Abstract
Background:
Milk is considered as complete food and an important part of human diet throughout the world including India. Bacterial contamination of milk such as Escherichia coli due to unhygienic condition and poor udder health can cause infections, especially in infants and elders or in immunocompromised persons. Possession of antimicrobial resistance genes by commensal bacteria present in milk makes the issue more serious.
Aim:
The study was aimed to isolate and characterize extended-spectrum beta-lactamase (ESBL)-producing E. coli from milk samples collected from different parts of West Bengal, India, to assess the potential risk associated with the food.
Materials and Methods:
Around 182 milk samples were collected from apparently healthy cows reared by organized dairy farms in West Bengal. E. coli was isolated from collected samples as per standard methods followed by serotyping. The detection of ESBL-producing E. coli was done both phenotypically and genotypically by detecting the presence of blaCTX-M gene. Antibiogram of the ESBL-positive isolates was done using common 12 antibiotics by disc diffusion method.
Results:
A total of 22 (12.1%) samples were found to be positive for E. coli in this study. Different serotypes such as O11, O20, O22, O34, O35, O128, O149, and UT were isolated from the collected samples. 12 (54.5%) E. coli strains showed the capability of producing ESBL, both phenotypically and genotypically with the presence of blaCTX-M gene. Antibiogram of these ESBL-positive isolates revealed the drugs such as colistin (100%), levofloxacin (83.33%), and imipenem (66.67%) to be highly sensitive against this pathogen but drugs such as cefotaxime (100%), ceftazidime (91.67%), amoxicillin/clavulanic acid (83.33%), tetracycline (75.00%), and gentamicin (58.33%) to be very much resistant.
Conclusion:
More than 50% of the E. coli strains prevalent in the bovine milk samples were positive for ESBL production and are resistant to most of the common antimicrobials which may be alarming for human health.
Keywords: antibiogram, blaCTX-M, bovine milk, extended-spectrum beta-lactamase, Escherichia coli
Introduction
India is one of the largest milk producing countries in the world with dairy industry playing an important role in the rural economy [1] generating huge self-employment. Bovine milk is generally considered to be a good source of protein and vitamins to human beings, particularly to the infants. However, due to faulty handling and storage of milk and poor management of the animal, milk may get spoiled due to rapid multiplication of bacteria due to milk’s high nutritive value [2]. Escherichia coli is one dreadful pathogen, especially the “enterohemorrhagic E. coli” strains, causing infection through milk which has a great effect on human health [3,4].
The prevalence of extended-spectrum beta-lactamase (ESBL)-producing E. coli is increasing in the globe including India. These pathogens pose a major challenge for the treatment of general infections and cause a problem with the extensive use of second- or third-generation antibiotics for the treatment of bacterial infections [5]. ESBL E. coli is mostly insensitive to lots of commonly used antibiotics causing an increase in the use of last-resort antimicrobial drugs (i.e., carbapenems) during treatment. Again, E. coli strains carrying the resistance genes can easily transfer those genes to other pathogens leading to the spread of drug resistance [6]. Hence, the presence of ESBL-producing E. coli in the food processing chain or in the food of our daily consumption which is possibly coming from these healthy farm animals is the fact which has to be appropriately studied.
The present study was aimed for the detection and characterization of ESBL-producing E. coli from raw milk samples (by detecting blaCTX-M gene in the isolates) from different dairy farms followed by further characterization and to know their antibiotic resistance patterns in vitro.
Materials and Methods
Ethical approval
As per the guidelines of the Committee for the Purpose of Control and Supervision of Experiments on Animals, this study does not require any ethical approval from the University Animal Ethics Committee.
Collection of samples
Bovine milk samples (n=182) were collected from different unorganized dairy farms of West Bengal aseptically in sterile plastic containers (Table-1) during the period of April-June, 2018. Milk samples were taken at 15 ml (approximately) in sterile plastic containers directly from the teats of the cows. The cows were selected on the basis of the history of illness/with decreased milk yield. The samples containers were kept in sample flask under ice and cooling pad cover and were transported through shortest route (for maximum 5-6 h approximately) to the Department of Veterinary Microbiology, Mohanpur, Nadia, for further study. All the collected samples were studied on the same day of receiving at the laboratory.
Table-1.
