Abstract
Pneumonia is an important issue for sheep production, leading to reduced growth rate and a predisposition to pleurisy. The objective of this study was to identify loci associated with pneumonic lesions and pleurisy in New Zealand progeny test lambs. The lungs from 3,572 progeny-test lambs were scored for presence and severity of pneumonic lesions and pleurisy at slaughter. Animals were genotyped using the Illumina Ovine Infinium HD SNP BeadChip (606,006 markers). The heritability of lung lesion score and pleurisy were calculated using the genomic relationship matrix, and genome-wide association analyses were conducted using EMMAX and haplotype trend regression. At slaughter, 35% of lambs had pneumonic lesions, with 9% showing lesions on more than half of any individual lobe. The number of lambs recorded as having pleurisy by the processing plants was 9%. Heritability estimates for pneumonic lesions and pleurisy scores adjusted for heteroscedasticity (CPSa and PLEURa) were 0.16 (± 0.03) and 0.05 (± 0.02), respectively. Five single-nucleotide polymorphisms (SNPs) were significantly associated with pneumonic lesions at the genome-wide level, and additional 37 SNPs were suggestively significant. Four SNPs were significantly associated with pleurisy, with an additional 11 SNPs reaching the suggestive level of significance. There were no regions that overlapped between the 2 traits. Multiple SNPs were in regions that contained genes involved in either the DNA damage response or the innate immune response, including several that had previously been reported to have associations with respiratory disease. Both EMMAX and HTR analyses of pleurisy data showed a significant peak on chromosome 2, located downstream from the transcription factor SP3. SP3 activates or suppresses the expression of numerous genes, including several genes with known functions in the immune system. This study identified several SNPs associated with genes involved in both the innate immune response and the response to DNA damage that are associated with pneumonic lesions and pleurisy in lambs at slaughter. Additionally, the identification in sheep of several SNPs within genes that have previously been associated with the respiratory system in cattle, pigs, rats, and mice indicates that there may be common pathways that underlie the response to invasion by respiratory pathogens in multiple species.
Keywords: disease, genomics, genome-wide association study, pneumonia, sheep
INTRODUCTION
Chronic nonprogressive pneumonia is the most common form of ovine pneumonia in New Zealand, and is an important issue for sheep production, leading to reduced growth rate (Kirton et al., 1976; Alley, 1987; Goodwin et al., 2004; McRae et al., 2016) and a predisposition to pleurisy (Alley, 2002). There is well-documented evidence for between-animal variation in the ability of livestock to resist multiple diseases of economic importance, including respiratory disease (Bishop and Morris, 2007; Davies et al., 2009). Previous work has established that the heritability of pneumonic lesions at slaughter in New Zealand mixed breed progeny tested lambs is 0.07 ± 0.02 (McRae et al., 2016), which is comparable to estimates of respiratory disease in cattle (Snowder, 2009). With growing pressure to reduce the use of antibiotics and drugs in agriculture, these heritable differences mean that improvement of animal health through genetic selection for enhanced resistance can be used as a complementary approach to current methods for disease control (Goddard, 2012). More fundamentally, genomics, through tools such as genome-wide association studies (GWAS), can also be used to further increase our understanding of the genetic mechanisms underlying the host response to disease, and compare these mechanisms between breeds or species. Discovering regions of the genome associated with resistance or susceptibility may also lead to the development of new diagnostic tools and alternative treatments. The aim of this study was to utilize genotype data to identify regions of the ovine genome associated with pneumonic lesions and pleurisy in New Zealand lambs.
MATERIALS AND METHODS
Animals were managed in accordance with the provisions of the New Zealand Animal Welfare Act 1999, and the New Zealand Codes of Welfare developed under sections 68–79 of the Act.
Animals
The lungs from a total of 3,572 ewe and ram lambs from 4 flocks were scored for the presence and severity of pneumonic lesions. Lambs were from 3 South Island (Flocks A, B, and C) and one North Island (Flock D) progeny test flocks. All flocks were fixed-date slaughters, which took place when lambs were between 4 and 8 mo of age. Dams were composites of the main dual-purpose sheep breeds used in New Zealand, including Romney, Coopworth, Perendale, and Texel. Sires were a mixture of dual-purpose and terminal sire composites.
Phenotypic Measurements
The methodology for scoring pneumonic lesions has been previously described (Baird et al., 2012; McRae et al., 2016). Briefly, lungs were scored at chain speed postslaughter at the processing plant. The “consolidated pneumonia score” (CPS) system has a range from 0 to 2, where 0 = no lesions present; 1 = any individual lobe with up to 50% of the lobe affected and 2 = any individual lobe with greater than 50% of the lobe affected. Pneumonic lesions were defined as compacted, dark purple-red areas of the lung that were firm to touch. Information on lamb carcasses that were identified as having pleurisy and detained for trimming was obtained from the processing plants.
