Abstract
Background
Kid (kinesin-like DNA binding protein), a member of microtubule-dependent molecular motor proteins, also known as KIF22, is reported to be associated with carcinogenesis and cancer progression in different types of malignant tumor, but the biologic behavior and clinical outcome of KIF22 in prostate cancer (PCa) has not been well studied. This study aimed to analyze the association between KIF22 and clinical outcome in PCa patients.
Material/Methods
The expression of KIF22 in tumor specimens compared with paired paracancerous tissue from 114 patients undergoing radical prostatectomy was detected by immunohistochemistry; results were verified using The Cancer Genome Atlas (TCGA) database. Subsequently, the relationship between KIF22 expression and clinical prognosis of PCa patients was then statistically analyzed.
Results
Both immunohistochemistry and database analysis showed that KIF22 was obviously overexpressed in PCa tissues compared with paracancerous tissue. The overexpression of KIF22 at the protein level was significantly related to higher clinical stage (P=0.025), Gleason score (P=0.002), seminal vesicle invasion (P=0.007), and lymph node metastasis (P=0.009). Furthermore, with the overexpression of KIF22 mRNA level in PCa patients, the oncological prognosis of PCa patients was much poorer.
Conclusions
High-level expression of KIF22 was related to both tumor progression and adverse clinical outcome. For this reason, KIF22 may become a potential prognostic factor for PCa.
MeSH Keywords: Immunochemistry, Carcinogenesis, DNA-Binding Proteins, Prostatic Neoplasms, Survival Analysis
Background
Prostate cancer (PCa) is a common cancer in developed countries with high morbidity and mortality [1,2]. In 2018, there will be 161 360 estimated new diagnosed cases of PCa in the United States and 26 730 of these patients will die from this disease [3,4]. Simultaneously, the incidence rate of PCa in China has continued to increase in recent years. Although PCa is an indolent solid tumor and patients with early PCa obtain a benefit from radical prostatectomy [5], the clinical outcome of PCa patients has not been improved significantly [6,7]. The underlying molecular mechanism of carcinogenesis, progression, and recurrence of PCa is not fully understood [8]. Hence, there is a great demand to find an effective prognostic factor to understand PCa better and to guide urologists to improve the efficacy of PCa treatment.
Kinesin-like DNA binding protein (Kid), a member of the kinesin-10 family [9,10], participates in regulating microtubule stability, synaptic development, and cytoskeletal dynamics [11,12]. As a motor protein located in the nucleus in normal cells [13], its functions have been well defined during mitosis. Kid is known to be overexpressed in many cancers, such as breast cancer, cervical cancer, ovarian cancer, and lung cancer [14]. Moreover, it has been demonstrated that phosphorylation of KIF22 plays a vital role in the cell division cycle 25C (CDC25C) protein’s transcriptional regulatory, leading to delayed mitotic exit in breast cancer [14]. Besides, a recent study has shown that delays in epidermal growth factor receptor (EGFR) internalization, enhancement of EGFR signaling and the coxsackievirus and adenovirus receptor (CAR) dynamics cell-cell junctions are regulated by KIF22-dependent microtubule dynamics in lung cancer [12]. So, it indicated that the Kid played an essential role during oncogenesis and cancer progression. However, the expression of Kid on the protein level in PCa has not been analyzed, and its potential value in PCa is unclear.
In this study, we found that the mRNA level of Kid is differentially expressed between PCa tissues and paracancerous prostate tissues in The Cancer Genome Atlas (TCGA) dataset and the Genotype-Tissue Expression (GTEx) dataset by bioinformatics analysis. Then we used immunohistochemistry to determine the expression of Kid at the protein level in PCa tissues and paracancerous prostate tissues and investigated the relation between elevated Kid expression and the clinical features of PCa patients. More importantly, we use bioinformatics to evaluate the effect of Kid expression on biochemical recurrence (BCR)-free survival and overall survival in PCa patients.