Details of milk sample collection from different districts of West Bengal, India.
| Name of the districts | Number of dairy farms covered | Total number collected samples |
|---|---|---|
| Purba Bardhaman | 04 | 32 |
| Paschim Bardhaman | 05 | 47 |
| Nadia | 03 | 29 |
| Hooghly | 11 | 74 |
| Total | 23 | 182 |
Isolation and characterization of E. coli
The collected milk samples were enriched adding sterile nutrient broth at 37°C for 6-8 h followed by streaking on to sterile MacConkey’s agar (HiMedia, India) plates. The plates were incubated aerobically at 37°C for 10-12 h. The tentative pinkish single colonies (i.e., lactose fermenting) were selected for selective isolation by further streaking on sterile Eosin Methylene Blue (EMB) agar (HiMedia, India) plates followed by 10-12 h incubation again at 37°C. The single colonies showing characteristic greenish “metallic sheen” were picked up and were stored using sterile nutrient agar (HiMedia, India) slants for further morphological study by Gram’s staining method and biochemical characterization with tests such as indole, methyl red, citrate utilization, Voges–Proskauer, catalase, and nitrate reduction [7,8]. All positive isolates showing typically to be E. coli were further confirmed for ESBL positivity later.
Serotyping of the E. coli isolates
All positive E. coli isolates were sent to the National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, Himachal Pradesh, India, for serotyping. All E. coli strains were subcultured in small sterile glass vials and were properly packed in hardboard box under cotton cover followed by sending to the NSEC, Kasauli, by registered post.
Phenotypic detection of ESBL production in E. coli isolates
Phenotypic detection of the presence of ESBL in E. coli isolates was done in vitro by disc diffusion method [9] using both cefotaxime (30 µg) and ceftazidime disks (30 µg) with and without clavulanate (10 µg) as per the CLSI methods by Patel et al. [10]. A difference of >5 mm between the zone diameters of each disk and their respective clavulanate disk is measured to phenotypically confirm the ESBL production by the E. coli isolates under study [10].
Molecular detection of ESBL production in E. coli isolates by polymerase chain reaction (PCR)
Bacterial culture lysate preparation
Selective E. coli strains were inoculated into nutrient broth (HiMedia, India) followed by 18 h incubation at 37°C. 1 ml of young broth culture of each sample was taken in a sterile 1.5 ml microcentrifuge tube (Tarsons, India) followed by centrifugation at 6000 rpm for 5 min [11]. The obtained pellet was washed 3 times with TE buffer and was suspended again in TE buffer (1 ml). The microcentrifuge tube with culture was then boiled in water for 10 min followed by chilling in ice. Again each tube was centrifuged at 5000 rpm for 5 min followed by removal of cell debris and the supernatant with crude DNA was collected and stored at −20°C for further use as a template in PCR [11].
Detection of blaCTX-M gene (540 bp) in E. coli isolates
All phenotypically ESBL-positive E. coli isolates were considered for confirmation by PCR detection of the blaCTX-M gene in them as per the protocol followed by Weill et al. [12] with slight modifications. All standard reagents and primers (GCC Biotech, India) were used in this process. Amplification reaction mixture containing 3 µl DNA templates, 50 pmol the primer set [540 bp] (forwardCTX-M-F 5’-CAATGTGCAGCACCAGTAA-3’ and reverseCTX-M-R 5’-CGCGATATCATTGGTGGTG-3’), 1U GoTaq DNA polymerase (Promega, USA), 200 mM deoxynucleoside triphosphate, 10% dimethyl sulfoxide, and 2 mM MgCl2 was prepared in a 25 µl reaction mixture and used in PCR amplification conducted in a thermocycler (Eppendorf, Germany). The PCR amplification was done in the following cycle condition with an initial denaturation at 94°C for 10 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 53°C for 30 s, and elongation at 72°C for 60 s with a 10 min final extension period at 72°C. The amplified PCR products were loaded onto a 1.5% w/v agarose gel (SRL, India), with ethidium bromide (0.5 µg/ml) (SRL, India) followed by agarose gel electrophoresis and were visualized by gel documentation system (UVP, UK). One ESBL-producing E. coli strain (O2) which is the departmental isolate and one Pseudomonas aeruginosa (ATCC 27853) were used as positive and negative controls in PCR assays.
