Abstract
CCCTC-binding factor (CTCF) sites are enriched at the boundaries of topologically associated domains (TADs), but their function within TADs is unclear. Removal of sub-TAD CTCF sites adjacent to the α-globin enhancers is now shown to result in inappropriate activation of neighbouring genes. Intra-TAD enhancer insulation might be broadly important for tissue specificity of enhancers.
Topologically associated domains (TADs) are defined as chromosomal regions of approximately 1 Mb that contain sequences that interact with each other more frequently than they do with sequences throughout the rest of the genome1,2. TADs are mostly conserved among different cell types and even across species, suggesting that they form a fundamental layer of chromosome folding in the nucleus. One of the defining features of TAD boundaries is enrichment of CCCTC-binding factor (CTCF) and cohesin complex shared sites. These boundary sites typically interact to create a looped domain that might favour interactions within TAD boundaries3. Consistent with this, enhancers almost always interact with genes in the same TAD. Conceptually, this boundary function of CTCF fits well with its function as an architectural protein, but also as an insulator of enhancer activity. Several studies have shown that deletion of TAD boundary elements or specifically TAD boundary CTCF sites resulted in inappropriate contacts of enhancers with neigh-bouring genes and the subsequent expression of these genes, termed ‘enhancer adoption’4–6.
However, despite enrichment at TAD boundaries, most CTCF sites (85%) occur within TADs, and how they function is largely unknown. They are thought to define smaller domains of interaction termed sub-TADs7, but 80% of enhancer–gene long-range contacts skip over intervening CTCF sites within TADs, suggesting that the insulator activity of sub-TAD CTCF is context-dependent8. One possible explanation is that interaction patterns of intra-TAD CTCF–cohesin sites might have a tissue-specific character. In this issue of Nature Cell Biology, Hanssen et al.9 reveal that a series of CTCF–cohesin sites surrounding the α-globin genes and their complex enhancer define a sub-TAD domain that forms in mouse primary erythroid cells, but not in embryonic stem cells (ESCs). A small deletion removing critical CTCF–cohesin sites resulted in enlargement of this looped domain and allowed the enhancers to inappropriately activate newly encompassed nearby, non-erythroid genes. Although the basis for the tissue specificity of the CTCF–cohesin loop remains unknown, these results show that intra-TAD CTCF–cohesin sites are likely to play an important role in keeping enhancers faithful to correct target genes.
To address the function of intra-TAD CTCF–cohesin sites in regulation of gene expression, Hanssen et al. used a genome locus containing several sub-TADs including one with the syntenic α-globin gene cluster as the main component. A wealth of information is available detailing the chromatin structure and transcriptional activity of the duplicated α-globin genes (Hba-a1 and Hba-a2) and their enhancers (R1–R4, and Rm that is mouse-specific) that contribute to α-globin gene transcriptional activity exclusively in erythroid cells (Fig. 1a)10. Interestingly, the enhancers are almost all within the introns of the widely expressed gene Nprl3 that is the nearest upstream neighbour to the α-globin genes.
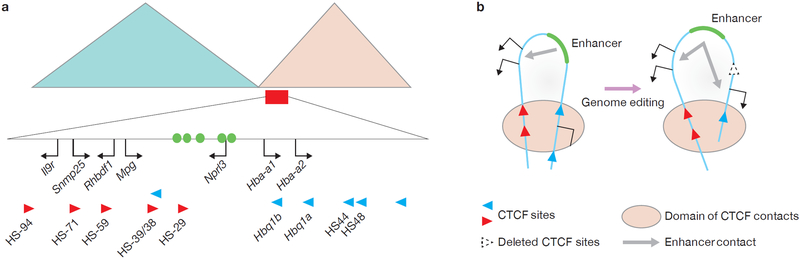
Figure 1.
The α-globin cluster in mouse ESCs and primary erythroid cells. (a) The large triangles represent Hi-C results showing two adjacent TADs in mouse ESCs with a boundary between them1. The domain within the red rectangle includes the α-globin genes and their enhancers (green circles), and upstream widely expressed genes. Red and blue arrowheads denote DNase I hypersensitive sites (HSs) bound by CTCF. The directionality of the arrowhead indicates the orientation of the underlying CTCF-binding motif. (b) A simplified model depicting interaction of convergently arranged CTCF sites to form a ‘hairpin’ structure described by Hanssen et al.9. The head of the pin is a loop that encloses an enhancer and its target genes but excludes an unrelated gene. When genome editing is used to remove the CTCF site at one end of the loop, the loop still forms but is enlarged by interaction of the next appropriate CTCF site. This results in incorporation of a new gene that then becomes a target of the enhancer.
What provides the specificity that defines the interaction between the α-globin enhancers and genes? Applying high-throughput sequencing for DNase I hypersensitive sites (DNase-seq), transposase-accessible chromatin (ATAC-seq) and chromatin immunoprecipitation (ChIP-seq), Hanssen et al. comprehensively demonstrated that sites of open chromatin surrounding the α-globin cluster correspond to CTCF–cohesin sites. Taking into account the insulator role of CTCF, the authors proposed that intra-TAD CTCF loops might be responsible for determining α-globin enhancer–gene interaction patterns. Based on the recent observation that CTCF loops are preferentially formed between CTCF-occupied sites in a convergent orientation11, the authors screened sequences up-and downstream of the α-globin genes and their enhancers and discovered clusters of convergently oriented (with one exception) CTCF–cohesin sites that are occupied by these proteins in both ESCs and primary erythroid cells.
