Abstract
Hereditary tyrosinemia type 1 (HT1) is an autosomal recessive disorder caused by deficiency of fumarylacetoacetate hydrolase (FAH). It has been previously shown that ex vivo hepatocyte-directed gene therapy using an integrating lentiviral vector to replace the defective Fah gene can cure liver disease in small- and large-animal models of HT1. This study hypothesized that ex vivo hepatocyte-directed gene editing using CRISPR/Cas9 could be used to correct a mouse model of HT1, in which a single point mutation results in loss of FAH function. To achieve high transduction efficiencies of primary hepatocytes, this study utilized a lentiviral vector (LV) to deliver both the Streptococcus pyogenes Cas9 nuclease and target guide RNA (LV-Cas9) and an adeno-associated virus (AAV) vector to deliver a 1.2 kb homology template (AAV-HT). Cells were isolated from Fah−/− mice and cultured in the presence of LV and AAV vectors. Transduction of cells with LV-Cas9 induced significant indels at the target locus, and correction of the point mutation in Fah−/− cells ex vivo using AAV-HT was completely dependent on LV-Cas9. Next, hepatocytes transduced ex vivo by LV-Cas9 and AAV-HT were transplanted into syngeneic Fah−/− mice that had undergone a two-thirds partial hepatectomy or sham hepatectomy. Mice were cycled on/off the protective drug 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) to stimulate expansion of corrected cells. All transplanted mice became weight stable off NTBC. However, a significant improvement was observed in weight stability off NTBC in animals that received partial hepatectomy. After 6 months, mice were euthanized, and thorough biochemical and histological examinations were performed. Biochemical markers of liver injury were significantly improved over non-transplanted controls. Histological examination of mice revealed normal tissue architecture, while immunohistochemistry showed robust repopulation of recipient animals with FAH+ cells. In summary, this is the first report of ex vivo hepatocyte-directed gene repair using CRISPR/Cas9 to demonstrate curative therapy in an animal model of liver disease.
Keywords: : hepatocytes, CRISPR/Cas9, hereditary tyrosinemia type 1, metabolic liver disease, gene therapy
Introduction
Hereditary tyrosinemia type I (HT1) is an autosomal recessive inborn error of metabolism of the liver caused by a deficiency in fumarylacetoacetate hydrolase (FAH), the final enzyme in the catabolism of tyrosine. The buildup of the toxic metabolites fumarylacetoacetate and succinylacetoacetate in hepatocytes leads to oxidative stress, resulting in apoptotic cell death or disrupted gene expression.1 Affecting about 1/100,000 live births worldwide, HT1 is fatal if untreated.2 Without treatment, children present with cirrhosis, renal tubular injury, neurological crises, hepatocellular carcinoma, and, eventually, liver failure early in life.3,4 Liver transplant remains the only curative treatment but comes with its own difficulties, including organ shortage and the need for lifelong immunosuppression. The protective drug 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) ameliorates the disease by inhibiting an earlier enzyme in the tyrosine catabolism pathway and preventing the accumulation of toxic metabolites. When combined with a diet low in phenylalanine and tyrosine, NTBC can mitigate the need for liver transplant in 90% of cases.2 However, even responsive patients remain at increased risk for the development of hepatocellular carcinoma and neurological degeneration.5 The severity of these complications prompts the need for a new curative therapy beyond that of transplant.
Recent advances in gene therapy have made it possible to develop targeted treatments for a variety of disorders. Not only can gene addition therapies restore phenotype in diseases that lack a functional gene or protein product, but gene correction is becoming more precise and efficient.6 Using a guide RNA and a Cas9 protein to induce a double-stranded break at a specific genomic target sequence, it is now possible to use CRISPR/Cas9 to produce indels through a non-homologous end-joining (NHEJ) repair pathway or to introduce a homology sequence to be incorporated via homologous recombination.7 HT1 provides an excellent opportunity for gene therapy, as there is a strong selective advantage for corrected cells in vivo in the absence of NTBC.8 When targeting a particular organ or cell type, ex vivo protocols—wherein cells are collected, edited, and re-transplanted—further increase the correction efficacy and safety of gene editing.9 However, primary hepatocytes, the main cell type affected by HT1, are difficult cells to target using ex vivo gene therapy, as they are thought to be quiescent and have a limited life-span in vitro.10 Nevertheless, several studies have had success transducing hepatocytes ex vivo using self-inactivating lentiviral (LV) vectors, including previous work by the authors' group correcting a pig model of HT1 through an ex vivo LV gene addition protocol.11–14 LV vectors are strong candidates for gene therapy in hepatocytes, as they are known to transfect nondividing cells, have relatively safe integration profiles, and can carry large amounts of genetic information.15
The goal of this study was to develop a targeted and effective gene-editing treatment for HT1 by delivering both the Cas9 protein and a homology template to cells ex vivo prior to transplantation. Using a LV to deliver the Cas9 gRNA and protein (herein referenced as LV-Cas9), as well as an adeno-associated virus (AAV) to deliver the homology template (AAV-HT), it was possible to demonstrate a curative ex vivo gene-correction protocol in a murine model of HT1.
Materials and Methods
Plasmid and vector construction
Guides targeting the point mutation in the Fah gene were designed using software from Benchling.com using previously published algorithms for active guide design.16,17 Oligos containing BsmBI-clonable overhangs were ligated downstream of a U6 promoter in a LV vector co-expressing Cas9 (S. pyogenes) and green fluorescent protein (GFP; pL-CRISPR.EFS.GFP was a gift from Benjamin Ebert; Addgene plasmid # 57818).18 A control single-guide RNA targeted against the LacZ gene (ACCCGAGTGTATCTGGTCGC) was also cloned into a LV expressing Cas9 (LV-LacZ). LV-Cas9 was generated and harvested using a three-plasmid construct, as described previously.19 Vector titers, expressed in the manuscript as LV particles (LPs), were determined by p24 enzyme-linked immunosorbent assay (Clontech, Mountain View, CA).