Material and Methods
Patients and tissue samples
Following Institutional Review Board approval, informed consent was acquired from all of the patients undergoing radical prostatectomy in the Tianjin Institute of Urology, Tianjin, China between July 2016 and October 2017. For immunohistochemical analysis, tissues from 114 patients were assembled and stored at –80°C. All the samples used were analyzed by 2 experienced pathologists to ensure they were appropriately diagnosed as prostate adenocarcinoma. All patients did exhibit signs suggestive of distant tumor metastasis as examined by emission computed tomography. Patients were classified according to the 2017 Union for International Cancer Control (UICC) TNM staging as well as in compliance with the 2016 World Health Organization/International Society of Urological Pathology (WHO/ISUP) classifications. All patients had no history of preoperative androgen deprivation treatment, chemotherapy, or radiation therapy. The recorded clinic pathologic variables included age, preoperative prostate-specific antigen (PSA), clinical stage, Gleason score, lymph node metastasis, and seminal vesicle invasion.
Bioinformatics analysis
Data for differential genetic analysis were obtained from TCGA public database (including 498 PCa tissues and 52 non-cancerous prostate tissues) and GTEx database (including 100 non-cancerous prostate tissues). Samples from all patients were frozen soon after surgery to prevent degradation of the RNA and DNA. FFPE (formalin fixed paraffin embedded) samples were not used because of potential changes to the RNA and DNA that may arise from the fixation process. Second-generation sequencing platforms were used for genomic analysis include detection of KIF22 expression at the mRNA level. This project began in 2006, and the patients were followed for over 13 years. The method for differential analysis was one-way ANOVA, using tissues from different locations (tumor or non-cancerous prostate tissues) as variable for calculating differential expression. Genes with higher log2FC than pre-set thresholds (pre-set threshold=0.7) were considered differentially expressed genes. The biochemical recurrence (BCR)-free survival time and overall survival time were obtained from TCGA public database was collected to analyze the association between the expression of KIF22 at mRNA level and the patients’ survival time. The analysis is presented in Figures 1 and 2.
Figure 1.
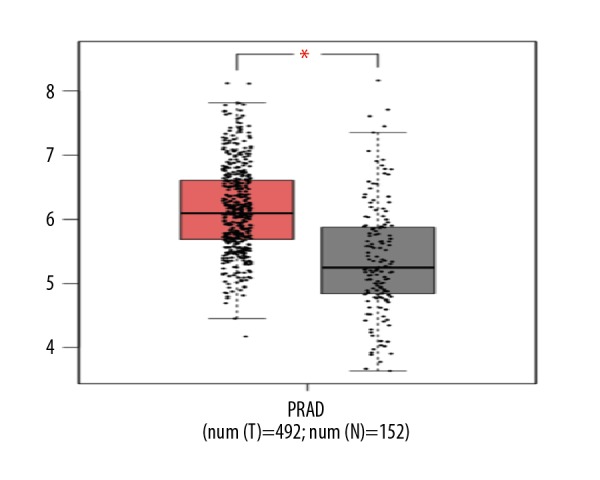
Compared with normal prostate tissues, the expression of KIF22 mRNA level was higher in prostate cancer tissues.
Figure 2.
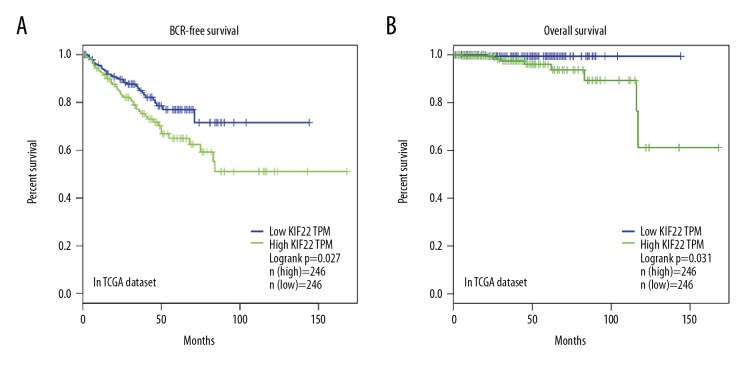
Kaplan-Meier survival analysis of BCR-free survival (A) and overall survival (B) for KIF22 expression in prostate cancer. BCR – biochemical recurrence.
Immunohistochemistry and scoring
Standard immunoperoxidase staining procedure was performed in observing KIF22 expression at the protein level. Briefly, 5-μm sections were obtained on the FFPE samples. Slides were dewaxed in xylene and rehydrated in different concentrations of ethanol. Subsequently, slides were immersed in 0.01M citrate buffer to repair the antigen, and heated for 15 minutes. Then slides were incubation with 3% hydrogen peroxide and placed in at 37°C for 15 minutes. Tissues were incubated with primary rabbit anti-human monoclonal antibody KIF22 antibody overnight. After washing with phosphate buffered saline (PBS), the tissues were incubated with goat anti-rabbit IgG as the secondary antibody. Then it was dripped with 3,3′-diaminobenzidine (DAB) to provide color development. After that, tissues were counterstained with hematoxylin for 90 seconds, dehydrated in ethanol, and sealed with coverslips. Negative controls sections were stained as described but with the primary antibody omitted.