Data cleaning consisted of removal of records with 1) missing values, and 2) contemporary groups (CG) containing less than 5 observations. CG was defined as flock, birth year, sex, weaning mob, and slaughter date; animals needed to have all of these in common to be considered in the same CG. Weaning mob was obtained from Sheep Improvement Limited (SIL), the New Zealand sheep genetic evaluation database. To adjust for heteroscedasticity, CPS (initially scored as 0, 1, or 2) was scaled using the formula CPSa = CPS/SQRT[CPSm*(2-CPSm)], where m is the mean incidence rate within the CG where phenotypic score is being adjusted. Pleurisy (initially coded as 0 or 1) values were also transformed using the formula PLEURa = PLEUR/SQRT[PLEURm*(1-PLEURm)].
Genotypes and Quality Control
Genomic DNA was extracted from ear tissue samples collected from lambs at tailing, using a high-throughput DNA extraction method (Clarke et al., 2014). Animals were genotyped with the Illumina Ovine Infinium HD SNP BeadChip (606,006 markers) according to the manufacturer’s protocol. Genome coordinates of each single-nucleotide polymorphism (SNP) were based on the OARv3.1 ovine genome assembly (Jiang et al., 2014). Quality control checks excluded markers that appeared nonautosomal (including pseudoautosomal), had a call rate below 90%, and/or had a minor allele frequency (MAF) ≤0.01. Individuals were excluded from the analysis if there was more than 5% genotyping failure. After quality control measures, 3,546 phenotyped animals were available, with 537,117 of the initial 606,006 SNPs utilized for analysis.
Heritability
Variance components were estimated using restricted maximum likelihood (REML) procedures fitting an animal model in ASReml (Gilmour et al., 2015), with the genomic relationship matrix (GRM) estimated in GenABEL (Aulchenko et al., 2007) using HD genotypes. Heritabilities were obtained by running a univariate analysis on the respective traits. Data analysis models for both CPSa and PLEURa, included CG as a fixed effect (McRae et al., 2016).
Genome-wide Association Analyses
Pneumonic lesion and pleurisy data were analyzed using values adjusted for heteroscedasticity (CPSa and PLEURa). Pneumonic lesion data was also analyzed by only including animals with no lesions and those with severe lesions [CPSa (0&2)]. Genome-wide association analyses were performed using SNP & Variation Suite v8.4.0 (Golden Helix, Inc., Bozeman, MT, www.goldenhelix.com) using 2 of the following approaches: 1) Efficient Mixed-Model Association eXpedited (EMMAX) using identity-by-state (IBS), and 2) haplotype trend regression (HTR) with a 3-SNP sliding window. Analyses were performed on adjusted values, with CG fitted as a covariate. Genome browse software was used to visualize results with an added track of Ovis aries genes from Ensembl 84. After Bonferonni correction, thresholds were 9.31 × 10−8 and 1.86 × 10−6 for genome-wide significance (P < 0.05) and suggestive significance (P < 0.1), respectively.
RESULTS
Incidence of Pneumonia and Pleurisy
In total, 3,572 lungs were scored for pneumonic lesions from lambs born between 2013 and 2015 (Table 1). Of these, 1,234 (35%) had lesions, with 329 (9%) showing lesions on more than 50% of any individual lobe (CPS of 2). The number of lambs recorded as having pleurisy by the processing plants was 310 (9%). The incidence of pneumonia was significantly higher in 2014-born lambs than those born in 2013 or 2015 (P < 0.001). Of the 310 animals recorded as having pleurisy, 118 (38%) had a CPS of 0, 71 (23%) had a score of 1, 60 (19%) had a score of 1, and 61 (20%) were unable to be scored due to the lungs being retained in the carcass.
Table 1.
Incidence of pneumonic lesions and pleurisy by flock and year of birth
| Flock | Year born | Lungs scored | CPSa > 0 | CPSa = 2 | Pleurisy |
|---|---|---|---|---|---|
| A | 2013 | 292 | 52 (18%) | 20 (7%) | 25 (9%) |
| 2014 | 483 | 194 (40%) | 57 (12%) | 70 (14%) | |
| 2015 | 467 | 98 (21%) | 29 (6%) | 77 (16%) | |
| B | 2014 | 766 | 334 (44%) | 80 (10%) | 59 (8%) |
| 2015 | 292 | 46 (16%) | 11 (4%) | 6 (2%) | |
| C | 2015 | 56 | 14 (25%) | 3 (5%) | 13 (23%) |
| D | 2014 | 1,216 | 496 (41%) | 129 (11%) | 60 (5%) |
| Total | 3,572 | 1,234 (35%) | 329 (9%) | 310 (9%) |
aCPS = Consolidated Pneumonia Score, where 0 = no lesions present; 1 = individual lobes with up to 50% of the lobe affected and 2 = individual lobes with greater than 50% of the lobe affected.
