SUMMARY
The molecular foundations of Hürthle cell carcinoma (HCC) are poorly understood. Here, we describe a comprehensive genomic characterization of 56 primary HCC tumors that span the spectrum of tumor behavior. We elucidate the mutational profile and driver mutations and show that they exhibit a wide range of recurrent mutations. Notably, we report an extremely high number of disruptive mutations to both protein-coding and tRNA-encoding regions of the mitochondrial genome. We reveal unique chromosomal landscapes that involve whole-chromosomal duplications of chromosomes 5 and 7 and widespread loss of heterozygosity arising from haploidization and copy number–neutral uniparental disomy. We also identify fusion genes and disrupted signaling pathways that may drive disease pathogenesis.
In Brief
Ganly et al. elucidate recurrent mutations impacting the RTK/RAS/AKT/mTOR pathway, DNA damage/repair, epigenetic modifiers, TERT promoter and the mitochondrial genome in Hürthle cell carcinoma (HCC). HCCs also display prevalent chromosome 5 and 7 duplications, loss of heterozygosity, and in-frame gene fusions.
INTRODUCTION
Hürthle cell carcinomas (HCCs) account for 5% of all malignancies arising from thyroid follicular cells and are, in general, understudied (Hundahl et al., 1998). The most noteworthy characteristic of HCC cells is their remarkable abundance of dysfunctional mitochondria (> 75% of cell volume) (Máximo et al., 2016). HCCs are categorized on the basis of their degree of vascular invasion—those with < 4 foci of vascular invasion are categorized as minimally invasive (HMIN), whereas those with ≥ 4 foci are categorized as widely invasive (HWIDE) (Ghossein et al, 2006). The HWIDE phenotype is aggressive, is prone to metastasize early, and accounts for the majority of deaths attributable to HCCs (Ghossein et al., 2006; Shaha et al., 1996; Grossman and Clark, 1997; Lopez-Penabad et al., 2003). Little is known about the genomic drivers of either subclass of HCC.
Metastatic HWIDE tumors are refractory to radioactive iodine and unresponsive to chemotherapeutic agents (Besic et al., 2003; Carcangiu et al., 1991; Kushchayeva et al., 2004). Clinical outcomes for these tumors are inferior to those for differentiated thyroid cancers. HCCs were not included in The Cancer Genome Atlas (TCGA) thyroid cancer study, which focused solely on papillary thyroid carcinomas (Cancer Genome Atlas Network, 2014). Here, we report a comprehensive analysis of the genomic landscape of HCCs.
RESULTS
Clinical and pathological features of the HCC cohort
Tumor samples were available from 56 patients: 32 HWIDE tumors, and 24 HMIN tumors (Figure S1A). DNA from pairs of tumor and normal tissue samples was characterized by hybrid capture, followed by whole-exome sequencing (WES). The transcriptomes of RNA samples were profiled by RNA sequencing (RNA-Seq). Clinicopathological characteristics (including age, sex, stage, and pathological subtype), treatment characteristics (surgery and use of radioactive iodine), and outcomes (locoregional recurrence, distant recurrence, and survival) are described in Figure 1A and S1A-S1C. Compared with patients with HMIN HCC, patients with HWIDE HCC had larger tumors (T3/T4) with extrathyroidal extension (p = 0.016), presented more frequently with stage III or IV disease (p = 0.014), and were more likely to have locoregional (p = 0.018) and distant (p = 0.034) metastatic recurrences.
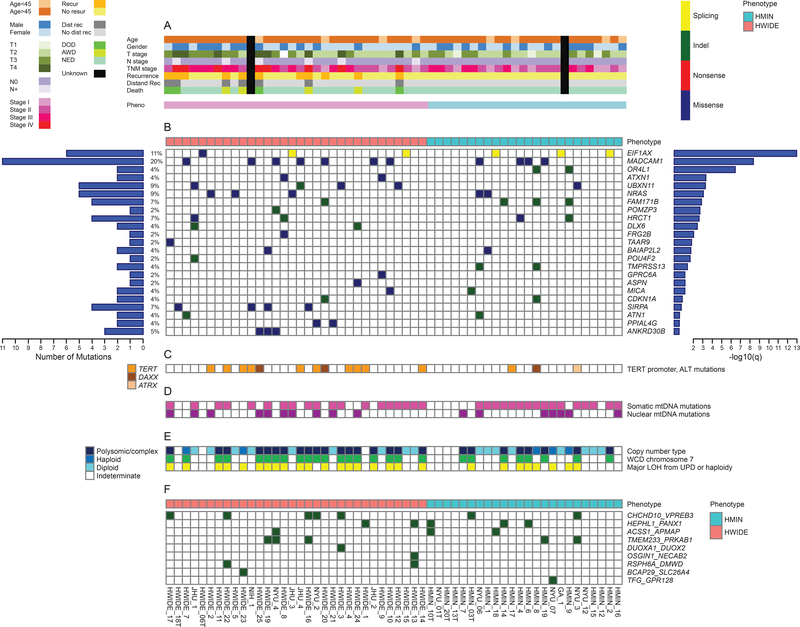
Figure 1. Somatic genomic alterations in HCCs.
(A) Summary of clinical characteristics stratified by histological phenotype. (B) Significantly mutated genes, as determined by the MutSig algorithm. Left histogram: Overall count for each gene. Right histogram: Significance level of mutations, as determined by log10 transformation of the MutSig q value. (C) TERT mutations. (D) Mitochondrial mutations. (E) Copy number alterations as determined by FACETS and FISH (F) In-frame structural variant fusion genes detected by RNA-Seq. See also Figures S1-S4 and S6 and Tables S1 and S6.
Mutational landscape of HCC
To identify somatic mutations, massively parallel sequencing was performed on tumors from 56 patients. Across all tumors, 4,293 somatic mutations were identified (2,347 in HWIDE and 1,946 in HMIN tumors) (Figure S2A). Among these mutations were 3,932 somatic single-nucleotide variants (SNVs) and 361 small insertions and deletions (indels). The SNVs included 3,621 missense, 162 nonsense, and 149 splice-site mutations. All mutations and indels are listed in Table S1. Figure 1B–1F shows a summary of the genes mutated most frequently in HCCs. The mean mutation burden per tumor for HCC was 2.6/Mb, which is comparable to that seen for ovarian cancer and glioblastoma, and far greater than that seen for other papillary thyroid cancers (0.41/Mb).
Candidate driver mutations were identified using the MutSig algorithm
(http://software.broadinstitute.org/cancer/software/genepattern/modules/docs/MutSigCV). A total of 23 genes were identified as genes with a significant frequency of mutation –log (q value) > 0.5 (Figure 1B); of these, 11 had –log (q value) > 2.0 [q < 0.01]. To validate mutation calls, a sample of 500 SNVs and indels detected by WES were subjected to validation by targeted resequencing using Ion Torrent next-generation sequencing (AmpliSeq, https://www.thermofisher.com /order/catalog/product/4475345). Ninety-nine percent of SNVs and indels were validated.
The frequency of recurrent gene mutations, together with the corresponding MutSig significance values, is shown in Figure 1B. The top 11 mutated genes, with –log (q value) > 2.0 [q < 0.01], were EIF1AX, MADCAM1, OR4L1, ATXN1, UBXN11, NRAS, FAM171B, POMZP3, HRCT1, DLX6, and FRG2B. Details of each mutation are shown in Figure S2B. The mutated genes are involved in protein translation, signaling, cytoskeletal dynamics, and other processes. For example, EIF1AX was mutated in 11% of HCC tumors. This gene encodes a protein that is a component of the translation preinitiation complex and has recently been reported to be mutated in 48% of uveal melanomas (Martin et al., 2013) and 1% of thyroid papillary carcinomas (Cancer Genome Atlas Research Network, 2014). Moreover, 11% of poorly differentiated thyroid cancers (PDTCs) and 9% of anaplastic thyroid cancers (ATCs) have EIF1AX mutations (Landa et al., 2016), which were associated with NRAS mutations in these cancers. The cooccurrence of EIF1AX and NRAS mutations was not observed in HCC. The EIF1AX mutations were located in 3 general regions of the gene (Figure S2C): in the N-terminal domain (position G15N; also observed in uveal melanomas (Castellana et al., 2017), at a unique splice acceptor site between exons 5 and 6 (p. A113 splice; observed in PDTCs and ATCs (Landa et al., 2016), and at a second splice acceptor site (p143splice) in 1 of 6 tumors. MADCAM1 (mutated in 20% of tumors [7 HWIDE and 4 HMIN]) encodes mucosal vascular addressin, a 58- to 66-kD glycoprotein adhesion receptor for lymphocytes that directs leukocytes into mucosal and inflamed tissue. Overexpression of MADCAM1 was recently found to increase AKT phosphorylation and protein translation (Wang et al, 2015). UBXN11 (mutated in 9% of tumors [4 HWIDE and 1 HMIN]) is involved in the reorganization of actin cytoskeleton mediated by RND1, RND2, and RND3. NRAS (mutated in 9% of tumors [3 HWIDE and 2 HMIN]) is the most common member of the RAS family and is mutated in all forms of thyroid cancer, as well as in other cancers (Cancer Genome Atlas Research Network, 2014; Cercek et al., 2017; Haas et al., 2017). SIRPA (mutated in 7% of tumors [4 HWIDE]) encodes a regulatory membrane glycoprotein (SIRPA) that is expressed mainly by myeloid cells, as well as by stem cells and neurons. SIRPA acts as an inhibitory receptor and interacts with the broadly expressed transmembrane protein CD47, which has also been called the “don’t eat me” signal (Willingham et al., 2012). This interaction negatively controls the effector function of innate immune cells, such as host cell phagocytosis.
The MutSig algorithm has also been used to identify potential driver mutations in over 21 different tumor types, with 247 genes identified (Lawrence et al., 2014). Of these 247 genes, 79 were identified in the mutational landscape of HCC, implicating them as potential drivers in HCC as well (Figure S3). Most HWIDE (28/32) and HMIN (23/24) tumors contained 1 or more of these mutations. These 79 identified genes included ERBB2 (11%), HLA-A (9%), NF1 (9%), ALKBH7 (7%), FAT1 (7%), NBPF1 (7%), and TP53 (7%).
The mutational profile was very different to that of other types of well differentiated thyroid cancer such as papillary and follicular thyroid cancer (Cancer Genome Atlas Research Network, 2014). There were no BRAF mutations (present in 62% of papillary thyroid cancer) and the frequency of NRAS was much lower than that of follicular thyroid cancer. Genes important in poorly differentiated and ATC (Landa et al., 2016) such as TP53, TERT promoter, PTEN, PIK3CA and ATM were identified in our HCC cohort but at a lower frequency (TP53: 7% vs 73%; TERT: 22% vs 73%; ATM: 5% vs 9%; PIK3CA: 0% vs 18%; PTEN: 4% vs 15%).
