Abstract
The last decade has seen impressive progress in human embryonic stem cell-derived cardiomyocytes (hESC-CMs) that makes them ideal tools to repair injured hearts. To achieve an optimal outcome, advanced molecular imaging methods are essential to accurately track these transplanted cells in the heart. Herein, we demonstrate for the first time that a class of photoacoustic nanoparticles (PANPs) incorporating semiconducting polymers (SPs) as contrast agents can be used in the photoacoustic imaging (PAI) of transplanted hESC-CMs in living mouse hearts. This is achieved by virtue of two benefits of PANPs. First, strong PA signals and specific spectral features of SPs allow PAI to sensitively detect and distinguish a small number of PANP-labeled cells (2,000) from background tissues in vivo. Second, the PANPs show a high efficiency for hESC-CM labeling without adverse effects on cell structure, function, and gene expression. Assisted by ultrasound imaging, the delivery and engraftment of hESC-CMs in living mouse hearts can be assessed by PANP-based PAI with high spatial resolution (~100 μm). In summary, this study explores and validates a novel application of SPs as a PA contrast agent to track labeled cells with high sensitivity and accuracy in vivo, highlighting the advantages of integrating PAI and PANPs to advance cardiac regenerative therapies.
Keywords: cardiac regenerative therapy, photoacoustic imaging, semiconducting polymer, cardiac cell tracking
Graphical Abstract
Ultrasensitive semiconducting polymer nanoparticles (SPNs) for photoacoustic imaging of transplantation and engraftment of human embryonic stem cell-derived cardiomyocytes (hESC-Cs) were developed. The SPNs have strong and stable PA signals as well as specific PA spectrum, which facilitate real-time monitoring of delivery and localization of hESC-CMs in mouse hearts for cardiac regenerative therapy.
1. Introduction
Ischemic heart disease is a leading cause of death worldwide [1]. It causes a permanent loss of cardiomyocytes that, if left untreated, will ultimately result in heart failure or sudden death. Restoring lost cardiomyocytes is still a challenge in current clinical practice due to the limited regenerative capability of adult hearts. Recent studies show that the transplantation of exogenously generated human embryonic stem cell-derived cardiomyocytes (hESC-CMs) has significant potential to repair injured hearts [2]. Effective delivery and engraftment of hESC-CMs are crucial to achieving optimal outcomes for these treatments [3]. To this end, molecular imaging has played an indispensable role. Various exogenous and endogenous imaging probes, including nanoparticles (NPs) and reporter genes, have been used to assist the tracking of transplanted cells in vivo [4]. However, current molecular imaging techniques for tracking hESC-CMs have limitations. For instance, positron emission tomography (PET) and magnetic resonance imaging (MRI) have been used to track transplanted hESC-CMs in hearts [5], but they are time-consuming and expensive, and have limited time or spatial resolution. Optical imaging techniques such as fluorescence imaging (FI) and bioluminescence imaging (BLI) encounter problems in accurately locating the engraftment of hESC-CMs because of their limited penetration and spatial resolution [4b]. Thus, an alternative approach that overcomes these imaging limitations to accurately track transplanted cells in vivo is needed, particularly for preclinical studies.
Photoacoustic imaging (PAI) techniques, which combine the high sensitivity and contrast of optical imaging with the desired penetration depth and resolution of ultrasound, show great potential to address the aforementioned problems while maintaining low cost and ease of operation [6]. To ensure high imaging contrast, the PAI of transplanted cells is enhanced by exogenous PA contrast agents [7], such as small-molecule dyes [8], carbon materials [9], and metallic nanoparticles [10]. However, these contrast agents suffer from several drawbacks, including intrinsic poor photostability unless they are protected by a shell, limited sensitivity to detect small cell numbers in vivo, and non-specific broad PA spectra that are difficult to be distinguished from the background signals [11]. These barriers particularly obstruct the application of PAI in tracking hESC-CMs in hearts, because the heart motion and cell death cause a majority of cell loss post transplantation, and the intracardial environment generates complex background signals. Therefore, a PAI contrast agent that has strong signals and specific spectral features would be ideal to track and discriminate the transplanted hESC-CMs from host tissues.
Recently, near-infrared light absorbing semiconducting polymers (SPs) have been shown to generate stronger PA signals compared with other PA nanoagents on a per mass basis [7,11b], highlighting their potential as next generation PAI contrast agents for stem cell imaging in deeper tissues [12]. Notably, SPs usually have specific and narrow PA spectra that are significantly distinguishable from those of host tissues in the background [11b,12f,13]. Thus, encapsulating SPs into nanoparticles provides a promising solution to enable the PAI of hESC-CMs in hearts. In addition, unlike other cell types, hESC-CMs have low endocytosis activity leading to a low efficiency for the passive internalization of NPs [14]. It thus requires an efficient approach to deliver NPs into hESC-CMs to avoid the NP dilution caused by cell proliferation and/or cell death, which is a common problem in current approaches that usually involve a delivery of NPs into stem cells before taking several weeks to induce them into terminally differentiated cardiomyocytes [14–15].
In this study, we developed a class of photoacoustic nanoparticles (PANPs) to enable the PAI of transplanted hESC-CMs in living mouse hearts. The PANPs were synthesized using SPs, poly[2, 6-(4,4-bis-(2-ethylhexyl)-4H-cyclopenta[2,l-b;3,4-b’]dithiophene)-alt-4,7(2,1,3 benzothiadia-zole)] (PCPDTBT), as PA contrast agents and a FDA-approved lipid-based copolymer as the encapsulation matrix. Specifically, the PANP surfaces were immobilized with cell penetrating peptides (CPPs) to facilitate effective labeling of hESC-CMs through a simple incubation for a short period. Following labeling, we characterized the properties of PANPs in vitro and the detection limit of labeled cells in vivo. We also evaluated the effects of PANPs on cell functions and gene expression of hESC-CMs in vitro. We further verified the feasibility of using PAI to track the delivery and engraftment of PANP-labeled hESC-CMs in living mouse hearts. These in vivo results were validated by histological analysis.
2. Results
2.1. Preparation and Characterization of PANPs
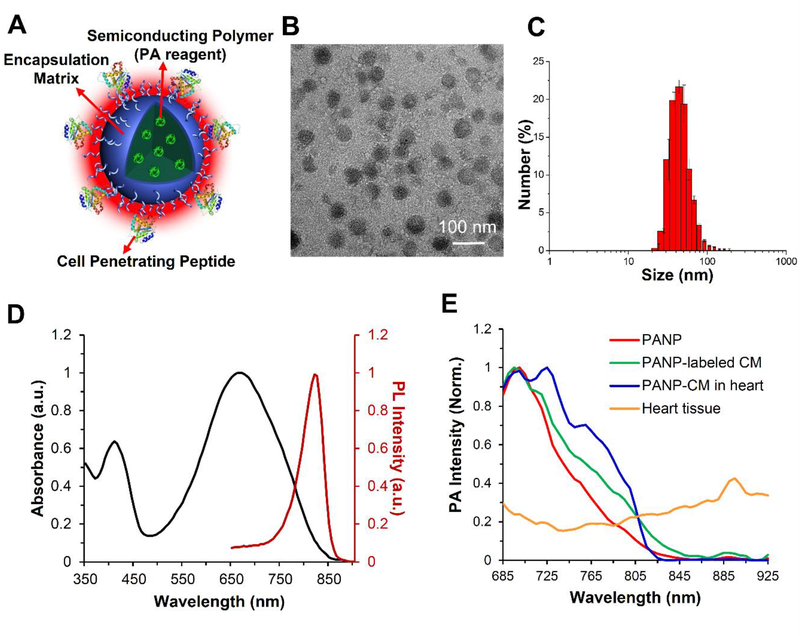
We synthesized a class of photoacoustic nanoparticles (PANPs) through a nanoprecipitation method for hESC-CM imaging [16]. As illustrated in Figure 1, the PANPs are composed of three components: SPs as the PA contrast agent, polymer lipids as the encapsulation matrix, and CPPs as the labeling enhancer. The morphology of synthesized PANPs was studied by transmission electron microscopy (TEM), which showed they were in spherical shape with uniformly distributed sizes (Figure 1C). Dynamic light scattering results revealed that the average size of PANPs was 48.6 ± 1.2 nm in water suspension (Figure 1C). Notably, the PANPs kept these stable sizes (50.8 ± 2.5 nm) even after being stored at 4°C for 1 year without obvious change in the profile of size distribution (Figure S1A in the Supporting Information). The optical properties of PANPs showed a UV-visible absorption peak at 670 nm and a photoluminescence spectral peak at 820 nm (Figure 1D). Following multi-spectral excitation of near-infrared lasers (680–970 nm), PANPs showed a specific PA spectrum that reached a maximal peak at 705 nm and gradually decreased to zero after 850 nm (Figure 1E). Importantly, this specificity of PA spectrum was preserved for PANP-labeled hESC-CMs, even when they were embedded in cardiac tissues. Moreover, this spectrum had a distinct pattern and a much higher signal intensity compared to the one of local cardiac milieu (Figure 1E). This feature ensured a specific identification of PANP-labeled hESC-CMs from surrounding tissues by unmixing the PA signals. The near-infrared excitation of PANPs made PAI capable of imaging deep tissues (>10 mm) in small animals such as mouse hearts in the present study.
