1823–1832. doi: 10.1097/PAS.0b013e3181b6da19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, et al. Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 16.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz Z, Aguilar-Mahecha A, Paquet ER, Basik M, Orain M, Camlioglu E, et al. Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine. Mod Pathol. 2013;26:1413–1424. doi: 10.1038/modpathol.2013.81. [DOI] [PubMed] [Google Scholar]
- 18.Walsh MD, Clendenning M, Williamson E, Pearson SA, Walters RJ, Nagler B, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26:1642–1656. doi: 10.1038/modpathol.2013.101. [DOI] [PubMed] [Google Scholar]
- 19.Khaidakov M, Lai KK, Roudachevski D, Sargsyan J, Goyne HE, Pai RK, et al. Gastric proteins MUC5AC and TFF1 as potential diagnostic markers of colonic sessile serrated adenomas/polyps. Am J Clin Pathol. 2016;146:530–537. doi: 10.1093/ajcp/aqw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban S, Mitomi H, Horiguchi H, Sato H, Shimizu M. Adenocarcinoma arising in small sessile serrated adenoma/polyp (SSA/P) of the colon: clinicopathological study of eight lesions. Pathol Int. 2014;64:123–132. doi: 10.1111/pin.12147. [DOI] [PubMed] [Google Scholar]
- 21.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 22.Bettington M, Walker N, Rosty C, Brown I, Clouston A, McKeone D, et al. Clinicopathological and molecular features of sessile serrated adenomas with dysplasia or carcinoma. Gut. 2017;66:97–106. doi: 10.1136/gutjnl-2015-310456. [DOI] [PubMed] [Google Scholar]
- 23.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Day FL, Jorissen RN, Lipton L, Mouradov D, Sakthianandeswaren A, Christie M, et al. PIK3CA and PTEN gene and exon mutation-specific clinicopathologic and molecular associations in colorectal cancer. Clin Cancer Res. 2013;19:3285–3296. doi: 10.1158/1078-0432.CCR-12-3614. [DOI] [PubMed] [Google Scholar]
- 26.Spruck CH, Strohmaier H, Sangfelt O, Müller HM, Hubalek M, Müller-Holzner E, et al. Hcdc4 gene mutations in endometrial cancer. Cancer Res. 2002;62:4535–4539. [PubMed] [Google Scholar]
- 27.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 28.Nateri AS, Riera-sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 29.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF (FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 34.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC: CDK8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Tetzlaff MT, Yu W, Li M, Zhang P, Finegold M, Mahon K, et al. Defective cardiovascular development and elevated cyclin e and notch proteins in mice lacking the fbw7 f-box protein. Proc Natl Acad Sci U S A. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, et al. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- 37.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 38.Kwon YW, Kim IJ, Wu D, Lu J, Stock WA, Jr, Liu Y, et al. Pten regulates Aurora-a and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol Cancer Res. 2012;10:834–844. doi: 10.1158/1541-7786.MCR-12-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R, Wang Y, Liu N, Ren C, Jiang C, Zhang K, et al. FBW7 regulates endothelial functions by targeting KLF2 for ubiquitination and degradation. Cell Res. 2013;23:803–819. doi: 10.1038/cr.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bengoechea-Alonso MT, Ericsson J. Tumor suppressor Fbxw7 regulates TGFβ signaling by targeting TGIF1 for degradation. Oncogene. 2010;29:5322–5328. doi: 10.1038/onc.2010.278. [DOI] [PubMed] [Google Scholar]
- 42.Cao J, Ge MH, Ling ZQ. Fbxw7 tumor suppressor: a vital regulator contributes to human tumorigenesis. Medicine (Baltimore) 2016;95:e2496. doi: 10.1097/MD.0000000000002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 44.Malapelle U, Pisapia P, Sgariglia R, Vigliar E, Biglietto M, Carlomagno C, et al. Less frequently mutated genes in colorectal cancer: evidences from next-generation sequencing of 653 routine cases. J Clin Pathol. 2016;69:767–771. doi: 10.1136/jclinpath-2015-203403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CC, Lin HH, Lin JK, Lin CC, Lan YT, Wang HS, et al. FBXW7 mutation analysis and its correlation with clinicopathological features and prognosis in colorectal cancer patients. Int J Biol Markers. 2015;30:e88–e95. doi: 10.5301/jbm.5000125. [DOI] [PubMed] [Google Scholar]
- 46.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 47.O'Brien MJ, Zhao Q, Yang S. Colorectal serrated pathway cancers and precursors. Histopathology. 2015;66:49–65. doi: 10.1111/his.12564. [DOI] [PubMed] [Google Scholar]
- 48.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 49.Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 50.Cottu PH, Muzeau F, Estreicher A, Fléjou JF, Iggo R, Thomas G, et al. Inverse correlation between RER+ status and p53 mutation in colorectal cancer cell lines. Oncogene. 1996;13:2727–2730. [PubMed] [Google Scholar]
- 51.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36:744–752. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the conclusions are included in the article. Raw data are available upon request.