TERT promoter mutations
Altogether, 22% of HCC tumors (11/50) had TERT promoter mutations (Figure 1C): 9 C228T mutations and 2 C250T mutations. These mutations were more common in HWIDE (32% [10/31]) than HMIN (5% [1/19]) tumors (p < 0.03) (Figure S4A-S4C). In addition, 3 tumors had a DAXX mutation and 1 tumor had an ATRX mutation, which could result in alternative telomere elongation (Amorim et al., 2016).
Somatic mutations of cancer pathways in HCC
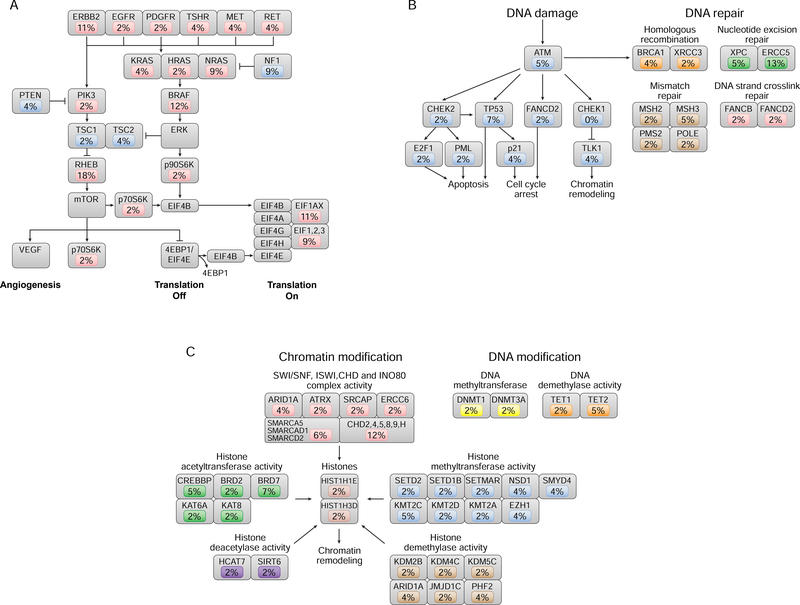
Figure 2A–2C shows mutations most commonly altered in cancer-related pathways in HCCs. The RAS/RAF/MAPK and PI3K/AKT/mTOR pathways (Samatar and Poulikakos, 2014; McCubrey et al., 2012) were found to be dysregulated in 55% of tumors (Figure 2A). As shown, at least 1 receptor tyrosine kinase was mutated in 20% of HCC tumors overall: EGFR (2%), ERBB2 (11%), PDGFR (2%), TSHR (4%), MET (4%), and RET (4%). PIK3CA mutations were found in 2% of HCC tumors and were mutually exclusive with PTEN mutations (4%). TSC1/2 mutations occurred in 6% of tumors. NF1 was deleted or mutated in 9% of tumors. Mutations in NRAS, HRAS, or KRAS occurred in 15% of tumors (NRAS [9%], HRAS [2%], and KRAS [4%]).
Figure 2. Somatic mutations of the canonical signal transduction and tumor suppressor pathways in HCC.
(A) RAS/RAF/MAPK and PI3K/AKT/mTOR pathway mutations occurred in 55% of tumors, and the incidence of mutation in each component of the pathway is indicated. Oncogenic alterations are shown in red; tumor suppressor alterations are shown in blue. All alterations were somatic mutations, with the exception of amplification events for RHEB and BRAF and deletion events for NF1. (B) DNA damage and DNA repair pathways were mutated in 38% of tumors, and the incidence of mutation in each component of the pathway is indicated. Gene mutations involved in DNA damage are shown in blue. Mutations in DNA repair pathways occurred in homologous recombination (orange), nucleotide excision repair (green), mismatch repair (brown), and DNA strand cross-link repair (red). (C) Epigenetic-modifying gene mutations occurred in 33 tumors (59%), by either chromatin modification (55%) or DNA modification (9%); the incidence of mutation in each component of the pathway is indicated. Chromatin-modifying mutations occurred in chromatin-modifying complexes, such as the SWI/SNF complex (red), histone acetyltransferases (green), methyltransferases (blue), histone deacetylases (purple), and demethylases (brown), as well as in histones themselves. DNAmodifying mutations occurred in DNA methyltransferases (yellow) and demethylases (orange).
Mutations in either p70S6K or p90S6K occurred in 4% of tumors. Mutations in EIF1AX occurred in 11% of tumors, and mutations in other EIF1, 2, or 3 genes occurred in 9% of tumors. In addition to these mutations, we also show genes which are overexpressed due to WCD of chromosome 7. These include BRAF (overexpressed in 12% tumors) and RHEB (overexpressed in 18%).
DNA damage and DNA repair pathways (Roos et al., 2016; Hustedt and Durocher, 2016) were altered in 38% of tumors (Figure 2B). In the DNA damage pathway, mutations occurred in ATM (5%), TP53 (7%), CHEK2 (2%), CDKN1A (p21) (4%), TLK1 (4%), E2F1 (2%), and PML (2%). Concurrently, 2% of tumors had RB1 mutation. In the DNA repair pathway, mutations were found in genes involved in nucleotide excision repair (XPC [5%] and ERCC5 [13%]), homologous recombination (BRCA1 [4%] and XRCC3 [2%]), mismatch repair (MSH2 [2%], MSH3 [5%], PMS2 [2%], and POLE [2%]), and DNA strand cross-link repair (FANCB [2%] and FANCD2 [2%]).
Epigenetic modification mutations (Baxter et al., 2014; Clapier and Cairns, 2009; Plass et al., 2013) were common (59% of tumors [33/56]) (Figure 2C) in genes encoding chromatin modifiers (55% [31/56 tumors]) or DNA modifiers (9% [6/56 tumors]). Mutations occurred in chromatin-modifying complexes, such as the SWI/SNF, ISWI/CHD, and INO80 family of complexes (ARID1A [4%], CHD2, 4, 5, 8, 9, and H [12%]), histone acetyltransferases (CREBBP [5%] and BRD7 [7%]), histone methyltransferases (KMT2C [5%], NSD1 [4%], and EZH1 [4%]), histone deacetylases (HCAT7 [2%] and SIRT6 [2%]), histone demethylases (PHF2 [4%] and KDM2B, 4C, and 5C [6%]), and histones (HIST1H1E [2%] and HIST1H3D [2%]). Mutations in DNA modification occurred in DNA methyltransferases (DNMT1 [2%] and DNMT3A [2%]) and DNA demethylases (TET1 [2%] and TET2 [5%]). All mutations occurred in a mutually exclusive fashion.
Mutations in the ephrin genes, important in angiogenesis (Kullander and Klein, 2002), occurred in 16% of tumors (9/56), mainly in the HWIDE subgroup (7/32 [22%]). Mutations occurred in EPHA1 (2%), EPHA2 (2%), EPHA6 (4%), EPHA7 (2%), EPHA10 (2%), EPHA8 (2%), EPHB2 (2%), and EPHB4 (2%), all in a mutually exclusive fashion.
Mitochondrial DNA (mtDNA) mutations
HCCs have a high mitochondrial load and high levels of mitochondrial dysfunction. The basis of this phenotype is unknown. Therefore, we examined the mutational landscape of the mitochondria in HCC tumors (Figure 1D). We surveyed reads aligning with the mitochondrial genome (mtDNA: mean coverage, 2160x in tumors, 240x in normal tissue) in 49 tumors which had a matching normal sample (Table S2). In total, 35 of 49 HCC tumors (71%) harbored a nonsilent mtDNA mutation (excluding rRNA mutations, Figure 3A). Interestingly, 17 of 49 tumors (24%) harbored ≥ 2 nonsilent mutations, suggesting an excess mtDNA mutation load in HCC tumors. Unlike in other cancer types with mtDNA mutations (e.g. chromophobe renal cell carcinomas), in HCCs we found no association between increased mtDNA copy number and mtDNA mutations.
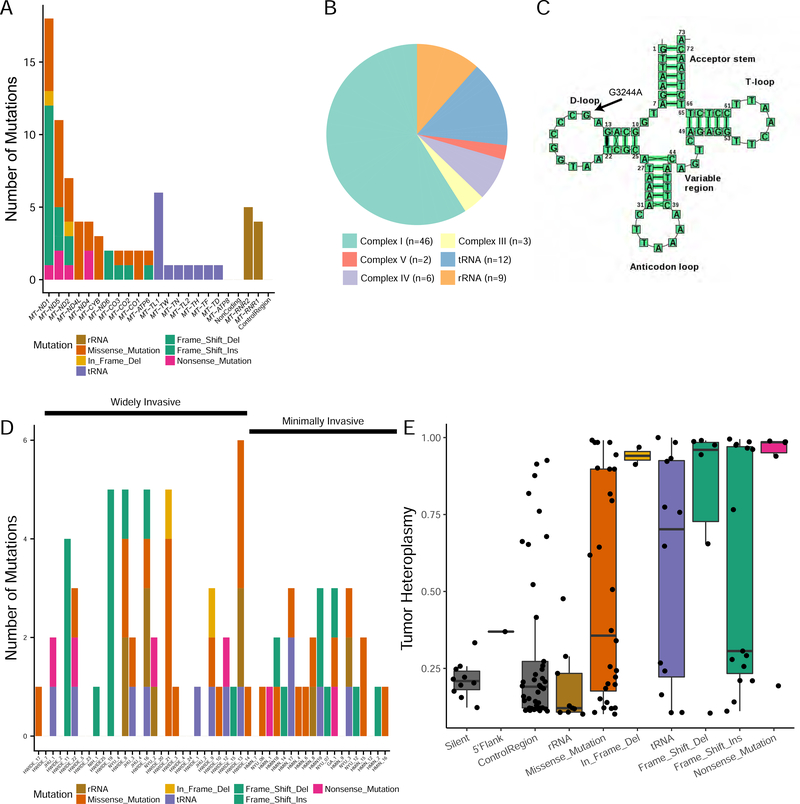
Figure 3. Mitochondrial mutations in HCC tumors.
(A) Types and frequency of mutations observed in mtDNA. B) Mitochondrial mutations categorized by the genes encoding the mitochondrial complexes the mutations alter. C) Location of the recurrent leucine tRNA MTTL1 (G3244A) mutation. D) Type and frequency of mitochondrial mutations categorized for each tumor sample. E) Tumor heteroplasmy categorized by the type of mitochondrial mutation. Boxplots showing the degree of heteroplasmy are shown for each type of mitochondrial mutation. For each boxplot, the horizontal line inside the box is the median and the upper and lower horizontal lines of the box represent the interquartile range (25th-75th percentile). The line above the box shows tumors from the 75th to 100th percentile and the line below shows tumors from 0–25th percentile. See also Table S2.