A 1,192 base pair (bp) fragment of homology to the Fah exon 8 locus was synthesized into a gBLOCK (IDT, Coralville, IA) and cloned into an AAV vector that, after ligation, had a total length of 3,622 bp between the inverted terminal repeats. AAV-HT was created by standard triple plasmid transfection of HEK-293T cells20 using Fugene6 (Promega, Madison, WI). Cells were incubated for 72 h and collected, and the AAV particles were purified, aliquoted, and stored at −80°C. Vectors titers were determined by quantitative polymerase chain reaction (PCR) using Luna Universal qPCR Mix (NEB, Ipswich, MA) with the following primers: 5′-TTGCATATACGATACAAGGCTGTT, 5′-AAAACTGCAAACTACCCAAGAAA. Vector titers are expressed throughout as vector genome copies (GC).
Animals and animal care
All animals received humane care in compliance with the regulations of the institutional animal care and use committee at Mayo Clinic. Fah5981SB mice, which bear a single-point mutation at the exon 8 locus as a result of N-ethyl-N-nitrosourea-mediated mutation, were a generous gift from Dr. Markus Grompe (OHSU, Portland, OR). These mice are referred to as Fah−/− mice throughout this article. In order to encourage positive selection for FAH+ hepatocyte proliferation, NTBC was withheld from Fah−/− transplant recipient mice until they showed signs of weight decline, at which point it was administered in their drinking water at 8 mg/L for 5–7 days before it was withdrawn again.
Cell culture
In vitro experiments were conducted on Fah−/− mouse fibroblasts. Fibroblast cell lines were derived from neonatal Fah−/− tissue and were immortalized using a LV vector expressing the small and large SV40 T antigens. Fibroblasts were kept in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Waltham, MA) containing 10% heat-inactivated fetal bovine serum (FBS; Corning, Herndon, VA) and 1% penicillin/streptomycin (Corning). Cells were kept at 37°C and 5% CO2 and were passaged using 0.05% trypsin/EDTA as needed (Thermo Fisher Scientific).
PCR and T7 endonuclease analysis
Total cells from in vitro assays were collected and processed using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). For PCR amplification of uncorrected, disrupted sequence, the following two primers were used: 5′-GGAAAGGGCTCATGTGAGTAG, 5′-TCCATCCTTCCACTTATGAGAA. PCR conditions using Phusion polymerase (NEB) were as follows: 98°C for 3 min; 35 cycles of 98°C for 10 s, 60°C for 20 s, 72°C for 30 s; 72°C for 5 min. Generated PCR products were 452 bp in length. PCR products were processed using a T7 endonuclease Alt-R® Genome Editing Detection Kit (Integrated DNA Technologies, Coralville, IA), electrophoresed on a 2% TAE agarose gel, and visualized with ethidium bromide staining. For selective PCR amplification of corrected sequence, the following two primers were used: 5′-TGGAGCGGTAATGCCTCC, 5′-AAAATGCAGGATCCACCAAG. The first primer binds selectively to the modified protospacer adjacent motif (PAM) sequence in corrected DNA, while the second binds outside of the homology template; only DNA that has successfully integrated the AAV-HT will amplify. PCR conditions were as follows: 98°C for 3 min; 35 cycles of 98°C for 10 s, 61°C for 20 s, 72°C for 45 s; 72°C for 5 min. The expected product length is 859 bp. PCR products were electrophoresed on a 2% TAE agarose gel and visualized with ethidium bromide staining.
Tracking of Indels by DEcomposition and sequencing analysis
PCR products that successfully showed sequence disruption using a T7 endonuclease assay were sequenced using Sanger sequencing. Chromatogram results were analyzed using the free online tool, Tracking of Indels by DEcomposition (TIDE).21 This program uses decomposition algorithms in order to compare mixed pool and control chromatograms and to determine the probability of various indels in the experimental group. For next-generation sequencing (NGS) of amplicons, PCR purified products using the primers 5′-CCAACTTTCTCCATGGCAGG and 5′-ACCCCTGTAGTACTTAGGCC were submitted to GENEWIZ for NGS-Amplicon-EZ platform sequencing (GENEWIZ, South Plainfield, NJ).
Flow cytometry
Various AAV vector serotypes expressing GFP under control of the ubiquitous promoter cytomegalovirus (Vigene Biosciences, Rockville, MD) were transduced into primary mouse hepatocytes at a multiplicity of infection (MOI) of 50,000 GC. The medium was changed daily, and cells were analyzed for GFP expression by flow cytometry after 4 days. To dissociate cells into a single cell suspension, 0.05% Trypsin/EDTA was used. Cells were washed and fixed in 1% paraformaldehyde for 15 min prior to analysis on a FACSCalibur (BD Biosciences, San Jose, CA). Data were analyzed using FlowJo (Treestar, Ashland, OR).
