Abstract
Background
In patients with human immunodeficiency virus (HIV) and hepatitis C virus (HCV) co-infection, HIV can modulate HCV replication and immune response as well as accelerate liver fibrosis. The role of miRNA in HIV/HCV co-infection is not fully elucidated. The aim of this study was to examine the differential expression of miRNAs in the liver.
Material/Methods
Thirteen patients who had undergone a liver transplant (7 HCV-infected and 6 HIV/HCV-co-infected patients) were examined using a miRNA array containing 1347 human miRNAs. To confirm the microarray results, data for 20 patients (10 HCV-infected and 10 HIV/HCV-co-infected) were validated using real-time polymerase chain reaction probing for miR101b, miR149, and miR200c. This miRNA was selected based on microarray results and its biological significance in liver fibrosis.
Results
Microarray analysis revealed 22 miRNAs that were differentially expressed in the HIV/HCV-co-infected group compared to the HCV-infected group (p<0.05). The expression of miR-101b and miR149 was significantly decreased in the HIV/HCV-co-infected group compared to that in the HCV-infected group (miR101b, 0.103±0.09 vs. 0.0157±0.0093, p=0.007; miR149, 0.152±0.159 vs. 0.0192±0.015, p=0.025).
Conclusions
HIV/HCV co-infection may promote liver fibrosis by modulating miRNA expression.
MeSH Keywords: Hepatitis C Antibodies; Hepatitis, Viral, Human; Liver Cirrhosis
Background
Co-infection of hepatitis C virus (HCV) and human immunodeficiency virus (HIV) is frequently seen due to their common routes of transmission [1,2]. With the development of highly active antiretroviral therapy (HAART), prognosis of patients with HIV has improved, allowing for a life expectancy comparable to that of the general population [3,4]. However, HCV-HIV-co-infected patients show rapid progression of fibrosis and a higher mortality rate than patients infected with HIV or HCV alone [5–7]. However, it remains unclear how these 2 viruses interact with each other.
MicroRNAs (miRNAs) are small-sized RNAs (21 to 25 bp in length) that regulate protein-coding genes by translational suppression or transcript degradation [8]. A recent study demonstrated that miRNAs influence the infection, replication, and host immune response to HCV [9]. MiR-122, the most abundant miRNA found in the liver, positively regulates HCV viral replication. We previously reported that miR-122 is associated with the interferon (IFN) response and could be a useful marker to predict the outcome of IFN therapy [10]. Moreover, miR-27-a is elevated in the HCV-infected liver and can inhibit HCV replication by regulating lipid metabolism [11].
With regard to HIV infection alone, miR-223 and miR-150 have been reported to inhibit HIV replication [12]. Several previous studies have investigated miRNAs in patients infected with HIV or HCV alone. However, no reports are available regarding the difference between end-stage liver miRNA expression in HIV/HCV co-infected and only HCV-infected groups. In this study, we examined liver miRNA profiles in HIV/HCV-co-infected patients who underwent liver transplantation.
Material and Methods
Patients
A total of 10 HIV/HCV-co-infected patients and 10 HCV-infected patients who underwent liver transplantation in 1 of 4 Japanese institutions involved in this study, were enrolled (Table 1). Thirteen (7 HCV-infected and 6 HIV/HCV-co-infected patients) of 20 patients were examined using a miRNA array. No patients had hepatocellular carcinoma. The liver function of both groups was similar as per the Child-Pugh score (HCV-infected group: 11.9±1.9; HCV/HIV-co-infected group: 11.2±1.4, p=0.332). All HIV/HCV co-infected patients were treated with HAART (9 patients with nucleoside reverse transcriptase inhibitor [NRTI] and 1 patient with non-NRTI [NNTRI]). The Institutional Review Board or the independent Ethics Committee at each institution approved this study.
Table 1.