The mitochondrial genome is composed of 13 protein-coding genes that encode 4 of the 5 complexes of the mitochondrial respiratory chain. Of these 13 genes, the 6 that encode complex I subunits were enriched for mutations (46/69 nonsilent mtDNA mutations were complex I [67%]) (Figure 3B). Nevertheless, we observed at least 1 mutation in each mtDNA protein-coding gene, with the exception of ND6 and ATP8. We also found 12 somatic mutations to mitochondrial tRNAs, 1 of which (G12183A, rs121434473) has been reported to cause pigmentary retinopathy and sensorineural deafness in a male patient (Crimi et al., 2003). Of the remaining tRNA mutations, 6 occurred at the same position in the leucine tRNA MT-TL1 (G3244A) (Figure 3C). Samples from patients with MELAS syndrome (a progressive neurodegenerative disorder caused by mitochondrial dysfunction) with the G3244A mutation have been found to be deficient in a taurine-containing modification at the anticodon wobble position (Kirino et al., 2005). The type and frequency of mitochondrial mutations categorized for each tumor sample are shown in Figure 3D. Most interestingly, 18 of 49 HCC tumors (37%) harbored a frameshift or nonsense mutation to mtDNA, suggesting potent inactivation of mitochondrial respiration in these samples. A number of these mutations were nearly homoplastic, suggesting that an uncharacterized positive selection is driving the accumulation and fixation of loss-of-function mtDNA variants (Figure 3E).
We further examined whether HCC tumors harbor mutations in nuclear DNA–encoded genes that are essential for mtDNA maintenance. Using a manually curated list of such genes, we identified several mutated genes (e.g. POLG, NDUFV3, DNA2, and SPG7) that may potentially affect mtDNA maintenance (Table S2). These mutations occurred in 17 tumors, 11 of which also had a somatic mtDNA mutation. Overall, we found that mutations in nuclear DNA–encoded genes were insufficient to explain mtDNA dysfunction in mtDNA wild-type HCC samples. We observed no association between mitochondrial mutations and tumor aggressiveness,
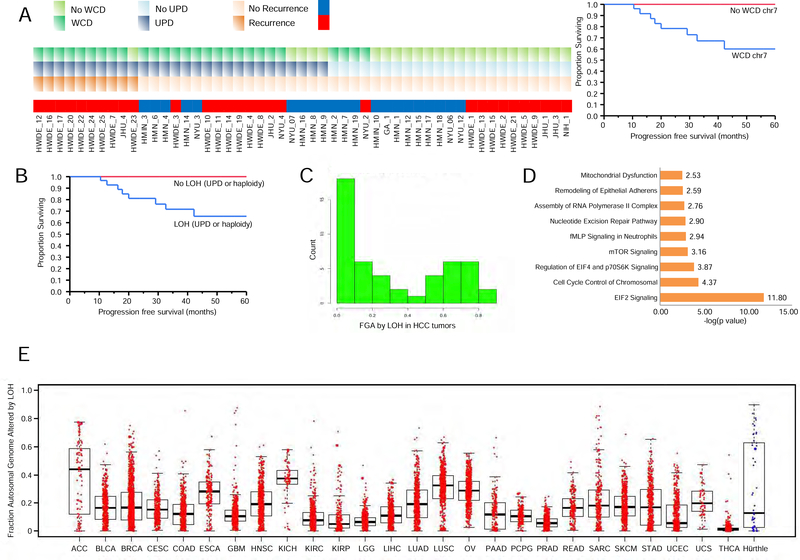
Whole chromosome duplication (WCD) and loss of heterozygosity (LOH) by haploidy or uniparental disomy (UPD) in HCC
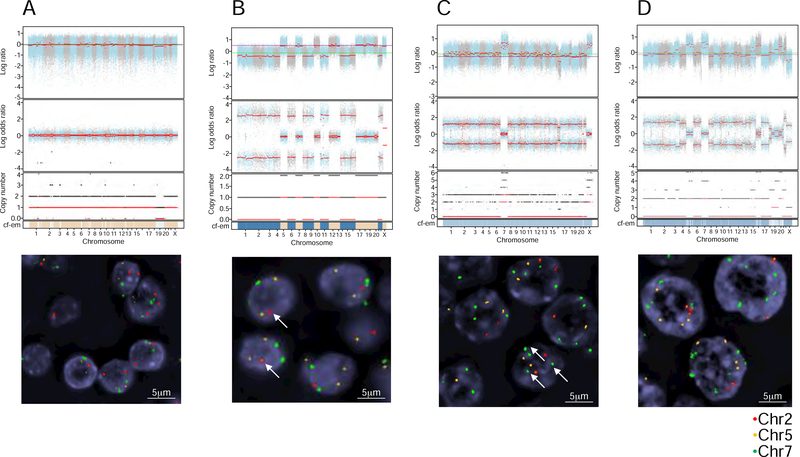
We examined the copy number landscape of HCCs using a combination of computational fraction and allele-specific copy number estimates from tumor sequencing (FACETS) and fluorescence in situ hybridization (FISH) analysis (Figure 1E). A summary of the FISH analysis and the combined copy number analysis using FACETS and FISH is shown in Table S3. Several striking features were apparent from this analysis. The majority of the HMIN tumors were diploid, with a small number of near-haploid and polysomic tumors. In contrast, the majority of HWIDE tumors were polysomic. In the polysomic tumors, there was universal duplication of chromosome 7, with 4 or more copies, as well as occasional duplication of chromosomes 5 and 12. The remaining chromosomes had UPD, suggesting that loss of a whole chromosome was subsequently followed by duplication of the remaining chromosome, yielding a diploid configuration. HMIN 18 is an example of a tumor with a predominant diploid cell type (Figure 4A). HMIN 9 has a haploid phenotype, with 1 copy of chromosome 2 but 2 copies of chromosomes 5 and 7 (Figure 4B). HWIDE 17 and HWIDE 16 are examples of polysomic tumors. HWIDE 17 has extensive UPD involving virtually all chromosomes, with WCD of chromosome 7 (Figure 4C). Using FISH probes to chromosomes 2, 5, and 7, we found that each tumor cell had 2 copies of chromosomes 2 and 5 but multiple copies of chromosome 7. HWIDE 16, again, has extensive UPD involving multiple chromosomes, with WCD of chromosomes 5, 7, and 16 (Figure 4D). Validation with FISH found that chromosome 2 was diploid but chromosomes 5 and 7 had multiple copies.
Figure 4. Example of cytogenic subtypes, characterized using genomic copy number analysis and FISH.
(A–C) FACETS plots show the GC-corrected normalized log ratio of tumor to normal read depths at a set of single-nucleotide polymorphism (SNP) loci; the log odds ratio was determined from cross-tabulating the tumor and normal reads into alleles for loci that are heterozygous in the germline; the total (black) and minor (red) integer copy number assignment for the segments is indicated; the final band shows the cellular fractions, where dark blue represents 1, lighter shades represent lower numbers, and beige represents no copy number change. FISH validation was performed for chromosomes 2, 5, and 7. These analyses are provided for 4 representative patients: HMIN 18 (A), HMIN 9 (B), HWIDE 17 (C), and HWIDE 16 (D). The arrows in HMIN 9(B) show 1 copy of chromosome 2 (haploid phenotype). The arrows in HWIDE 17(C) show multiple copies of chromosome 7 (WCD chromosome 7) See also Figure S5 and Table S3 and S5
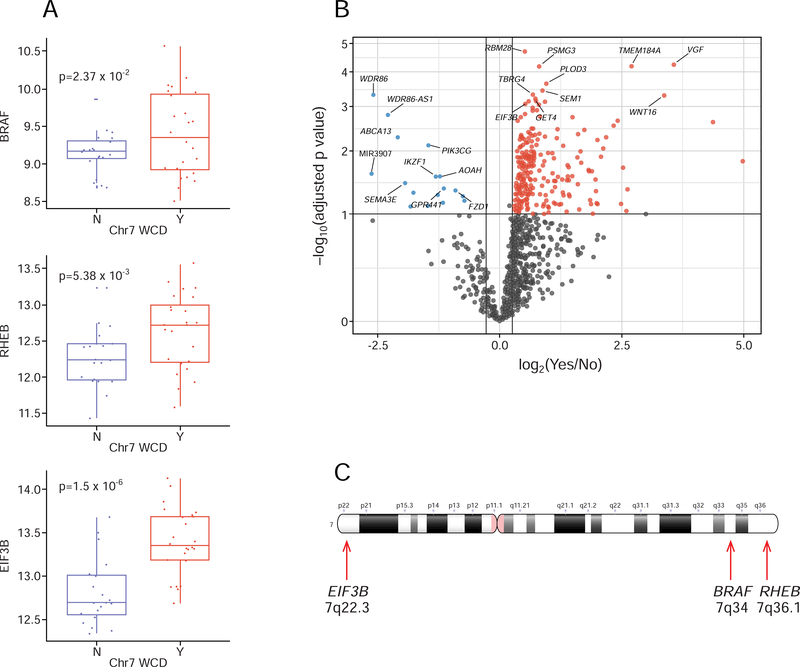
Of the 49 tumors with matched normal genomic DNA, 27 had WCD of chromosome 7 (19 HWIDE and 8 HMIN) and 6 had near-haploidy (2 HWIDE and 4 HMIN) (Table S3). There was a statistically significant association between WCD of chromosome 7 and tumor recurrence (9/10 patients with recurrence had WCD of chromosome 7; p = 0.002). Patients with WCD of chromosome 7 were also more likely to experience progression (5-year progression-free survival, 60% vs 100%; p = 0.02 Log-rank test) (Figure 5A). Polysomic tumors had UPD of the remaining chromosomes, resulting in widespread LOH. Tumors that were near-haploid but that had 2 copies of chromosome 7 also had widespread LOH. Kaplan-Meier analysis showed that tumors with widespread LOH were associated with significantly worse outcomes (5-year progression-free survival, 66% vs. 100%; p = 0.02 Log-rank test) (Figure 5B). Figure 5C quantifies HCC tumors by the fraction of the genome altered by LOH. HCCs appear to fall into a bimodal distribution, with a subset of tumors with widespread LOH due to UPD or haploidy. In total, 28% of tumors had > 0.6 of the genome altered by LOH, and 16% of tumors had >0.7 of the genome altered by LOH. Pathway analysis, which differentiated tumors with widespread LOH from those with lower levels of LOH, showed that EIF2 signaling, cell-cycle control of chromosomal replication, EIF4 and p70S6K signaling, and mTOR signaling are enriched in tumors with major LOH (Table S4 and Figure 5D). Genes identified in cell-cycle control of chromosomal replication included PCNA, RPA3, POLA1, CDK5, CDK6, CDC7, ORC5, and DBF4. A pan-cancer analysis of LOH using data from TCGA (Figure 5E) showed that, other than HCC, only glioblastoma multiforme (GBM) and sarcoma (SARC) had > 0.6 of the genome altered by LOH, highlighting the significance of the high levels of LOH observed in HCCs. The tumors with WCD of chromosomes 5, 7, and 12 contained several genes involved in the RAS/RAF/MAPK pathway, as well as the PI3K/AKT/mTOR pathway: chromosome 5, RICTOR and GOLPH3; chromosome 7, EGFR/MET/CDK6/BRAF/RHEB; and chromosome 12, KRAS, CDK4, and MDM2. Tumors with WCD of chromosome 7 had a statistically significant overexpression of several genes involved in mTOR signaling and protein translation (Table S4), including BRAF (p = 0.0237), RHEB (p = 0.005) and EIB3B (p = 1.5 × 10−6) (Figure 6A). Other genes overexpressed on chromosome 7 are shown in the top right panel of the volcano plot in Figure 6B. The chromosome location of these 3 genes is shown in Figure 6C. Overexpression was not observed in all tumors with chromosome duplication. The relationship between overexpression and copy number gain has been well-documented. Amplification of BRAF, rather than activating mutations, has been reported as a potential mechanism of activation (Ciampi et al., 2005). Amplification of BRAF has also recently been identified as a potential mechanism of resistance of BRAF-mutant melanoma cell lines to vemurafenib (Shi et al., 2012). Of the 20 tumors with WCD of chromosome 5, 9 had mRNA overexpression of RICTOR (45%), and 5 had mRNA overexpression of GOLPH3 (25%) which could also potentiate mTOR signaling. Of the 11 tumors with WCD of chromosome 12, 3 had mRNA overexpression of MDM2 (27%).