The expression of KIF22 at protein levels was assessed semi-quantitatively according to the sum of the scores of the proportion of positive-stained cells and staining intensity. The proportion of positive stained cells was made as follows: 0, less than 5% positive staining; 1, 5% to 50% positive staining; 2, more than 50% positive staining. The staining intensity was made as follows: 0, negative/weak; 1, moderate; and 2, strong. The sum of 2 parameters represented the expression levels: 0~2 was low expression; 3~4 was high expression [15]. Independent score was estimated by 2 pathologists, the means of the scores was used as the final immunostaining score. The controversial cases were re-examined by the original pathologist and a senior pathologist using other areas on the slide until an agreement was reached.
Statistical analysis
The SPSS edition 22.0 software for analyzing data. We use SPSS edition 22.0 software to analyze the data (SPSS, Chicago, IL, USA). The chi-square test was performed for variables. GEPIA website [16] (http://gepia.cancer-pku.cn/) was used for survival analysis (Kaplan-Meier method). The Cox proportional hazard model was conducted to determine the prognostic effect of KIF22 expression for the BCR-free survival and overall survival. Two side values of P<0.05 were considered significant.
Results
KIF22 protein level was overexpressed in PCa tissues by immunohistochemical staining
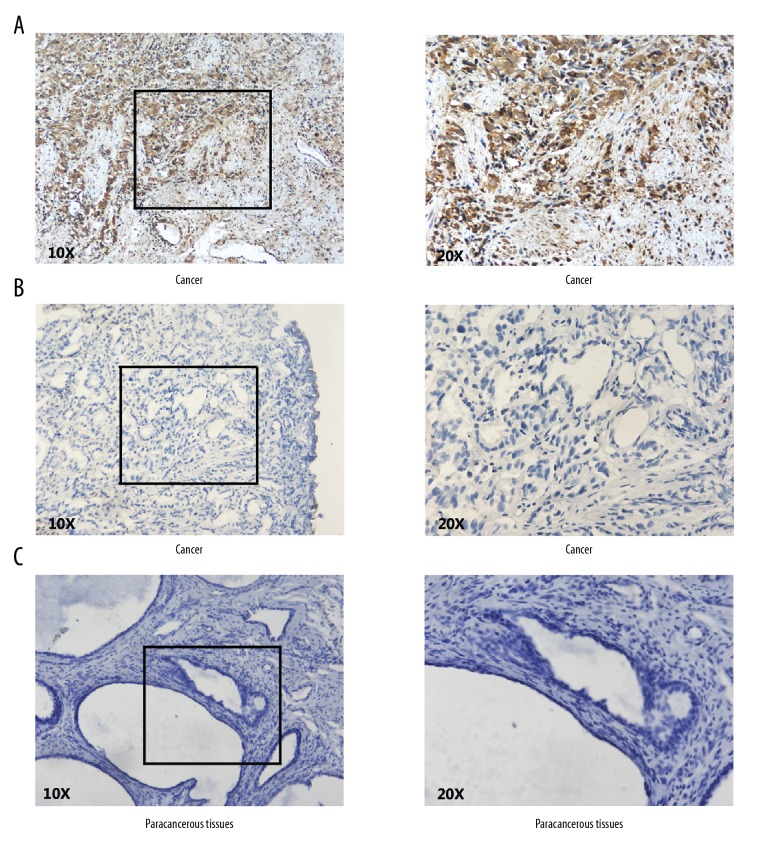
The expression of KIF22 at protein level was detected in surgical specimen from 114 PCa patients in our institution. As shown in Figure 3, KIF22 expression was located in the cytoplasm and nucleus of prostate tumor cells. All the specimens were examined; we found that KIF22 expression was elevated in 74 out of 114 patients (64.9%) and expression was low in 40 out of 114 patients (35.1%), but was almost negatively expressed in paracancerous tissues. The expression of KIF22 in tumor tissues was apparently higher than in paracancerous tissues as shown by using statistical analysis (Table 1, P<0.001).