Figure 1. Characterization of semiconducting polymer (SP)-based photoacoustic nanoparticles (PANPs).
(A) The PANPs were composed of three components: SPs (Poly[2, 6-(4,4-bis-(2-ethylhexyl)-4H-cyclopenta[2,l-b;3,4-b’]dithiophene)-alt-4,7(2,l,3 benzothiadia-zole)]) as the PA contrast agent, polymer lipids as the encapsulant, and cell-penetrating peptides as the labeling enhancer. (B) The morphology was studied by transmission electron microscopy (TEM). (C) Particle size distribution of PANPs in water determined by a dynamic light scattering analyzer. (D) The maximum peaks of UV-visible absorption and photoluminescence spectrum for PANPs were at 670 nm and 820 nm, respectively. (E) Following multi-spectral excitations of near-infrared lasers (680–970 nm), pure PANPs showed a specific PA spectrum that reached a maximal peak at 705 nm and gradually decreased to zero after 850 nm (red). This narrow spectrum was consistent with PANP-labeled hESC-CMs (green), even when they were injected into the heart (blue). This spectrum had a largely different pattern and a much higher signal peak compared to the one from cardiac tissues (yellow).
2.2. In Vitro Evaluation of PANPs in hESC-CM Labeling
2.2.1. Quantification of Cell Labeling Efficiency of PANPs
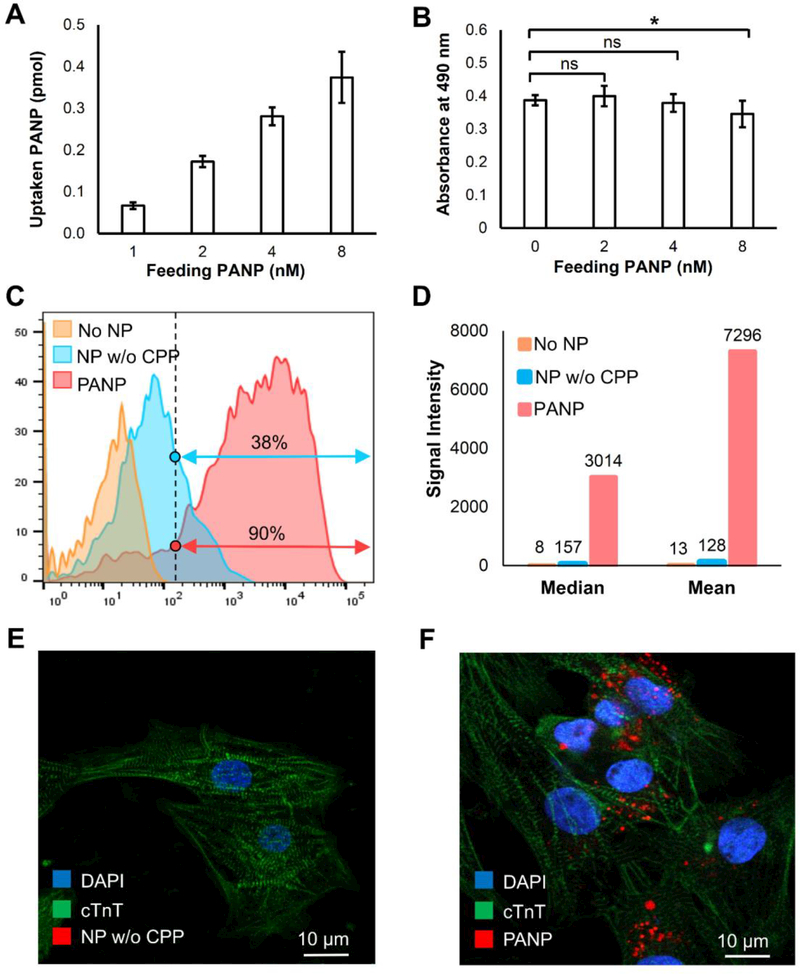
We then evaluated the direct labeling efficiency of PANPs to hESC-CMs through in vitro studies. PANP dose for the hESC-CM labeling was optimized based on both cellular uptake and toxicity assays. Although the increased feeding dose of PANPs from 1 nM to 8 nM led to enhanced uptake efficiency for cells (Figure 2A), we chose 4 nM as the optimal labeling dose because the results from cell viability assays indicated potential cell toxicity when the PANP dose exceeded 4 nM (Figure 2B). Moreover, the release ratio of PANPs from the labeled cells were all measured at below 5% in 24 hours post labeling (Figure S1B in the Supporting Information). Taking advantage of the near-infrared fluorescence of PANPs, fluorescence-activated cell sorting (FACS) was used to quantitatively evaluate the PANP labeling efficiency (4 nM) for hESC-CMs after overnight incubation at 37 °C (Fig. 2C). Using NPs without CPP-functionalization as a benchmark, we showed that PANPs significantly increased the labeling efficiency of hESC-CMs from 38% to 90% as measured by the percentage of fluorescent cells. Moreover, we were able to achieve a ~60-fold increase of PANP loading dose per cell as measured by the averaged fluorescent intensity (Figure 2D). These FACS results were also confirmed by confocal images. Bright fluorescence (red) from internalized PANPs was observed in hESC-CMs in Figure 2F, while no fluorescence from NPs without CPPs could be detected in hESC-CMs in Figure 2E, due to their ultra-low internalization efficiency from endocytosis. Notably, direct labeling of hESC-CMs avoided the major problem of NP dilution in stem cell labeling because hESC-CMs had very limited proliferation compared to stem cells. Additionally, the PANP labeling did not interfere with the striated sarcomeric structure of hESC-CMs, as indicated by cardiac α-actinin in the confocal images of Figure 2F. Therefore, our strategy provided a promising solution for direct labeling of hESC-CMs with PANPs.
Figure 2. Assessing the uptake, toxicity, and efficiency of hESC-CM labeling with PANPs in vitro.
We first compared the uptake ratio and toxicity of hESC-CM labeling under different feeding PANP doses. (A) A higher feeding PANP dose from 1 nM to 8 nM led to a higher uptake rate for cells. (B) The PANPs showed significant toxicity to the labeled hESC-CMs when the feeding dose of PANPs exceeded 4 nM. Using feeding PANP dose of 4 nM, we further compared two approaches for direct labeling of hESC-CMs: our proposed PANP-based labeling versus the routine endocytosis-based labeling that used the nanoparticles without CPPs (NP w/o CPP). (C) Following an overnight labeling, FACS-based quantification indicated that the percentage of hESC-CMs labeled by PANPs was 2.4 times more than that labeled by NPs w/o CPP. (D) The fluorescence intensity analysis further indicated that the loaded dose of PA contrast agents with PANP labeling was almost 60 times higher than that labeled by NPs w/o CPP. (E-F) Confocal images of delivered NP w/o CPP and PANP (red) in hESC-CMs, which revealed no fluorescence from NP w/o CPP in cells (left) due to the ultra-low internalization efficiency through endocytosis.
