Abstract
Genetic connectedness assesses the extent to which estimated breeding values can be fairly compared across management units. Ranking of individuals across units based on best linear unbiased prediction (BLUP) is reliable when there is a sufficient level of connectedness due to a better disentangling of genetic signal from noise. Connectedness arises from genetic relationships among individuals. Although a recent study showed that genomic relatedness strengthens the estimates of connectedness across management units compared with that of pedigree, the relationship between connectedness measures and prediction accuracies only has been explored to a limited extent. In this study, we examined whether increased measures of connectedness led to higher prediction accuracies evaluated by a cross-validation (CV) based on computer simulations. We applied prediction error variance of the difference, coefficient of determination (CD), and BLUP-type prediction models to data simulated under various scenarios. We found that a greater extent of connectedness enhanced accuracy of whole-genome prediction. The impact of genomics was more marked when large numbers of markers were used to infer connectedness and evaluate prediction accuracy. Connectedness across units increased with the proportion of connecting individuals and this increase was associated with improved accuracy of prediction. The use of genomic information resulted in increased estimates of connectedness and improved prediction accuracies compared with those of pedigree-based models when there were enough markers to capture variation due to QTL signals.
Keywords: genomic connectedness, genomic prediction, relatedness
INTRODUCTION
Genetic connectedness quantifies the extent of risk associated with the comparisons of estimated breeding values (EBV) across management units (Foulley et al., 1990). Best linear unbiased prediction (BLUP) of EBV can be fairly compared across units in the presence of a sufficient level of connectedness. On the other hand, an insufficient level of connectedness increases the risk of uncertainty in EBV comparisons when selecting individuals across units due to imperfect uncoupling of genetic signal from noise. A number of studies have shown that increasing pedigree-based connectedness through exchange of common reference sires can result in more accurate comparisons of genetic values of individuals from different management units (Foulley et al., 1983; Hanocq et al., 1996; Kuehn et al., 2008). The magnitude of estimates of connectedness is a function of genetic relatedness or relationships among individuals. Despite the critical importance of connectedness towards enabling genetic evaluations, the impact of genomic information on the degree of connectedness relative to pedigree only has been explored to a limited extent.
Use of genomics can affect genetic evaluations in 2 related but different contexts. One is related to determining whether EBV can be safely compared across management units and the other is related to enhancing the reliability of EBV. In the former context, Yu et al. (2017) employed 3 measures of connectedness to examine the extent to which genomic information increases the estimates of connectedness. They found that the use of genomic relatedness improved genetic connectedness measures across management units compared with the use of pedigree relationships.
However, it remains an open question as to whether increased connectedness observed by genomic relatedness also leads to increased prediction accuracy of genetic values across management units. Although improving the quality of breeding value comparisons and improving the accuracy of genomic prediction have been discussed in different contexts historically, it is worth investigating how these 2 items are related to each other. The objectives of this study were to examine how choice of relationship matrices and connectedness statistics affect the estimates of connectedness under various simulated scenarios and to assess the relationship between connectedness level and genome-enabled prediction accuracy. In addition, a guideline with respect to a sufficient level of connectedness is discussed.
MATERIALS AND METHODS
Data Simulation
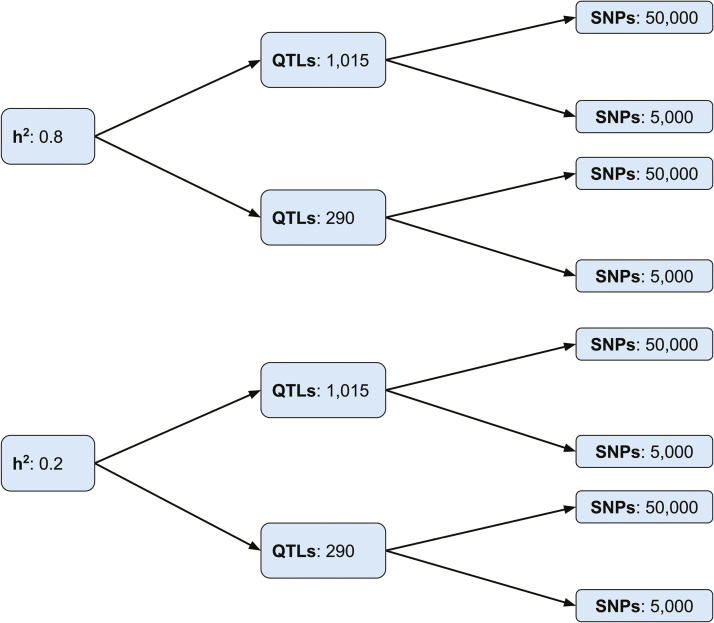
Ten replicates of genotypes and phenotypes were simulated using the QMSim software (Sargolzaei and Schenkel, 2009) with details summarized in Figure 1. One single historical population with 1,100 generations was simulated with the forward-in-time approach to create the initial linkage disequilibrium (LD) and mutation-drift equilibrium. The mating system was based on the random union of gametes sampled from sires and dams and the only evolutionary forces simulated were mutation and drift. The first 1,000 historical generations had a constant size of 1,000 per generation and then linearly decreased from 1,000 to 320 in the last hundred historical generations to account for population bottlenecks. The numbers of individuals from each sex were equal across the historical generations except the last historical generation which included a random sample of 20 males and 300 females (generation 0).