Heritability
The heritability estimated for CPSa was 0.16 (± 0.03) and for PLEURa was 0.05 (± 0.02). This is slightly higher than the previously published estimates of 0.07 ± 0.02 and 0.02 ± 0.01, respectively (McRae et al., 2016). This is likely to be due to the use of a GRM rather than recorded pedigree information in estimating the heritability in the current analysis; the 1,216 lambs from Flock D were included in both studies, however only sire information is recorded for these animals, therefore using a GRM rather than pedigree is a more accurate estimation of relatedness. The genetic correlation between the 2 traits was 0.58 (± 0.16), and the phenotypic correlation was (0.15 ± 0.02), which was in line with previous estimates.
Genome-wide Association Analyses
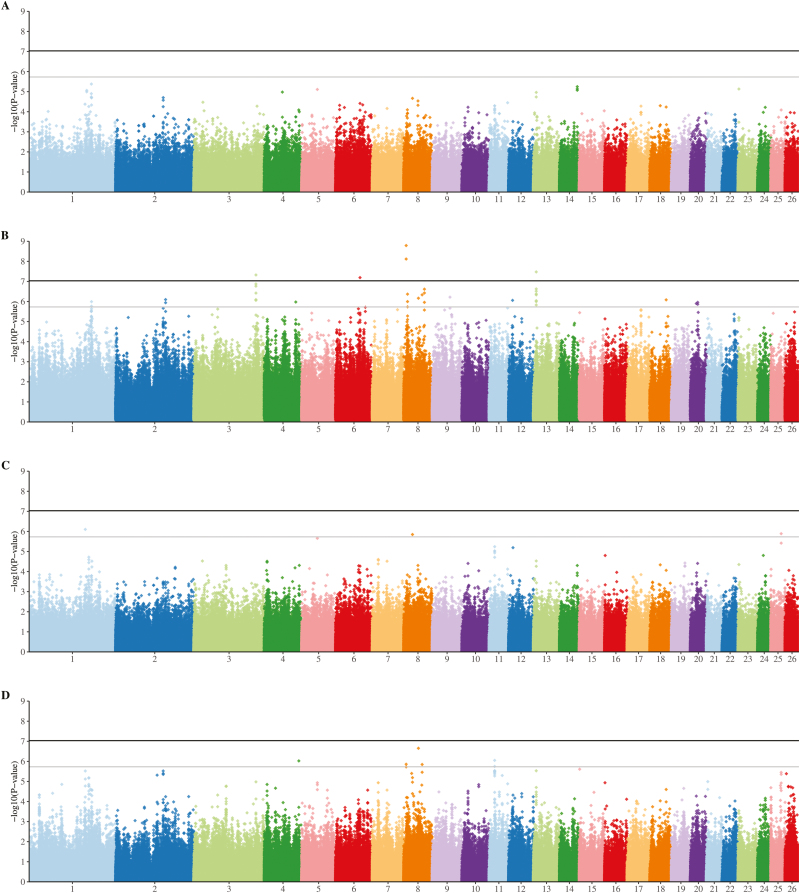
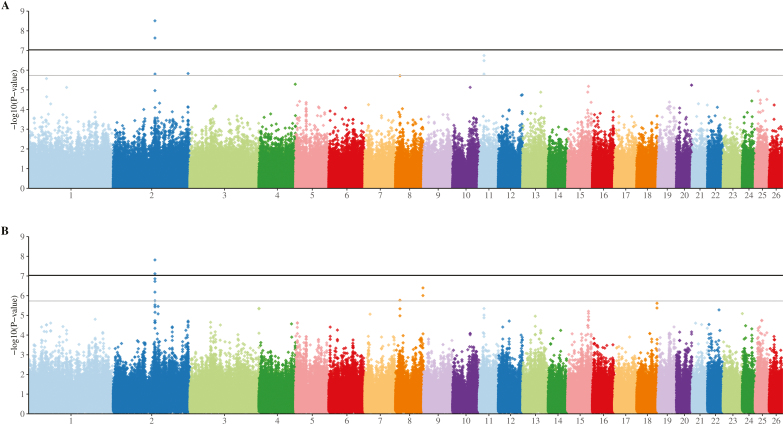
When adjusted pneumonic lesion (CPSa) information from all animals was included, there were no SNPs that passed the threshold for suggestive significance in the EMMAX analysis (Fig. 1A). In the HTR analysis, however, 4 regions, on chromosomes 3, 6, 8, and 13, were significant (Fig. 1B), with a further 31 SNPs passing the level for suggestive significance (Table 2). When only the extreme animals were included (i.e., animals with no lesions compared to those with severe lesions; CPS of 0 vs. CPS of 2), several SNPs in each analysis were of suggestive significance, although none reached the genome-wide level of significance (Fig. 1C and D). The top 3 SNPs in the EMMAX analysis were all intronic variants, in the LSAMP, PPIL6, and KCNMA1 genes (Table 2). The top SNPs in the HTR analysis included the same SNP in EYA4 that reached the suggestive significance level in the CPSa HTR analysis, along with 2 missense variants in exon 2 of ATAD5. Both EMMAX and HTR analyses of pleurisy data showed a significant peak on chromosome 2 (Fig. 2). Additionally, there were multiple suggestively significant intergenic SNPs on chromosomes 8 and 11 (Table 2).