Hepatocyte transplantation
Hepatocytes were harvested from an anesthetized donor mouse using a standard in situ perfusion of the liver using a mix of Collagenase NB (Serva, Heidelberg, Germany) and Thermolysin (Sigma–Aldrich, St. Louis, MO). Cell viability was determined via trypan blue exclusion assay. Cells were plated into six-well Primaria culture plates (BD Biosciences) containing the following media: DMEM, 10% FBS, 10 mM of HEPES, 10 μM of dexamethasone (Sigma–Aldrich), 7 mg/L of NTBC, 10 ng/mL of murine epidermal growth factor (Peprotech, Rocky Hill, NJ), and 1% penicillin/streptomycin. Fresh media were added 100 min later. LV-Cas9 was added at this time at the indicated MOI. AAV-HT was also added at this time at a MOI between 10,000 and 20,000, as indicated. Twenty-four hours post transduction, Hepatocytes were harvested with 0.05% trypsin/EDTA, centrifuged, and re-suspended for transplantation in 200 μL of media containing 2 μg/mL of DNaseI (Sigma–Aldrich). The cells were then injected into recipient mice via intra-splenic injection, as described previously.22 A two-thirds partial hepatectomy was performed on one cohort of mice, as previously described.23
Histology and biochemical analysis
For histological analysis, tissue samples were fixed in 10% neutral-buffered formalin (Protocol; Thermo Fisher Scientific, Pittsburgh, PA) and processed for paraffin embedding and sectioning. For hematoxylin and eosin (H&E) staining, slides were prepared with standard protocols. A blinded histopathological analysis was performed by a trained veterinarian pathologist. FAH immunohistochemistry using a polyclonal rabbit anti-FAH primary antibody24 was performed with a Bond III automatic stainer (Leica, Buffalo Grove, IL) with a 20 min antigen retrieval step using Bond Epitope Retrieval Solution 2 (Leica), and stained with diaminobenzidine (Leica). GFP immunohistochemistry using a monoclonal rabbit anti-GFP primary antibody (GFP #2956; Cell Signaling Technology, Danvers, MA) was also performed with a Bond III automatic stainer with a 20 min antigen retrieval step using Bond Epitope Retrieval Solution 2, and stained with diaminobenzidine. For biochemical analysis of alkaline phosphatase (ALP), alanine aminotransferase (ALT), aspartate aminotransferase (AST), albumin, and total bilirubin (TBIL), plasma was analyzed with the VetScan VS2 benchtop analyzer (Mammalian Liver Profile; Abaxis, Union City, CA) according to the manufacturer's instructions. Tyrosine values were determined using tandem mass spectrometry and chromatography via Mayo Clinic's internal biochemical PKU test. Quantification of FAH-positive cells was obtained using a cytoplasmic stain algorithm in Aperio ImageScope. Fifteen rectangular areas totaling 954,829.4 μm2 were randomly selected and analyzed. Reported results are total percentage of cytoplasmic FAH positivity among the cells selected.
Statistical analysis
Data were analyzed using GraphPad Prism v7 (GraphPad Software, Inc., San Diego, CA). Variances were determined by the F-test. Experimental groups with equal variances were compared using an unpaired two-tailed Student's t-test. Experimental groups with unequal variances were compared using a two-tailed Welch's t-test. Differences between multiple groups were compared using one-way analysis of variance followed by Tukey's multiple comparisons test. Due to the inability to keep untreated Fah−/− mice alive off of NTBC for the duration of the experiment, historical controls were used for the plasma data at time of sacrifice. A p-value of <0.05 was considered statistically significant.
Results
LV-Cas9.sgFah can robustly edit Fah−/− cells at the target locus
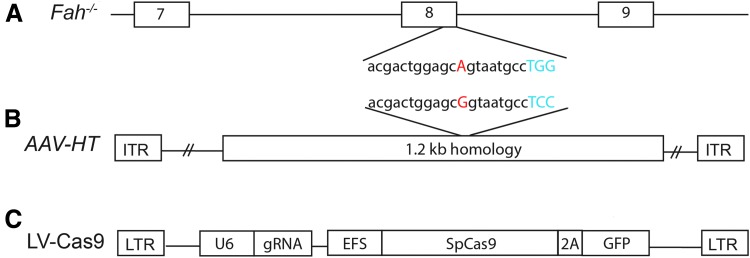
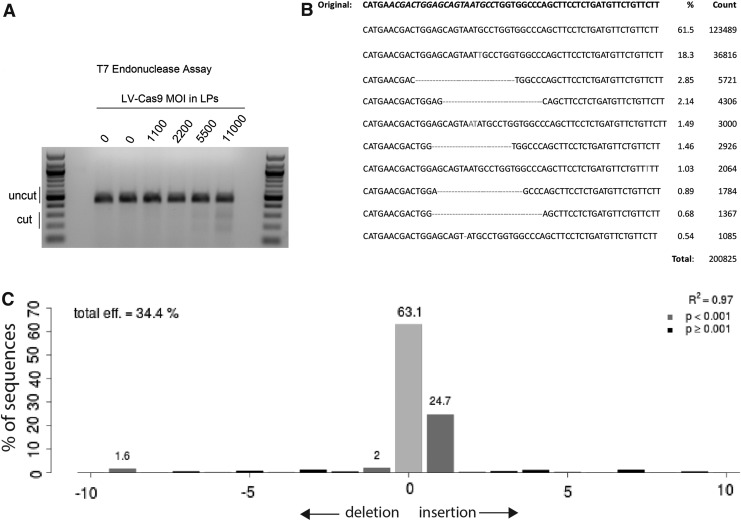
For this series of experiments, Fah−/− cells from a mouse model of HT1 were utilized. These Fah−/− mice carry a G → A point mutation in the last nucleotide of exon 8 (Fig. 1A), leading to a splicing defect and a truncated, unstable, and non-functioning FAH protein.25 A single LV vector was generated that expressed S. pyogenes Cas9 nuclease and a guide RNA targeted to the point mutation surrounding exon 8 of the Fah gene (LV-Cas9; Fig. 1C). Fibroblasts from Fah−/− mice were transduced to determine the efficiency and spectrum of mutations caused by CRISPR/Cas9 at the targeted locus using LV-Cas9. Generation of double-strand breaks was confirmed by T7 endonuclease analysis (Fig. 2A). As expected, a range of mutations were identified using both NGS of amplicons and TIDE analyses of Sanger sequencing results (Fig. 2B and C).
Figure 1.
Schematics of Fah gene and viral vectors used. (A) Fah−/− mouse gene locus. The hereditary tyrosinemia type 1 (HT1) mutation is on the last nucleotide of exon 8, as shown in red. The protospacer adjacent motif (PAM) sequence for the indicated guide RNA is highlighted in blue. (B) Adeno-associated virus (AAV) vector homology template with 1.2 kb homology fragment flanked by inverted terminal repeats. The corrected Fah gene sequence is shown in red, and the PAM sequence, which is modified to prevent re-cutting, is shown in blue. (C) The LV-Cas9 vector contains a single-guide RNA (gRNA) under the control of the U6 promoter. Cas9 from Streptococcus pyogenes is co-expressed with green fluorescent protein (GFP) under the control of the EFS promoter by means of a P2A cleavage site.
Figure 2.