Figure 1.
Genomic data simulation parameters. SNPs, QTLs, and represent total single nucleotide polymorphisms, quantitative trait loci, and trait heritability, respectively. Simulations were carried out across 2 different (0.8 and 0.2), 2 different numbers of QTLs (1,015 and 290), and 2 different SNP densities (50,000 and 5,000).
Using the 20 males and 300 females as founder animals, the population size was expanded by simulating 7 generations (genreations 1 to 7) with the total population size approximately equal to 2,210. Each dam had 1 or 2 progenies within each generation with the probability of 0.95 and 0.05, respectively. As with the historical population, the mating was at random without selection and proportion of male progeny was 50%. The replacement rates of sires and dams were 0.6 and 0.2, respectively. Phenotypes with heritability levels of 0.2 and 0.8 were simulated with phenotypic variance of 1.0, where the overall heritability was accounted for by the variance of QTL additive genetic effects assuming no extra polygenic effect. Allelic effects of QTLs were sampled from a gamma distribution with a shape parameter of 0.4 and a corresponding scale parameter to ensure that the sum of QTLs variances was equal to the predefined QTL variances. The residual effects were randomly sampled from a Gaussian distribution with a mean of 0 and variance equal to heritability. The overall phenotypic effects were the sum of QTL effects and residual effects.
Pedigree information was recorded in the recent population from generations 0 to 7. Genotypic data were simulated for individuals (n = 2,210) in generations 1 to 7 coupled with 5,000 or 50,000 biallelic single nucleotide polymorphisms (SNPs) markers evenly distributed across 29 pairs of autosomes with each chromosome length of 100 cM. The number of autosomes and total chromosome length followed those of the bovine genome. Additionally, 290 or 1,015 randomly distributed QTLs were simulated: the former is equivalent to 10 QTLs per chromosome and the latter corresponds to 35 QTLs per chromosome. Markers and QTLs were simulated with a starting allele frequency of 0.5 and a recurrent mutation rate of 2.5 × 10−5 was used to create mutation-drift equilibrium in historical generations. In generation 1,100, markers and QTLs with minor allele frequency greater than 0.05 were randomly drawn from the segregating loci. Only SNPs but not QTLs were used to infer measures of connectedness and to assess accuracy of prediction.
Management Units Simulation
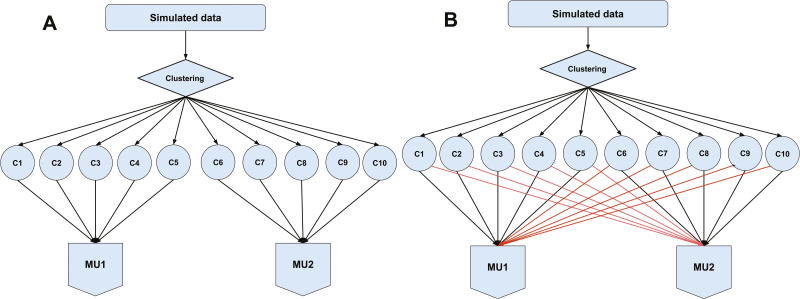
The management units were simulated in 2 steps following Yu et al. (2017): 1) individuals were classified into clusters and 2) clusters were assigned to management units (Figure 2). First, 10 individuals were chosen to represent medoids and then 10 distinctive groups were formed by assigning the remaining individuals to the closest medoid using the k-medoid algorithm (Kaufman and Rousseeuw, 1990). The size of 10 distinctive groups ranged from 91 to 590, varying slightly between replications. A dissimilarity matrix was created from the A (numerator relationship) matrix by calculating the distance between highest similarity and each similarity coefficient such that the largest similarity coefficient becomes zero. Clustering based on the k-medoid algorithm coupled with the dissimilarity matrix resulted in higher relationship coefficients within a cluster than between clusters.
Figure 2.