2.2.2. Impact of PANPs on Cell Function and Gene Expression of hESC-CMs
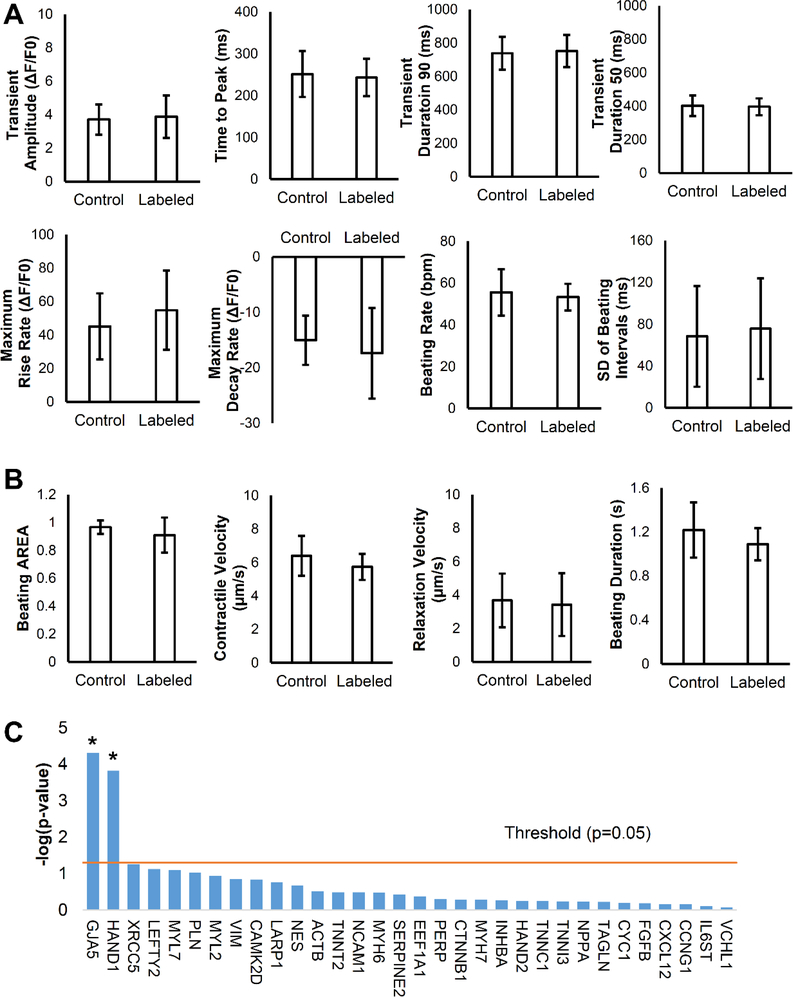
Post hESC-CM labeling, we evaluated the cardiotoxicity of PANPs to the labeled cells. Because calcium handling and contractility are essential functions of cardiomyocytes, we quantitatively compared these parameters between PANP-labeled cells and unlabeled control cells in vitro (Figure S2A and S2B in the Supporting Information). The calcium handling was comprehensively assessed by Fluo-4 AM confocal calcium imaging with eight major parameters and showed no significant differences between labeled and control cells (Figure 3A). Similarly, the cell contractility was quantified by an image motion-tracking method with four parameters, which revealed comparable results between the two groups (Figure 3B). We next assessed the effect of PANP internalization on gene expression patterns of hESC-CMs following PANP labeling. We used quantitative polymerase chain reaction (qPCR) to examine genes related to cardiomyocyte structures and functions and found no adverse effects of PANP labeling on most genes (Figure S2C in the Supporting Information). Interestingly, the expression of two beneficial genes, GJA5 and HAND1, were up-regulated following PANP labeling (Figure 3C). The expression levels of IL-6 and IL-8 were also up-regulated, which could be due to increased inflammation effect upon labeling (Figure S3 in the Supporting Information). In addition, although most intravenously injected PANPs were quickly accumulated in the liver [17], blood tests showed no liver abnormalities of mice caused by the injected PANPs compared to control mice with PBS injection (Figure S4 in the Supporting Information). Overall, these results demonstrated that cell function and transcriptome of hESC-CMs were not compromised by PANP labeling.
Figure 3. Assessment of cell functions and gene expression for PANP-labeled hESC-CMs in vitro.
(A) Eight parameters regarding calcium handling were comprehensively compared between PANP-labeled cells and unlabeled control cells and all of them showed no significant difference (P > 0.05). (B) Four parameters regarding contractility were compared between PANP-labeled cells and unlabeled control cells, and none of them showed a significant difference (P > 0.05). These results indicate that cell functions of hESC-CMs are minimally affected by PANP labeling. (C) Negative Log-transformed P-values for each gene are plotted as bar graph with threshold (P = 0.05, -log10[0.05] = 1.301) plotted as a straight line. Except the upregulation of GJA5 and HAND1 following PANP labeling, the remaining 30 genes showed no significant differences between control and labelled cells. P < 0.05 stands for significance. N = 17 for each group.
2.3. In Vivo Monitoring of PANP-labeled hESC-CM
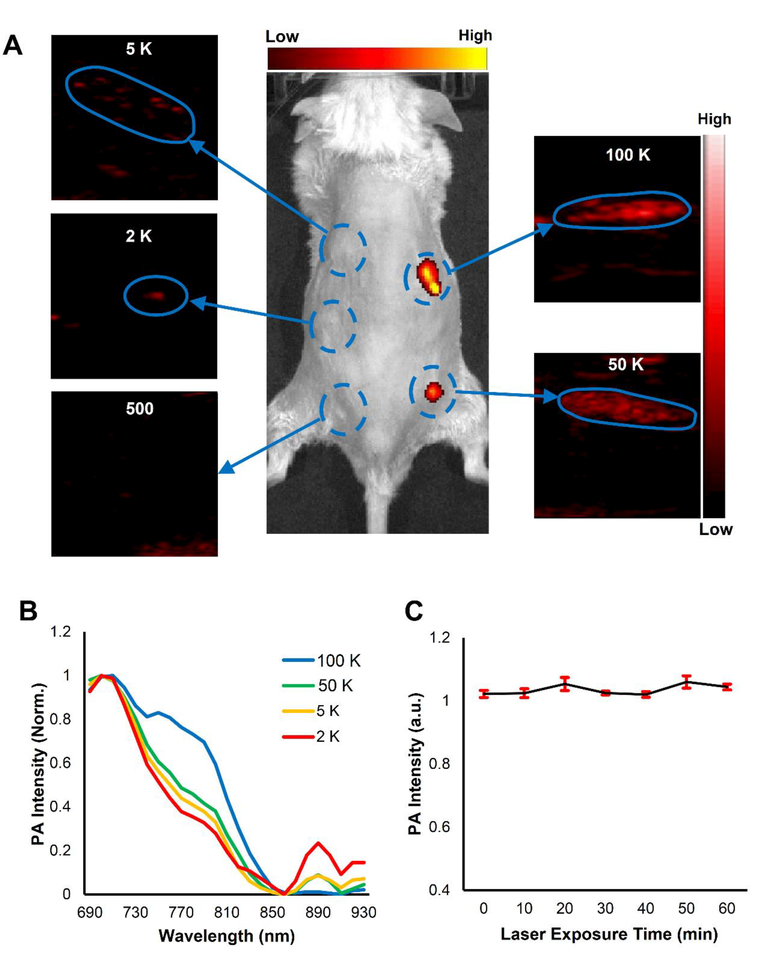
Notably, we also evaluated the detection limit of PANP-labeled cells by PAI in vivo. Five groups with different numbers of PANP-labeled hESC-CMs were mixed with 10 μL Matrigel. Following subcutaneous injections, these cells were imaged by FI and PAI in vivo. The results from Figure 4A showed that the FI was significantly less sensitive for the detection of injected cells when their numbers were less than 50,000. By contrast, PAI was able to detect the cell clusters with a minimal cell number of 2,000 (Figure 4A), comparable to a recent report [12i]. The high sensitivity of PAI was mainly due to the strong PA signal and specific PA spectrum of PANPs. For instance, Figure 4B indicated that the in vivo PA spectra did not differ with different number of PANP-labeled cells, enabling us to accurately identify the injected cells from their surrounding host tissues through PAI. Moreover, PA signals from PANPs were consistently robust without obvious photobleaching even after repetitive laser exposure for up to 1 hour (Figure 4C).