LV-Cas9-mediated gene editing at the Fah−/− locus in fibroblasts. (A) A 452 base pair (bp) region around the HT1 single nucleotide polymorphism was amplified using polymerase chain reaction (PCR), re-annealed, and analyzed using a T7 endonuclease assay, which cleaves DNA at mismatched sequences. The lower bands in the gel indicated that LV-Cas9 did disrupt the Fah locus. (B) Gene disruption was confirmed by next-generation sequencing of amplicons. The top line is the non-modified mutant sequence, and the lines below indicate the frequency of each modified sequence at this locus. The guide sequence is italicized in the top line. (C) Representative Tracking of Indels by DEcomposition analysis of cells treated with LV-Cas9 at a multiplicity of infection of 8,000 lentiviral (LV) particles. The Cas9 efficiency in this sample was 34.4%, with the most frequent indel being a +1 insertion (24.7%).
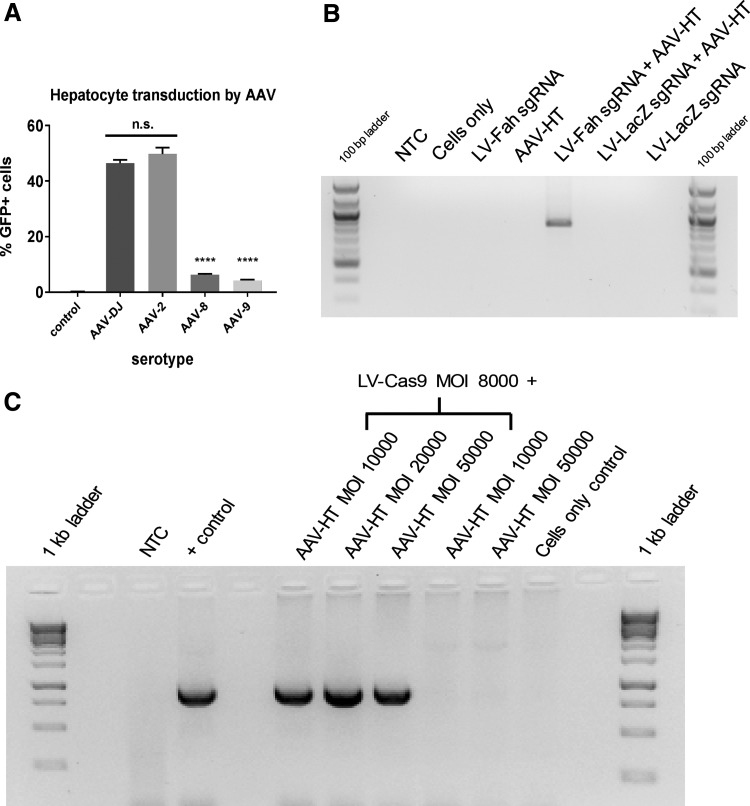
Correction of Fah using AAV is dependent on CRISPR/Cas9
Initially, a pilot experiment was performed in primary mouse hepatocytes to determine the efficiency of common AAV serotypes to transduce hepatocytes. Using a GFP vector, the serotypes 2 and DJ gave significantly better transduction than serotypes 8 and 9 (Fig. 3A). Based on these data, and previous use of the DJ serotype to perform gene editing in pig fibroblasts for somatic cell nuclear transfer,26 the DJ serotype was utilized for the remainder of the experiments. The AAV-targeting vector contained approximately 1.2 kb of homology that contained the correct G for proper Fah splicing, in addition to a modified sequence downstream of the corrected point mutation that would disrupt future cutting at the target site by modifying the PAM sequence (Fig. 1B). The hypothesis was tested that Cas9 expression is critical for gene correction at the Fah locus by comparing gene correction in the presence of AAV-HT alone or combined with LV-Cas9 (with a sgRNA Fah guide) or with LV-LacZ (a control sgRNA targeted against the LacZ gene; Fig. 3B). A positive PCR signal was only detected in cells receiving both LV-Cas9 against Fah and AAV-HT. Next, Fah−/− fibroblasts were transduced with various MOIs of AAV with or without LV-Cas9. After 72 h, PCR analysis using specific primers that can only amplify repaired Fah confirmed correction of the target site at various MOIs of AAV (Fig. 3C).
Figure 3.
Fah−/− gene correction using AAV is dependent on LV-Cas9. (A) Hepatocytes were transduced with different serotypes of AAV vector expressing GFP, and the percentage of cells transduced was measured by flow cytometry. AAV-2 and AAV-DJ yielded statistically significant more GFP+ cells compared to control, AAV-8, and AAV-9 (****p < 0.001) but not from each other. AAV-DJ was used for the duration of the experiments. (B) Fah−/− fibroblasts were transduced, as indicated with a combination of a LV vector carrying Cas9 and a guide for the Fah locus (LV-Fah sgRNA), a LV vector carrying Cas9 and a guide for the LacZ locus (LV-LacZ sgRNA), and AAV-HT for the Fah locus. Using PCR primers that selectively amplify corrected sequence, only cells that were transduced with both LV-Fah sgRNA and AAV-HT showed correction. The expected product size is 859 bp. (C) Fah−/− fibroblasts were transduced as indicated. Using the same PCR primers as (B), only cells transduced by LV-Cas9 and AAV-HT together resulted in gene correction.