Figure 3.
Immunostaining of KIF22 in prostate cancer and adjacent benign prostate tissues. (A) Immunostaining showed positive KIF22 in cytoplasm of prostate cancer cells. (B) Immunostaining showed negative KIF22 in prostate cancer tissues. (C) KIF22 was negative or weakly expressed in adjacent benign prostate tissues.
Table 1.
Expression of KID in prostate specimens.
| Groups | KIF22 expression | P value# | ||
|---|---|---|---|---|
| n | High-expression | % | ||
| Non-cancerous | 114 | 9 | 7.89% | <0.001* |
| PCa | 114 | 74 | 64.91% | |
P value was analyzed by chi-square test;
indicates P<0.05 with statistical significance.
KIF22 mRNA level was elevated in PCa tissues by bioinformatics analysis of the TCGA dataset
To validate the accuracy of our data, we used bioinformatics to analyze the expression of KIF22 mRNA in PCa and normal tissues in TCGA database and the GTEx database. The KIF22 mRNA expression level of KIF22 in PCa tissues was 1.7 times higher than normal tissues based on the 2 datasets (KIF22 with higher log2FC than pre-set thresholds are considered differentially expressed genes, in TCGA and GTEx data) as shown in Figure 1. In our clinical specimens and TCGA database and the GTEx database, it was confirmed that KIF22 was overexpressed in PCa tissues when compared with normal tissues.
Associations between KIF22 expression and clinicopathological parameters
The relationship between KIF22 expression and commonly used clinicopathological characteristics in PCa are presented in Table 2. Based on patient data and immunohistochemical findings from our clinical specimens, we found that strong intensity of KIF22 expression was related with higher clinical stage (P=0.025), Gleason score (P=0.002), seminal vesicle invasion (P=0.007), and lymph node metastasis (P=0.009). Nevertheless, no significant correlation was found between KIF22 expression and age (P=0.541) or preoperative PSA (P=0.682).
Table 2.
Clinicopathologic variables and KIF22 expression in 114 prostate cancer patients.
| Variable | Group | KIF22 expression | P value# | ||
|---|---|---|---|---|---|
| n | High | Low | |||
| Age | <70 | 64 | 40 (62.5%) | 24 (37.5%) | 0.541 |
| ≥70 | 50 | 34 (68.0%) | 16 (32.0%) | ||
| Perioperative PSA | ≤10 | 49 | 16 (51.5%) | 10 (38.5%) | 0.682 |
| >10 | 65 | 58 (65.9%) | 30 (34.1%) | ||
| Clinical stage | T1 | 55 | 30 (54.5%) | 25 (45.5%) | 0.025* |
| T2–3 | 59 | 44 (74.6%) | 15 (25.4%) | ||
| Gleason score | <7 | 63 | 33 (52.4%) | 30 (47.6%) | 0.002* |
| ≥7 | 51 | 41 (80.4%) | 10 (19.6%) | ||
| Seminal vesicle invasion | Absence | 24 | 10 (41.7%) | 14 (58.3%) | 0.007* |
| Presence | 90 | 64 (71.1%) | 26 (28.9%) | ||
| Lymph node metastasis | Absence | 39 | 19 (48.7%) | 20 (51.3%) | 0.009* |
| Presence | 75 | 55 (73.3%) | 20 (26.7%) | ||
P value was analyzed by chi-square test;
indicates P<0.05 with statistical significance.
KIF22 was a poor prognostic indicator after radical prostatectomy
BCR-free survival and overall survival are vital parameters of most concerned for PCa patients after radical prostatectomy [17]. To explore whether KIF22 mRNA expression level is associated with BCR-free survival and overall survival in patients with PCa, we used the Kaplan-Meier curve method to analyze the relationship between them by using TCGA dataset. During the follow-up time of 165 months, there were 91 out of 498 cases (18.3%) of biochemical recurrence, and 10 deaths out of 498 cases (2%). The median BCR-free time and overall time were 18 and 30 months, respectively. The 5-year BCR-free survival and overall survival rate were 77.5% and 98.7%, respectively. The mRNA expression of the KIF22 was classified as low (n=246, in TCGA dataset) or high (n=246, in TCGA dataset) in relation to median KIF22 mRNA expression as the cutoff point. As shown in Figure 2, we found that the BCR-free survival (P=0.027, in TCGA dataset) and overall survival (P=0.031, in TCGA dataset) was significantly different between the KIF22 high group and the KIF22 low group.