Figure 4. PANPs enabled PAI to detect a small number of labeled cells in vivo.
Five groups with different numbers of PANP-labeled hESC-CMs (500/2,000/5,000/50,000 /100,000) were mixed with 10 μL Matrigel, respectively, and subcutaneously injected into mice. (A) While fluorescent imaging only detected the cell numbers of 50,000 or more, PAI was able to detect the cell numbers as few as 2,000 by virtue of the strong PA signals and specific PA spectrum of PANPs. (B) Different numbers of PANP-labeled cells showed a similar PA spectral pattern in vivo, which was a notable feature to distinguish these cells from background tissues. (C) The excited PA signals from PANPs were quite stable even under a continuous PAI for up to 1 hour.
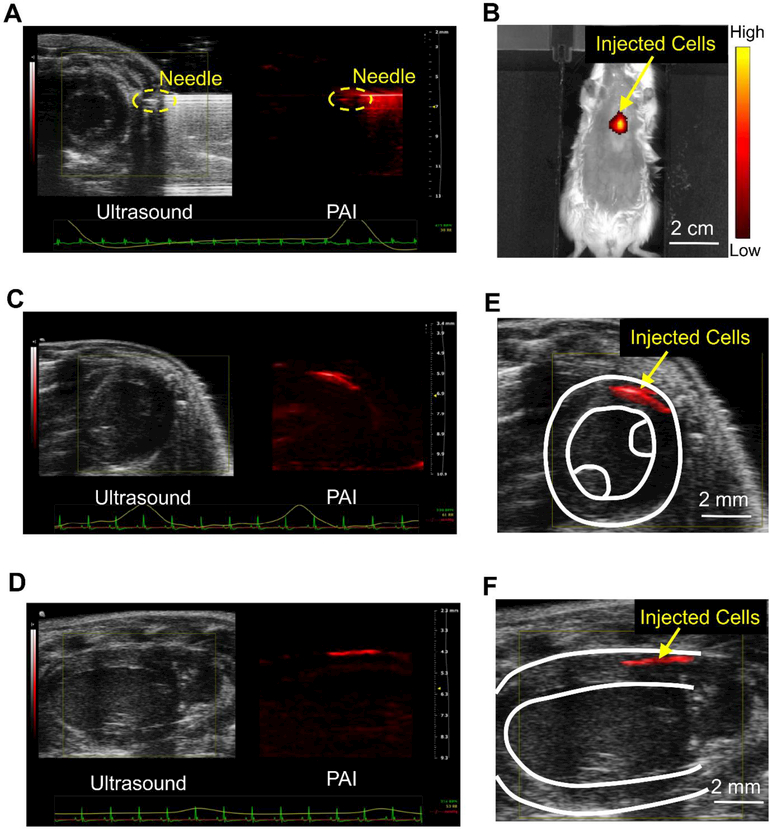
The specific PA spectrum of PANPs enables PAI to track the transplanted hESC-CMs in mouse heart at high resolution. To demonstrate this, we next intramyocardially injected the PANP-labeled hESC-CMs into mouse hearts under a real-time guidance of B-mode ultrasound and PAI. This injection was assisted by a previously reported engineered hydrogel to enhance cell retention [18]. Figure 5A shows an ultrasound image of cardiac structures in a short-axis view, in which the injecting needle is difficult to be differentiated from the ultrasound image due to its duplicated artifacts and similar intensity to the tissues. By contrast, the needle is clearly identified from the PA image because the surrounding tissues have much lower PA signals. Hence, the integration of PAI and ultrasound imaging provided improved guidance for cell injection by positioning both cardiac structures and needle locations. Post-cell delivery, PAI and ultrasound imaging were also performed to visualize the engraftment of transplanted cells in vivo. Using an electrocardiogram (ECG) and respiratory coupling, B-mode ultrasound images and multi-spectral PA images were acquired from short-axis (Figure 5C) and long-axis views (Figure 5E). Based on the specific spectrum of PANPs, the transplanted hESC-CMs were identified from PA images by an unmixing approach with the linear mixing model [19], whereas the host myocardium was outlined from the B-mode ultrasound images. Their merged images in Figure 5D and 5F showed the 3D spatial relationship between the transplanted cells and the host myocardium at a high resolution (~100 μm). In addition, the PANP-labeled cells also emitted a near-infrared fluorescence upon laser excitation (peak at 820 nm), which facilitated FI to further confirm the cell transplantation in vivo (Figure 5B). However, compared to PAI, FI was unable to provide accurate spatial information of the injected cells due to its low spatial resolution and limited depth information. This drawback rendered FI unable to confirm failed cell injections, whereas PAI successfully detected the leaked cells (Figure S6 in the Supporting Information).
Figure 5. PAI of the injection and engraftment of the PANP-labeled hESC-CMs in mouse hearts in vivo.
(A) Labeled hESC-CMs were intramyocardially injected into a mouse heart with an engineered hydrogel under a real-time guidance with PAI and B-mode ultrasound. The ultrasound image provides cardiac structures in a short-axis view and the PA image provides needle position simultaneously. (B) The transplanted cells were imaged by fluorescence imaging (FI) in vivo using the emitted near-infrared fluorescence (peak 820 nm) from PANPs. (C-D) Using an electrocardiogram (ECG) and respiratory coupling, B-mode ultrasound and multi-spectral PA imaging were performed to image the engraftment of the PANP-labeled hESC-CMs from short-axis and long-axis views, respectively. (E-F) Merged ultrasound and PA images showed the 3D spatial relationship between the transplanted cells and the host myocardium at a high resolution (~100 μm).
2.4. Ex Vitro Analysis
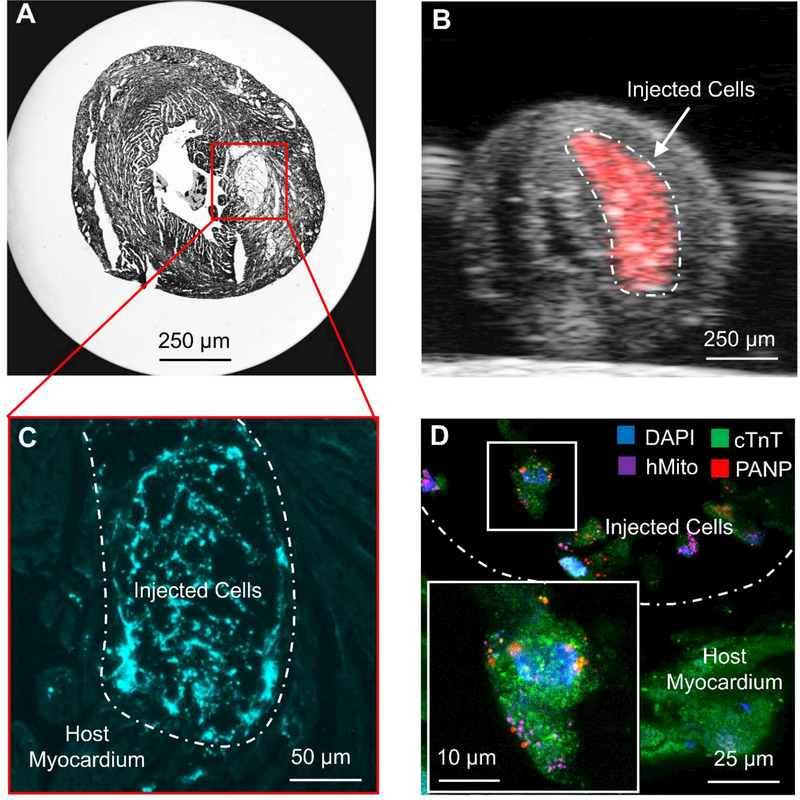
In addition to in vivo imaging, the PANPs were also used to detect the engrafted hESC-CMs from the host myocardium ex vivo (Figure 6). Figure 6B demonstrates that PAI enables detection of PANP-labeled cells from the dissected and fixed heart. Following histological sectioning, FI facilitates direct discrimination of the engrafted hESC-CMs from the host myocardium through the internalized PANPs in cells without any immunostaining (Figure 6C). This engraftment was confirmed by a follow-up immunofluorescence staining with multiple markers as shown in Figure 6D. In this confocal image, DAPI (blue) and cTnT (green) are demonstrated in both host myocardium and engrafted cardiomyocytes, whereas human mitochondria antibody (purple) and PANP fluorescence (red) distinguish the engrafted cells from the host myocardium. Thus, these results highlighted another potential application of PANPs to identify the labeled cells in histological analysis ex vivo.