Figure 1.
Manhattan plot of genome-wide association analysis for consolidated pneumonia score (CPS) in New Zealand lambs. Analyses were performed on all animals (A and B), or only including animals with scores of 0 or 2 (C and D). Genome-wide association analyses were conducted using 2 approaches: 1) Efficient Mixed-Model Association eXpedited (EMMAX) using identity-by-state (IBS) (A and C), and 2) haplotype trend regression (HTR) with a 3-SNP sliding window (B and D). Analyses were performed on pneumonic lesion scores after adjustment for heteroscedasticity, with contemporary group fitted as a covariate.
Table 2.
SNPs suggestively and significantly associated with consolidated pneumonia score (CPS) and pleurisy in New Zealand lambs
| Testa | Analysisb | P-valuec | Chr | Position | RSID | Gened | Gene named | Variant consequence (impact)d |
|---|---|---|---|---|---|---|---|---|
| CPSa | HTR | 1.72E-06 | 1 | 198656040 | rs421794454 | ENSOARG00000020512 | RFC4 | Intron variant |
| HTR | 1.02E-06 | 1 | 198674451 | rs416081302 | Multiple small nucleolar RNAs | Upstream gene variant | ||
| HTR | 8.00E-07 | 2 | 158991503 | rs407273673 | ||||
| HTR | 1.15E-06 | 2 | 158998132 | rs417102378 | ||||
| HTR | 8.05E-07 | 3 | 197683811 | rs426850802 | ||||
| HTR | 8.74E-07 | 3 | 197705247 | rs405096150 | ||||
| HTR | 1.72E-07 | 3 | 197720146 | rs430716198 | ||||
| HTR | 1.34E-07 | 3 | 197720936 | rs412869687 | ||||
| HTR | 3.80E-07 | 3 | 197824787 | rs407726225 | ||||
| HTR | 4.75E-08* | 3 | 197825391 | rs424070250 | ||||
| HTR | 1.06E-06 | 4 | 100364303 | rs403394816 | ||||
| HTR | 6.45E-08* | 6 | 77843695 | rs399606595 | ||||
| HTR | 1.63E-09* | 8 | 7733798 | rs429357466 | ||||
| HTR | 7.65E-09* | 8 | 7743164 | rs400905064 | ||||
| HTR | 4.29E-07 | 8 | 12061122 | rs425423371 | ||||
| HTR | 1.01E-06 | 8 | 12061431 | rs402511423 | ||||
| HTR | 1.85E-06 | 8 | 12149272 | rs423436094 | ||||
| HTR | 6.85E-07 | 8 | 46753989 | rs422310670 | ||||
| HTR | 4.64E-07 | 8 | 58610312 | rs418966278 | ENSOARG00000014564 | EYA4 | Intron variant | |
| HTR | 1.10E-06 | 8 | 65817474 | rs419752214 | ||||
| HTR | 2.43E-07 | 8 | 65827376 | rs399425501 | ||||
| HTR | 3.76E-07 | 8 | 65834390 | rs429368446 | ||||
| HTR | 6.06E-07 | 9 | 56587375 | rs424700173 | ||||
| HTR | 8.75E-07 | 12 | 12552747 | rs408273790 | ||||
| HTR | 1.01E-06 | 13 | 8840940 | rs400804234 | ||||
| HTR | 3.36E-08* | 13 | 8848881 | rs417728121 | ||||
| HTR | 2.36E-07 | 13 | 8850719 | rs423025524 | ||||
| HTR | 4.67E-07 | 13 | 8855334 | rs416260513 | ||||
| HTR | 1.49E-06 | 13 | 8860196 | rs420406541 | ||||
| HTR | 3.06E-07 | 13 | 8864106 | rs428620400 | ||||
| HTR | 4.21E-07 | 13 | 8871693 | rs430812458 | ||||
| HTR | 9.09E-07 | 13 | 8872416 | rs404379883 | ||||
| HTR | 8.25E-07 | 18 | 51969325 | rs419274927 | ENSOARG00000026456 | Novel lincRNA | Noncoding transcript variant | |
| HTR | 1.26E-06 | 20 | 18894279 | rs401389671 | ||||
| HTR | 1.37E-06 | 20 | 23420251 | rs417356235 | ||||
| HTR | 1.10E-06 | 20 | 23423229 | rs399485900 | ||||
| CPSa (0&2) | EMMAX | 7.91E-07 | 1 | 178857472 | rs410655004 | ENSOARG00000019641 | LSAMP | Intron variant |
| EMMAX | 1.42E-06 | 8 | 27846793 | rs430024463 | ENSOARG00000010461 | PPIL6 | Intron variant | |
| EMMAX | 1.29E-06 | 25 | 32872254 | rs422854508 | ENSOARG00000009163 | KCNMA1 | Intron variant | |
| HTR | 9.49E-07 | 4 | 110104884 | rs425466808 | ||||
| HTR | 1.39E-06 | 8 | 7743164 | rs400905064 | ||||
| HTR | 2.23E-07 | 8 | 46753989 | rs422310670 | ||||