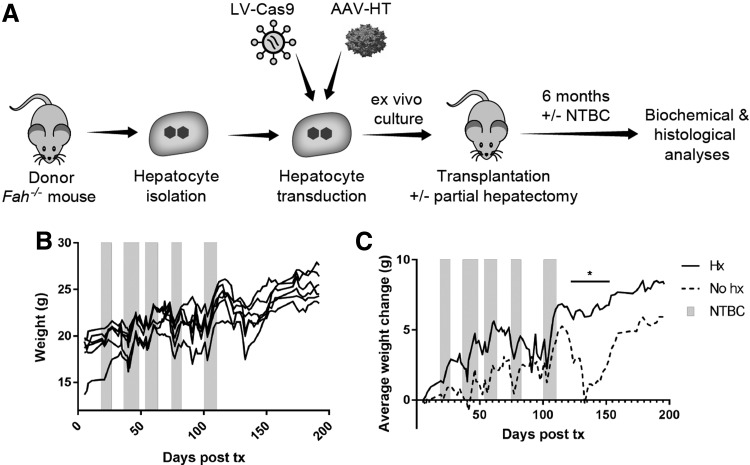
Transplantation of gene-edited hepatocytes can prevent liver failure in Fah−/− mice
Previous work with primary hepatocytes identified optimal conditions for high transduction with VSV-G-pseudotyped LV vectors.27 Using these culture conditions, hepatocytes from a donor Fah−/− mouse were isolated and cultured for <24 h in the presence of LV-Cas9 and AAV-HT. Hepatocytes were harvested and transplanted into syngeneic Fah−/− mice by intra-splenic injection. One group of recipient mice received a two-thirds partial hepatectomy immediately prior to transplantation to investigate whether regenerative cues would improve gene repair and proliferation in vivo (Fig. 4A). Recipient mice were cycled on/off NTBC to stimulate expansion of corrected hepatocytes in vivo. Interestingly, while both groups of animals became weight stable off NTBC (Fig. 4B), mice that received a partial hepatectomy became weight stable significantly sooner than animals that only received hepatocyte injections (Fig. 4C).
Figure 4.
Transplantation of ex vivo gene edited hepatocytes is curative. (A) Schematic depicting the ex vivo procedure. Fah−/− hepatocytes were isolated from a donor mouse, transduced with the LV-Cas9 and AAV-HT, cultured for <24 h, and transplanted into syngeneic Fah−/− mice via splenic injection. One cohort of mice (n = 3) received a partial hepatectomy at the time of transplant. A second cohort (n = 3) received splenic injection only. After 6 months of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC) cycling, biochemical and histological data were obtained. (B) Mice were cycled on and off NTBC (gray bars) following transplant until NTBC-independence was obtained. Weight data from all mice are shown. Mice were NTBC-independent for 92 days before sacrifice. (C) Change in average weights (n = 3 for each group) from day 5 post transplant indicates that mice that received a partial hepatectomy (Hx) became weight stable off NTBC more quickly than mice that did not receive a partial hepatectomy (No hx).
Transplantation of gene-edited hepatocytes can cure metabolic disease
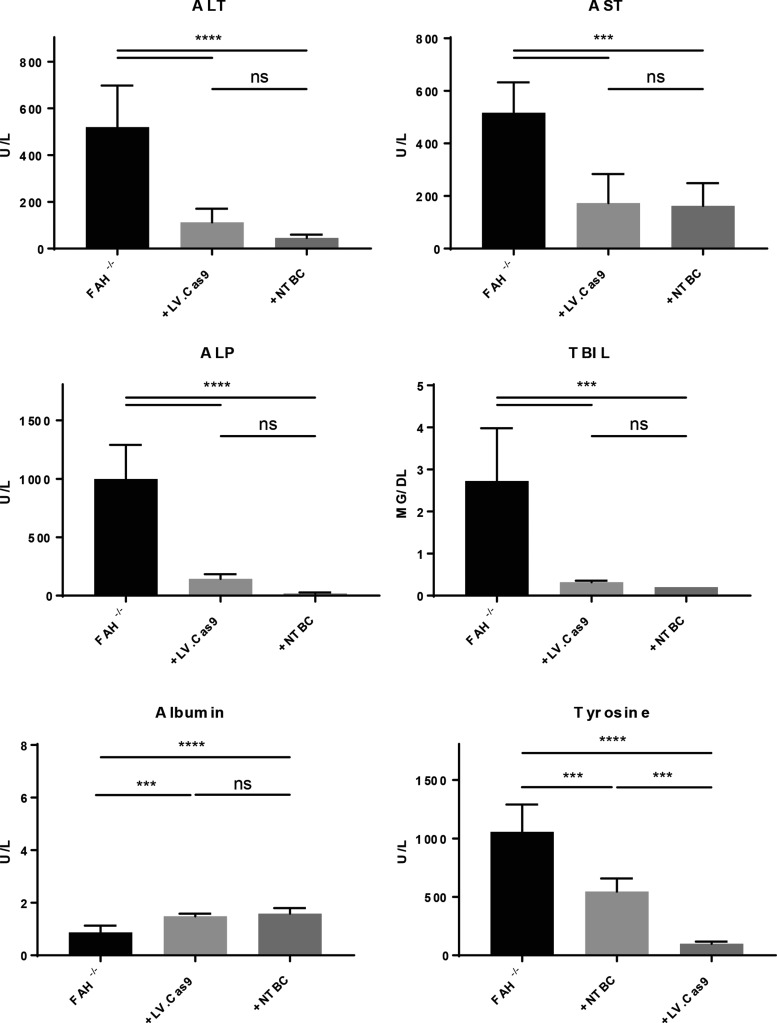
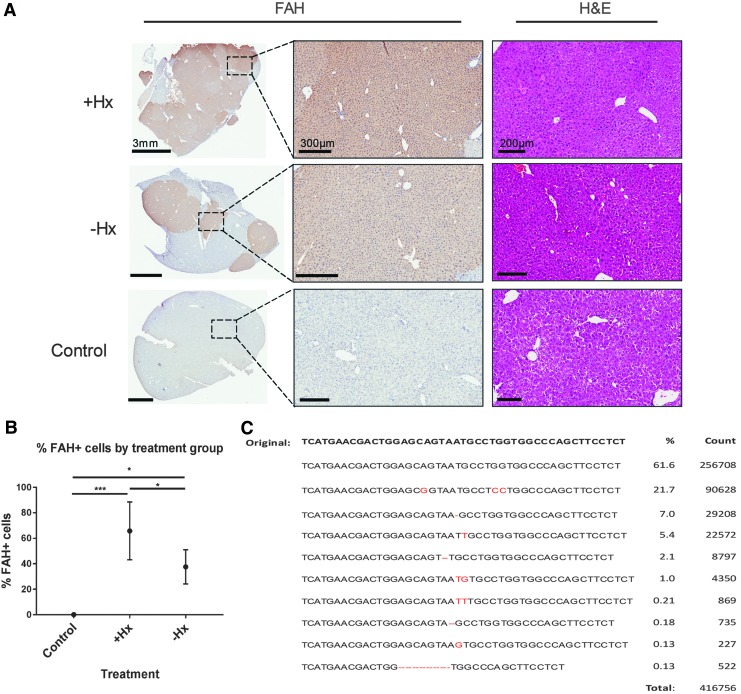
Six months after transplantation, all mice were euthanized, and a thorough analysis of biochemical and histological markers of disease was performed. As expected based on the weight data, all mice had significant reductions in biochemical makers of liver injury, including ALT, AST, ALP, and TBIL (Fig. 5). Albumin levels were also significantly higher in transplanted mice compared to controls. Specific to HT1, tyrosine levels in the blood were reduced to normal levels, indicating complete amelioration of aberrant tyrosine catabolism in these mice. Next, the histology of the livers was analyzed using FAH (Fig. 6A and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hum), GFP (co-expressed with Cas9 by the LV; Supplementary Fig. S2), and H&E (Fig. 6A). All mice displayed significant repopulation of the Fah−/− mouse liver with corrected hepatocytes, as indicated by FAH staining. In congruence with the weight data, mice that received partial hepatectomy had significantly increased numbers of FAH+ cells, as determined by digital quantification of FAH+ cells in transplanted livers (Fig. 6B). Finally, using NGS of amplicons, the molecular status of the corrected allele was characterized in all six mice (Fig. 6C). The mean percentage of corrected alleles in transplanted mice was calculated to be 21.7%. Based on the hepatocyte cell number in the liver and the natural polyploidy status of hepatocytes, these data correlate with the FAH histology showing robust hepatocyte expansion of corrected cells in vivo. Interestingly, a number of other mutations were also detected at high frequency at the edited locus, indicating the potential for CRISPR/Cas9 to cause unwanted changes in the genome when delivered by integrating vectors (Fig. 6C).