Figure 6. Imaging the engraftment of hESC-CMs ex vivo.
(A) A sectioned slide of the heart under bright field microscopy (×2). (B) PAI detected the engrafted hESC-CMs from a fixed heart before sectioning ex vivo. (C) Fluorescent microscopy directly identified the engrafted cells (brighter blue) from the host myocardium without any immunostaining. (D) Confocal images of the engrafted cells and the host myocardium with immunostaining to distinguish human cells from mouse cells. DAPI (blue) and cTnT (green) marked both host and engrafted cardiomyocytes, whereas human mitochondria antibody (purple) and PANP fluorescence (red) distinguished the engrafted hESC-CMs from the host cells.
3. Discussion
This study reports an innovative cell tracking approach using PANPs as contrast agents. The PANPs consisting of an organic semiconductor core, a lipid shell, and cell penetration peptides were demonstrated to facilitate the PA imaging of transplanted hESC-CMs with high sensitivity. Specifically, the in vivo results highlighted the advantage of encapsulating SPs as PA contrast agents for hESC-CM imaging. SPs is a class of new-generation PA contrast agents that can generate higher PA signals compared to routine PA nanoagents on a per mass basis. They also have a specific PA spectrum that enables PAI to detect a trace number of labeled cells (~2,000) following a subcutaneous injection. These advantages of SPs allowed us for the first time to perform the PAI of transplanted hESC-CMs in living mouse hearts. To our knowledge, this is also the first study to assess the impact of SP-generated NPs on the functions and behaviors of hESC-CMs.
Another advantage of PANPs is the high labeling efficiency for hESC-CMs. Because hESC-CMs are a terminally differentiated cell type with limited endocytosis activity, they are difficult to label by typical NP-labeling approaches. Thus, a previously reported approach was to use an indirect way to label hESC-CMs through delivering NPs into stem cells first before differentiating them into cardiomyocytes [14]. Besides being time-consuming, this approach was suboptimal because of NP dilution from cell proliferation and/or cell death during differentiation. By contrast, our PANPs with CPPs can be directly introduced into hESC-CMs through simple incubation and had minimal effects on the cell proliferation and excytosis. Its high labeling efficiency further helps the sensitive detection of labeled cells.
The PANPs showed no negative effects on the labeled cells. We evaluated the cell morphology, functions, and gene expression of the labeled hESC-CMs in vitro. Confocal images showed that the cytoskeleton of hESC-CMs was not affected by the loaded PANPs. Calcium imaging showed similar calcium handling patterns between labeled and control cell groups. Similarly, contractility measurement also indicated negligible differences in contractile function between both groups. In addition, compared to control cells, PANP-labeled hESC-CMs had minimal alterations of their gene expression. Most notably, the key genes related to cardiac structural proteins (MYH7, MYL7, and TNNT2) and cardiac stress markers (NPPA) were unaffected. Interestingly, we found that gap junction protein alpha 5 (GJA5) was up-regulated after PANP labeling, which was consistent with a recent report [20]. GJA5 is part of the cardiac gap junction proteins that are involved in cell-cell interactions. The up-regulation of GJA5 may increase the engraftment efficiency, which will be pursued in future studies. Similarly, we found IL-6 and IL-8 were up-regulated after PANP labeling as previously reported[10a]. Finally, the PANPs were synthesized with organic materials and did not cause any toxicity to liver function.
The PANPs enabled us to track the labeled hESC-CMs in mouse hearts in vivo and ex vivo. We first performed PAI and ultrasound imaging to image PANP-labeled hESC-CMs in mouse hearts in vivo. With cardiorespiratory coupling, this dual-imaging method provided precise 3D spatial information of both host myocardium and hESC-CMs to accurately assess the delivery and engraftment of these cells. Compared to ultrasound, PAI detected a specific and high-contrast spectrum from PANP-labeled cells, which can sensitively identify the transplanted cells from the host tissues. Compared to FI, PAI provided a higher spatial resolution as well as depth information, which enabled the detection of the failed cell injections that were undetectable for FI. In addition to in vivo imaging, the PANPs were also used to assess the engraftment of transplanted cells ex vivo. Taking advantage of the intense fluorescence of SPs in near-infrared region (emission peak 820 nm), the labeled cells were directly marked by PANPs in histological slides (~10 μm thickness) without immunostaining.
This study has several limitations. Like most NP-based labeling methods [21], currently the PANPs can only monitor the cell delivery and engraftment rather than more comprehensive cell fates (e.g., apoptosis). Thus, future work will focus on broadening its application by incorporating PANPs with other molecules to create “smart” nanoplatforms that are sensitive to cell fate outcomes such as apoptosis [22]. Another limitation arises from the imaging modality. Although PAI can detect PANP-labeled cells, it is less sensitive in imaging the host myocardium. However, ultrasound imaging can be used as a complementary modality to provide details on specific myocardial structures [23].
4. Conclusion
In summary, we developed SP-based PANPs to image transplanted hESC-CMs in living mouse hearts. The strong and stable PA signals as well as specific PA spectrum of SPs enabled a sensitive detection of as few as 2,000 labeled cells. The PANPs also provided a facile and efficient way to directly label hESC-CMs while avoiding the problem of NP dilution caused by stem cell proliferation and death in previous labeling approaches. In vitro assessments showed that PANP-based labeling had no adverse effects on hESC-CMs regarding cell structure, function, or gene expression. Moreover, the labeled PANPs allowed us to assess the delivery and engraftment of hESC-CMs in mouse hearts by photoacoustic imaging in vivo or by fluorescence imaging ex vivo. Collectively, these results suggest that PANPs together with photoacoustic imaging provide a convenient and sensitive imaging technique to facilitate and advance the field of cardiac regenerative therapy.