| HTR | 1.43E-06 | 8 | 58610312 | rs418966278 | ENSOARG00000014564 | EYA4 | Intron variant | |
| HTR | 8.91E-07 | 11 | 17720321 | rs419581914 | ENSOARG00000012322 | ATAD5 | Missense variant (moderate) | |
| HTR | 1.79E-06 | 11 | 17720415 | rs400520703 | ENSOARG00000012322 | ATAD5 | Missense variant (moderate) | |
| PLEURa | EMMAX | 3.10E-09* | 2 | 134984962 | rs398681238 | |||
| EMMAX | 2.32E-08* | 2 | 134985148 | rs424471052 | ||||
| EMMAX | 1.57E-06 | 2 | 135163372 | rs415671617 | ||||
| EMMAX | 1.48E-06 | 2 | 242723012 | rs421193149 | ||||
| EMMAX | 1.58E-06 | 11 | 15261261 | rs420254502 | ||||
| EMMAX | 3.29E-07 | 11 | 15262540 | rs409974296 | ||||
| EMMAX | 1.80E-07 | 11 | 15265356 | rs417033802 | ||||
| HTR | 1.38E-07 | 2 | 134976058 | rs404285802 | ENSOARG00000000469 | SP3 | Downstream gene variant | |
| HTR | 7.66E-08* | 2 | 134979525 | rs428634189 | ||||
| HTR | 1.53E-08* | 2 | 134984962 | rs398681238 | ||||
| HTR | 6.59E-07 | 2 | 134985148 | rs424471052 | ||||
| HTR | 1.88E-07 | 2 | 134998369 | rs414115266 | ||||
| HTR | 1.82E-06 | 2 | 135006264 | rs412779979 | ||||
| HTR | 1.72E-06 | 8 | 13863996 | rs414046873 | ||||
| HTR | 9.86E-07 | 8 | 88651287 | rs412134993 | ||||
| HTR | 4.03E-07 | 8 | 88659717 | rs398705894 |
aAnalyses were performed on consolidated pneumonia and pleurisy scores after adjustment for heteroscedasticity (CPSa and PLEURa, respectively). For CPSa data, analyses were performed using all animals, or only including animals with scores of 0 or 2 [CPSa (0&2)].
bGenome-wide association analyses were conducted using 2 approaches: 1) Efficient Mixed-Model Association eXpedited (EMMAX) using identity-by-state (IBS), and 2) haplotype trend regression (HTR) with a 3-SNP sliding window. Contemporary group (sex, birth year, flock, weaning mob, and kill date) was fitted as a covariate in all analyses.
cAfter Bonferonni correction, thresholds were 9.31 × 10−8 and 1.86 × 10−6 for genome-wide significance (P < 0.05*) and suggestive significance (P < 0.1), respectively.
dGene names and variant consequences are based on Ensembl Release 84.
Figure 2.
Manhattan plot of genome-wide association analysis for pleurisy in New Zealand lambs. Genome-wide association analyses were conducted using 2 approaches: 1) Efficient Mixed-Model Association eXpedited (EMMAX) using identity-by-state (IBS) (A), and 2) haplotype trend regression (HTR) with a 3-SNP sliding window (B). Analyses were performed on pleurisy scores after adjustment for heteroscedasticity, with contemporary group fitted as a covariate.
DISCUSSION
In sheep, as with other ruminants, respiratory disease such as pneumonia is etiologically complex, resulting from a complex interaction between multiple infectious agents and the host, which is often compromised by physical and physiological stress. GWAS help provide an understanding of the genes and pathways involved in the response to disease. GWAS in both dairy (Neibergs et al., 2014) and beef (Keele et al., 2015) cattle have identified multiple loci associated with bovine respiratory disease complex (BRDC). Neibergs et al. (2014) discovered candidate loci involved in viral susceptibility, viral entry into cells, and modulation of inflammation in a case–control analysis of preweaned Holstein calves. A GWAS of lung lesions in beef cattle identified SNPs near candidate genes involved in functions such as tissue repair and regeneration, cell proliferation, apoptosis, and immunity (Keele et al., 2015).