Figure 5. WCD and major LOH from UPD or haploidy in HCC tumors.
A) The correlation between WCD, major LOH, and recurrence is shown for each tumor (left). Kaplan-Meier plot showing progression-free survival in patients based on WCD of chromosome 7 (right). B) Kaplan-Meier plot showing progression-free survival in patients based on LOH from UPD and haploidy. C) Number of tumors with the indicated fraction of the genome altered by LOH from UPD or haploidy in HCC tumors is shown. D) Results of Ingenuity pathway analysis for tumors enriched with UPD. Pathways are shown along the y-axis and statistical significance (-log10 p value) along the x-axis. E) A pan-cancer analysis of LOH comparing HCC (blue) to data from TCGA for other cancer types (red). Fraction of the genome altered (FGA) by LOH is shown on the y-axis. Tumor types are shown on the x-axis. Boxplots show the fraction of genome altered for each tumor type. For each boxplot, the horizontal line inside the box is the median and the upper and lower horizontal lines of the box represent the interquartile range (25th–75th percentile). The line above the box shows tumors from the 75th to 100th percentile and the line below shows tumors from 0–25th percentile. See also Table S4.
Figure 6. Correlation of WCD of chromosome 7 with gene overexpression.
A) Boxplots showing the distribution of expression of 3 genes involved in mTOR signaling and translation in tumors with (Y) and without (N) chromosome 7 amplification. For each boxplot, the horizontal line inside the box is the median and the upper and lower horizontal lines of the box represent the interquartile range (25th–75th percentile). The line above the box shows tumors from the 75th to 100th percentile and the line below shows tumors from 0–25th percentile. B) Volcano plot showing differentially expressed genes in tumors with chromosome 7 amplification. Magnitude fold changes are shown on the x-axis and statistical significance (log10 of p value) on the y-axis. Genes with a fold change less than 2 (log2 = 1) are shown in grey. Genes in the top right colored red are overexpressed with large statistically significant fold changes. Genes in the top left colored blue are underexpressed with large statistically significant fold changes. C) Ideogram of chromosome 7 showing the location BRAF, RHEB and EIF3B genes.
Interestingly, mutations in the RAS family of genes (NRAS, HRAS, KRAS), EIF1AX, TSC1 and RET were unique to tumors which had a diploid phenotype. Of the 17 tumors which were diploid, 13 had either a mutation in a RAS family member, EIF1AX, TSC1 or RET suggesting these were driving the RAS/PIK3/AKT/mTOR pathway in diploid tumors whereas this pathway was driven by the chromosomal alterations in the polysomic group of tumors.
As well as arm level changes involving WCD and UPD, we also identified focal copy number alterations. Figure S5A shows the significant focal copy number changes as quantified by GISTIC analysis. These focal events appeared to be somewhat more common in HWIDE tumors (Figures S5B and S5C). Table S5 shows the significant focal amplifications and deletions with their associated genes detected.
Recurrent structural variants
We identified 200 candidate somatic rearrangements, including 52 unique rearrangements (Figure S6A and Table S6). The number of events per sample ranged from 0 to 11 (median = 1). Eighteen samples had no rearrangements detected. Of the 52 unique rearrangements, 43 were noncoding, and 9 were in-frame coding rearrangements. The 9 in-frame coding rearrangements were all intrachromosomal events, including 5 that were recurrent (Figures 1F and S6A). The inframe coding rearrangements were as follows: CHCHD10_VPREB3 (n = 7, chromosome 22), HEPHL1_PANX1 (n = 5, chromosome 11), TMEM233_PRKAB1 (n = 5, chromosome 12), ACSS1_APMAP (n = 3, chromosome 20), RSPH6A_DMWD (n = 2, chromosome 19), DUOXA1_DUOX2 (n = 1, chromosome 15), OSGIN1_NECAB2 (n = 1, chromosome 16), BCAP29_SLC26A4 (n = 1, chromosome 7), and TFG_GPR128 (n = 1, chromosome 3). We validated 4 rearrangements by PCR in cDNAs from the index tumors. Detailed schematic figures showing the fusion points in the 4 validated structural variants, with Sanger sequencing validation, are shown in Figure S6B.
The TMEM233_PRKAB1 rearrangement is a fusion between exon 1 of TMEM233 and exon 2 of PRKAB1. TMEM233 is a transmembrane protein of unknown function. PRKAB1 is the regulatory subunit of the AMP-activated protein kinase AMPK, which is an important energy-sensing enzyme that monitors cellular energy status. The fusion gene was found in 5 tumors (3 HWIDE and 2 HMIN). Transfection of TMEM233_PRKAB1 cDNA into Nthy-ori cells increased their proliferation, compared with empty vector control, suggesting that this fusion may result in a gain of function (Figure S6C).
Molecular pathways in HCCs defined by transcriptome analysis
Supervised clustering of mRNA expression profiles showed marked differences in expression between HWIDE and HMIN tumors (Figure S7A). Ingenuity pathway analysis showed that these differences were attributable to alterations in tRNA splicing, cAMP-mediated signaling, Gprotein receptor signaling, angiopoietin signaling, and p53 signaling, among other processes (Table S7).
Supervised clustering between HWIDE and normal thyroid tissue, as well as HMIN and normal thyroid tissue, is shown in Figures S7B and S7C. Ingenuity pathway analysis showed that these differences were dominated by oxidative phosphorylation, mitochondrial dysfunction, and tricarboxylic acid cycle (Table S7), which are consistent with the abundance of dysfunctional mitochondria that is a hallmark of HCCs.
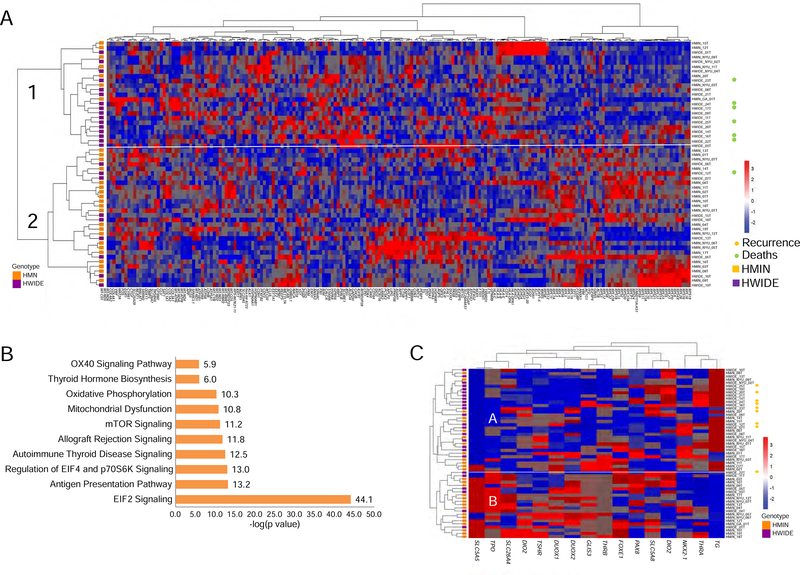
Unsupervised clustering of the 200 most variant genes revealed 2 main groups (Figure 7A). Most tumors in group 1 (16/22) were HWIDE tumors. The majority of recurrences and deaths occurred in group 1. The principal pathway alterations that differentiated the 2 groups were EIF2 signaling, EIF4 and p70S6K signaling, mTOR signaling, and alterations in mitochondrial dysfunction and oxidative phosphorylation (Figure 7B).
Figure 7. Transcriptome profiles of HCC tumors.
A) Unsupervised clustering of the 200 most variant genes. B) Pathway analysis of the most significant pathways altered between the clusters in panel A. C) Unsupervised clustering using a 16-gene thyroid differentiation score shows 2 main groups A and B. Group A is over represented by widely invasive HCC with greater loss of thyroid differentiation. All patients with recurrence, shown by orange sympos, are in group A. (SLC5A5, TPO, SLC26A4, DIO2, TSHR, DUOX1, DUOX2, GLIS3, THRB, FOXE1, PAX8, SLC5A8, DIO1, NKX-2, THRA, and TG). See also Figure S7 and Table S7.
Unsupervised clustering using a 16-gene thyroid differentiation score (Cancer Genome Atlas Research Network, 2014) (16 genes important in thyroid differentiation: SLC5A5, TPO, SLC26A4, DIO2, TSHR, DUOX1, DUOX2, GLIS3, THRB, FOXE1, PAX8, SLC5A8, DIO1, NKX2, THRA, and TG) identified 2 main groups (Figure 7C). Slightly more than half of the tumors in group A (19/32) were HWIDE tumors, with greater loss of thyroid differentiation. All patients who experienced recurrence were in group A. This indicates that although the more aggressive widely invasive Hürthle cancer phenotype lacked the histological features of poorly differentiated thyroid cancer (i.e. presence of mitosis and necrosis), these cancers did have loss of thyroid differentiation as compared to the minimally invasive phenotype.