Clinical features of patients.
| HIV/HCV co-infection | HCV-infection | p-value | |
|---|---|---|---|
| Age (years) | 34.1±7.4 | 50.2±6.9 | 0.001 |
| Chid-Pugh score | 11.9±1.5 | 11.2±1.4 | 0.332 |
| MELD score | 25.9±10.4 | 19.9±11.1 | 0.230 |
| AST | 80.5±30.2 | 57.7±35.5 | 0.140 |
| ALT | 44.1±15.1 | 37.1±36.8 | 0.589 |
| Protronbin time (INR) | 3.24±2.21 | 1.95±0.56 | 0.087 |
| Albumin | 2.8±0.4 | 2.7±0.3 | 0.540 |
| Prior treatment | 2 patients were treated with interferon | 6 patients were treated with interferon | |
| HCV genotype | 3 patients: 1a; 2 patients: 1b 1 patient: 1a+1b, 1 patient: 2a 1 patient: 2a+2b; 2 patients: 1b+3a |
10 patients: 1b | |
| HCV RNA (log IU/mL) | 5.0±0.7 | 5.1±1.4 | 0.861 |
| HIV load (copy/ml) | 2.7±1.9 | No data | |
| CD4 count (/μl) | 273±231 | No data |
MELD – Model For End-Stage Liver Disease, HCV – hepatitis C virus, HIV – human immunodeficiency virus, INR – international normalized ratio.
RNA isolation
RNA was extracted from explant liver specimens. Total RNA containing miRNA was isolated from formalin-fixed paraffin-embedded (FFPE) liver specimens using the Recover All Total Nucleic Acid Isolation Kit for FFPE (Ambion) according to the manufacturer’s protocol.
miRNA microarray
miRNA array analysis containing 1347 human miRNAs was performed with the miRNA Complete Labeling and Hub kit (Agilent, Santa Clara, CA). An aliquot of 100 μg of total RNA was taken from each specimen and treated with calf intestine phosphatase, denatured using DMSO, and directly labeled with Cy3 using T4 ligase. Labeled samples were hybridized to the miRNA array 8×15 k (G4878A) platform and washed with the supplied buffer (Agilent, Santa Clara, CA). To identify miRNAs that were significantly differentially expressed between the HCV-infected (n=7) and HIV/HCV-co-infected (n=6) groups, only those samples for which the total RNA extracted was adequate for microarray analysis were used. A difference in miRNA expression between the 2 groups was considered significant if the fold-change of expression values was >2.0 and the p-value was <0.05 using the Student’s t test. The correlations were analyzed using Spearman’s correlation coefficient. We clustered the miRNA without multiple testing and performed qPCR to validate microarray data, because only 2 miRNAs passed multiple testing.
Quantitative reverse transcription polymerase chain reaction
We validated the miRNAs identified using a miRNA array in all 20 patients. Based on the microarray results (fold-change of >20) and their biological significance in liver disease, miR-101, miR194, and miR-200c were selected for amplification via real-time PCR to confirm the results of microarray analysis.
miR-101b, miR194, and miR-200c, obtained by quantitative reverse transcription polymerase chain reaction, were analyzed using TaqMan MicroRNA assays (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. The expression of these miRNAs was calculated by the relative standard curve method and normalized to RNU6 levels. Both groups were compared using Student’s t test.
Results
Identification of differentially expressed miRNAs
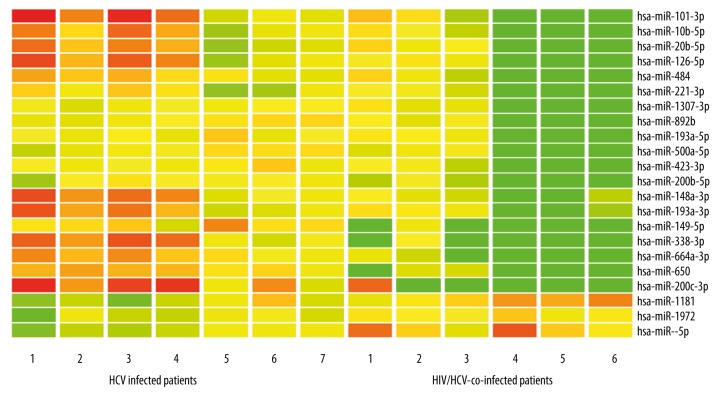
Comparison of 6 HIV/HCV-co-infected patients and 7 HCV-infected patients revealed that 22 miRNAs were significantly differently expressed, as per the miRNA array. Among these, 19 miRNAs were significantly decreased in HIV/HCV-co-infected patients as compared to HCV-infected patients (Figure 1). Moreover, 5 miRNAs showed the highest fold-change (>20-fold) (Table 2).