Management unit (MU) simulation scenarios. (A) Scenario 1 (least connected design). Individuals within clusters 1 to 5 were assigned to MU1 and clusters 6 to 10 were assigned to MU2. (B) Scenarios 2 to 6 (partially connected to connected). The degree of connectedness was gradually increased by exchanging 10% (Scenario 2), 20% (Scenario 3), 30% (Scenario 4), 40% (Scenario 5), and 50% (Scenario 6) of randomly sampled individuals between MU1 and MU2. Scenario 6 corresponds to the connected design.
Two management units were simulated with individuals within clusters assigned to a management unit in 6 ways. In Scenario 1, a least connected design was simulated by assigning individuals within clusters 1 to 5 into management unit 1 (MU1) and clusters 6 to 10 into management unit 2 (MU2). In Scenarios 2 to 6, the degree of genetic link was gradually increased by exchanging 10%, 20%, 30%, 40%, and 50% of randomly sampled individuals between MU1 and MU2.
Prediction Error Variance
Prediction error variance (PEV) can be derived from a linear mixed model,
where , , , and refer to a vector of phenotypes, fixed effects, random additive genetic effects, and residuals, respectively. The incidence matrices and connect fixed effects and random additive genetic effects with phenotypes. The joint distribution of random effects is as follows:
where is the additive genetic variance, is the residual variance, and represents a relationship matrix, which will be defined in a later section. Following the mixed model equation of Henderson (1984),
| (1) |
where is a ratio of variance components which equals to . BLUP of is given by
where is the absorption matrix for fixed effects. Then, the PEV of is given by (Henderson, 1984)
where denotes the lower right quadrant of the inverse of coefficient matrix in equation 1.
Genetic Connectedness
Two statistics applied in Yu et al. (2017) were used to measure connectedness in this study. The first one is the prediction error variance of the differences (PEVD) of EBV between individuals from different management units (Kennedy and Trus, 1993). A pair-wise comparison between th and th individuals is given by the variance of
where and refer to the diagonal elements of the matrix corresponding to th and th individuals, respectively, and denotes the off-diagonal elements of matrix. The summary connectedness of PEVD across all pairs of comparisons in a contrast notation is defined as follows (Laloë, 1993):
where the sum of elements in a contrast vector is zero. For instance, a pair-wise comparison between th and th management units with and individuals, the contrast vector will be set as and 0 corresponding to individual belonging to th, th, and remaining units. The boundary of PEVD is not restricted, with a lower value indicating stronger connectedness. To express connectedness independent of unit of measurement, PEVD was scaled by additive genetic variance (Kuehn et al., 2008; Yu et al., 2017).
The generalized CD measures the precision of EBV (Laloë, 1993). Different from PEVD, CD penalizes connectedness measurements if the genetic variability is too small across populations,
where denotes a pair-wise comparison between th and th individuals. A summary CD of contrast between any management unit is defined as follows (Laloë et al., 1996):
where is the vector of contrast defined earlier. This statistic ranges from 0 to 1 and measures the accuracy of the design. A larger value suggests a stronger estimate of connectedness among management units.
Relationship Matrix
Any kind of (semi)-positive definite relationship matrices can be used to define (Morota and Gianola, 2014). We used 3 types of in this study constructed from different sources. The numerator relationship matrix ( = ) measures the expected additive genetic relationship coefficient between individuals on the basis of pedigree information. The diagonal elements are , where represents inbreeding coefficient and off-diagonal elements are equal to twice the kinship coefficients. The construction of the matrix was based on tracing all individuals extending over 8 generations to account for historical information and animals from generations 1 to 7 were used for analysis. This matrix expresses relationships as identical by descent (IBD) as it measures the probability of alleles inherited from the same ancestor by tracing pedigree (Wright, 1922).
In contrast, a genomic relationship matrix ( = ) measures the molecular similarity among individuals. A typical matrix is obtained as a function of the gene content matrix () including elements of 0, 1, and 2 corresponding to the number of reference alleles. The distribution of th marker follows the binomial distribution of , where is the allele frequency of th marker. The matrix of VanRaden (2008) is obtained as follows:
where is the standardized gene content equal to and is the total number of markers.
One item that needs to be addressed when the and matrices are compared is that they are not on the same scale. For instance, the matrix represents relationships among individuals and inbreeding level as deviations from the unrelated base population; conversely the matrix expresses those relationships relative to the allele frequencies in the current generation. The following = matrix rescales to the same base population as in by adjusting the inbreeding coefficient level in similar to that of ,
where and refer to the average inbreeding coefficient of whole population in the matrix and the n × n square matrix filled with 1, respectively (Powell et al., 2010).
