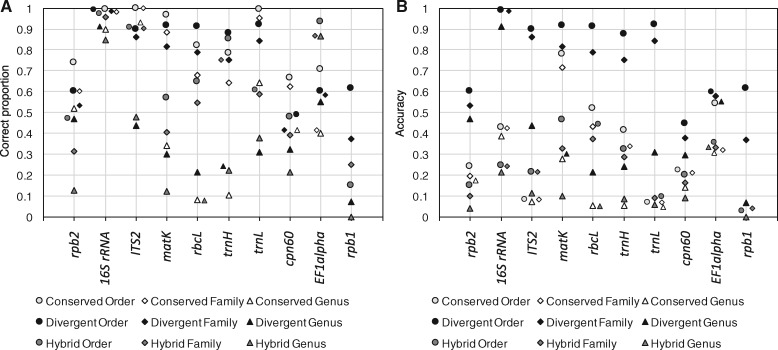
Fig. 1.
Self-evaluated performance of the Metaxa2 Database Builder. Evaluation was performed in all operating modes (conserved, divergent and hybrid) on 10 different DNA barcoding regions. (A) Proportion of assigned sequences classified to the correct order (circles), family (diamonds) and genera (triangles). (B) Accuracy, i.e. proportion of correctly assigned sequences multiplied with the proportion of sequences included in the final classification databases (see Supplementary Fig. S2). The ATP9-NAD9 genetic marker is not shown, because it only had relevant taxonomic differences at the species level