Figure 5.
Biochemical analyses confirmed amelioration of metabolic disease in transplanted mice. Plasma from the time of sacrifice (6 months post transplantation) in all transplanted mice (n = 6) was compared to plasma of Fah−/− untreated controls off NTBC (–NTBC; n = 5) and untreated Fah−/− mice on NTBC (+NTBC; n = 5). ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; TBIL, total bilirubin. ***p < 0.001; ****p < 0.0001.
Figure 6.
(A) Histology and immunohistochemistry slides from treated and untreated control mice. Low-magnification fumarylacetoacetate hydrolase (FAH) images are on the left; higher-magnification view of dashed box is on the right. (B) Average percentage of FAH+ cells in untreated controls (n = 3), +Hx mice (n = 3), and −Hx mice (n = 3). *p < 0.05; ***p < 0.0001. (C) Combined sequencing data from the time of autopsy detailing frequency of targeted and untargeted modifications at the Fah locus.
Discussion
This study aimed to genetically edit and phenotypically cure a mouse model of HT1 through the use of a viral vector–delivered Cas9 protein and homology template using an ex vivo protocol. After delivering the LV-Cas9 and the AAV-HT to purified hepatocytes and transplanting them via intra-splenic injection, recipient mice were cycled on and off NTBC to encourage proliferation of corrected cells until NTBC-independent weight gain was achieved. As shown by weight profiles, biochemical data, and histological imaging, the treatment was successful, demonstrating for the first time proof-of-concept for ex vivo hepatocyte-directed gene editing using CRISPR/Cas9.
Although allogeneic hepatocyte transplantation has demonstrated safety and partial efficacy in preclinical and clinical settings, its widespread use is severely limited by a shortage of donor hepatocytes and the immune response against the allogeneic cells.28,29 Ex vivo autologous hepatocyte-directed gene therapy using integrating vectors has been previously demonstrated in small- and large-animal models of metabolic liver disease. Initially utilizing gamma retroviral vectors to deliver and integrate the missing gene randomly into the host genome, a number of preclinical studies demonstrated efficacy of an ex vivo approach for metabolic liver diseases.30–33 These preliminary data were sufficient for a clinical trial to occur in five patients with familial hypercholesterolemia in which a gamma-retroviral vector containing the LDLR gene was delivered to cultured hepatocytes after resection, and transduced autologous cells were re-transplanted.34,35 While only some therapeutic efficacy was shown, this important study provided clinical proof-of-concept for future ex vivo hepatocyte-directed gene therapy approaches. With the advent of self-inactivating LV vectors, and their encouraging success in clinical trials with hematopoietic stem cells for treating primary immunodeficiencies,36,37 a number of preclinical studies have shown that ex vivo transduction of primary hepatocytes with LV vectors can ameliorate metabolic liver disease in rodent and pig models.38,39 Therefore, this group of studies provided the foundation to continue to pursue the development of ex vivo hepatocyte-directed CRISPR/Cas9-mediated gene therapy as an alternative to orthotopic liver transplantation for metabolic liver disease.