Whole-Genome Prediction Model
The relationship between connectedness and prediction accuracy was investigated with a standard BLUP model,
| (2) |
where , , , and refer to a vector of observed phenotypes, intercept, random additive genetic effects, and residuals, respectively. The model was treated under a Bayesian framework, where was set as a flat prior, with the prior distributions for genetic and residual effects,
where is 1 of 3 (semi)-positive definite relationship matrices described earlier and refers to the identity matrix. The variance components and represent variance of additive genetic effects and residual variance, respectively. The scaled inverse distribution was assigned to and by setting the degrees of freedom () equal to 5 and choosing the scale parameter by equating the mode of scaled inverse distribution to the quantity of , where is the expected proportion of phenotypic variance () explained by the regression and refers to the average sum squares of the genotypes (Pérez and de los Campos, 2014). Here was set to 0.5 according to Pérez and de los Campos (2014).
The prediction accuracy was evaluated by 2-fold CV, where the 2 management units were treated as the training and testing sets instead of randomly partitioning all individuals into 2 sets. The variance components were inferred from the data and the predictive ability of the model was calculated as the Pearson correlation between predicted genetic values and true genetic values in the testing set. Throughout this study, the BGLR R package was used to fit equation 2. A Gibbs sampler was run for 10,000 iterations, where the first 2,000 samples were discarded as burn-in. A total of 8,000 samples coupled with a thinning rate of 5 were used to infer posterior means.
Criterion for Connectedness Measures
The challenge with discussing connectedness is that there is no clear standard or benchmark for true connectedness. Although zero connectedness may be an indicator of possible bias, this issue has been discussed since Foulley et al. (1990). In this respect, Kuehn et al. (2008) proposed threshold values for moderate and strong levels of connectedness based on the relationship between prediction error correlation and model-based mean squared error. In this study, we provide a guideline for connectedness measures in terms of whole-genome prediction by performing CV. Note that prediction accuracy may simply increase as PEVD continues to decrease no matter how individuals across management units become genetically alike. On the other hand, measures of CD start to decrease as in Yu et al. (2017) when across management units include individuals that are too genetically similar. CD is suited for deriving a criterion because there is no point in enhancing prediction accuracy by simply reducing relatedness variability. Therefore, we explored the approximate threshold of CD that yields a reasonable prediction accuracy while maintaining genetic diversity in a population (Laloë, 1993; Laloë et al., 1996).
RESULTS
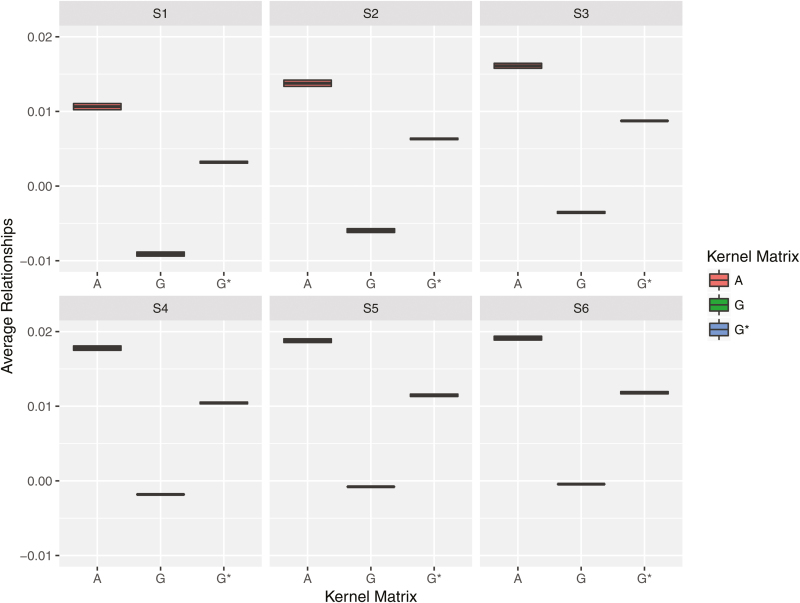
Figure 3 displays relationships between 2 management units with 5,000 markers used to compute 3 relationship matrices (, , and ) according to 6 simulated management unit scenarios. For each scenario, average relationships were the highest for and the smallest for , and produced relationships somewhere between and . Relationships increased when more individuals were exchanged between the 2 units. This increasing relationship pattern was observed regardless of relationship matrices used. A similar tendency was shown when the number of markers was equal to 50,000 (result not shown).
Figure 3.
Average relationship coefficients across management units with 5,000 markers over 2 heritability levels and 2 different numbers of quantitative trait loci. S1 to S6 denotes management unit simulation scenarios 1, 2, 3, 4, 5, and 6, respectively. The magnitude of connectedness level steadily increased from S1 to S6. We compared pedigree-based , genome-based , and rescaled genome-based relationship kernel matrices.