5. Experimental Section
Materials:
Poly[2,6-(4,4-bis-(2-ethylhexyl)-4H-cyclopenta,[2,l-b;3,4-b’]dithiophene)-alt-4,7(2,1,3-benzothiadia-zole)] (PCPDTBT) and tetrahydrofuran (THF) were purchased from Sigma-Aldrich. DSPE-PEG-Maleimide (Mw = 3,400) is a commercial product from Laysan Bio, Inc. Cell penetrating peptide (CCP), RKKRRQRRRC, was customized by GenicBio, China. Qiagen miRNeasy Mini Kit was purchased from Qiagen Sciences, Inc., USA. The optimal cutting temperature (OCT) embedding compound was from Tissue-Tek, Sakura Finetek, USA. The anti-human mitochondria antibody (clone 113–1, Alexa Fluor® 488) was purchased from EMD Millipore, USA. The anti-cardiac troponin T antibody (ab45932) was purchased from Abcam, USA. The donkey anti-rabbit IgG (H+L) secondary antibody (Alexa Fluor® 488) was purchased from Thermo Fisher Scientific, USA. Matrigel was purchased from Corning, USA. The engineered hydrogel was synthesized and purified as previously described [18,24]
Synthesis of Photoacoustic Nanoparticles (PANPs):
Semiconducting polymer (PCPDTBT, 1 mg) and DSPE-PEG-Maleimide (Mw = 3,400, 5 mg) were dissolved in 1 mL of tetrahydrofuran (THF) to form a homogeneous solution. The organic solution was quickly injected in 9 mL of deionized (DI) water, followed by continuous sonification using a probe sonicator at 50 W output for 90 seconds (Misonix Incorporated, NY). The obtained suspension was then washed with DI water for 5 times using an Amicon Ultra-15 Centrifugal Filter Unit to eliminate THF and then suspended in 5 mL of 1× PBS buffer. CCP (10 μmol) was mixed with the suspension for reaction overnight under gentle stirring at room temperature. The solution was then dialyzed against DI water for 3 days to eliminate the excess peptide and the final product (PANPs) was concentrated using Amicon Ultra-15 Centrifugal Filter Unit to 1 mL stock solution for further use.
Estimation of PANP Concentration:
We can first estimate the average volume of each PANP based on the average size of PANPs in water (~48.6 nm). Considering the fact that the PANP water suspension is stable, the density of NPs in suspension should be close to that of water. With the assumption of the density of PANPs is ~1 g/cm3, the concentration of PANPs in stock suspension can be calculated from the following equations:
The number of PANPs in stock solution:
The concentration of PANPs in 1 mL of stock solution was then calculated as following:
Characterizations:
The ultraviolet-visible (UV-vis) spectrum of PANP solution was recorded on a Shimadzu UV-1700 spectrometer. The fluorescence spectrum was measured using a fluorometer (LS-55, Perkin Elmer, USA). The average particle size and size distribution were determined by a Zetasizer Nano ZS equipment at room temperature. The morphology of PANPs was studied by a FEI Tecnai G2 F20 X-TWIN Transmission Electron Microscope.
Agarose Phantom for PA Spectrum:
The 2% (w/v%) Agarose gel was prepared through melting 8 g of agarose powder into 400 mL of DI water in the casting tray. PANP solution was injected into the prepared agarose phantom and the PA signals were recorded by a Vevo LAZR imaging system in Nanostepper mode with alternating acquisition from 680 nm to 970 nm. It is equipped with a MS-550D linear array transducer (40 MHz, 70%−6 dB two-way bandwidth, 256 elements) to detect US and PA signals, and a tunable Nd:YAG laser system (OPOTEK Inc., Carlsbad, CA, 680–950 nm, 20 Hz repetition rate, 5 ns pulse width, 50 mJ pulse peak energy) was used to trigger the system acquisition, and excite the samples with optical pulses to generate the PA effect. PA signal intensities (average pixel intensities) were measured using region of interest (ROI) manager tool in the Vevo LAZR imaging system software to acquire PA spectra. The PA spectra were calibrated with PA signal intensities of graphite.
hESC Culture, Differentiation and Labeling:
Human embryonic stem cell line WA07 (H7) and cardiomyocyte differentiation using small molecules were based on protocols described in previous studies [25]. Before labeling, hESC-CMs were dissociated and re-seeded into a new Matrigel-coated plate to form a single monolayer of cells. To label cells, PANPs diluted in cell culture media with a final concentration of 4 nM were added directly to hESC-CMs for incubation at 37°C. After 6 hours or overnight, the labeling efficiency was measured by both Flow Cytometry (BD FACSAria II, BD Biosciences, San Jose, CA) and a Leica SP8 White Light Confocal (Leica, Wetzlar, Germany).
Cell Viability:
5 × 103 of hESC-CMs were seeded in a 96-well plate. After 48 hours of recovery, 100 μL of culture medium with PANP at concentrations of 0, 2, 4, 8 nM in each well were used for incubation with hESC-CMs for 24 hours, then fresh culture medium was changed for another 24 hours. The terazolium reduction based cell viability assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS), Promega) was applied to evaluate the cytotoxicity on the hESC-CMs with varied PANP labeling concentrations. In brief, 100 μL of cell culture medium and 20 μL of MTS solution were pre-mixed, then added to each well of hESC-CMs. After 1 hour of incubation in the cell culture incubator at 37 °C and 5% CO2, the intensities of absorbance at 490 nm were record by a plate reader (Biotek). The final intensity was obtained with background subtraction, and each group had its own background based on the absorbance of culture medium with NP treated hESC-CMs at the corresponding concentration without adding MTS solution. The higher intensity of absorbance suggests better cell viability. Each group at varied PANP concentrations had 12 replicates (n = 12).
PANP Uptake Assay:
5 × 103 of hESC-CMs were seeded in a 96-well plate. After 48 hours recovery of hESC-CMs, 100 μL of culture medium with PANP at labeling concentrations of 0, 1, 2, 4, 8 nM in each well were used to label hESC-CMs. After 24 hours, the supernatant from each well was collected. Then the absorbance of PANP solutions at 670 nm was measured by a plate reader (Biotek). The percentage of PANP uptake at each PANP labeling concentration is then calculated: % uptake = (fresh culture medium with PANP - supernatant of culture medium at 24 hours) / fresh culture medium with PANP. Each group at varied PANP concentrations had 12 replicates (n = 8).
PANP Release Assay:
Following the collection of supernatant from each well after incubation with PANPs for 24 hours in the uptake assay, 100 μL of fresh medium was added to each well for further incubation of another 24 hours (assigned as supernatant at 48 hours). The supernatant from each well was then collected, and the absorbance at 670 nm was measured by a plate reader (Biotek). The percentage of PANP released from cells at each PANP labeling concentration is then calculated: % release = supernatant of culture medium at 48 hours / (fresh culture medium with PANP - supernatant of culture medium at 24 hours). Each group at varied PANP concentrations had 12 replicates (n = 8).
Cell Function Assessments:
The cardiac cell functions were assessed through (i) calcium imaging, (ii) contractility assay, and (iii) quantitative RT-PCR.
Prior to calcium imaging, both PANP-labeled and non-labeled hESC-CMs (control group) were replated in 8-well imaging chambers (Thermo Fisher Scientific Inc.) and recovered for 4 days. Then they were loaded with 5 μM of Fluo-4 AM in Tyrode’s solution at 37 °C for 5–10 min. Following a previously published protocol [26], the spontaneous calcium signals of hESC-CMs were sampled by confocal microscopy (Carl Zeiss, LSM 510 Meta, Göttingen, Germany) with a 63× oil immersed objective (Plan-Apochromat 63×/1.40 Oil DIC M27) and analyzed by a customized script using IDL (Interactive digital language).
Contractility of hESC-CMs was measured by a Sony SI8000 Live Cell Imaging System (Sony Biotechnology, San Jose, CA) that quantified cellular motion in a noninvasive label-free environment. Falcon 6-well plates seeded with both PANP-labeled and non-labeled beating hESC-CMs were first placed in the CO2-incubator of the system at 37 °C. Focus and light conditions of the phase contrast microscope were automated through the SI8000 software. When acquiring data, a high-speed charge-coupled device (CCD) video camera was used to capture cell activities at a high frame rate (up to 150 fps). Post image acquisition, displacements and magnitudes of cellular motions were calculated using a motion detection algorithm developed by Sony.
Total RNA was isolated using Qiagen RNeasy Mini Kit (QIAGEN, Germantown, MD) and reverse transcribed into cDNA using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Inc., Hercules, CA). cDNA template generated from 1 μg of total RNA was pre-amplified using TaqMan PreAmp Master Mix (Thermo Fisher Scientific Inc., Waltham, MA) based on protocol (PN 100–5876) from Fluidigm Corporation (South San Francisco, CA). Preamplified cDNA and Taqman Primer sets (Thermo Fisher Scientific Inc., Waltham, MA) were loaded into 48.48 Dynamic Array™ integrated fluidic circuit using IFC Controller MX (Fluidigm). Quantitative PCR was run using Biomark system (Fluidigm). Data were analyzed using Fluidigm Real-Time PCR Analysis software (Quality threshold: 0.65; Baseline correction method: Linear [Derivative]; Ct threshold method: Auto [Detectors]), normalized to 18S rRNA of each sample (Ctgene — Ct18S +, where is the average of Ct18S from all samples) and visualized using SINGULAR™ Analysis Toolset in R environment (3.1.1) (Limit of Detection or LoD: 24).