The majority of SNPs associated with pneumonic lesions in this study were in intergenic regions of the sheep genome. Intergenic variants within RFC4, EYA4, and a novel lincRNA were suggestively associated with pneumonic lesions when including all the data and variants within LSAMP, PPIL6, and KCNMA1 reached suggestive significance when only including the extreme animals. Additionally, 2 missense variants in exon 2 of ATAD5 also reached the suggestively significant level in the analysis of the extreme animals.
EYA4, ATAD5, and RFC4 all have roles in the response to DNA damage. Eyes Absent (EYA) proteins are implicated in a diverse range of processes, including DNA damage repair and innate immunity (Tadjuidje and Hegde, 2013). EYA4 has been shown to enhance the innate immune response to viruses through stimulating the interferon regulatory factor 3 (IRF3)-mediated transcription of IFN-β and CXCL10 in response to undigested DNA (Okabe et al., 2009). EYA4 has been associated with familial lung cancer risk (Wilson et al., 2014), and a SNP located within 15 kb of EYA4 has been significantly associated with lung lesions in commercial beef cattle (Keele et al., 2015).
The replication factor C (RFC) complex, composed of subunits RFC1-5, also plays an essential role in DNA replication and repair in eukaryotes (Kim and MacNeill, 2003). Additionally, several RFC-like complexes (RLC), made up of RFC2-5 and an alternative subunit that replaces RFC1, have been reported, including ATAD5-RLC (Ben-Aroya et al., 2003). Atad5+/− mice show high levels of genomic instability (Bell et al., 2011), and delayed DNA replication and cell division, leading to an altered adaptive immune response though reduced immunoglobulin class switching (Zanotti et al., 2015). The identification of 2 suggestively significant SNPs within genes that form ATAD5-RLC highlights the potential importance of this complex in the host response to respiratory challenge.
Although not their primary role, both PPIL6 and KCNMA1 have previously been associated with the respiratory system. A QTL containing the cyclophilin-like PPIL6 was associated with the variability of immune response in a crossbred swine population postinfluenza vaccination (Zanella et al., 2015). The potassium channel gene KCNMA1 was expressed at significantly higher levels in the lungs of asthmatic rats compared to those of control rats (Yin et al., 2008), and is differentially methylated during normal development in the mouse and human lung (Cuna et al., 2015).
The significant peak on chromosome 2 associated with pleurisy was detected using 2 independent methods. This peak is located downstream from the transcription factor SP3, which is involved in the activation or suppression the expression of numerous genes, including the interferon regulatory factor IRF3 and IL-10, an anti-inflammatory cytokine (Tone et al., 2000). Of interest is that Sp3 knockout mice die at birth of respiratory failure, although only minor structural abnormalities are observed in the lungs (Bouwman et al., 2000). As mentioned above, IRF3 is involved in the innate response to viral infection (Xu et al., 2012), and several bovine viral pathogens including bovine herpesvirus 1 (BHV-1) and bovine diarrhoea virus (BVDV) target IRF3 activity, halting the interferon response (Srikumaran et al., 2007). As with other farmed ruminants, in sheep, pneumonia is etiologically complex. While Mannheimia haemolytica is considered to be the predominant agent responsible for lung damage, multiple viruses (Davies et al., 1977; Davies et al., 1982; Davies and Jones, 1985) have also been shown to play a role through compromising the respiratory system, allowing secondary invasion by bacteria (Brogden et al., 1998). An enhanced immune response to viruses could therefore result in a reduced chance of developing lung damage.
As previously discussed, pneumonia can arise through a combination of a variety of environmental and pathogenic factors. Despite the complex nature of this disease, previous research in both sheep and cattle has shown that there is an underlying genetic component in the variation observed between animals in their susceptibility to pneumonia (Snowder, 2009; McRae et al., 2016). This indicates that it is possible to select for animals with the ability to withstand and/or recover from infection that can be the result of multiple causative factors. This study identified several SNPs associated with genes involved in both the innate immune response and the response to DNA damage that are associated with pneumonic lesions and pleurisy in lambs at slaughter. Additionally, the identification in sheep of several SNPs within genes that had previously been reported to be involved in the respiratory system in cattle, pigs, rats, and mice indicates that there may be common genetic pathways underlying the response to respiratory disease in multiple mammalian species.
Footnotes
This work was supported by FarmIQ, AgResearch Core, and Beef + Lamb New Zealand Genetics funding. The flocks involved in this study were from FarmIQ and Pastoral Greenhouse Gas Research Consortium (PGgRc) funded progeny test programs. The authors would like to acknowledge the AgResearch Animal Genomics field staff and Silver Fern Farms staff from the Takapau and Finegand processing plants for their help in data collection. The Illumina Ovine Infinium® HD SNP BeadChip was used with the kind permission of the International Sheep Genomics Consortium (www.sheephapmap.org).