Prediction Error Variance of the Difference
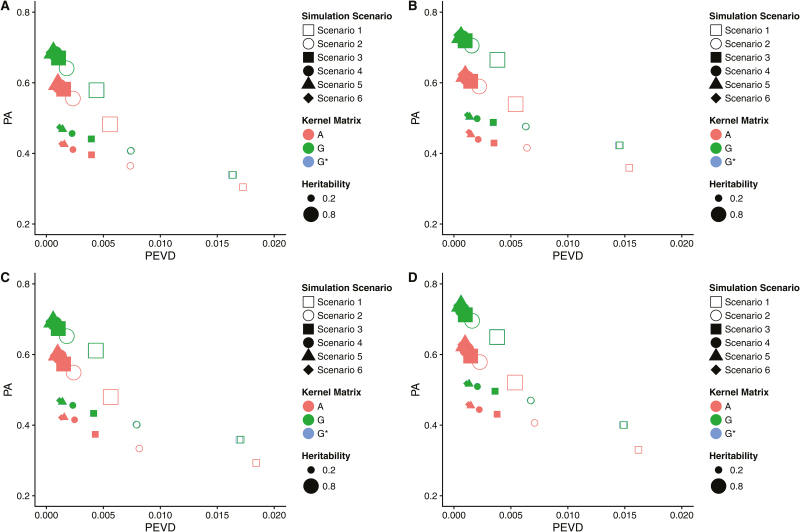
The relationships between measures of connectedness and prediction accuracies obtained from the Bayesian BLUP model are shown in Figures 4 and 5. The prediction accuracies in Figures 4 and 5 are identical as they are based on the same simulations. Figure 4 depicts connectedness measured as PEVD of contrast with smaller values inferring increased connectedness. Generally, increased connectedness measures and prediction accuracies were observed as more individuals from the same clusters were shared between management units, regardless of levels, type of kernel matrices, the number of QTLs, and marker density. Similarly, standard errors of estimates over 10 replicates ranged from 0.008 to 0.068 for prediction accuracy, and from 0.001 to 0.002 for PEVD, regardless of levels, type of kernel matrices, the number of QTLs, and marker density. In Figure 4A with 290 QTLs and 5,000 markers, the and matrices delivered similar or stronger connectedness measures and higher prediction accuracies than those of the matrix. The results from strongly resembled those of in terms of measures of connectedness and prediction accuracies. When marker density increased to 50,000, with the same number of QTLs, slightly improved prediction accuracies and increased estimates of connectedness were observed (Figure 4B). Stronger connectedness and higher prediction accuracy were shown with and than The pattern in Figure 4C with 1,015 QTLs and 5,000 markers resembled that of Figure 4A; however, we observed marginally decreased genomic prediction accuracies. Figure 4D with 1,015 QTLs and 50,000 markers presented the clearest pattern: the and matrices consistently produced stronger estimates of connectedness and higher prediction accuracies than those of the regardless of simulation scenarios and levels.
Figure 4.
Relationship between connectedness and prediction accuracy. PEVD and PA denote prediction error variance of the differences and prediction accuracy, respectively. PA was defined as the correlation between phenotypes and estimated breeding values Connectedness of pedigree-based genome-based and rescaled genome-based within 6 management units simulation scenarios across 2 heritabilities were compared with their prediction accuracies in each graph. (A) 290 QTLs and 5,000 markers. (B) 290 QTLs and 50,000 markers. (C) 1,015 QTLs and 5,000 markers. (D) 1,015 QTLs and 50,000 markers.
Figure 5.
Relationship between connectedness and prediction accuracy. CD and PA denote coefficient of determination and prediction accuracy, respectively. PA was defined as the correlation between phenotypes and estimated breeding values Connectedness of pedigree-based genome-based and rescaled genome-based within 6 management units simulation scenarios across 2 heritabilities were compared with their prediction accuracies in each graph. (A) 290 QTLs and 5,000 markers. (B) 290 QTLs and 50,000 markers. (C) 1,015 QTLs and 5,000 markers. (D) 1,015 QTLs and 50,000 markers.
Coefficient of Determination
The change of prediction accuracies with the increasing proportion of linked individuals quantified with CD of contrast is shown in Figure 5, where larger CD values suggest stronger connectedness. The standard errors of estimates for CD through 10 replicates varied from 0.004 to 0.057, regardless of levels, type of kernel matrices, the number of QTLs, and marker density. In general, the prediction accuracy improved when more individuals from the same clusters were assigned across units. Within each scenario, the estimates of CD increased up to Scenario 3 and decreased at Scenario 4 because CD penalized connectedness measures for reduced genetic variability. This corresponded to 20% exchange rate.