Animals:
All animal procedures were performed in accordance with the protocol of Stanford’s Administrative Panel on Laboratory Animal Care. Female NOD SCID mice (n = 3) aged 8 to 16 weeks old (Charles River Laboratories, Wilmington, MA) were used for the subcutaneous cell injection. Mice were anesthetized with 1–2% (v/v) isoflurane in oxygen, placed in supine position on an imaging paddle, and kept at 37 ± 0.4 °C via a heating system of the paddle. Hair removal creams were performed to clean hairs from the mouse backs. Different numbers of hESC-CMs were mixed with 10 μL of Matrigel, respectively. Then, each of them was subcutaneously injected into the mouse back via a 30-gauge needle at the tip of the syringe. Another five NOD SCID mice were used for intramyocardial cell injection. Mice were anesthetized with the same procedures. Hair removal creams were performed to clean the left side of mouse chests. A 10 μL of pellet containing 1 × 106 hESC-CMs was mixed with an equivalent volume of ice-cold hydrogel. The mixture was loaded into a 0.5 mL insulin syringe at room temperature and delivered via a 30-gauge needle at the tip of the syringe.
In Vivo Imaging:
Photoacoustic and ultrasound imaging was performed with a Vevo LAZR system (Visualsonics, Toronto, Canada). It used a 256-element transducer with broadband frequency (22 MHz-55 MHz) to receive tissue-generated high-resolution signals (axial resolution 44 μm; lateral resolution 140 μm), which enabled fast and real-time acquisitions. The excited laser was generated from a flash lamp pumped Q-switched Nd: YAG laser system with optical parametric oscillator (OPO) and second harmonic generator. Its electrical specifications included frequency at 20 Hz, wavelength between 680 and 970 nm with a step size of 2 nm, pulse duration about 4–6 ns, peak energy of 45±5 mJ, and spot size of 1 mm × 24 mm. Its imaging parameters were dynamic range of 70 dB, signal-to-noise-ratio of 20±10 dB, and data acquisition time of 0.2 seconds. PAI and ultrasound imaging system was also integrated with an advanced physiological monitoring unit to record physiological traces, including electrocardiogram and respiration signals. PAI was performed with 40 MHz frequency, 100% power, 40 dB gain, and multi-spectrum from 680 nm to 925 nm with a step size of 5 nm. Using the recorded cardio-respiratory coupling, the spectral images of the mice heart at the same cardiac phase were reconstructed. Based on the recorded reference spectrums of PANPs and current image intensities, the transplanted cells were identified from the image series using an in-house developed unmixing method with Matlab (Mathworks, MA). The IVIS Spectrum in vivo imaging system (PerkinElmer, Waltham, MA) was used to perform high-sensitivity in vivo fluorescence imaging. It was equipped with 10 narrow band excitation filters (30 nm bandwidth) and 18 narrow band emission filters (20 nm bandwidth) to reduce autofluorescence. It had a 23-cm field of view and multiple filters with a wavelength ranging from 430 nm to 850 nm. This system also supported spectral unmixing applications. The excitation filter of 675 nm and the emission filter of 820 nm were selected for PANP imaging.
Histological Analysis:
Dissected mouse hearts were immediately fixed in 4% paraformaldehyde (PFA) for 24 hours and then kept in 30% sucrose solution for 48 hours. Following fixation, the fixed hearts were embedded in OCT and kept in −80 °C freezer. Under the temperature of −20 °C to −30 °C, the frozen hearts were serially sliced with a thickness of 10 μm. The sections were stained for human mitochondria (Alexa Fluor® 488 conjugated, EMD Millipore, USA), cardiac troponin T (Abcam), and DAPI (ProLong® gold antifade mountant, Thermo Fisher Scientific) following previously published procedures [2d]. Immunofluorescence was analyzed with a Leica SP8 White Light Laser Confocal Microscope (Leica, Wetzlar, Germany) and a Revolve Microscope (Echo laboratories, San Diego, CA).
Liver Function Analysis:
Mice (n = 3) was injected with 50 μL of diluted PANPs solution (4 mmol/mL) via intravenous injection. Equivalent volume of PBS was also injected into the control group (n = 3). One week post-injection, 1 mL of blood was collected from each mouse following euthanasia. These blood samples were processed to quantify the amounts of biomarkers for liver functions, including AST, ALT, alkaline phosphatase, total bilirubin, total protein, and albumin.
Statistical Analysis:
Results are presented as mean ± SD. Student’s t-tests were used to compare the PANP uptake, exocytosis, toxicity, as well as cellular functions including calcium handling and contractility between PANP-labeled and control cells. One-way analysis of variance (ANOVA) test of normalized Ct value was used for the comparison of gene expression changes in PANP-labeled and control cells. A Student’s t-test was used to compare the liver functions of mice in PANP-treated and PBS-treated groups. The statistical analysis was performed using SPSS Statistics software (IBM, Armonk, NY). A value of P < 0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Stanford Center for Innovation and In Vivo Imaging (NIH S10 RR026917), Stanford Veterinary Service Center, Stanford Cell Sciences Imaging Facility, Stanford Neuroscience Microscopy Service, and FACS Core of Stanford Institute for Stem Cell Biology for their infrastructure supports. This work was supported by National Institutes of Health (NIH) R01 HL132875, R01 HL133272, T32 EB009035 (J.C.W.), PCBC HL099997 (X.Q., H.C., H. Wu), and R21 EB020235 (S.C.H.); California Institute of Regenerative Medicine (CIRM) RT3–07798 and DR2A-05391 (J.C.W.) and RT3–07948 (S.C.H.); Stanford TRAM Program (X.Q.); Kodak Fellowship (H. Wang); Agency for Science, Technology and Research (A*STAR, Singapore) (K.L.); and National Natural Science Foundation of China (NSFC) 31571011 and 51622305 (D.D).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Xulei Qin, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Dr. Haodong Chen, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Dr. Huaxiao Yang, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Dr. Haodi Wu, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Dr. Xin Zhao, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Dr. Huiyuan Wang, Department of Materials Science and Engineering, Stanford, CA, 94305, United States
Mr. Tony Chour, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Dr. Evgenios Neofytou, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States
Prof. Dan Ding, State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, China, 300071
Prof. Heike Daldrup-Link, Department of Radiology, Stanford, CA, 94305, United States
Prof. Sarah C. Heilshorn, Department of Materials Science and Engineering, Stanford, CA, 94305, United States
Dr. Kai Li, Department of Radiology, Stanford, CA, 94305, United States; Institute of Materials Science and Engineering, A*STAR, Singapore, 138634.
Prof. Joseph C. Wu, Stanford Cardiovascular Institute, Stanford, CA, 94305, United States; Department of Medicine, Division of Cardiology, Stanford, CA, 94305, United States..