Integration of genome and transcriptome data
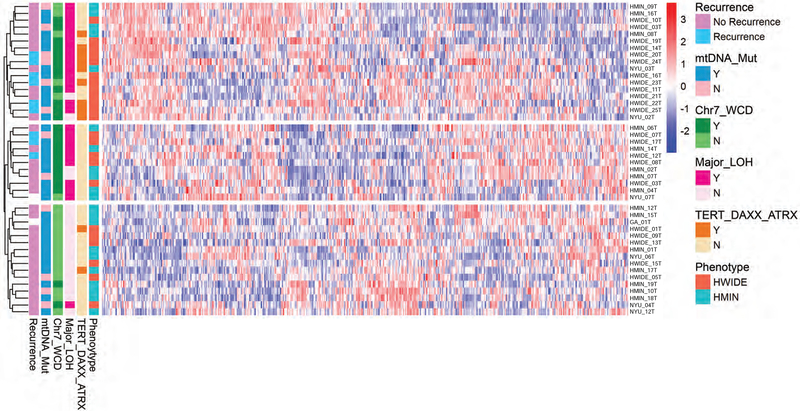
We carried out unsupervised clustering of the 500 most differentially expressed genes, and integrated these data with the major genomic findings of WCD, LOH, TERT mutation status and mitochondrial mutation status (Figure 8). The unsupervised clustering indicated there are, at least, 3 subtypes of HCCs. The first (top) group is enriched with HWIDE; these tumors are enriched with TERT alterations, major LOH, and chromosome 7 WCD HCCs. The second (middle) group is enriched with non-TERT alterations, major LOH, and chromosome 7 WCD HCCs. The third (bottom) group is enriched with HMIN and non-recurrent HCCs; these tumors lack TERT alterations, major LOH, and chromosome 7 WCD.
Figure 8. Heatmap showing integration of genomic and transcriptomic data.
The unsupervised clustering of the 500 most differentially expressed genes integrated with the major genomic findings of WCD, LOH, TERT mutation status and mitochondrial mutation status in HCC tumors.
DISCUSSION
The molecular foundations of HCCs are poorly defined (DeLellis et al., 2017; Ganly et al., 2013; Jun et al., 2012; Li and Durbin, 2010) and were not studied by the TCGA project. The present study is an integrated genomic analysis of HCCs, and with the availability of detailed clinical information, including treatment and survival outcomes, for nearly the entire cohort, this rich data set has enabled the identification of genomics-based biomarkers.
Perhaps most strikingly, we found that a large fraction of HCCs harbor extensive polysomy, characterized by WCD of chromosomes 5 and 7 and extensive UPD of the remaining chromosomes (Lapunzina and Monk, 2011; Makishima and Maciejewski, 2011; Tuna et al., 2009). These chromosomal alterations were not identified in other aggressive forms of thyroid cancer, such as poorly differentiated or ATC (Landa et al., 2016). Although UPD has been observed in several cancers, all of these previous cases involved only single chromosomes (Torabi et al., 2015; Tuna et al., 2015; Walsch et al., 2008). The extent of UPD seen in some HCC tumors is among the highest observed in human cancers. In some cases, almost the entire genome is affected by UPD. This high level of UPD results in widespread LOH, potentially promoting inactivation of many tumor suppressor genes. Furthermore, WCD is common and occurs in a nonrandom fashion involving specific chromosomes, particularly chromosomes 5 and 7, which indicates that these chromosomes consistently avoid LOH. This could result in a coordinated increase in expression of several genes that may provide clonal advantage (Boot et al., 2016). This appears to be particularly important for several genes in the RAS/RAF/MAPK pathway, as well as the PI3K/AKT/mTOR pathway. Importantly, polysomic tumors with WCD and UPD mark cancers that are aggressive, more likely to be widely invasive, less differentiated, and associated with a greater risk of thyroid cancer–related recurrence and death. In addition to polysomy, we identified haploidy in some of the tumors. Haploidization would also result in generalized LOH. Haploid tumors consistently retained 2 copies of chromosome 7 but had single copies of the remaining chromosomes. It has been postulated that HCCs first undergo haploidization, with retention of chromosome 7, during cell division and then undergo endoduplication, resulting in UPD and WCD of chromosome 7 (Corver et al., 2012; Corver et al., 2014). Interestingly, tumors with high levels of LOH appear to be present in a number of different cancer types, most notably glioma and sarcoma. There is a clear bimodal distribution of LOH in HCCs: HCCs have either high (due to UPD or haploidization) or low levels of LOH. It is likely that these high-LOH tumors represent a subset of human tumors with common operative mechanisms generating genetic instability. High-LOH tumors have hyperactivity of cyclindependent kinase–related signaling, and we hypothesize that agents that inhibit cyclin-dependent kinases may have efficacy against these tumors.
The mutational landscape reveals several important features of HCCs. The RTK/RAS/RAF/MAPK and PI3K/mTOR/AKT pathways are among the most frequently altered pathways in human cancers (Samatar and Poulikakos, 2014; McCubrey et al., 2012). Mutations and transcriptional alterations converge to activate these pathways in HCCs. This may provide a rationale to use agents targeting these pathways, such as mTOR inhibitors, in patients with HCC. Other genes that are affected in HCCs include those involved in DNA repair and DNA damage response pathways (Roos et al., 2016; Hustedt and Durocher, 2016), as well as a very large number of epigenetic modification–related factors involving chromatin modification and DNA modification (Baxter et al., 2014; Clapier and Cairns, 2009; Plass et al, 2013). Many of these genes are widely mutated in different human malignancies (Cancer Genome Atlas Research Network, 2014; Cancer Genome Atlas Network, 2012a; Cancer Genome Atlas Research Network, 2012b; Cancer Genome Atlas Research Network, 2012c; Cancer Genome Atlas Research Network, 2011; Cancer Genome Atlas Research Network, 2008), highlighting the central role that these pathways play in the oncogenesis of many human cancer types, including HCCs. Mutations in the TERT promoter gene, as well as mutations in other genes that cause alternate telomere elongation, such as DAXX and ATRX (Amorim et al., 2016), are common in more-aggressive forms of thyroid cancer, such as PDTC (40%) and ATC (70%) (Lott et al., 2013). These mutations also appear to be important in the more-aggressive, HWIDE form of HCC (32%).
Interestingly, HCCs were at first thought to be a form of follicular thyroid carcinoma. Our findings suggest that this is not the case. Instead, it would appear that HCCs are a distinct class of thyroid cancer. Although some mutations found in HCCs are also observed in other forms of thyroid cancer, the mutational landscape of HCC is on the whole distinct from that of both papillary and follicular thyroid carcinoma (Cancer Genome Atlas Research Network, 2014). In particular, the extensive UPD observed in HCCs is not observed in papillary and follicular thyroid cancer (Cancer Genome Atlas Research Network, 2008; Caria and Vanni, 2010; Vu-Phan and Koenig, 2014). A number of genes that are frequently mutated in HCCs are rarely mutated in other thyroid cancers, such as MADCAM1, UBXN11, SIRPA, XPC, and ERCC5.
HCCs are characterized by a high rate of mitochondrial mutations, which is interesting given the high frequency of mitochondrial dysfunction in these tumors. HCC mitochondrial mutations frequently affect complex I subunits. Interestingly, of 12 somatic mutations in mitochondrial tRNAs, 6 occurred at the same position in the leucine tRNA MT-TL1 (G3244A). One could postulate that mutations at this position indirectly affect leucine metabolism, which could affect leucine/glutamine transport and indirectly affect bioenergetics. Importantly, many of the mtDNA mutations were homoplastic—again illustrating the importance of these mutations in HCCs. The extent of mitochondrial dysfunction observed in HCCs is not seen in any other type of thyroid cancer. We analyzed TCGA data on both the classical and follicular variants of papillary thyroid carcinoma (PTC). Nonsynonymous and loss of function mtDNA mutations were identified in 26.8% and 14.8% of classical variant PTC, respectively, and 25.4% and 12.7% of follicular variant PTC, respectively. We also analyzed IMPACT data from Memorial Sloan Kettering Cancer Center (MSKCC) on more aggressive forms of thyroid cancer. Nonsynonymous and loss of function mtDNA mutations were identified in 4% and 0% of medullary thyroid cancers, 10.8% and 2.1% of PDTC, and 20.4% and 9% of ATC. Compared with the high rates that we identified in HCC (71.4% and 36.7%), our data illustrates the unique role that mitochondrial mutations may play in HCCs. We observed no association between mitochondrial mutations and tumor aggressiveness, suggesting that mitochondrial mutations are involved in the establishment, rather than the progression, of HCCs. This could be by favoring a bioenergetic shift from oxidative phosphorylation to aerobic glycolysis as a main source of ATP supply (Warburg effect) (Warburg, 1956). Indeed, HCC cells have high glucose uptake and excessive lactate production but low levels of respiration activity. They are extremely hypermetabolic on FDGPET imaging, which is consistent with a dependence on high levels of glucose uptake. In addition, HCCs have upregulation of reactive oxygen species formation—creating oxidative stress and activating signaling pathways that favor oncogenic transformation and malignant progression, as well as the development of antitumor drug and radiation resistance. Alterations in mtDNA have also been shown to initiate epigenetic modifications in the nuclear genome that are probably involved in tumor formation (Xie et al., 2007) and that may account for the excessive epigenetic alterations we observed in our study. In addition to induction of epigenetic alterations, mtDNA mutations can result in genetic changes in the nuclear genome (Ma et al., 2010). However, we have not been able to assess the frequency of mitochondrial DNA mutations in benign thyroid conditions with oncocytic change such as in Hashimotos thyroiditis and in more indolent forms of thyroid cancer such as oncocytic follicular variant of PTC. Therefore, our data does not definitively show that mitochondrial mutations contribute to HCC, only that they occur at a high frequency. Future work is needed to determine the frequency and type of mitochondrial mutations in these more benign conditions.
Our findings indicate that the regulation of translation plays an important role in the pathogenesis of HCC. The high incidence of EIF1AX mutations, at a frequency similar to that seen in PDTC and ATC, illustrates the importance of this particular gene. We identified other mutations in EIF genes which initiate translation. Mutations in several genes in the mTOR signaling pathway together with chromosomal duplication of chromosome 7 and 5 with corresponding mRNA overexpression of genes driving mTOR signaling, such as BRAF, RHEB and EIF3B also drive translation. Eukaryotic initiation factor 3 (EIF3) complex is essential for initiation of protein synthesis, and is closely associated with mTOR and S6K binding (Holz et al., 2005). It consists of 13 subunits (EIF3A to M), among which EIF3B serves as a major scaffolding subunit. EIF3B is elevated in several cancers, including human bladder cancer and prostate cancer (Wang et al., 2013), esophageal cancer (Xu et al., 2016), and glioblastoma (Liang et al., 2012). Elevated levels of EIF3B are associated with tumor depth, lymph node metastasis, and advanced tumor, node, metastasis stage (Wang et al., 2013). The differential gene expression profile also highlighted that EIF2, EIF4, and mTOR signaling pathways are especially important in the more-aggressive forms of HCCs.