In Figure 5A with 290 QTLs and 5,000 markers, similar or stronger connectedness and higher prediction accuracies were observed by the matrix than those using for all scenarios. An analogous tendency was identified in Figure 5C with 1,015 QTLs and 5,000 markers, except that marginal reduction of genomic prediction accuracies was observed. With 290 QTLs and an increased number of markers (50,000), both genomic prediction accuracies and estimates of connectedness increased slightly (Figure 5B). Overall, and presented stronger estimates of connectedness and higher prediction accuracies than those of . Clearer differences were observed when increasing the number of QTLs to 1,015 (Figure 5D). The matrix clearly yielded higher estimates of connectedness and higher prediction accuracies when compared with . The performances of were very similar to those of in CD across all cases.
DISCUSSION
The concept of connectedness dates back to estimability in experimental design in the sense of all-or-none connectedness (Weeks and Williams, 1964; Eccleston and Hedayat, 1974). A dataset can be seen as connected if merging cells in a cross-table are possible such that all filled cells are connected (Searle, 1986). It was later extended to a random effect model or BLUP genetic evaluation known as reference sire progeny testing schemes by Foulley et al. (1983, 1990) and Miraei Ashtiani and James (1991). The central idea is when sires from 1 management unit are compared against sires in another unit, at least 1 sire should be tested in both units. Such common sires are known as link sires or reference sires. These authors investigated the efficient strategy of reference sire used to minimize PEVD between EBV by identifying the optimal number of progeny. Since then connectedness based on pedigree information has taken center stage in both theoretical development and real data applications (e.g., Laloë (1993), Hanocq and Boichard (1999), and Kuehn et al. (2008)). In addition, non-PEV-based genetic connectedness metrics have been developed (e.g., Foulley et al. (1992)). Connectedness is often used as an indicator of the robustness of genetic evaluation comparisons, where a higher level of connectedness suggests more reliable comparison of EBV across units. Past studies found that BLUP evaluations correctly yielded the likely ranking of individuals distributed across units when connectedness was present. Although research in pedigree-based connectedness is still critical, as shown in Yu et al. (2017) and in the current study, availability of genomic information now offers an opportunity to revisit a number of critical questions related to connectedness, such as how prediction accuracy is influenced given the level of connectedness between management units.
The extent of connectedness level boils down to the ability of to capture relationships among individuals. Connectedness increases with stronger across unit genetic relationship and it decreases with stronger within unit relationship (Kennedy and Trus, 1993). Advantages of genomic over pedigree relationships are as follows: 1) genomic measures relatedness arising from more distant ancestors than those included in a pedigree and 2) genomic captures the variation in realized kinship arising from the stochastic effects of Mendelian sampling and recombination. We tested 3 types of to capture the relationship among individuals in this study. The 2 matrices and mainly differ in 1) the distinction between IBD and IBS and 2) the relationships are relative to the baseline population vs. current population. The relationship matrix helps us to put and on a similar scale. Although those factors contributed to the improved quality of genetic evaluation design with the increased proportion of connecting individuals as shown in Yu et al. (2017), the relationship between connectedness level and CV-derived prediction accuracy has been yet-to-be answered. The present study aimed to bridge this gap by applying PEVD and CD of contrasts to simulated phenotypes, pedigrees, genomics, and management units. Note that the magnitude of the differences in results may be observed when applied to real data compared with the simulation results shown in this study.
Relationship Between Connectedness and Prediction Accuracy
We used contrasts of PEVD and CD to investigate the relationship between connectedness and prediction accuracy. We found prediction accuracy improved with increased capturing of connectedness between units. This suggests that increase in the accuracy of the EBV comparison is positively associated with an increase in accuracy of CV-based prediction. In general, genomic prediction accuracy improved as more markers were used to infer a genomic relationship matrix and as more QTLs contributed to the genetic variation given plenty of markers. These can be attributed to the fact that 1) the greater the number of markers, the better capturing of QTL relationships among individuals (Ober et al., 2012) and 2) genomic best linear unbiased prediction (GBLUP) performs better when the number of QTLs is large, because of its infinitesimal model assumption (Daetwyler et al., 2010). This result may change when an alternative whole-genome prediction model is used instead of GBLUP. For instance, a BayesB type of model performs well when the number of QTLs is small (Daetwyler et al., 2010). Measures of connectedness increased as more markers were used to characterize connectedness. When more markers were used, genomic information captures more variation in relationships which results in increased measures of connectedness.
Across 6 management unit scenarios, the extent of connectedness measured by PEVD and prediction accuracy from BLUP were higher as the proportion of individuals exchanged between the 2 units increased. The measurement of PEVD decreases when the number of markers increase regardless of QTL numbers and levels. This was not always the case in CD because this statistic penalizes connectedness estimates when the amount of genetic variability across units was small.