Figure 1.
Differentially expressed miRNAs in HIV/HCV-co-infected and HCV-infected patients. Red indicates higher miRNA expression and green indicates lower miRNA expression.
Table 2.
Fold-change of miRNA in HIV/HCV-co-infected versus HCV-infected patients.
| P value | Fold change | ||
|---|---|---|---|
| hsa-miR-101 | 0.011822 | Down | 20.36468 |
| hsa-miR-10b-5p | 0.019522 | Down | 10.90453 |
| hsa-miR-1181 | 0.010273 | Up | 3.131051 |
| hsa-miR-126-5p | 0.021261 | Down | 11.55823 |
| hsa-miR-1307-3p | 0.034467 | Down | 4.957585 |
| hsa-miR-148a-3p | 0.023237 | Down | 14.62462 |
| hsa-miR-149-5p | 0.00021 | Down | 21.23182 |
| hsa-miR-193a-3p | 0.042631 | Down | 8.882698 |
| hsa-miR-193a-5p | 0.038456 | Down | 8.148207 |
| hsa-miR-1972 | 0.038585 | Up | 2.010676 |
| hsa-miR-200b-5p | 0.011813 | Down | 6.402043 |
| hsa-miR-200c-3p | 0.001608 | Down | 44.20504 |
| hsa-miR-20b-5p | 0.036423 | Down | 12.50701 |
| hsa-miR-221-3p | 0.046994 | Down | 6.563181 |
| hsa-miR-338-3p | 0.000317 | Down | 29.83419 |
| hsa-miR-423-3p | 0.016273 | Down | 6.016482 |
| hsa-miR-484 | 0.014421 | Down | 8.84227 |
| hsa-miR-500a-5p | 0.02642 | Down | 7.400069 |
| hsa-miR-574-5p | 0.006449 | Up | 3.266655 |
| hsa-miR-650 | 0.000849 | Down | 20.01017 |
| hsa-miR-664a-3p | 0.00115 | Down | 19.64892 |
| hsa-miR-892b | 0.026285 | Down | 8.342825 |
PCR validation of significantly differentially expressed miRNAs
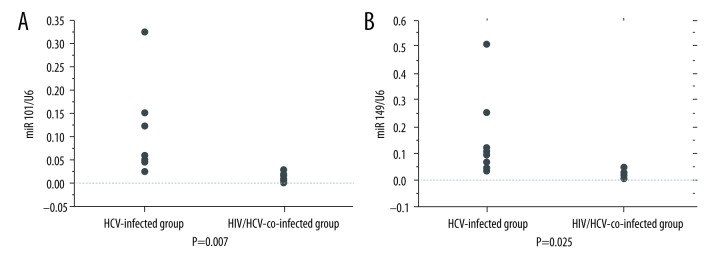
The expression of miR-101b and miR-149 was significantly decreased in the HIV/HCV-co-infected group compared to that in the HCV-infected group (miR 101-b 0.103±0.09 vs. 0.0157±0.0093, p=0.007, miR149 0.152±0.159 vs. 0.0192±0.015, p=0.025) (Figure 2A, 2B). However, miR-101 did not show a significant correlation with the HIV (r=0.553, p=0.097) and HCV (r=0.090, p=0.707) viral load. Moreover, miR-149 showed a correlation with HCV viral load (r=−0.644, p=0.04) but not with HIV viral load (r=−0.354, p=0.350). No significant difference was found for miR-101 and miR-194 expression in each treatment group.
Figure 2.