To further discuss the prognostic effect of KIF22 in patients with PCa, we used univariate and multivariate Cox proportional hazards regression to verify the clinical prognostic impact of KIF22 in TCGA dataset (Tables 3, 4). We found that KIF22 (P=0.042), Gleason score (P<0.001), and pT-stage (P<0.001) were identified as prognostic factors for BCR-free survival univariate analysis. Next, multivariate Cox analysis identified Gleason score (P<0.001) as an independent prognostic factor for BCR-free survival. However, both univariate analysis and multivariate analysis showed that KIF22 was not significantly related to overall survival.
Table 3.
Prognostic value of KIF22 mRNA expression level for the BCR-free survival via Cox proportional hazards model.
| Covariant | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Exp(B) | 95% CI | P value# | Exp(B) | 95% CI | P value | |
| KIF22 | 1.59 | 1.018–2.482 | 0.042* | 1.51 | 0.966–2.371 | 0.071 |
| Age | 1.029 | 0.996–1.064 | 0.085 | 1.009 | 0.975–1.044 | 0.616 |
| Gleason | 2.266 | 0.805–2.845 | <0.001* | 2.028 | 1.584–2.597 | <0.001* |
| pT-stage | 2.669 | 1.715–4.151 | <0.001* | 1.621 | 0.950–2.767 | 0.077 |
P value was analyzed by chi-square test;
indicates P<0.05 with statistical significance.
Table 4.
Prognostic value of KIF22 mRNA expression level for the overall survival via Cox proportional hazards model.
| Covariant | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Exp(B) | 95% CI | P value | Exp(B) | 95% CI | P value | |
| KIF22 | 4.265 | 0.927–19.623 | 0.063 | 3.751 | 0.801–17.571 | 0.093 |
| Age | 1.097 | 1.002–1.202 | 0.045* | 1.059 | 0.958–1.172 | 0.264 |
| Gleason | 3.269 | 1.593–6.707 | 0.001* | 2.423 | 1.076–5.457 | 0.033* |
| pT-stage | 5.126 | 1.550–16.949 | 0.007* | 2.158 | 0.417–11.181 | 0.359 |
Indicates P<0.05 with statistical significance.
Discussion
The benefit of the popularity of PSA screening is that PCa can be detected at an early stage that led to a decrease in mortality rates [18]. However, PCa has an unpredictable clinical outcome, such as variable biochemical recurrence time, because it is a biologically heterogeneous and multifocal disease [19–22]. Meanwhile, the underlying mechanism of its carcinogenesis and cancer progression have not been totally explored. Many experts argue that PSA screening has failed to complete the clinical requirement of discriminating high- and low-risk PCa [7,23,24]. Consequently, there is a great demand to find effectual prognostic factors for better understanding PCa that can guide urologists to improve the diagnostic and therapeutic understanding of PCa [25–27].
Previous studies have shown KIF22 is abnormally overexpressed in breast cancer, cervical cancer, ovarian cancer, and lung cancer [14]. To our knowledge, this is the first study that explores the role of KIF22 on the progression of PCa. First, we found a significant differential expressed in KIF22 mRNA level between PCa tissues and paracancerous tissues from TCGA dataset and the GTEx dataset by bioinformatics analysis. Then we analyzed KIF22 expression at the protein level by immunohistochemistry using 114 surgical specimens of PCa patients. Subsequently, we assessed the correlation between the differential expression of KIF22 and clinical pathological characteristics of PCa patients. We confirmed that high KIF22 expression was significantly related to higher Gleason score, clinical stage, seminal vesicle invasion, and lymph node metastasis, but not with age and preoperative PSA level. Moreover, we used GEPIA to analyze the BCR-free time and overall survival of 492 PCa patients from TCGA database by Kaplan-Meier method; the data showed that high KIF22 expression was associated with poor BCR-free survival and overall survival.