In vitro antibiotic sensitivity test of ESBL E. coli isolates
Antibiogram of the ESBL-positive E. coli isolates was performed using 12 antimicrobials, i.e., amikacin, amoxicillin/clavulanic acid, azithromycin, colistin, cotrimoxazole, cefotaxime, ceftazidime, gentamicin, imipenem, levofloxacin, piperacillin-tazobactam, and tetracycline by Kirby–Bauer disc diffusion method [9]. Young broth cultures of all the ESBL-positive isolates were produced for the test. Separate and sterile Mueller-Hinton agar (HiMedia, India) plates were used for uniform spreading of each broth culture using sterile L-spreader, and standard discs (HiMedia, India) were placed with sterile forceps. All the plates were incubated at 37°C for 10-12 h, and the results were interpreted by measuring the inhibition zone diameter and comparing those with the standard chart [10].
Results
Out of 182 bovine milk samples tested, 22 (12.08%) samples were found to be identified as E. coli in this study. All the positive isolates showed typical characteristics during cultural, i.e., produced typical “metallic sheen” when grown on EMB agar plates (Figure-1) and morphological examinations (pink rods after Gram’s staining). All showed typical results during their biochemical characterization, i.e., positive to indole, methyl red, catalase, and nitrate reduction whereas negative to VP and citrate utilization.
Figure-1.
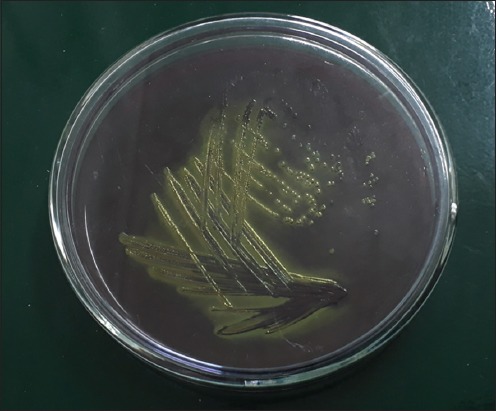
Characteristic “metallic sheen” produced by positive Escherichia coli isolated from bovine milk samples on Eosin Methylene Blue agar plate.
Serotyping of all 22 E. coli isolates was done at the National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, HP, India, to get the following 8 different serotypes, i.e., O11, O20, O22, O34, O35, O128, O149, and UT (Table-2).
Table-2.
Frequency of E. coli serotypes isolated from bovine milk samples.
| Sl. No. | E. coli serotypes | Frequency of prevalence (%) |
|---|---|---|
| 1 | O11 | 3 (13.63) |
| 2 | O20 | 7 (31.82) |
| 3 | O149 | 3 (13.63) |
| 4 | O22 | 5 (22.72) |
| 5 | O35 | 1 (4.55) |
| 6 | O34 | 1 (4.55) |
| 7 | O128 | 1 (4.55) |
| 8 | UT | 1 (4.55) |
| Total | 22 (100.00) |
E. coli=Escherichia coli
During ESBL detection, a total of 12 (54.54%) E. coli isolates were found to be phenotypically positive as ESBL producers by double disc method in this study. All phenotypically ESBL-positive E. coli isolates were detected to have the blaCTX-M gene (540 bp) by PCR (Figure-2). The ESBL-producing strains belonged to O11[2 nos.], O20 [4 nos.], O22 [5 nos.], and O128 [1 no.]. Samples from Purba Bardhaman district showed the highest positivity in comparison to other districts (Table-3).
Figure-2.
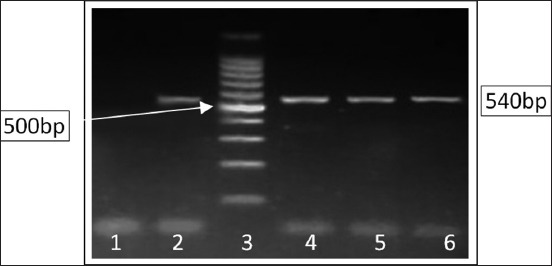
Polymerase chain reaction detection of blaCTX-M gene (540 bp) in extended-spectrum beta-lactamase-producing Escherichia coli strains isolated from bovine milk samples in West Bengal, India. Lane 1: Negative control, Lane 2: Positive control, Lane 3: 100 bp ladder, Lanes 4-6: Test samples.