(A) Data are presented as mean ± standard deviation (SD). The expression of miR-101 was significantly decreased in the HIV/HCV-co-infected group (n=10) as compared to that in the HCV-infected group (n=10) (0.103±0.09 vs. 0.0157±0.0093, p=0.007, Student’s t test.). (B) Data are presented as mean ±SD. The expression of miR-149 decreased significantly in the HIV/HCV-co-infected group (n=10) as compared to that in the HCV-infected group (n=10) (0.152±0.159 vs. 0.0192±0.015, p=0.025, Student’s t test).
Discussion
This is first study to examine the difference in miRNA expression during end-stage liver fibrosis in patients with HIV/HCV co-infection. In a previous study, miRNA expression in peripheral blood mononuclear cells (PBMCs) from patients with HIV/HCV co-infection was examined [13]. HIV/HCV-co-infected patients were shown to develop liver fibrosis more rapidly and had a higher mortality due to more aggressive fibrosis. Therefore, it seems plausible that the miRNA profile can be used to target the liver of HIV/HCV-co-infected patients to develop novel treatment strategies.
The unique feature of this study is the comparison of miRNA profiles between patients with HIV/HCV co-infection and HCV-infected patients. All patients enrolled in this study were diagnosed with end-stage liver disease and underwent liver transplantation. In fact, the liver function of both groups was similar as per the Child-Pugh score. Therefore, the miRNA profile is not influenced by liver function. The microarray analysis revealed that 22 miRNAs were significantly differentially expressed between the HCV-infected and HIV/HCV-co-infected groups. Of these miRNAs, miR-101 and miR-194 was selected for further analysis because of their biological significance in liver disease. HIV does not infect hepatocytes, but it can replicate in hepatic stellate cells, macrophages, and CD4+ T lymphocytes [14]. The HIV viral protein gp120 can activate cell signaling pathways in hepatic stellate cells and certain immune cells [15,16]. Therefore, HIV infection may modulate miRNA expression and promote the progression of liver fibrosis.
Previous studies have demonstrated different cytokine expression levels in the livers of HIV/HCV-co-infected patients and HCV-infected patients [17,18]. These studies showed that intrahepatic mRNA levels of tumor necrosis factor-α, interleukin (IL)-8, and IL-10 were increased and that of tumor growth factor (TGF)-β was decreased in HIV/HCV-co-infected as compared to HCV-infected patients. Of these, TGF-β is a potent inducer of fibrosis in the effector cells of hepatic fibrosis. A previous in vitro study found that the co-culture of hepatic stellate cells with HIV and HCV resulted in increased expression level of TGF-β [19]. In a recent report, miR-101 promoted the reversal of activated HSCs to a quiescent state by suppressing TGF-β signaling. Moreover, in hepatocytes, miR-101 inhibits TGF-β signaling and suppresses the upregulation of profibrogenic cytokines [20], whereas miR-101 is downregulated in activated cells when compared to that in quiescent hepatic stellate cells [21].
Taken together, these results suggest that HIV/HCV co-infection might promote liver fibrosis by modulating the miR-101-TGF-β signal pathway.
A previous study has shown that miR-101 expression is lower in PBMCs of HIV/HCV-co-infected patients compared with that in HCV-mono-infected patients [13]. Thus, HIV may modulate miR-101 expression in the liver. Taking these results together, suppression of HIV virus by therapy could improve liver fibrosis by modulating miR-101 expression.
In a previous in vitro study, inhibition of miR-149 increased HCV entry and HCV RNA replication. In our study, miR-149 showed a significant correlation with HCV viral load. Therefore, HIV co-infection might affect HCV entry and replication. Moreover, mir-149 was associated with inflammation by modulating pro-inflammatory cytokines such as TNFα, IL1β, and IL-6.
Previous studies have shown that the expression of miR-122, miR-22, miR34a, mir-29b, and miR-124 are correlated with liver injury in patients with HIV [22,23]. However, in our study, expression of these miRNAs was not significantly different. This discrepancy could be attributed to the patient groups studied. Previous studies included patients with mild fibrosis, whereas patients with advanced liver disease only were enrolled in our study. Moreover, previous studies analyzed miRNA in PBMCs, which may be expressed differently in the liver, as analyzed in this study.