Recently, many studies have confirmed that KIF22 was a unique role as an oncogene in cancer carcinogenesis and cancer progression. The identified underlying mechanism demonstrated that KIF22 might influence normal mitotic and promote cell proliferation implicated in various human cancers. Germani et al. [28] suggested that SIAH-1 which binds ubiquitin-conjugating enzymes, affected the correct proceeding of mitosis by promoting KIF22 ubiquitination. Heriberto et al. [29] further confirmed that the association between KIF22 and SIAH-1 might suggest an additional regulation step which was more than an interaction among protein-protein and regulation stability of the protein in their analysis of the KIF22 and SIAH-1 mRNA variations between normal and tumor tissues in breast cancer. Interestingly, they found KIF22 mRNA level was decreased in some of the breast tumor cases compared to normal tissue. But in our study, we found that both KIF22 mRNA and protein level was higher than normal tissue. Yu et al. [14] found that KIF22 was overexpressed in the breast cancer and deletion of KIF22 may delay mitotic exit by upregulation of its direct transcriptional target CDC25C, resulting in suppressing cancer cell proliferation. These data were in agreement with our data of KIF22 expression in tumor compared with paired tissues adjacent to cancers. Both studies consider KIF22 as an oncogene promoting carcinogenesis. Manning et al. [30] identified that KIF22 and KIF2C play a significant role in EZH2-dependent melanoma invasion and lung colonization. Pike et al. [12] demonstrated that chromokinesin KIF22 was coordinated with CAR and EGFR, 2 key plasma membrane receptors that facilitate cancer cell division, resulting in promoting CAR- and EGFR-dependent tumorigenesis in lung cancer. Meanwhile, our study data concluded that there was an association between high KIF22 expression and advanced clinicopathological features and poor clinical outcome in PCa patients.
Conclusions
We demonstrated that KIF22 was overexpressed in patients with PCa. We further confirmed that high KIF22 expression level was correlated with higher Gleason score, clinical stage, seminal vesicle invasion, and lymph node metastasis. Finally, we found that upregulation of KIF22 predicted shorter BCR-free survival and overall survival in patients who had undergone radical prostatectomy in TCGA dataset. This study had some limitations. The sample size from our institution was not large enough, and we did not analyze the BCR-free survival and overall survival of these patients due to the short follow-up time. A randomized study and a long-term follow-up should be conducted to further confirm KIF22 as a novel prognostic factor in PCa patients.
Footnotes
Source of support: This work was supported by the Science & Technology Development Fund of Tianjin Education Commission for Higher Education (NO: 2016YD13)
Conflict of interest
None.
References
- 1.Punnen S, Cooperberg MR. The epidemiology of high-risk prostate cancer. Curr Opin Urol. 2013;23(4):331–36. doi: 10.1097/MOU.0b013e328361d48e. [DOI] [PubMed] [Google Scholar]
- 2.Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: An important clinical entity. Nat Rev Urol. 2014;11(6):317–23. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet. 2016;387(10013):70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 5.Molitierno J, Evans A, Mohler JL, et al. Characterization of biochemical recurrence after radical prostatectomy. Urol Int. 2006;77(2):130–34. doi: 10.1159/000093906. [DOI] [PubMed] [Google Scholar]
- 6.Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: A review. JAMA. 2017;317(24):2532–42. doi: 10.1001/jama.2017.7248. [DOI] [PubMed] [Google Scholar]
- 7.Pezaro C, Woo HH, Davis ID. Prostate cancer: measuring PSA. Intern Med J. 2014;44(5):433–40. doi: 10.1111/imj.12407. [DOI] [PubMed] [Google Scholar]
- 8.Rodrigues DN, Boysen G, Sumanasuriya S, et al. The molecular underpinnings of prostate cancer: impacts on management and pathology practice. J Pathol. 2017;241(2):173–82. doi: 10.1002/path.4826. [DOI] [PubMed] [Google Scholar]
- 9.Yu Y, Feng YM. The role of kinesin family proteins in tumorigenesis and progression: Potential biomarkers and molecular targets for cancer therapy. Cancer. 2010;116(22):5150–60. doi: 10.1002/cncr.25461. [DOI] [PubMed] [Google Scholar]