To investigate the mechanisms by which CTCF sites might influence the tissue specificity of gene expression, they applied next generation Capture-C technology using mouse ESCs and primary erythroid cells12. The authors detected broad, diffuse interactions across the α-globin locus in ESCs but much stronger interactions in erythroid cells including specific contacts between the enhancers and genes. The locus flanking convergent CTCF sites make contact with each other but, for the most part, do not contact the α-globin enhancers or genes. Moreover, the CTCF–CTCF contacts are considerably stronger in erythroid cells than in ESCs. The observations suggest that the α-globin cluster-encompassing CTCF looping interactions constitute a sub-TAD and function to constrain α-globin enhancer activity in a tissue-specific manner. Consistent with this model, expression of Nprl3, located inside the CTCF loops, is substantially increased in erythroid cells compared to ESCs. However, genes located in the upstream, neighbouring sub-TAD are expressed similarly in ESCs and erythroid cells (Mpg, Snmp25 and Il9r) or are silenced (Rhbdf1).
To provide in vivo functional evidence supporting the model, the authors applied TALEN and CRISPR–Cas9 genome editing to generate mice with deletions of CTCF-binding sites located just upstream of the α-globin enhancers (HS-38, HS-39 and HS-29) (Fig. 1a). Simultaneous deletion of the HS-38 and HS-39 sites completely abolished CTCF occupancy at the sites and, strikingly, resulted in an enlarged sub-TAD domain delineated by the same downstream CTCF sites and new CTCF sites further upstream from HS-38/HS-39 in primary erythroid cells. Within this enlarged loop, the α-globin enhancers and the non-erythroid genes Mpg, Rhbdf1 and Snmp25, formerly excluded from the α-globin sub-TAD, interact. RNA sequencing experiments confirmed that de novo looping between α-globin enhancers and these ectopic genes substantially increased their expression, although to varying extents. Expression of Il9r, which remains outside the newly enlarged sub-TAD loop, was not affected.
Not surprisingly, there was an increase in the H3K4me3 histone mark and in RNA polymerase II occupancy, accompanied by an increase in chromatin accessibility at the newly activated gene promoters. Interestingly, ectopic activation of Rhbdf1, which is normally silenced in erythroid cells, was not accompanied by loss of H3K27me3 or PRC2 complex occupancy at the gene promoter, suggesting that insulation of Rhbdf1 from the α-globin enhancers is required for polycomb-mediated repression of gene expression. Deletion of the HS-38 site alone had a more moderate effect compared with the double deletion on activation of gene expression, while deletion of only HS-39 did not result in gene expression changes. The results suggest the two divergently oriented CTCF sites work together to form a functional insulator. Independent deletion of the HS-29 CTCF site that is between two of the enhancers increased looping of the enhancers to the α-globin genes, but did not measurably affect the already very high transcription level.
Overall, the results of Hanssen et al. show that convergent, interacting CTCF sites surrounding the α-globin cluster act as a boundary to ensure erythroid-specific activation of the α-globin genes but not their non-erythroid neighbours (Fig. 1b). Enlargement of the region constrained by CTCF looping to include the non-erythroid Mpg, Rhbdf1 and Snmp25 genes and to allow their inappropriate activation by the α-globin enhancers is a satisfying mechanistic dissection of insulator antagonism of enhancers in vivo. Still, questions remain. What attributes of CTCF-occupied sites in chromatin underlie the erythroid-specific looping around the α-globin cluster observed in this work? The authors speculate that there might be additional cohesin recruitment to the locus in erythroid cells that could contribute to stabilization of CTCF–CTCF interactions but this remains to be demonstrated. Why are the ectopically activated gene promoters receptive to the α-globin enhancers? The basis for the new long-range enhancer communication with non-erythroid genes, which is thought to depend on the binding of compatible transcription factors to each13,14, is unclear.
Other uncertainties remain. For example, what is the basis for the functional distinction among CTCF sites HS-29, HS-38 and HS-39 revealed by the differing effects of their deletion on chromatin structure and gene expression? Also, how can the α-globin enhancers skip over a CTCF site near Hbq1b to activate the Hba-a2 gene and how can these enhancers activate the NME4 gene, 400 kb from the human α-globin cluster, despite intervening CTCF sites/sub-TAD boundaries15? Overall, there is a pressing need to understand what makes CTCF sites different from one another.
It seems clear from this work that enclosure within a sub-TAD limits an enhancer’s choice of target genes. But does the sub-TAD organization actually favour enhancer–gene interactions? In these experiments, loss of at least one sub-TAD boundary, albeit with formation of an enlarged sub-TAD, had no deleterious effect on enhancer–α-globin interaction or activity, suggesting the answer might be negative. Nor did inclusion in the enlarged loop of additional genes responsive to the enhancers seem to affect α-globin expression by competition. However, since the α-globin genes are expressed at such high levels, any diminution of their activity after HS-38/39 deletion might be difficult to observe. Finally, the approaches illustrated in the work of Hanssen et al. will be important to address other questions in the field, such as, does the larger TAD encompassing the α-globin cluster change when sub-TAD boundaries and enhancer–gene interactions within it are changed? This kind of question begins to address the issue of whether form follows function or the other way around.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Dixon JR et al. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nora EP et al. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao SS et al. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupianez DG et al. Cell 161, 1012–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hnisz D et al. Science 351, 1454–1458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narendra V, Bulajic M, Dekker J, Mazzoni EO & Reinberg D Genes Dev 30, 2657–2662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phillips-Cremins JE et al. Cell 153, 1281–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li G et al. Cell 148, 84–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanssen LLP et al. Nat. Cell Biol 19, 952–961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay D et al. Nat. Genet 48, 895–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Y et al. Cell 162, 900–910 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies JO et al. Nat. Methods 13, 74–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noordermeer D et al. Nat. Cell Biol 13, 944–951 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plank JL & Dean A Mol. Cell 55, 5–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lower KM et al. Proc. Natl Acad. Sci. USA 106, 21771–21776 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]