LITERATURE CITED
- Alley M. R. 1987. The effect of chronic non-progressive pneumonia on weight gain of pasture fed lambs. N Z Vet J. 35:163–166. doi: 10.1080/00480169.1987.35429 [DOI] [PubMed] [Google Scholar]
- Alley M. R. 2002. Pneumonia in sheep in New Zealand: an overview. N Z Vet J. 50(3 Suppl):99–101. doi: 10.1080/00480169.2002.36281 [DOI] [PubMed] [Google Scholar]
- Aulchenko Y. S., Ripke S., Isaacs A., and van Duijn C. M.. 2007. Genabel: an R library for genome-wide association analysis. Bioinformatics. 23:1294–1296. doi: 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- Baird H. J., Clarke S. M., and Johnson P. L.. 2012. Brief communication: development of a visual scoring system for ovine pneumonia at the processing plant. Proc N Z Soc Anim. 72:169–171. [Google Scholar]
- Bell D. W., Sikdar N., Lee K. Y., Price J. C., Chatterjee R., Park H. D., Fox J., Ishiai M., Rudd M. L., Pollock L. M., et al. ; NISC Comparative Sequencing Program. 2011. Predisposition to cancer caused by genetic and functional defects of mammalian atad5. Plos Genet. 7:e1002245. doi: 10.1371/journal.pgen.1002245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S., Koren A., Liefshitz B., Steinlauf R., and Kupiec M.. 2003. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA. 100:9906–9911. doi: 10.1073/pnas.1633757100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop S. C., and Morris C. A.. 2007. Genetics of disease resistance in sheep and goats. Small Ruminant Res. 70:48–59. doi: 10.1016/j.smallrumres.2007.01.006 [DOI] [Google Scholar]
- Bouwman P., Göllner H., Elsässer H. P., Eckhoff G., Karis A., Grosveld F., Philipsen S., and Suske G.. 2000. Transcription factor sp3 is essential for post-natal survival and late tooth development. Embo J. 19:655–661. doi: 10.1093/emboj/19.4.655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A., Lehmkuhl H. D., and Cutlip R. C.. 1998. Pasteurella haemolytica complicated respiratory infections in sheep and goats. Vet. Res. 29:233–254. [PubMed] [Google Scholar]
- Clarke S. M., Henry H. M., Dodds K. G., Jowett T. W., Manley T. R., Anderson R. M., and McEwan J. C.. 2014. A high throughput single nucleotide polymorphism multiplex assay for parentage assignment in New Zealand sheep. Plos One. 9:e93392. doi: 10.1371/journal.pone.0093392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuna A., Halloran B., Faye-Petersen O., Kelly D., Crossman D. K., Cui X., Pandit K., Kaminski N., Bhattacharya S., Ahmad A., et al. 2015. Alterations in gene expression and DNA methylation during murine and human lung alveolar septation. Am. J. Respir. Cell Mol. Biol. 53:60–73. doi: 10.1165/rcmb.2014-0160OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. H., Dungworth D. L., Humphreys S., and Johnson A. J.. 1977. Concurrent infection of lambs with parainfluenza virus type 3 and Pasteurella haemolytica. N. Z. Vet. J. 25:263–265. doi: 10.1080/00480169.1977.34425 [DOI] [PubMed] [Google Scholar]
- Davies G., Genini S., Bishop S. C., and Giuffra E.. 2009. An assessment of opportunities to dissect host genetic variation in resistance to infectious diseases in livestock. Animal. 3:415–436. doi: 10.1017/S1751731108003522 [DOI] [PubMed] [Google Scholar]
- Davies D. H., Herceg M., and Thurley D. C.. 1982. Experimental infection of lambs with an adenovirus followed by Pasteurella haemolytica. Vet. Microbiol. 7:369–381. doi: 10.1016/0378-1135(82)90017-7 [DOI] [PubMed] [Google Scholar]
- Davies D. H., and Jones S.. 1985. Serological evidence of respiratory syncytial virus infection in lambs. N. Z. Vet. J. 33:155–156. doi: 10.1080/00480169.1985.35211 [DOI] [PubMed] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Welham S. J., and Thompson R.. 2015. ASReml User Guide Release 4.1 Structural Specification. VSN International, Hemel Hempstead, HP1 1ES, UK. [Google Scholar]
- Goddard M. E. 2012. Uses of genomics in livestock agriculture. Anim Prod Sci. 52:73–77 (Review). doi: 10.1071/AN11180 [DOI] [Google Scholar]