- 10.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: Insights into structure and function. Trends Cell Biol. 2005;15(9):467–76. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Park SM, Littleton JT, Park HR, Lee JH. Drosophila homolog of human KIF22 at the autism-linked 16p11.2 loci influences synaptic connectivity at larval neuromuscular junctions. Exp Neurobiol. 2016;25(1):33–39. doi: 10.5607/en.2016.25.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pike R, Ortiz-Zapater E, Lumicisi B, Santis G, Parsons M. KIF22 coordinates CAR and EGFR dynamics to promote cancer cell proliferation. Sci Signal. 2018;11(515) doi: 10.1126/scisignal.aaq1060. pii: eaaq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tahara K, Takagi M, Ohsugi M, et al. Importin-beta and the small guanosine triphosphatase Ran mediate chromosome loading of the human chromokinesin Kid. J Cell Biol. 2008;180(3):493–506. doi: 10.1083/jcb.200708003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Wang XY, Sun L, et al. Inhibition of KIF22 suppresses cancer cell proliferation by delaying mitotic exit through upregulating CDC25C expression. Carcinogenesis. 2014;35(6):1416–25. doi: 10.1093/carcin/bgu065. [DOI] [PubMed] [Google Scholar]
- 15.Tan LD, Xu YY, Yu Y, et al. Serum HER2 level measured by dot blot: A valid and inexpensive assay for monitoring breast cancer progression. PLoS One. 2011;6(4):e18764. doi: 10.1371/journal.pone.0018764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abd Elmageed ZY, Yang Y, Thomas R, et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells. 2014;32(4):983–97. doi: 10.1002/stem.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabayoyong W, Abouassaly R. Prostate cancer screening and the associated controversy. Surg Clin North Am. 2015;95(5):1023–39. doi: 10.1016/j.suc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Morrison GJ, Goldkorn A. Development and application of liquid biopsies in metastatic prostate cancer. Curr Oncol Rep. 2018;20(4):35. doi: 10.1007/s11912-018-0683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardsen E, Andersen S, Al-Saad S, et al. Evaluation of the proliferation marker Ki-67 in a large prostatectomy cohort. PLoS One. 2017;12(11):e0186852. doi: 10.1371/journal.pone.0186852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valkenburg KC, De Marzo AM, Williams BO. Deletion of tumor suppressors adenomatous polyposis coli and Smad4 in murine luminal epithelial cells causes invasive prostate cancer and loss of androgen receptor expression. Oncotarget. 2017;8(46):80265–77. doi: 10.18632/oncotarget.17919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faramarzi S, Ghafouri-Fard S. Expression analysis of cancer-testis genes in prostate cancer reveals candidates for immunotherapy. Immunotherapy. 2017;9(12):1019–34. doi: 10.2217/imt-2017-0083. [DOI] [PubMed] [Google Scholar]
- 23.Bravaccini S, Puccetti M, Bocchini M, et al. PSMA expression: A potential ally for the pathologist in prostate cancer diagnosis. Sci Rep. 2018;8(1):4254. doi: 10.1038/s41598-018-22594-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin RM, Donovan JL, Turner EL, et al. Effect of a low-intensity PSA-based screening intervention on prostate cancer mortality: The cap randomized clinical trial. JAMA. 2018;319(9):883–95. doi: 10.1001/jama.2018.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panigrahi GK, Ramteke A, Birks D, et al. Exosomal microRNA profiling to identify hypoxia-related biomarkers in prostate cancer. Oncotarget. 2018;9(17):13894–910. doi: 10.18632/oncotarget.24532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cozar JM, Robles-Fernandez I, Martinez-Gonzalez LJ, et al. Genetic markers a landscape in prostate cancer. Mutat Res. 2018;775:1–10. doi: 10.1016/j.mrrev.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Khorasani M, Teimoori-Toolabi L, Farivar TN, et al. Aberrant expression of miR-141 and nuclear receptor small heterodimer partner in clinical samples of prostate cancer. Cancer Biomark. 2018;22(1):19–28. doi: 10.3233/CBM-170696. [DOI] [PubMed] [Google Scholar]
- 28.Germani A, Bruzzoni-Giovanelli H, Fellous A, et al. SIAH-1 interacts with alpha-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene. 2000;19(52):5997–6006. doi: 10.1038/sj.onc.1204002. [DOI] [PubMed] [Google Scholar]
- 29.Bruzzoni-Giovanelli H, Fernandez P, Veiga L, et al. Distinct expression patterns of the E3 ligase SIAH-1 and its partner Kid/KIF22 in normal tissues and in the breast tumoral processes. J Exp Clin Cancer Res. 2010;29:10. doi: 10.1186/1756-9966-29-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manning CS, Hooper S, Sahai EA. Intravital imaging of SRF and Notch signalling identifies a key role for EZH2 in invasive melanoma cells. Oncogene. 2015;34(33):4320–32. doi: 10.1038/onc.2014.362. [DOI] [PMC free article] [PubMed] [Google Scholar]