References
- [1].Moran AE, Forouzanfar MH, Roth GA, Mensah GA, Ezzati M, Flaxman A, Murray CJ, Naghavi M, Circulation 2014, 129, 1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE, Nat. Biotechnol 2007, 25, 1015. [DOI] [PubMed] [Google Scholar]; b) Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ, Gantz JA, Fugate JA, Muskheli V, Gough GM, Vogel KW, Astley CA, Hotchkiss CE, Baldessari A, Pabon L, Reinecke H, Gill EA, Nelson V, Kiem HP, Laflamme MA, Murry CE, Nature 2014, 510, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ong SG, Huber BC, Hee Lee W, Kodo K, Ebert AD, Ma Y, Nguyen PK, Diecke S, Chen WY, Wu JC, Circulation 2015, 132, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Riegler J, Tiburcy M, Ebert A, Tzatzalos E, Raaz U, Abilez OJ, Shen Q, Kooreman NG, Neofytou E, Chen V, Wang M, Meyer T, Tsao PS, Connolly AJ, Couture LA, Gold JD, Zimmermann WH, Wu JC, Circ. Res 2015, 17, 720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nguyen PK, Neofytou E, Rhee JW, Wu JC, JAMA Cardiol. 2016, 1, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Nguyen PK, Lan F, Wang Y, Wu JC, Circ. Res 2011, 109, 962. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nguyen PK, Riegler J, Wu JC, Cell Stem Cell 2014, 14, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jokerst JV, Khademi C, Gambhir SS, Sci. Transl. Med 2013, 5, 177ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang LV, Hu S, Science 2012, 335, 1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Weber J, Beard PC, Bohndiek SE, Nat. Methods 2016, 13, 639. [DOI] [PubMed] [Google Scholar]
- [8].a) Akers WJ, Kim C, Berezin M, Guo K, Fuhrhop R, Lanza GM, Fischer GM, Daltrozzo E, Zumbusch A, Cai X, Wang LV, Achilefu S, ACS nano 2011, 5, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Fan QL, Cheng K, Yang Z, Zhang RP, Yang M, Hu X, Ma XW, Bu LH, Lu XM, Xiong XX, Huang W, Zhao H, Cheng Z, Adv. Mater 2015, 27, 843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) De La Zerda A, Zavaleta C, Keren S, Vaithilingam S, Bodapati S, Liu Z, Levi J, Smith BR, Ma TJ, Oralkan O, Cheng Z, Chen XY, Dai HJ, Khuri-Yakub BT, Gambhir SS, Nat. Nanotechnol 2008, 3, 557. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim JW, Galanzha EI, Shashkov EV, Moon HM, Zharov VP, Nat. Nanotechnol 2009, 4, 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Jokerst JV, Thangaraj M, Kempen PJ, Sinclair R, Gambhir SS, ACS Nano 2012, 6, 5920. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nam SY, Ricles LM, Suggs LJ, Emelianov SY, PloS one 2012, 7, e37267. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cook JR, Frey W, Emelianov S, ACS Nano 2013, 7, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Homan KA, Souza M, Truby R, Luke GP, Green C, Vreeland E, Emelianov S, ACS Nano 2012, 6, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Comenge J, Fragueiro O, Sharkey J, Taylor A, Held M, Burton NC, Park BK, Wilm B, Murray P, Brust M, Levy R, ACS Nano 2016, 10, 7106. [DOI] [PubMed] [Google Scholar]; b) Pu KY, Shuhendler AJ, Jokerst JV, Mei JG, Gambhir SS, Bao ZN, Rao JH, Nat. Nanotechnol 2014, 9, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Li K, Liu B, Chem Soc Rev 2014, 43, 6570. [DOI] [PubMed] [Google Scholar]; b) Liu J, Geng JL, Liao LD, Thakor N, Gao XH, Liu B, Polym Chem-Uk 2014, 5, 2854 [Google Scholar]; c) Pu KY, Shuhendler AJ, Valta MP, Cui LN, Saar M, Peehl DM, Rao JH, Adv. Healthc. Mater 2014, 3, 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lyu Y, Pu K, Adv Sci (Weinh) 2017, 4, 1600481. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Lyu Y, Zhen X, Miao Y, Pu K, ACS Nano 2017, 11, 358. [DOI] [PubMed] [Google Scholar]; f) Xie C, Zhen X, Lei QL, Ni R, Pu KY, Adv Funct Mater 2017, 27 [Google Scholar]; g) Jiang YY, Pu KY, SMALL 2017, 13. [DOI] [PubMed] [Google Scholar]; h) Jiang YY, Upputuri PK, Xie C, Lyu Y, Zhang LL, Xiong QH, Pramanik M, Pu KY, Nano Lett 2017, 17, 4964. [DOI] [PubMed] [Google Scholar]; i) Kim T, Lemaster JE, Chen F, Li J, Jokerst JV, ACS Nano 2017, DOI: 10.1021/acsnano.7b03519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pu K, Mei J, Jokerst JV, Hong G, Antaris AL, Chattopadhyay N, Shuhendler AJ, Kurosawa T, Zhou Y, Gambhir SS, Bao Z, Rao J, Adv. Mater 2015, 27, 5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Castaneda RT, Boddington S, Henning TD, Wendland M, Mandrussow L, Liu S, Daldrup-Link H, Pediatric. Radiology 2011, 41, 1384. [DOI] [PubMed] [Google Scholar]
- [15].a) Mahmoudi M, Hosseinkhani H, Hosseinkhani M, Boutry S, Simchi A, Journeay WS, Subramani K, Laurent S, Chem. Rev 2011, 111, 253. [DOI] [PubMed] [Google Scholar]; b) Hachani R, Lowdell M, Birchall M, Thanh NTK, Nanoscale 2013, 5, 11362. [DOI] [PubMed] [Google Scholar]
- [16].Jin GR, Mao D, Cai PQ, Liu RR, Tomczak N, Liu J, Chen XD, Kong DL, Ding D, Liu B, Li K, Adv. Funct. Mater 2015, 25, 4263. [Google Scholar]
- [17].Zhao QL, Li K, Chen SJ, Qin AJ, Ding D, Zhang S, Liu Y, Liu B, Sun JZ, Tang B, J. Mater. Chem 2012, 22, 15128. [Google Scholar]
- [18].Wang H, Zhu D, Paul A, Cai L, Enejder A, Yang F, Heilshorn SC, Adv. Funct. Mater 2017, 27, 1605609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Keshava N, Mustard JF, IEEE. Signal. Proc. Mag 2002, 19, 44. [Google Scholar]
- [20].Han J, Kim B, Shin JY, Ryu S, Noh M, Woo J, Park JS, Lee Y, Lee N, Hyeon T, Choi D, Kim BS, ACS Nano 2015, 9, 2805. [DOI] [PubMed] [Google Scholar]
- [21].Naumova AV, Modo M, Moore A, Murry CE, Frank JA, Nat. Nanotechnol 2014, 32, 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].a) Kim K, Lee M, Park H, Kim JH, Kim S, Chung H, Choi K, Kim IS, Seong BL, Kwon IC, J. Am. Chem. Soc 2006, 128, 3490. [DOI] [PubMed] [Google Scholar]; b) Nejadnik H, Ye D, Lenkov OD, Donig JS, Martin JE, Castillo R, Derugin N, Sennino B, Rao J, Daldrup-Link H, ACS Nano 2015, 9, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].a) Piao Z, Ma T, Li J, Wiedmann MT, Huang S, Yu M, Kirk Shung K, Zhou Q, Kim CS, Chen Z, Appl. Phys. Lett 2015, 107, 083701. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xiang L, Wang B, Ji L, Jiang H, Sci. Rep 2013, 3, 1113. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wei CW, Nguyen TM, Xia J, Arnal B, Wong EY, Pelivanov IM, O’Donnell M, IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2015, 62, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cai L, Dewi RE, Heilshorn SC, Adv. Funct. Mater 2015, 25, 1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].a) Qin X, Riegler J, Tiburcy M, Zhao X, Chour T, Ndoye B, Nguyen M, Adams J, Ameen M, Denney TS Jr., Yang PC, Nguyen P, Zimmermann WH, Wu JC, Circ. Cardiovasc. Imaging 2016, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chen VC, Couture SM, Ye J, Lin Z, Hua G, Huang HI, Wu J, Hsu D, Carpenter MK, Couture LA, Stem Cell Res. 2012, 8, 388. [DOI] [PubMed] [Google Scholar]
- [26].Wu H, Lee J, Vincent LG, Wang Q, Gu M, Lan F, Churko JM, Sallam KI, Matsa E, Sharma A, Gold JD, Engler AJ, Xiang YK, Bers DM, Wu JC, Cell Stem Cell 2015, 17, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.