The advent of precision gene editing has changed the landscape for hepatocyte-directed gene therapy. A number of elegant studies have demonstrated the potential for in vivo delivery of endonucleases, including zinc finger nucleases and CRISPR/Cas9, combined with delivery of a repair template, to correct and cure disease in rodent models.40–43 While an in vivo approach to liver gene therapy is attractive for a number of reasons, ex vivo gene therapy provides many advantages for liver-directed gene therapy. First, the source of autologous cells is inherent in the therapy, and in addition to providing cells by means of liver resection, the procedure is strongly pro-regenerative,44 therefore providing a stimulus for transplanted cells to proliferate. Second, in contrast to an in vivo approach, this therapeutic method ensures specific targeting of the intended cell type—the hepatocyte—at much lower and safer vector doses than are needed with systemic administration of viral vectors. Third, although much more research is needed to make this feasible, ex vivo gene therapy opens up the possibility of correcting and expanding hepatocytes ex vivo, prior to re-transplantation into the patient.45
To the authors' knowledge, this study is the first to demonstrate the efficacy of combining CRISPR/Cas9-mediated gene editing and hepatocyte transplantation together to cure a metabolic liver disease. However, a number of challenges will need to be overcome prior to bringing this novel regenerative therapy to the clinic for the many metabolic liver diseases. First, the use of a LV vector to express the Cas9 and guide RNA would not be the optimal method to deliver these gene-editing components due to the risk of continued Cas9-mediated off-target gene editing. The present data demonstrate continued expression of GFP, which is co-expressed with Cas9, 6 months after transplantation. Therefore, future ex vivo gene-editing approaches should focus on the use of other non-integrating viral vectors (e.g., AAV) or, even preferably, the use of non-viral vectors for short-term expression of Cas9. Second, hepatocyte gene editing provides the formidable obstacle of cells not actively replicating and therefore predominantly repairing DNA damage of double-strand breaks by NHEJ. This obstacle has been previously highlighted with recent in vivo gene-editing approaches, with DNA repair efficiencies significantly lower in adult mice compared to neonatal mice.43 A partial hepatectomy was utilized as a clinically relevant method to improve gene editing in transplanted hepatocytes.29 Further work is needed to evaluate the therapeutic benefit of this approach in other models. Other methods that either stimulate hepatocyte replication or provide a selective advantage for transplanted hepatocyte proliferation46 could also be optimized for efficient gene editing in adult hepatocytes to occur. Third, while the Fah−/− mouse model provides an excellent proof-of-concept for gene-editing studies, the robustness and clinical relevance of ex vivo gene editing will need to be demonstrated in other models. The Fah−/− mouse model provides a massive repopulation advantage for FAH+ cells over host FAH– cells; previous studies have shown that as few as 1,000 FAH+ hepatocytes can correct metabolic deficiency in Fah−/− mice.8 Therefore, while other metabolic diseases may also provide some selective advantage for corrected cells (most notably alpha-1 antitrypsin deficiency47,48) improved gene-editing protocols will need to be optimized for this novel therapeutic strategy to be applied to other metabolic diseases where no apparent repopulation advantage is present. Finally, future work is needed to evaluate the ability of AAV to mediate robust gene editing in the absence of CRISPR/Cas9. The molecular status of cells ex vivo (Fig. 3B) and after 6 months in vivo (Fig. 6C) indicated significant indels at the Fah locus produced by CRISPR/Cas9. Further studies are needed to evaluate the on-target as well as off-target effects of CRISPR/Cas9, particularly in the context of an integrating vector.
In summary, the present data provide proof-of-concept for the application of hepatocyte-directed CRISPR/Cas9-mediated gene editing to metabolic liver diseases. The results demonstrated curative gene and cell therapy in a mouse model of HT1. Future work is needed to optimize this novel therapeutic approach, but these results lay the foundation for future endeavors to bring this therapy to the clinic for patients with metabolic liver disease who are currently only curable by whole-organ transplantation. In much the same way as has been demonstrated for patients with primary immunodeficiencies, 36,37,49 the potential for autologous gene therapy—using partial hepatectomy to isolate hepatocytes—is envisioned to be an alternative regenerative therapy for patients with metabolic liver disease.
Supplementary Material
Acknowledgments
We thank Yasuhiro Ikeda and Stephen Russell for thoughtful discussion. We thank Robert Kaiser for thoughtful review of the manuscript. We thank LouAnn Gross and Tony Blahnik for histology and immunohistochemistry support. R.D.H. was funded through a National Institutes of Health K01 DK106056 award and a Mayo Clinic Center for Regenerative Medicine Career Development Award.
Author Disclosure
The authors declare no competing financial interests exist.
References
- 1.Grompe M. The pathophysiology and treatment of hereditary tyrosinemia type 1. Semin Liver Dis 2001;21:563–571 [DOI] [PubMed] [Google Scholar]
- 2.Simoncelli M, Samson J, Bussieres JF, et al. . Cost–consequence analysis of nitisinone for treatment of tyrosinemia type I. Can J Hosp Pharm 2015;68:210–217 [PMC free article] [PubMed] [Google Scholar]
- 3.Russo P, O'Regan S. Visceral pathology of hereditary tyrosinemia type I. Am J Hum Genet 1990;47:317–324 [PMC free article] [PubMed] [Google Scholar]
- 4.Grompe M, Lindstedt S, al-Dhalimy M, et al. . Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet 1995;10:453–460 [DOI] [PubMed] [Google Scholar]
- 5.Sniderman King L, Trahms C, Scott CR. Tyrosinemia type I. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews®. Seattle, WA: University of Washington, 1993 [Google Scholar]
- 6.Salsman J, Dellaire G. Precision genome editing in the CRISPR era. Biochem Cell Biol 2017;95:187–201 [DOI] [PubMed] [Google Scholar]
- 7.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Overturf K, Al-Dhalimy M, Tanguay R, et al. . Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet 1996;12:266–273 [DOI] [PubMed] [Google Scholar]
- 9.Gregory-Evans K, Bashar AM, Tan M. Ex vivo gene therapy and vision. Curr Gene Ther 2012;12):103–115 [DOI] [PubMed] [Google Scholar]
- 10.Gao S, Seker E, Casali M, et al. . Ex vivo gene delivery to hepatocytes: techniques, challenges, and underlying mechanisms. Ann Biomed Eng 2012;40:1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen TH, Oberholzer J, Birraux J, et al. . Highly efficient lentiviral vector-mediated transduction of nondividing, fully reimplantable primary hepatocytes. Mol Ther 2002;6:199–209 [DOI] [PubMed] [Google Scholar]