Table-3.
District-wise Distribution of E. coli isolates with ESBL positivity.
| Name of the Districts | Number of samples studied | Number of E. coli strains isolated (%) | ESBL positivity in E. coli strains |
|---|---|---|---|
| Purba Bardhaman | 32 | 6 (18.75) | 5 |
| Paschim Bardhaman | 47 | 7 (14.89) | 4 |
| Nadia | 29 | 2 (6.90) | 1 |
| Hooghly | 74 | 7 (9.46) | 2 |
| Total | 182 | 22 (12.08) | 12 |
E. coli=Escherichia coli, ESBL=Extended-spectrum beta-lactamase
In vitro, antibiotic sensitivity assay of the ESBL-positive E. coli isolates showed high-level resistance to cefotaxime, ceftazidime, amoxicillin-clavulanic acid, tetracycline, gentamicin, amikacin, etc. (with the range of 60-100%). Piperacillin-tazobactam (83.33%) was detected to be intermediately sensitive to these isolates (Table-4), and drugs such as colistin, levofloxacin, and imipenem were found to be sensitive against these pathogens.
Table-4.
Antibiogram of 12 ESBL-producing E. coli strains isolated from bovine milk samples in West Bengal, India.
| Sl. No. | Antimicrobials (Conc. in µg) | Isolates sensitive | Isolates intermediately sensitive | Isolates resistant |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| 1. | Amikacin (30) | 2 (16.67) | 3 (25.00) | 7 (58.33) |
| 2. | Amoxicillin/Clavulanic acid (20/10) | 2 (16.67) | 0 (0) | 10 (83.33) |
| 3. | Colistin (10) | 12 (100) | 0 (0) | 0 (0) |
| 4. | Cotrimoxazole (25) | 5 (41.67) | 4 (33.33) | 3 (25.00) |
| 5. | Cefotaxime (30) | 0 (0) | 0 (0) | 12 (100) |
| 6. | Ceftazidime (30) | 0 (0) | 1 (8.33) | 11 (91.67) |
| 7. | Imipenem (10) | 8 (66.67) | 4 (33.33) | 0 (0) |
| 8. | Gentamicin (10) | 0 (0) | 5 (41.67) | 7 (58.33) |
| 9. | Levofloxacin (5) | 10 (83.33) | 2 (16.67) | 0 (0) |
| 10. | Pipercillin-Tazobactam (100/10) | 2 (16.67) | 10 (83.33) | 0 (0) |
| 11. | Azithromycin (30) | 4 (33.33) | 3 (25.00) | 5 (41.67) |
| 12. | Tetracycline (30) | 2 (16.67) | 1 (8.33) | 9 (75.00) |
ESBL=Extended-spectrum beta-lactamase, E. coli=Escherichia coli
Discussion
Approximately 12% of the total milk samples screened were found to yield E. coli isolates in this study which were also supported by Kamaruzzaman [13], Badri et al. [14], Geser et al. [15], and Ali et al. [16] who also reported 12.22-13.7% E. coli positivity in the bovine milk samples during their study. All positive E. coli isolates showed typical cultural, morphological, and biochemical nature in this study which was also supported by Carter and Wise [7], Samanta [17], and Quinn et al. [8].
The serotypes reported by the National Salmonella and Escherichia Centre, Central Research Institute, Kasauli, were also supported by Osman et al. [18] who detected E. coli serogroups O26, O86, O111, and O127 from cattle milk in their study. Al-Zogibi et al. [19] reported the prevalence of E. coli serogroups, namely O22, O111, O113, and O172, from bovine milk samples in their study which also supports the current findings.
Approximately, 54.54% E. coli isolates were both phenotypically and genotypically positive to produce ESBL in this study which was also supported by Geser et al. [15] and Ibrahim et al. [20]. Kamaruzzaman [13] reported a high prevalence of ESBL-producing E. coli in milk (66.7%) followed by farm environment (27.8%) and cattle (5.5%) in his work. Ali et al. [16] reported 36 (23.53%) and Badri et al. [14] reported 29.3% ESBL-positive E. coli strains from bovine milk samples which may be of great concern as these pathogens may be carried out to the human consumers as well as calves leading to the spread of the antibiotic-resistant pathogens over human and animal population. Sharma et al. [21] also reported ESBL-positive E. coli serotypes in their study, matching the current findings.