- Goodwin K. A., Jackson R., Brown C., Davies P. R., Morris R. S., and Perkins N. R.. 2004. Pneumonic lesions in lambs in New Zealand: patterns of prevalence and effects on production. N Z Vet J. 52:175–179. doi: 10.1080/00480169.2004.36425 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Xie M., Chen W., Talbot R. T., Maddox J. F., Faraut T., Wu C., Muzny D. M., Li Y., Zhang W.,. et al. 2014. The sheep genome illuminates biology of the rumen and lipid metabolism. Science. 344:1168–1173. doi: 10.1126/science.1252806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele J. W., Kuehn L. A., McDaneld T. G., Tait R. G., Jones S. A., Smith T. P., Shackelford S. D., King D. A., Wheeler T. L., Lindholm-Perry A. K., et al. 2015. Genomewide association study of lung lesions in cattle using sample pooling. J. Anim. Sci. 93:956–964. doi: 10.2527/jas.2014-8492 [DOI] [PubMed] [Google Scholar]
- Kim J., and MacNeill S. A.. 2003. Genome stability: a new member of the RFC family. Curr. Biol. 13:R873–R875. doi:10.1016/j.cub.2003.10.048 [DOI] [PubMed] [Google Scholar]
- Kirton A. H., O’Hara P. J., Shortridge E. H., and Cordes D. O.. 1976. Seasonal incidence of enzootic pneumonia and its effect on the growth of lambs. N Z Vet J. 24:59–64. doi: 10.1080/00480169.1976.34284 [DOI] [PubMed] [Google Scholar]
- McRae K. M., Baird H. J., Dodds K. G., Bixley M. J., and Clarke S. M.. 2016. Incidence and heritability of ovine pneumonia, and the relationship with production traits in New Zealand sheep. Small Ruminant Res. 145:136–141. doi: 10.1016/j.smallrumres.2016.11.003 [DOI] [Google Scholar]
- Neibergs H. L., Seabury C. M., Wojtowicz A. J., Wang Z., Scraggs E., Kiser J. N., Neupane M., Womack J. E., Van Eenennaam A., Hagevoort G. R., et al. ; Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team. 2014. Susceptibility loci revealed for bovine respiratory disease complex in pre-weaned Holstein calves. BMC Genomics. 15:1164. doi: 10.1186/1471-2164-15-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y., Sano T., and Nagata S.. 2009. Regulation of the innate immune response by threonine-phosphatase of eyes absent. Nature. 460:520–524. doi: 10.1038/nature08138 [DOI] [PubMed] [Google Scholar]
- Snowder G. 2009. Genetics, environment and bovine respiratory disease. Anim. Health Res. Rev. 10:117–119. doi: 10.1017/S1466252309990144 [DOI] [PubMed] [Google Scholar]
- Srikumaran S., Kelling C. L., and Ambagala A.. 2007. Immune evasion by pathogens of bovine respiratory disease complex. Anim. Health Res. Rev. 8:215–229. doi: 10.1017/S1466252307001326 [DOI] [PubMed] [Google Scholar]
- Tadjuidje E., and Hegde R. S.. 2013. The eyes absent proteins in development and disease. Cell. Mol. Life Sci. 70:1897–1913. doi: 10.1007/s00018-012-1144-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone M., Powell M. J., Tone Y., Thompson S. A., and Waldmann H.. 2000. IL-10 gene expression is controlled by the transcription factors sp1 and sp3. J. Immunol. 165:286–291. [DOI] [PubMed] [Google Scholar]
- Wilson I. M., Vucic E. A., Enfield K. S., Thu K. L., Zhang Y. A., Chari R., Lockwood W. W., Radulovich N., Starczynowski D. T., Banáth J. P., et al. 2014. EYA4 is inactivated biallelically at a high frequency in sporadic lung cancer and is associated with familial lung cancer risk. Oncogene. 33:4464–4473. doi: 10.1038/onc.2013.396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. G., Jin R., Ren W., Zou L., Wang Y., and Zhou G. P.. 2012. Transcription factors sp1 and sp3 regulate basal transcription of the human IRF-3 gene. Biochimie. 94:1390–1397. doi: 10.1016/j.biochi.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Yin L. M., Jiang G. H., Wang Y., Wang Y., Liu Y. Y., Jin W. R., Zhang Z., Xu Y. D., and Yang Y. Q.. 2008. Serial analysis of gene expression in a rat lung model of asthma. Respirology. 13:972–982. doi: 10.1111/j.1440-1843.2008.01398.x [DOI] [PubMed] [Google Scholar]
- Zanella R., Gava D., Peixoto J. d. e. O., Schaefer R., Ciacci-Zanella J. R., Biondo N., da Silva M. V., Cantão M. E., and Ledur M. C.. 2015. Unravelling the genetic components involved in the immune response of pigs vaccinated against influenza virus. Virus Res. 210:327–336. doi: 10.1016/j.virusres.2015.09.003 [DOI] [PubMed] [Google Scholar]
- Zanotti K. J., Maul R. W., Castiblanco D. P., Yang W., Choi Y. J., Fox J. T., Myung K., Saribasak H., and Gearhart P. J.. 2015. ATAD5 deficiency decreases B cell division and Igh recombination. J. Immunol. 194:35–42. doi: 10.4049/jimmunol.1401158 [DOI] [PMC free article] [PubMed] [Google Scholar]