The and matrices clearly outperformed that of in prediction and also produced increased measures of connectedness (Figures 4 and 5). Interestingly, although the average relationship of individuals across management units computed from the matrix was more similar with that of than (Figures 3), the results of connectedness estimates and prediction accuracies obtained from the matrix were more similar with those of (Figures 4 and 5). This is most likely because of the similar variation in relationships across management units captured by and , which play an important role in measures of connectedness and prediction accuracies. The effect of scaling to be more similar to was minimal for PEVD and CD as produced increased measure of connectedness compared with that of . This is in agreement with Yu et al. (2017) where they found that genome-based connectedness consistently increased estimates of connectedness in most cases regardless of rescaling to the level of .
In addition, we observed marginally decreased genomic prediction accuracies when the number of QTLs was increased while the number of SNPs remained constant (Figures 4A vs. 4C and 5A vs. 5C). This is because the number of parameters we need to accurately predict increased and a sufficient number of markers is required to establish a sufficient level of LD to capture QTL signals. With more QTL, more markers are needed for them to contribute to or enhance prediction accuracy. This observation can also be supported theoretically from interactive deterministic genomic prediction accuracy simulators (Morota, 2017).
What is the Sufficient Level of Connectedness?
The extent to which a design is genetically connected or not has been the subject of discussion in the literature (e.g., Petersen (1978) and Fernando et al. (1983)). These authors proposed statistical approaches to determine the presence or absence of connectedness. A related question is to find a desired or sufficient level of connectedness based on connectedness metrics as in Kuehn et al. (2008). Here CD statistic offers an important insight because it accounts for the reduction of connectedness due to reduced genetic variability between individuals under comparison. This pattern was also observed by using both pedigree and genome-based CD connectedness in Yu et al. (2017). From the perspective of designing a breeding program, increasing connectedness simply by making individuals genetically similar to each other should be avoided (Laloë, 1993). Thus, the use of CD allows us to identify an upper limit of sufficient CD value that gives a reasonable prediction accuracy while maintaining the variability of relatedness. The CD began to fall around 20% exchange rate and the threshold CD value was in the range of 0.7 to 0.9 across simulation scenarios. When the measures of CD exceeded this threshold, prediction accuracy continued to improve in a mild degree or stayed the same, whereas connectedness estimates started to decrease. Although this cutoff value slightly varies among different scenarios (Yu et al., 2017), the CD metric can be used to optimize selective genotyping and phenotyping along the lines of Rincent et al. (2012) and Isidro et al. (2015). In contrast, when connectedness was determined with PEVD, prediction accuracy and connectedness both continued to increase when shifting more individuals across management units, thereby increasing genetic similarity. Such is clearly not a desired property in designing a breeding program.
CONCLUSIONS
In general, connectedness measures and prediction accuracies increased as more individuals from the same clusters were shared across management units. We found prediction accuracy improved with increased capturing of connectedness across units suggesting that increase in the accuracy of the EBV comparison is positively associated with increase in accuracy of CV-based prediction. This was entirely true for PEVD and partly so for CD. The impact of genomics was more marked compared with pedigree when a sufficient number of markers was present to capture QTLs. Although there is a need to establish increased levels of connectedness, simply increasing connectedness results in rapid decrease of relatedness variability which may not be desired in a breeding program. Use of CD allows us to find a connectedness level that gives a reasonable prediction accuracy while maintaining genetic diversity in a population.
Footnotes
This work was supported in part by the University of Nebraska startup funds to G.M.