The high level of antibiotic resistance as shown in this report was also reported earlier by Kamaruzzaman [13], Ibrahim et al. [20], and Hinthong et al. [22]. Ali et al. [16] also found resistance against drugs such as ampicillin (86.11%), amoxicillin-clavulanic acid (63.89%), cefotaxime (100%), ceftazidime (66.67%), tetracycline (72.22%), and gentamicin (61.11%) by ESBL E. coli pathogens in their study. Faruk et al. [23] reported that ampicillin, cefotaxime, ceftazidime, and cefuroxime (all 100%) and tetracycline (93.54%) were highly resistant but imipenem (100%) to be highly sensitive to the ESBL E. coli strains isolated from cattle in their study which almost matches with the current findings.
Conclusion
The drug-resistant ESBL gene is significantly present in approximately 55% of the E. coli strains isolated from cattle milk samples which may be of great health concern for human beings. This drug resistance can easily be transferred between closely related pathogens in vivo which may result in risky and fatal health hazards due to unsuccessful treatment with common antimicrobials. Hence, proper care should be taken to combat these dreadful pathogens.
Author’s Contributions
KB and SD designed the study. AB and ADS collected the samples. KB, AB, and SP carried out the experiment. SNJ, SD, and IS analyzed the data. DPI, KB, and SD drafted the article. SNJ and IS revised the article. All authors read and approved the final manuscript.
Acknowledgments
The authors thank the University Authorities and the Faculties of the Department of Veterinary Microbiology, West Bengal University of Animal and Fishery Sciences, West Bengal, India, for providing necessary funds [ICAR Development Grant 2017-18 (2nd Installment), Order no. VCS/WBUAFS/1-18(Pt-1)/09(11)/10, dated. 24.11.17], research facilities, and support for this study. They are also thankful to the farm owners, dairy farmers, and local veterinary doctors of the concerned districts of West Bengal, India, for their technical and field support.
Competing Interests
The authors declare that they have no competing interests.
References
- 1.A Study of India's Dairy Sector 2017. The World's Largest Producer and Consumer - Research and Markets. [Last accessed on 18-07-2018]. Available from: https://www.businesswire.com/news/home/20180102005671/en/Study-Indias-Dairy-Sector-2017-Worlds-Largest .
- 2.Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implications. Foodborne Pathog. Dis. Summer. 2005;2(2):115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- 3.Hickey CD, Sheehan JJ, Wilkinson MG, Auty MAE. Growth and location of bacterial colonies within dairy foods using microscopy techniques: A review. Front. Microbiol. 2015;6:99. doi: 10.3389/fmicb.2015.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen Y, Sperandio V. Enterohemorrhagic E. coli(EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2012;2(90):1–7. doi: 10.3389/fcimb.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenover FC, Mohammed MJ, Gorton TS, Dembek ZF. Detection and reporting of organisms producing extended spectrum-beta-lactamases: Survey of laboratories in Connecticut. J. Clin. Microbiol. 1999;37(12):4065–4070. doi: 10.1128/jcm.37.12.4065-4070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Yang X, Li J, Lv N, Liu F, Wu J, Lin I.Y.C, Wu N, Weimer BC, Gao GF, Liu Y, Zhu B. The transfer network of bacterial mobile resistome connecting animal and human microbiome. Appl. Environ. Microbiol. 2016;82(22):66726681. doi: 10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter GR, Wise DJ. Essentials of Veterinary Bacteriology and Mycology. 6th ed. Iowa: Iowa State Press; 2004. pp. 129–134. [Google Scholar]
- 8.Quinn PJ, Markey BK, Leonard FC, Fitz P.E.S, Fanning S, Hartigan PJ. Veterinary Microbiology and Microbial Diseases. 2nd ed. Oxford: Blackwell Publishing Ltd; 2011. pp. 266–273. [Google Scholar]