A limitation of this study was the small sample size that included only patients with advanced liver disease. These micro RNAs in patients with middle stages may show different results. Second, we did not enroll an HIV-infected comparison group. Moreover, HIV/HCV co-infected patients were significantly younger compared to HCV-infected patients. Therefore, the miRNA profile could be influenced by age.
In addition, HCV genotype 3 infection is associated with higher steatosis scores than infection with other HCV genotypes (24). In our study, HIV/HCV co-infected patients had more varied HCV genotypes compared to the HCV-infected patients. Therefore, the higher diversity of HCV genotypes in the co-infection group may have influenced the miRNA profile. Nonetheless, this is one of the first studies comparing intrahepatic miRNA in HIV/HCV-co-infected and HCV-infected patients.
Conclusions
We found that 19 miRNAs were significantly downregulated and 3 miRNAs were significantly up-regulated in HIV/HCV-co-infected patients as compared to HCV-infected patients. Of all miRNAs studied, miR-101 and miR-149 were significantly downregulated in HIV/HCV-co-infected patients and thus could play an important role in the rapid progression of liver failure.
Acknowledgments
We thank the doctors listed below for their kind support in this study:
Dr. Yasuhiko Sugawara (Japanese Red Cross Medical Center, Tokyo, Japan), Dr. Hideaki Okajima (Kyoto University), Dr. Hidekazu Yamamoto (Kumamoto University), and Dr. Kohei Ishiyama (Hiroshima University).
Abbreviations
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- miRNA
microRNA
- PCR
polymerase chain reaction
- HAART
highly active antiretroviral
- IFN
interferon
- FFPE
formalin-fixed paraffin-embedded
- PBMC
peripheral blood mononuclear cells
Footnotes
Source of support: The authors were supported by a Grant-in-Aid for Research on HIV/AIDS from the Ministry of Health, Labor, and Welfare of Japan, the “Eguchi Project”
References
- 1.Alter MJ, Kruszon-Moran D, Nainan OV, et al. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N Engl J Med. 1999;341:556–62. doi: 10.1056/NEJM199908193410802. [DOI] [PubMed] [Google Scholar]
- 2.Sherman KE, Rouster SD, Chung RT, Rajicic N. Hepatitis C Virus prevalence among patients infected with Human Immunodeficiency Virus: A cross-sectional analysis of the US adult AIDS Clinical Trials Group. Clin Infect Dis. 2002;34:831–37. doi: 10.1086/339042. [DOI] [PubMed] [Google Scholar]
- 3.Lohse N, Hansen AB, Pedersen G, et al. Survival of persons with and without HIV infection in Denmark, 1995–2005. Ann Intern Med. 2007;146:87–95. doi: 10.7326/0003-4819-146-2-200701160-00003. [DOI] [PubMed] [Google Scholar]
- 4.Nakagawa F, May M, Phillips A. Life expectancy living with HIV: Recent estimates and future implications. Curr Opin Infect Dis. 2013;26:17–25. doi: 10.1097/QCO.0b013e32835ba6b1. [DOI] [PubMed] [Google Scholar]
- 5.de Ledinghen V, Barreiro P, Foucher J, et al. Liver fibrosis on account of chronic hepatitis C is more severe in HIV-positive than HIV-negative patients despite antiretroviral therapy. J Viral Hepat. 2008;15:427–33. doi: 10.1111/j.1365-2893.2007.00962.x. [DOI] [PubMed] [Google Scholar]
- 6.Soto B, Sanchez-Quijano A, Rodrigo L, et al. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J Hepatol. 1997;26:1–5. doi: 10.1016/s0168-8278(97)80001-3. [DOI] [PubMed] [Google Scholar]
- 7.Mohsen AH, Easterbrook PJ, Taylor C, et al. Impact of human immunodeficiency virus (HIV) infection on the progression of liver fibrosis in hepatitis C virus infected patients. Gut. 2003;52:1035–40. doi: 10.1136/gut.52.7.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen PY, Meister G. microRNA-guided posttranscriptional gene regulation. Biol Chem. 2005;386:1205–18. doi: 10.1515/BC.2005.139. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57–67. doi: 10.1016/j.virol.2009.11.036. [DOI] [PubMed] [Google Scholar]