Our analysis also revealed that in-frame gene fusions occur in HCC tumors. These include TMEM233_PRKAB1 and CHCHD10_VPREB3. A number of these fusions are recurrent and may contribute to tumor initiation or progression. The spectrum of fusions is also quite different from those that appear in other forms of thyroid cancer, again emphasizing the distinctiveness of HCCs (Cancer Genome Atlas Research Network, 2008; Caria and Vanni, 2010).
Overall, our study has revealed several insights into the molecular foundations of HCC. Through our analysis, we have identified the importance of the RTK/RAS/RAF/MAPK and PIK3/AKT/mTOR pathways in this disease. We have initiated a phase II randomized clinical trial of the multiple tyrosine kinase inhibitor sorafenib and the mTOR inhibitor everolimus and shown a significant response rate for these agents (Sherman et al., 2015). Our study has also identified the importance of WCD and UPD in the pathogenesis of this cancer, as well as the association between high levels of UPD and poor outcomes. Moreover, we have identified frequent mitochondrial mutations that may underlie the mitochondrial dysfunction that is a hallmark of this malignancy. We believe that our findings can help form a foundation for the development of diagnostic and treatment modalities for HCCs.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Timothy A. Chan (chant@mskcc.org).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Tumor samples
Tumor and matched normal (peripheral blood or nonneoplastic normal tissue) specimens were obtained from HCC patients treated at multiple institutions (Memorial Sloan Kettering Cancer Center, New York University, National Institutes of Health, Johns Hopkins Medical Center, and University of Georgia) after informed consent and Institutional Review Board approval was received from each participating institution. The demographic and clinical characteristics of the patients are shown in Figure S1. All tissue samples were snap-frozen in liquid nitrogen at the time of surgery and stored at −80°C. Hematoxylin and eosin–stained tumor sections were reevaluated by a head and neck pathologist (R.G.), confirming the diagnosis of HCC and the classification of either HMIN or HWIDE. We detail our exact definition of minimally and widely invasive as follows: Minimally invasive HCC (HMIN) was defined as encapsulated tumor harboring < 4 foci of vascular invasion (foci of vascular invasion that were closely adjacent to one another were counted as separate foci) and lacking both gross invasion as well as vascular invasion of extrathyroid vessels). Widely invasive HCC (HWIDE) was defined as a tumor with gross invasion/significant vascular invasion if the tumor was grossly invasive, had extrathyroid vascular invasion and/or was encapsulated with 4 or more foci of vascular invasion. The terms HMIN and HWIDE are abbreviations specific to our study to define “minimally invasive HCC” and “widely invasive HCC”.
Cell lines
The immortalized thyroid cell line NThy-Ori (Sigma-Aldrich; cat 90011609) was used for creation of the stable transfectant cell line with TMEM233_PRKAB1 fusion gene.
METHODS DETAILS
DNA and RNA extraction
DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen) and quantified with the PicoGreen assay (Thermo Fisher Scientific). RNA was extracted using the RNeasy Mini kit (Qiagen) and quantified with the RiboGreen assay (Thermo Fisher Scientific). DNA and RNA quality and integrity were characterized using a Bioanalyzer (Agilent Technologies) and Fragment Analyzer (Advanced Analytics). Samples were then prepared for WES of tumor and normal DNA, as well as whole-transcriptome sequencing.
WES libraries
To identify somatic mutations, WES of tumor and matched normal genomic DNA were performed for 49 tumors. For the remaining 7 tumors, from which no paired normal DNA was available, mutations were determined using haplotype caller from RNA-Seq analysis. WES libraries were prepared using the SureSelect XT library preparation kit (Agilent). DNA was sheared using a LE220 Focused-ultrasonicator (Covaris), and the fragments were end-repaired, adenylated, ligated to Illumina sequencing adapters, and amplified by PCR. Exome capture was performed using the SureSelect XT v4 51Mb capture probe set (Agilent), and captured exome libraries were enriched by PCR. Final libraries were quantified using the KAPA library quantification kit (KAPA Biosystems), Qubit fluorometer (Life Technologies), and 2100 Bioanalyzer (Agilent) and were sequenced on a HiSeq 2500 sequencer (Illumina) using 2× 125bp cycles with a depth of coverage of > 100x.
RNA-seq libraries
RNA-seq libraries were prepared using the KAPA Stranded RNA-Seq with RiboErase sample preparation kit (KAPA Biosystems). Total RNA (100 ng) was ribo-depleted and fragmented, followed by first and second strand synthesis, A tailing, adapter ligation, and PCR (using 11 cycles). Final libraries were quantified using the KAPA library quantification kit, Qubit fluorometer, and 2100 Bioanalyzer, and were sequenced on a HiSeq 2500 v4 chemistry sequencer using 2× 125-bp cycles with a depth of coverage of > 100x.
Mutation analysis
Matches between tumor and normal samples for each patient were confirmed with fingerprinting analysis using an in-house panel of 118 SNPs and with VerifyBamID (Jun et al., 2012) (Figure S8). Raw-sequencing data were aligned with the hg19 genome build using the Burrows-Wheeler Aligner (version 0.7.10) (Li and Durbin, 2010). Indel realignment, base quality score recalibration, and removal of duplicate reads were performed using the Genome Analysis Toolkit (version 3.2.2, broadinstitute.org/gatk), following the guidelines for raw read alignment (DePristo et al., 2011). SNVs were independently detected by 4 callers: MuTect (broadinstitute.org/cancer/cga/mutect), SomaticSniper (version 1.0.4.2, gmt.genome.wustl.edu/packages/somatic-sniper), Strelka (version 1, sites. google.com/site/strelkasomaticvariantcaller), and VarScan (version 2.3.8, varscan.sourceforge.net).
SNVs that were identified by at least 2 different callers, with > 10% variant allelic fraction and > 7x coverage in tumor and ≥ 15x coverage with > 97% normal allelic fraction in normal tissue, were considered high-confidence variants. SNVs that did not meet these criteria but that had ≥ 4x coverage, > 6% variant allelic fraction in tumor, and ≥ 4x normal coverage with > 97% normal allelic fraction in normal tissue were considered low-confidence variants. Each low-confidence variant was manually reviewed via Integrative Genomics Viewer (IGV; version 2.3, broadinstitute.org/igv) and selected individually if thought to be real. Insertions and deletions (indels) were detected by Strelka and VarScan. Variants that passed manual review in IGV, with ≥ 4x tumor allelic coverage, > 10% tumor allelic fraction, ≥ 4x normal DNA coverage, and > 97% normal allelic fraction, were considered to be potential indels.
Orthogonal validation of mutations
A random selection of 500 high-confidence SNVs were subjected to orthogonal validation using alternative Ion Torrent next-generation sequencing Ion AmpliSeq technology (Thermo Fisher Scientific). The sequencing and variant calling and annotation were performed by the Integrated Genomics Core at MSKCC, in accordance with the Ion AmpliSeq workflow (https://www.thermofisher.com/us/en/home/life-science/sequencing/next-generationsequencing/ion-torrent-next-generation-sequencing-workflow/ion-torrent-next-generationsequencing-select-targets/ampliseq-target-selection.html). The selected 500 SNVs demonstrated an excellent validation rate with resulting AmpliSeq calls, with 496 of 500 (99%) validated.
TERT mutation testing
TERT promoter mutations were identified from sequencing data of 50 tumors (19 HMIN and 31 HWIDE). In addition, the TERT proximal promoter was amplified from genomic DNA from 47 HCC tumors (25 of these had been reported in our original paper describing these alterations in thyroid cancers (Landa et al., 2013). We adopted either a regular or a nested PCR approach, using primers and conditions previously described (Horn et al., 2013) and subsequently sequenced on a 3730 capillary sequencer (Applied Biosystems).
Somatic mitochondrial mutations
Aligned reads were analyzed using a custom informatics pipeline for mtDNA analysis. Briefly, a pileup file was generated with samtools mpileup (Li, 2011; Li et al., 2009), with minimum mapping quality 10 and base alignment quality 10. Other options for mpileup were: region MTcount-orphans -- --ignore-RG --excl-flags UNMAP,SECONDARY,QCFAIL,DUP --BCF -output-tags DP,AD,ADF,ADR --gap-frac 0.005 --tandem-qual 80 --no-BAQ. Pileup files were generated for paired tumor and normal tissue samples and piped into BCFtools call with options --multiallelic-caller --ploidy GRCh37 --keep-alts. Mutation annotation format files for mtDNA variants were then generated using vcf2maf. Variants were further annotated with calls from MitImpact (including the APOGEE score (Castellana et al., 2017) and MitoTIP (Lott et al., 2013).
Filtering criteria were used to remove spurious variant calls and prioritize variants. Variants were retained when satisfying the following criteria: (1) at least 5 reads supporting the variant in the tumor and at least 2 reads in both forward and reverse orientation; and (2) > 10% heteroplasmy/allelic fraction for the variant in the tumor and < 5% heteroplasmy/allelic fraction in the normal. Silent variants and those located in the control region were filtered and not included in further analysis. The last condition ensures that frameshift/nonsense variants are retained even if they are observed at high allelic fraction in the germline. mtDNA copy number was calculated as previously described (Reznik et al., 2016; Reznik et al., 2017). Briefly, the ratio of the number of reads aligning with mtDNA to the number of reads aligning with nuclear DNA was calculated. Only high-quality reads (MAPQ > 30, properly paired, excluding reads that failed quality control or that were marked as duplicates) were used. This ratio was then corrected for changes in tumor purity and ploidy. Because exome sequencing is often characterized by changes in target enrichment efficiency from batch to batch, a batch correction was implemented using log10 mtDNA copy number estimates (Reznik et al., 2016). The log10-corrected copy number estimates were then correlated against mtDNA mutation calls.
Copy number analysis
To characterize allele-specific somatic DNA copy number alterations, we applied FACETS to tumor and normal tissue pairs of bam files (Shen and Seshan, 2016). Copy number alterations, tumor purity, ploidy, and cellular fractions were estimated and reported. We also generated seg files for IGV viewing and applied GISTIC (version 2.0, http://www.broadinstitute.org/cgibin/cancer/publications/pub_paper.cgi?mode=view&paper_id=216&p=t) to define regions of the genome that are significantly amplified or deleted across the set of samples.