- 9.Bauer AW, Kirby WM, Sherrris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966;45(4):493–496. [PubMed] [Google Scholar]
- 10.Patel JB, Cockerill FR, Bradford PA, Eliopoulos GM, Hindler JA, Jenkins SG, Lewis JS, Limbago B, Miller LA, Nicolau DP, Powell M, Swenson JM, Traczewski MM, Turnidge JD, Weinstein MP, Zimmer BL. Wayne, Pa, USA: CLSI; 2015. Clinical and Laboratory Standard Institute: Performance Standards for Antimicrobial Susceptibility Testing;Twenty-Fifth Informational Supplement. CLSI Document M100-S25; pp. 1–240. [Google Scholar]
- 11.Salehi TZ, Mahzounieh M, Saeedzadeh A. Detection of inv A gene in isolated salmonella from broilers by PCR method. Int. J. Poult. Sci. 2005;4(8):557–559. [Google Scholar]
- 12.Weill F, Claude J, Demartin CS, Grimont P. Characterization of ESBL (CTX-M-15) producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 2004;238(2):353–358. doi: 10.1016/j.femsle.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 13.Kamaruzzaman EA. Occurrence of Extended-Spectrum Beta-Lactamase Producing Escherichia coli in Dairy Cattle, Farm Environment and Milk. Masters Degree Thesis Submitted in the University of Putra, Malaysia. 2015 [Google Scholar]
- 14.Badri AM, Ibrahim IT, Mohamed SG, Garbi MI, Kabbashi AS, Arbab MH. Prevalence of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae isolated from raw milk samples in Al Jazirah State, Sudan. Mol. Biol. 2017;7(1):201. [Google Scholar]
- 15.Geser N, Stephan R, Hachler H. Occurrence and characteristics of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae in food-producing animals, minced meat and raw milk. BMC Vet. Res. 2012;8(1):21. doi: 10.1186/1746-6148-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali T, Ur Rahman S, Zhang L, Shahid M, Zhang S, Liu G, Gao J, Han B. ESBL-producing Escherichia coli from cows suffering mastitis in China contain clinical class 1 integrons with CTX-M linked to ISCR1. Front. Microbiol. 2016;7:1931. doi: 10.3389/fmicb.2016.01931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samanta I. In: Veterinary Bacteriology. Ch. 6. New Delhi: New India Publishing Agency; 2013. Enterobacteriaceae; pp. 119–135. [Google Scholar]
- 18.Osman KM, Mustafa AM, Aly MA, Abd EGS. Serotypes, virulence genes, and intimin types of Shiga toxin-producing Escherichia coli and enteropathogenic Escherichia coli isolated from mastitic milk relevant to human health in Egypt. Vector Borne Zoonotic Dis. 2012;12(4):297–305. doi: 10.1089/vbz.2010.0257. [DOI] [PubMed] [Google Scholar]
- 19.Al-Zogibi OG, Mohamed MI, Hessain AM, El-Jakee JK, Kabli SA. Molecular and serotyping characterization of Shiga toxogenic Escherichia coli associated with food collected from Saudi Arabia. Saudi J. Biol. Sci. 2015;22(4):438–442. doi: 10.1016/j.sjbs.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim DR, Dodd C.E.R, Stekel DJ, Ramsden SJ, Hobman JL. Multidrug-resistant, extended spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS Microbiol. Eco. 2016;92(4):fiw013. doi: 10.1093/femsec/fiw013. [DOI] [PubMed] [Google Scholar]
- 21.Sharma S, Kaur N, Malhotra S, Madan P, Ahmad W, Hans C. Serotyping and antimicrobial susceptibility pattern of Escherichia coli isolates from urinary tract infections in pediatric population in a tertiary care hospital. J. Pathog. 2016;2016(2548517):4. doi: 10.1155/2016/2548517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinthong W, Pumipuntu N, Santajit S, Kulpeanprasit S, Buranasinsup S, Sookrung N, Chaicumpa W, Pisinee AP, Indrawattana N. Detection and drug resistance profile of Escherichia coli from subclinical mastitis cows and water supply in dairy farms in Saraburi Province, Thailand. Peer J. 2017;5:e3431. doi: 10.7717/peerj.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faruk P, Hulya T, Dilek O, Hakan Y. Molecular characterization of ESBL-producing Escherichia coli isolated from healthy cattle and sheep. Acta Vet. Beogr. 2016;66(4):520–533. [Google Scholar]