- 12.Parouchev A, Nguyen TH, Dagher I, et al. . Efficient ex vivo gene transfer into non-human primate hepatocytes using HIV-1 derived lentiviral vectors. J Hepatol 2006;45:99–107 [DOI] [PubMed] [Google Scholar]
- 13.VandenDriessche T, Thorrez L, Naldini L, et al. . Lentiviral vectors containing the human immunodeficiency virus type-1 central polypurine tract can efficiently transduce nondividing hepatocytes and antigen-presenting cells in vivo. Blood 2002;100:813–822 [DOI] [PubMed] [Google Scholar]
- 14.Hickey RD, Mao SA, Glorioso J, et al. . Curative ex vivo liver-directed gene therapy in a pig model of hereditary tyrosinemia type 1. Sci Transl Med 2016;8:349ra399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakuma T, Barry MA, Ikeda Y. Lentiviral vectors: basic to translational. Biochem J 2012;443:603–618 [DOI] [PubMed] [Google Scholar]
- 16.Doench JG, Hartenian E, Graham DB, et al. . Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 2014;32:1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu PD, Scott DA, Weinstein JA, et al. . DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 2013;31:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heckl D, Kowalczyk MS, Yudovich D, et al. . Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 2014;32:941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hickey RD, Mao SA, Amiot B, et al. . Noninvasive 3-dimensional imaging of liver regeneration in a mouse model of hereditary tyrosinemia type 1 using the sodium iodide symporter gene. Liver Transpl 2015;21:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm D. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods 2002;28:146–157 [DOI] [PubMed] [Google Scholar]
- 21.Brinkman EK, Chen T, Amendola M, et al. . Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 2014;42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ponder KP, Gupta S, Leland F, et al. . Mouse hepatocytes migrate to liver parenchyma and function indefinitely after intrasplenic transplantation. Proc Natl Acad Sci U S A 1991;88:1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc 2008;3:1167–1170 [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Montini E, Al-Dhalimy M, et al. . Kinetics of liver repopulation after bone marrow transplantation. Am J Pathol 2002;161:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin H, Xue W, Chen S, et al. . Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickey RD, Lillegard JB, Fisher JE, et al. . Efficient production of Fah-null heterozygote pigs by chimeric adeno-associated virus-mediated gene knockout and somatic cell nuclear transfer. Hepatology 2011;54:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey RD, Mao SA, Amiot B, et al. . Noninvasive 3-dimensional imaging of liver regeneration in a mouse model of hereditary tyrosinemia type 1 using the sodium iodide symporter gene. Liver Transpl 2015;21:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhawan A, Puppi J, Hughes RD, et al. . Human hepatocyte transplantation: current experience and future challenges. Nat Rev Gastroenterol Hepatol 2010;7:288–298 [DOI] [PubMed] [Google Scholar]
- 29.Jorns C, Nowak G, Nemeth A, et al. . De novo donor-specific HLA antibody formation in two patients with Crigler–Najjar syndrome type I following human hepatocyte transplantation with partial hepatectomy preconditioning. Am J Transplant 2016;16:1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overturf K, Al-Dhalimy M, Manning K, et al. . Ex vivo hepatic gene therapy of a mouse model of hereditary tyrosinemia type I. Hum Gene Ther 1998;9:295–304 [DOI] [PubMed] [Google Scholar]
- 31.Chowdhury JR, Grossman M, Gupta S, et al. . Long-term improvement of hypercholesterolemia after ex vivo gene therapy in LDLR-deficient rabbits. Science 1991;254:1802–1805 [DOI] [PubMed] [Google Scholar]
- 32.Kay MA, Baley P, Rothenberg S, et al. . Expression of human alpha 1-antitrypsin in dogs after autologous transplantation of retroviral transduced hepatocytes. Proc Natl Acad Sci U S A 1992;89:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grompe M, Jones SN, Loulseged H, et al. . Retroviral-mediated gene transfer of human ornithine transcarbamylase into primary hepatocytes of spf and spf-ash mice. Hum Gene Ther 1992;3:35–44 [DOI] [PubMed] [Google Scholar]
- 34.Grossman M, Raper SE, Kozarsky K, et al. . Successful ex vivo gene therapy directed to liver in a patient with familial hypercholesterolaemia. Nat Genet 1994;6:335–341 [DOI] [PubMed] [Google Scholar]
- 35.Grossman M, Rader DJ, Muller DW, et al. . A pilot study of ex vivo gene therapy for homozygous familial hypercholesterolaemia. Nat Med 1995;1:1148–1154 [DOI] [PubMed] [Google Scholar]
- 36.Aiuti A, Biasco L, Scaramuzza S, et al. . Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott–Aldrich syndrome. Science 2013;341:1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biffi A, Montini E, Lorioli L, et al. . Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:1233158. [DOI] [PubMed] [Google Scholar]
- 38.Rittelmeyer I, Rothe M, Brugman MH, et al. . Hepatic lentiviral gene transfer is associated with clonal selection, but not with tumor formation in serially transplanted rodents. Hepatology 2013;58:397–408 [DOI] [PubMed] [Google Scholar]
- 39.Hickey RD, Mao SA, Glorioso J, et al. . Curative ex vivo liver-directed gene therapy in a pig model of hereditary tyrosinemia type 1. Sci Transl Med 2016;8:349ra399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin H, Xue W, Chen S, et al. . Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 2014;32:551–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Haurigot V, Doyon Y, et al. . In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 2011;475:217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin H, Song CQ, Dorkin JR, et al. . Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 2016;34:328–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Wang L, Bell P, et al. . A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 2016;34:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forbes SJ, Newsome PN. Liver regeneration—mechanisms and models to clinical application. Nat Rev Gastroenterol Hepatol 2016;13:473–485 [DOI] [PubMed] [Google Scholar]
- 45.Azuma H, Paulk N, Ranade A, et al. . Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Il2rg−/− mice. Nat Biotechnol 2007;25:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nygaard S, Barzel A, Haft A, et al. . A universal system to select gene-modified hepatocytes in vivo. Sci Transl Med 2016;8:342ra379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borel F, Tang QS, Gernoux G, et al. . Survival advantage of both human hepatocyte xenografts and genome-edited hepatocytes for treatment of alpha-1 antitrypsin deficiency. Mol Ther 2017;25:2477–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding J, Yannam GR, Roy-Chowdhury N, et al. . Spontaneous hepatic repopulation in transgenic mice expressing mutant human alpha1-antitrypsin by wild-type donor hepatocytes. J Clin Invest 2011;121:1930–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eichler F, Duncan C, Musolino PL, et al. . Hematopoietic stem-cell gene therapy for cerebral adrenoleukodystrophy. New Engl J Med 2017;377:1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.