LITERATURE CITED
- Daetwyler H. D., Pong-Wong R., Villanueva B., and Woolliams J. A.. 2010. The impact of genetic architecture on genome-wide evaluation methods. Genetics 185:1021–1031. doi: 10.1534/genetics.110.116855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston J., and Hedayat A.. 1974. On the theory of connected designs: characterization and optimality. Ann. Stat. 2:1238–1255. [Google Scholar]
- Fernando R., Gianola D., and Grossman M.. 1983. Identifying all connected subsets in a two-way classification without interaction. J. Dairy Sci. 66:1399–1402. [Google Scholar]
- Foulley J. L., Bouix J., Goffinet B., and Elsen M. J.. 1990. Connectedness in genetic evaluation. In: Gianola D., and Hammond K., editors.. Advances in statistical methods for genetic improvement of livestock. Springer Verlag, Heidelberg, Germany: p. 277–308. [Google Scholar]
- Foulley J. L., Hanocq E., and Boichard D.. 1992. A criterion for measuring the degree of connectedness in linear models of genetic evaluation. Genet. Sel. Evol. 24:315–330. [Google Scholar]
- Foulley J., Schaeffer L., Song H., and Wilton J.. 1983. Progeny group size in an organized progeny test program of ai beef bulls using reference sires. Can. J. Anim. Sci. 63:17–26. [Google Scholar]
- Hanocq E., and Boichard D.. 1999. Connectedness in the french holstein cattle population. Genet. Sel. Evol. 31:163. [Google Scholar]
- Hanocq E., Boichard D., and Foulley J. L.. 1996. A simulation study of the effect of connectedness on genetic trend. Genet. Sel. Evol. 28:67. [Google Scholar]
- Henderson C. R. 1984. Applications of linear models in animal breeding. 3rd ed. Schaeffer L. R., editor. Univ. of Guelph, Guelph. [Google Scholar]
- Isidro J., Jannink J. L., Akdemir D., Poland J., Heslot N., and Sorrells M. E.. 2015. Training set optimization under population structure in genomic selection. Theor. Appl. Genet. 128:145–158. doi: 10.1007/s00122-014-2418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L. and Rousseeuw P.. 1990. Finding groups in data: an introduction to cluster analysis. John Wiley and Sons, New York. [Google Scholar]
- Kennedy B. W., and Trus D.. 1993. Considerations on genetic connectedness between management units under an animal model. J. Anim. Sci. 71:2341–2352. [DOI] [PubMed] [Google Scholar]
- Kuehn L. A., Notter D. R., Nieuwhof G. J., and Lewis R. M.. 2008. Changes in connectedness over time in alternative sheep sire referencing schemes. J. Anim. Sci. 86:536–544. doi: 10.2527/jas.2007-0256 [DOI] [PubMed] [Google Scholar]
- Laloë D. 1993. Precision and information in linear models of genetic evaluation. Genet. Sel. Evol. 25:557. [Google Scholar]
- Laloë D., Phocas F., and Ménissier F.. 1996. Considerations on measures of precision and connectedness in mixed linear models of genetic evaluation. Genet. Sel. Evol. 28:359. [Google Scholar]
- Miraei Ashtiani S., and James J.. 1991. Efficient use of link rams in merino sire reference schemes. In: Proc. 9th Conf. Aust. Assoc. Anim. Breed. Genet University of Melbourne, Melbourne, Australia: p. 24–27. [Google Scholar]
- Morota G. 2017. Shinygpas: interactive genomic prediction accuracy simulator based on deterministic formulas. Genet. Sel. Evol. 49:91. doi: 10.1186/s12711-017-0368-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morota G., and Gianola D.. 2014. Kernel-based whole-genome prediction of complex traits: a review. Front. Genet. 5:363. doi: 10.3389/fgene.2014.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U., Ayroles J. F., Stone E. A., Richards S., Zhu D., Gibbs R. A., Stricker C., Gianola D., Schlather M., Mackay T. F.,. et al. 2012. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. Plos Genet. 8:e1002685. doi: 10.1371/journal.pgen.1002685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez P., and de los Campos G.. 2014. Genome-wide regression and prediction with the BGLR statistical package. Genetics 198:483–495. doi: 10.1534/genetics.114.164442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen P. 1978. A test for connectedness fitted for the two-way blup-sire evaluation. Acta Agric. Scand. 28:360–362. [Google Scholar]
- Powell J. E., Visscher P. M., and Goddard M. E.. 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 11:800–805. doi: 10.1038/nrg2865 [DOI] [PubMed] [Google Scholar]
- Rincent R., Laloë D., Nicolas S., Altmann T., Brunel D., Revilla P., Rodríguez V. M., Moreno-Gonzalez J., Melchinger A., Bauer E.,. et al. 2012. Maximizing the reliability of genomic selection by optimizing the calibration set of reference individuals: comparison of methods in two diverse groups of maize inbreds (Zea mays L.). Genetics 192:715–728. doi: 10.1534/genetics.112.141473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargolzaei M., and Schenkel F. S.. 2009. Qmsim: a large-scale genome simulator for livestock. Bioinformatics 25:680–681. doi: 10.1093/bioinformatics/btp045 [DOI] [PubMed] [Google Scholar]
- Searle S. 1986. Linear models. John Wiley & Sons, New York. [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Weeks D., and Williams D.. 1964. A note on the determination of connectedness in an n-way cross classification. Technometrics 6:319–324. [Google Scholar]
- Wright S. 1922. Coefficients of inbreeding and relationship. Am. Nat. 56:330–338. [Google Scholar]
- Yu H., Spangler M. L., Lewis R. M., and Morota G.. 2017. Genomic relatedness strengthens genetic connectedness across management units. G3 (Bethesda). 7:3543–3556. doi: 10.1534/g3.117.300151 [DOI] [PMC free article] [PubMed] [Google Scholar]