- 10.Kamo Y, Ichikawa T, Miyaaki H, et al. Significance of miRNA-122 in chronic hepatitis C patients with serotype 1 on interferon therapy. Hepatol Res. 2015;45:88–96. doi: 10.1111/hepr.12317. [DOI] [PubMed] [Google Scholar]
- 11.Singaravelu R, Chen R, Lyn RK, et al. Hepatitis C virus induced up-regulation of microRNA-27: A novel mechanism for hepatic steatosis. Hepatology. 2014;59:98–108. doi: 10.1002/hep.26634. [DOI] [PubMed] [Google Scholar]
- 12.Munshi SU, Panda H, Holla P, et al. MicroRNA-150 is a potential biomarker of HIV/AIDS disease progression and therapy. PLoS One. 2014;9:e95920. doi: 10.1371/journal.pone.0095920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta P, Liu B, Wu JQ, et al. Genome-wide mRNA and miRNA analysis of peripheral blood mononuclear cells (PBMC) reveals different miRNAs regulating HIV/HCV co-infection. Virology. 2014;450–451:336–49. doi: 10.1016/j.virol.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 14.Tuyama AC, Hong F, Saiman Y, et al. Human immunodeficiency virus (HIV)-1 infects human hepatic stellate cells and promotes collagen I and monocyte chemoattractant protein-1 expression: Implications for the pathogenesis of HIV/hepatitis C virus-induced liver fibrosis. Hepatology. 2010;52:612–22. doi: 10.1002/hep.23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruno R, Galastri S, Sacchi P, et al. gp120 modulates the biology of human hepatic stellate cells: A link between HIV infection and liver fibrogenesis. Gut. 2010;59:513–20. doi: 10.1136/gut.2008.163287. [DOI] [PubMed] [Google Scholar]
- 16.Hong F, Tuyama A, Lee TF, et al. Hepatic stellate cells express functional CXCR4: Role in stromal cell-derived factor-1alpha-mediated stellate cell activation. Hepatology. 2009;49:2055–67. doi: 10.1002/hep.22890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbate I, Cappiello G, Rosati S, et al. Intra-hepatic messenger RNA levels for interferons and related genes in hepatitis C virus/HIV co-infected patients. AIDS. 2004;18:691–92. doi: 10.1097/00002030-200403050-00015. [DOI] [PubMed] [Google Scholar]
- 18.Blackard JT, Kang M, Sherman KE, et al. Effects of HCV treatment on cytokine expression during HCV/HIV coinfection. J Interferon Cytokine Res. 2006;26:834–38. doi: 10.1089/jir.2006.26.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi L, Qin E, Zhou J, et al. HIV and HCV co-culture promotes profibrogenic gene expression through an epimorphin-mediated ERK signaling pathway in hepatic stellate cells. PLoS One. 2016;11:e0158386. doi: 10.1371/journal.pone.0158386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu X, Zhang H, Zhang J, et al. MicroRNA-101 suppresses liver fibrosis by targeting the TGFbeta signalling pathway. J Pathol. 2014;234:46–59. doi: 10.1002/path.4373. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, Wu CQ, Zhang ZQ, et al. Loss of expression of miR-335 is implicated in hepatic stellate cell migration and activation. Exp Cell Res. 2011;317:1714–25. doi: 10.1016/j.yexcr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Anadol E, Schierwagen R, Elfimova N, et al. Circulating microRNAs as a marker for liver injury in human immunodeficiency virus patients. Hepatology. 2015;61:46–55. doi: 10.1002/hep.27369. [DOI] [PubMed] [Google Scholar]
- 23.Jansen C, Reiberger T, Huang J, et al. Circulating miRNA-122 levels are associated with hepatic necroinflammation and portal hypertension in HIV/HCV coinfection. PLoS One. 2015;10:e0116768. doi: 10.1371/journal.pone.0116768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asselah T, Rubbia-Brandt L, Marcellin P, Negro F. Steatosis in chronic hepatitis C: why does it really matter? Gut. 2006;55:123–30. doi: 10.1136/gut.2005.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]