FISH analysis
FISH analysis was performed on formalin-fixed, paraffin-embedded sections using a home-brew 3-color probe designed to confirm the copy number changes of chromosomes 2, 5, and 7 detected by FACETS. Chromosome 2 served as the control for ploidy. The clones used in the probe mix were as follows: 5q11 (BAC clone RP11–412L4 and RP11–364C6; labeled with Orange dUTP), Centromere 7 (PAC clone p7t1; labeled with Green dUTP), and 2q11 (BAC clones RP11–708D7, RP11–468G5, and RP11–468G5; labeled with Red dUTP). All BAC clones were purchased from Roswell Park Cancer Institute Genomics Shared Resource (Buffalo, NY); the PAC clone is from the Memorial Sloan Kettering Cancer Center Molecular Cytogenetics Core Facility. Probe labeling, tissue processing, hybridization, post hybridization washing, and fluorescence detection were performed in accordance with standard laboratory procedures. Slides were scanned using a Zeiss Axioplan 2i epifluorescence microscope equipped with a megapixel CCD camera (CV-M4+CL, JAI) controlled by Isis imaging software (version 5.5.9, MetaSystems, Waltham, MA). The Metafer and VSlide modules were used to generate the virtual image of hematoxylin and eosin– and DAPI-stained sections.
To assess the quality of hybridization and possible intratumoral heterogeneity, the entire section was scanned under 63x or 100x objectives. Following initial scan, representative regions were imaged through the depth of the tissue (compressed/merged stack of 12 z-section images taken at 0.5-micron intervals), and at least 10 images per representative region were captured. Signal counts (scoring) were performed on captured images. For each case, a minimum of 3 distinct regions (representative of major signal pattern) were selected, and 100 discrete nuclei were scored. To minimize truncation artifacts, only nuclei with at least 1 signal, for 2 of the 3 chromosomes, were selected. Five normal thyroid tissue sections were also analyzed, and for each case, 100 nuclei were scored to derive the cut-off values (false-positive). The cut-off value for each probe was calculated as the mean of the false-positive + 3x the standard deviation and set at 50% for loss and 10% for gain. A tumor was considered to exhibit true loss if > 50% of the cells showed ≤ 1 copy of chromosomes 2, 5, or 7 and true gain if > 10% of the cells showed ≥ 3 copies of chromosomes 2, 5, or 7. Additionally, cells and tumors with signal pattern 1–2-2 (chromosome 2: ≤ 1 copy; chromosome 5: 2 copies; chromosome 7: 2 copies) were considered to be near-haploid, and cells/tumors with signal pattern 2–4-4 (chromosome 2: 2 copies; chromosome 5: 4 copies; chromosome 7: 4 copies) were considered to represent endoreduplication of the near-haploid clone and to be polysomic.
Copy number alterations integrative analysis
The FACETS results were integrated with the FISH results to determine the copy number alterations in each tumor. Tumors were categorized into 3 types: diploid, near-haploid, and polysomic. Diploid tumors had 2 copies of each chromosome, with or without focal alterations in individual chromosomes. Near-haploid tumors had single copies of the majority of chromosomes, with the exception of chromosome 7, which was always diploid, and chromosome 5, which was diploid in some cases. Polysomic tumors had whole-chromosome gains of chromosome 7 (usually WCD, resulting in 4 copies), as well as, in some cases, chromosome 5 and chromosome 12.
Fractional genome alteration analysis
For TCGA tumors, across 26 pan-cancer types, the corresponding Affymetrix SNP 6 array data were processed and genotyped together with Affymetrix power tools and the Birdseed algorithm, respectively. PennCNV was used to obtain the log R ratio and B allele frequencies for each sample. We subsequently ran the ASCAT package to generate the allele-specific copy number calls and to determine LOH status per tumor. For HCCs, the FACETS software was used to analogously obtain allele-specific copy number and LOH calls. Finally, a customized Ruby algorithm was used to generate the fractional genome alteration on a per sample basis. Only regions of the (autosomal) genome that had either a hemizygous deletion (loss of 1 allele) or a copy number–neutral LOH call were considered to be altered.
Gene expression analysis
For the RNA-Seq data from the HCC samples, raw FASTQ files were aligned with the hg19 genome using the STAR aligner with default parameters (Dobin et al., 2013). Aligned fragments were counted with Rsamtools (version 3.2) and annotated using the TxDb.Hsapiens.UCSC.hg19.knownGene (version 3.2) transcript database. Regularizedlogarithm transformation of the matrix was obtained with the rlog function of DESeq2 (version 1.10.1). The gene expression count data were used for clustering analysis. Unsupervised hierarchial clustering was performed in Partek (version 6.6) using batch correction and quantile normalization. Batch correction was incorporated to account for the multiple data sites. The 200 most variant genes across all samples were clustered using the Ward distance. Integrated pathway analysis of the 200 most differentially expressed genes across the HMIN and HWIDE samples was used to identify pathways characteristic of HCCs.
Fusion gene analysis
Two software packages were used for gene fusion discovery: deFuse (http://compbio.bccrc.ca/software/defuse/) and FusionCatcher (https://github.com/ndaniel/fusioncatcher). We ran both packages, following the best practices recommended by the developers. Results from both callers were combined, and overlapping calls were used for the downstream analysis. We also filtered out the fusions annotated as adjacent (fusions between adjacent genes), altsplice (fusions likely the product of alternative splicing between adjacent genes), and read-through (fusions involving adjacent genes potentially resulting from cotranscription, rather than genome rearrangement).
For validation of fusion genes, cDNA was synthesized from tumor RNA using the Clonetech RNA to cDNA EcoDry kit, amplified by PCR, and visualized on a 1.5% agarose gel. In-frame fusions were verified by designing PCR primers around the predicted fusion site. The resulting PCR products were purified, and the fusion site was then confirmed by Sanger sequencing. All primer sequences are as follows: TMEM233_VPREB3 (forward CGTCTCGTGTTTTTGCCCTG, reverse CGTCCACTGACCATCCACAA); CHCHD10_VPREB3 (forward CTCATGGCTCAGATGGCGAC, reverse CCGAGAATCGATCGGGGATG); ACSS1_APMAP (forward TCAAGGCCTACCCAGGCTAT, reverse TCCAGGCATGTTCTCCACAA); BCAP29_SLC26A4 (forward GAGAAGAGCTCCACCAGCAG, reverse GCACTGGCAATCAGGACTCT); DUOXA1_DUOX2 (forward TGTGGATGAAGACCCCATGC, reverse GGTAGCCAAAGAAGACCCCC).
Creation of stable fusion cell line TMEM233_PRKAB1 NTHY-ori
NTHY-ori 3.1 cells, an immortalized thyroid epithelial cell line, were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, L-glutamine, penicillin and streptomycin. Cells were incubated at 37°C and maintained at 5% CO2. HA-tagged TMEM233_PRKAB1 fusion gene was cloned into pCDF1-Puro vector by BamH1 and Not I enzymes. Transfection was performed with 1 ug plasmids using Lipofectamine™ reagent (Invitrogen). pCDF1 transfected cells were used as control for cell proliferation assays. For lentivirus infection, 6 μg TMEM233_PRKAB1 fusion expressing constructs and control vector were transfected along with packaging and envelop plasmids into Lenti-X™ 293T Cells (Clontech). Viral supernatants were collected 48 hr after transfection. Filtered and target cells were infected in the presence of 8μg/ml polybrene. Twenty-four hr later, virus medium was removed and replaced with normal culture medium. Three days after transduction, cells were selected with 3 μg/ml puromycin.
Proliferation assay in vitro of stable fusion cell line TMEM233_PRKAB1 NTHY-ori
The xCELLigence real time proliferation system (ACEA Biosciences Inc.) was used to detect changes in proliferation of fusion and control cells over a 120 hr period in real time. Briefly, NTHY-ori cells, an immortalized thyroid epithelial cell line was transfected with fusion gene TMEM233_PRKAB1 and control vector and trypsinized and counted after 48 hr. E-plate VIEW16 xCELLigence plates were prepared as manufacturer’s recommendations and transfected cells added to wells in groups of 4 wells per treatment. Cell proliferation was measured by a calculated cell index using the xCELLigence device.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was carried out using SPSS (version 21, IBM Corporation, Armonk, NY). Clinical and pathological variables were compared within HMIN and HWIDE groups using Pearson’s chi-squared test. Survival outcomes were analyzed using the Kaplan-Meier method. Outcomes data were calculated at 5 years. The p values are presented in figure legends where p < 5 x E-2 was considered statistically significant. Details of the statistical methodology for somatic mutation calls, mitochondrial DNA mutations calls, FACETS algorithm for copy number calls and mRNA expression profiling are described in detail in each relevant section.
Supplementary Material
SIGNIFICANCE
This is a comprehensive analysis of the molecular landscape of Hürthle cell cancer. Our study lays the foundations for many mechanistic studies to investigate the unique chromosomal alterations identified, frequent and recurrent somatic mitochondrial mutations, fusion genes and a large number of somatic mutations driving protein translation including the RTK/RAS/AKT/mTOR pathway. Taken together, our findings can help guide the development of treatments for one of the most aggressive types of thyroid cancer.
Highlights
HCCs have a high number of mutations, several of which regulate translation
HCCs exhibit a high number of somatic recurrent mitochondrial mutations
HCCs have a unique chromosomal landscape, which predicts aggressive behavior
Several recurrent fusion genes, including TMEM233_PRKAB1, are identified
Acknowledgments:
We thank Susan Weil for graphic design and Jessica Massler for editorial assistance. We thank the Chan lab for helpful discussions. This work was supported by Hutchison Shaw Foundation, Pechter Foundation, NIH SPORE grant P50 CA172012–01A1, and NIH/NCI Cancer Center Support Grant P30 CA008748.
Footnotes
Declaration of Interests: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AND SOFTWARE AVAILABILITY
Whole exome sequencing data has been deposited in ENS’s Sequence Read Archive. RNA Sequencing data has been deposited in SRA and is available at https://www.ncbi.nlm.nih.gov/sra/SRP136351
Supplementary Material
Table S1, related to Figure 1
Table S2, related to Figure 3
Table S3, related to Figure 4
Table S4, related to Figure 5
Table S5, related to Figure 4
Table S6, related to Figure 1
Table S7, related to Figure 7
REFERENCES
- Amorim JP, Santos G, Vinagre J, and Soares P (2016). The Role of ATRX in the alternative lengthening of telomeres (ALT) phenotype. Genes (Basel) 7, E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter E, Windloch K, Gannon F, and Lee JS (2014). Epigenetic regulation in cancer progression. Cell Biosci 4, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besic N, Vidergar-Kralj B, Frkovic-Grazio S, Movrin-Stanovnik T, and Auersperg M (2003). The role of radioactive iodine in the treatment of Hürthle cell carcinoma of the thyroid. Thyroid 13, 577–584. [DOI] [PubMed] [Google Scholar]
- Boot A, Oosting J, de Miranda NF, Zhang Y, Corver WE, van de Water B, Morreau H, and van Wezel T (2016). Imprinted survival genes preclude loss of heterozygosity of chromosome 7 in cancer cells. J Pathol 240, 72–83. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. (2012a). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2012b). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2012c). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2011). Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. (2014). Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcangiu ML, Bianchi S, Savino D, Voynick IM, and Rosai J (1991). Follicular Hürthle cell tumors of the thyroid gland. Cancer 68, 1944–1953. [DOI] [PubMed] [Google Scholar]
- Caria P and Vanni R (2010). Cytogenetic and molecular events in adenoma and welldifferentiated thyroid follicular-cell neoplasia. Cancer Genet Cytogenet 203, 21–29. [DOI] [PubMed] [Google Scholar]
- Castellana S, Fusilli C, Mazzoccoli G, Biagini T, Capocefalo D, Carella M, Vescovi AL, and Mazza T (2017). High-confidence assessment of functional impact of human mitochondrial non-synonymous genome variations by APOGEE. PLoS Comput Biol 13, e1005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercek A Braghiroli MI, Choi JF, Hechtman JF, Kemeny N, Saltz L, Capanu M, and Yaeger R(2017). Clinical features and outcomes of patients with colorectal cancers harboring NRAS mutations. Clin Cancer Res 23, 4753–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi R, Zhu Z, and Nikiforov YE (2005). BRAF copy number gains in thyroid tumors detected by fluorescence in situ hybridization. Endocr Pathol 16, 99–105. [DOI] [PubMed] [Google Scholar]
- Clapier CR and Cairns BR (2009). The biology of chromatin remodeling complexes. Annu Rev Biochem 78, 273–304. [DOI] [PubMed] [Google Scholar]
- Corver WE, Ruano D, Weijers K, den Hartog WC, van Nieuwenhuizen MP, de Miranda N, van Eijk R, Middeldorp A, Jordanova ES, Oosting J, et al. (2012). Genome haploidisation with chromosome 7 retention in oncocytic follicular thyroid carcinoma. PLoS One 7, e38287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corver WE, van Wezel T, Molenaar K, Schrumpf M, van den Akker B, van Eijk R, Ruano Neto D, Oosting J, and Morreau H (2014). Near-haploidization significantly associates with oncocytic adrenocortical, thyroid, and parathyroid tumors but not with mitochondrial DNA mutations. Genes Chromosomes Cancer 53, 833–844. [DOI] [PubMed] [Google Scholar]
- Crimi M, Galbati S, Perini MP, Bordoni A, Malferrari G, Sciacco M, Biunno I, Strazzer S, Moggio M, Bresolin N, et al. (2003). A mitochondrial tRNA(His) gene mutation causing pigmentary retinopathy and neurosensorial deafness. Neurology 60, 1200–1203. [DOI] [PubMed] [Google Scholar]
- DeLellis RA, Lloyd RV, and Heitz PU (2017). World Health Organization Classification of Tumors Pathology and Genetics Tumors of Endocrine Organs Ch. 2 (Lyon, France: IARC Press; ), pp. 65–142. [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Harti C, Philppakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganly I Ricarte Filho J, Eng S, Ghossein R, Morris LG, Liang Y, Socci N, Kannan K, Mo Q, Fagin JA, et al. (2013). Genomic dissection of Hürthle cell carcinoma reveals a unique class of thyroid malignancy. J Clin Endocrinol Metab 98, E962–E972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossein RA, Hiltzik DH, Carlson DL, Patel S, Shaha A, Shah JP, Tuttle RM, and Singh B (2006). Prognostic factors of recurrence in encapsulated Hürthle cell carcinoma of the thyroid gland: a clinicopathologic study of 50 cases. Cancer 106, 1669–1676. [DOI] [PubMed] [Google Scholar]
- Grossman RF and Clark OH (1997). Hürthle cell carcinoma. Cancer Control 4, 13–17. [DOI] [PubMed] [Google Scholar]
- Haas M, Ormann S, Baechmann S, Remold A, Kruger S, Westphalen CB, Siveke JT, Wenzel P, Schlitter AM, and Esposito I (2017). Extended RAS analysis and correlation with overall survival in advanced pancreatic cancer. Br J Cancer 116, 1462–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, and Blenis J (2005). mTOR and S6K1 Mediate Assembly of the Translation Preinitiation Complex through Dynamic Protein Interchange and Ordered Phosphorylation Events. Cell 123, 569–580. [DOI] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. (2013). TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961. [DOI] [PubMed] [Google Scholar]
- Hustedt N and Durocher D (2016). The control of DNA repair by the cell cycle. Nat Cell Biol 19, 1–9. [DOI] [PubMed] [Google Scholar]
- Hundahl SA, Fleming ID, Fremgen AM, and Menck HR (1998). A National Cancer Data Base report on 58,856 cases of thyroid carcinoma treated in the U.S., 1985–1995. Cancer 83, 2638–2648. [DOI] [PubMed] [Google Scholar]
- Jun G, Flickinger M, Hetrick KN, Romm JM, Doheny KF, Abecasis GR, Boehnke M, and Kang HM (2012). Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet 91, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Goto Y, Campos Y, Arenas J, and Suzuki T (2005). Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc Natl Acad Sci U S A 102, 7127–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullander K and Klein R (2002). Mechanisms and functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol 3, 475–486. [DOI] [PubMed] [Google Scholar]
- Kushchayeva Y, Duh QY, Kebebew E, and Clark OH (2004). Prognostic indications for Hürthle cell cancer. World J Surg 28, 1266–1270. [DOI] [PubMed] [Google Scholar]
- Landa I, Ganly I, Chan TA, Mitsutake N, Matsuse M, Ibrahimpasic T, Ghossein RA, and Fagin JA (2013). Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab 98, E1562–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, RicarteFilho JC, Krishnamoorthy GP, Xu B, et al. (2016). Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 126, 1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapunzina P and Monk D (2011). The consequences of uniparental disomy and copy number neutral loss-of-heterozygosity during human development and cancer. Biol Cell 103, 303–317. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, and Getz G (2014). Discovery and saturation analysis of cancer genes across 21 tumor types. Nature 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H and Durbin R (2010). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Ding X, Zhou C, Zhang Y, Xu M, Zhang C, and Xu L (2012). Knockdown of eukaryotic translation initiation factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in glioblastoma cells. Neurol Sci. 33, 1057–1062. [DOI] [PubMed] [Google Scholar]
- Lopez-Penabad L, Chiu AC, Hoff AO, Schultz P, Gaztambide S, Ordoñez NG, and Sherman SI (2003). Prognostic factors in patients with Hürthle cell neoplasms of the thyroid. Cancer 97, 1186–1194. [DOI] [PubMed] [Google Scholar]
- Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, and Wallace DC (2013). mtDNA variation and analysis using mitomap and mitomaster. Curr Protoc Bioinformatics 44, 1.23.1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bai RK, Trieu R, and Wong LJ (2010). Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta 1797, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H and Maciejewski JP (2011). Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res 17, 3913–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Maẞhöfer L, Temming P, Rahmann S, Metz C, Bornfeld N, van de Nes J, Klein-Hitpass L, Hinnebusch AG, Horsthemke B, et al. (2013). Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 45, 933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Máximo V, Lima J, Prazeres H, Soares P, and Sobrinho-Simões M (2016). The biology and the genetics of Hürthle cell tumors of the thyroid. Endocr Relat Cancer 23, X2. [DOI] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Montalto G, Cervello M, Nicoletti F, Fagone P, Malaponte G, Mazzarino MC, et al. (2012). Mutations and deregulation of Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR cascades which alter therapy response. Oncotarget 3, 954–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, and Lichter P (2013). Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet 14, 765–780. [DOI] [PubMed] [Google Scholar]
- Reznik E, Miller ML, Şenbabaoğlu Y, Riaz N, Sarungbam J, Tickoo SK, Al-Ahmadie HA, Lee W, Seshan VE, Hakimi AA, et al. (2016). Mitochondrial DNA copy number variation across human cancers. Elife 5, e10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik E, Wang Q, La K, Schultz N, and Sander C (2017). Mitochondrial respiratory gene expression is suppressed in many cancers. Elife 6, e21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos WP, Thoma AD, and Kaina B (2016). DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer 16, 20–33. [DOI] [PubMed] [Google Scholar]
- Samatar AA and Poulikakos PI (2014). Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov 13, 928–942. [DOI] [PubMed] [Google Scholar]
- Shaha AR, Shah JP, and Loree TR (1996). Patterns of nodal and distant metastasis based on histologic varieties in differentiated carcinoma of the thyroid. Am J Surg 172, 692–694. [DOI] [PubMed] [Google Scholar]
- Shen R and Seshan VE (2016). FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44, e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman EJ, Ho AL, Fury MG, Baxi SS, Dunn L, Lee JS, Lipson BL, and Pfister DG (2015). Combination of everolimus and sorafenib in the treatment of thyroid cancer: update on phase II study. J Clin Oncol 33, 6069. [Google Scholar]
- Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, et al. (2012). Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification mediated acquired B-RAF inhibitor resistance. Nature Commun 3, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi K, Miró R, Fernández-Jiménez N, Quintanilla I, Ramos L, Prat E, del Rey J, Pujol N, Killian JK, Meltzer PS, et al. (2015). Patterns of somatic uniparental disomy identify novel tumor suppressor genes in colorectal cancer. Carcinogenesis 36, 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuna M, Ju Z, Smid M, Amos CI, and Mills GB (2015). Prognostic relevance of acquired uniparental disomy in serous ovarian cancer. Mol Cancer 14, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuna M, Knuutila S, and Mills GB (2009). Uniparental disomy in cancer. Trends Mol Med 15, 120–128. [DOI] [PubMed] [Google Scholar]
- Vu-Phan D and Koenig RJ (2014). Genetics and epigenetics of sporadic thyroid cancer. Mol Cell Endocrinol 386, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CS, Ogawa S, Scoles DR, Miller CW, Kawamata N, Narod SA, Koeffler HP, and Karlan BY (2008). Genome-wide loss of heterozygosity and uniparental disomy in BRCA1/2-associated ovarian carcinomas. Clin Cancer Res 14, 7645–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Ma L, Tang X, Zhang X, Qiao Y, Shi Y, Xu Y, Wang Z, Yu Y, and Sun F (2015). Doxorubicin induces apoptosis by targeting Madcam1 and AKT and inhibiting protein translation initiation in hepatocellular carcinoma cells. Oncotarget 6, 24075–24091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ru Y, Sanchez-Carbayo M, Wang X, Kieft JS, and Theodorescu D (2013). Translation initiation factor eIF3b expression in human cancer and its role in tumor growth and lung colonization. Clin Cancer Res. 19, 2850–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956). On the origin of cancer cells. Science 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Miltra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, et al. (2012). The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A 109, 6662–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie CH, Naito A, Mizumachi T, Evans TT, Douglas MG, Cooney CA, Fan CY, and Higuchi M (2007). Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun 364, 656–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Xu CZ, Gu J, Liu X, Huang E, Yuan Y, Zhao G, Jiang J, Xu C, et al. (2016). Eukaryotic translation initiation factor 3B accelerates the progression of esophageal squamous cell carcinoma by activating β-catenin signaling pathway. Oncotarget 7, 43401–43411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.