Abstract
Background
Gene therapy can be an intriguing therapeutic option in wide-ranging neurological disorders. Though nonviral gene carriers represent a safer delivery system to their viral counterparts, a thorough design of such vehicles is crucial to enhance their transfection properties.
Purpose
This study evaluated the effects of combined use of two nonionic surfactants, poloxamer 188 (P) and polysorbate 80 (P80) into nanovesicles – based on 2,3-di(tetradecyloxy)propan-1-amine cationic lipid (D) – destined for gene delivery to central nervous system cells.
Methods
Niosome formulations without and with poloxamer 188 (DP80 and DPP80, respectively) were prepared by the reverse-phase evaporation technique and characterized in terms of size, surface charge, and morphology. After the addition of pCMS-EGFP plasmid, the binding efficiency to the niosomes was evaluated in agarose gel electrophoresis assays. Additionally, transfection efficiency of complexes was also evaluated in in vitro and in vivo conditions.
Results
In vitro experiments on NT2 cells revealed that the complexes based on a surfactant combination (DPP80) enhanced cellular uptake and viability when compared with the DP80 counterparts. Interestingly, DPP80 complexes showed protein expression in glial cells after administration into the cerebral cortices of rats.
Conclusion
These data provide new insights for glia-centered approach for gene therapy of nervous system disorders using cationic nanovesicles, where nonionic surfactants play a pivotal role.
Keywords: gene therapy, nonviral gene vectors, poloxamer 188, niosomes, cationic lipids
Background
The genetic base of many central nervous system (CNS) diseases along with the recent advances in the nanotechnology fields makes gene therapy an attractive therapeutic option.1 Gene therapy can improve the quality of life in patients with major brain diseases, including Parkinson, Alzheimer, or epilepsy.2–4 Successful delivery of DNA–vector hybrids that can correct such CNS disorders was probed.5–7 Although many phase-I clinical trials of CNS gene therapy are documented, only a few have reached phase-II.5 Such modest improvement is due mainly to the lack of optimal vehicles able to deliver the genetic material in an efficient way.8
Being easier in large-scale production with lower production cost, non-immunogenic and non-oncogenic, nonviral approach offers several advantages in delivering pDNA and siRNA.9,10 However, their main drawback is the lower transfection efficiency compared with virus-based vectors.11 Therefore, further research in the design of such formulations is necessary before they become a realistic medical option to treat CNS disorders.
Among the different available gene delivery systems such as liposomes or micelles, niosomes have attracted a great amount of attention.12
Nonionic surfactant lipid vesicles, such as niosomes, are synthetic vesicles with high drug-delivery potential.13 Niosomes can deliver both water- and lipid-soluble molecules. In addition, and depending on their chemical structure, they can also control the release of such molecules.14 However, niosomes are more stable and cheaper systems, compared with liposomes.15 Among several factors, the nonionic surfactants are known to provide stability to the formulation. Other components, such as cationic lipids, can interact with negatively charged genetic material to form niocomplexes, in gene delivery context.16,17
Recently, some encouraging results have been obtained with niosomes based on the hydrochloride salt of the cationic lipid 2,3-di(tetradecyloxy)propan-1-amine (DTPA-Cl) and polysorbate 80 (P80) nonionic surfactant to transfect retinal cells.16 DTPA-Cl is a fairly water-soluble lipid that can efficiently condense DNA. In addition, other cationic niosome formulations have been able to transfect neuronal cells after intracranial administration in the brain of rats.18 Considering such results, we explored the impact of adding poloxamer 188 nonionic surfactant (P) to niosomes that contained the hydrochloride salt of cationic lipid DTPA (D) and P80 to obtain two kind of niosomes, with only one surfactant (DP80 formulation) or with a mixture of them (DPP80 formulation). Previous studies showed that poloxamer 188, a biocompatible copolymer, could be well tolerated by cells.19 Moreover, P has been successfully used in several gene-delivery studies when combined with different lipid components.20,21 In addition, other reports in the literature highlight the advantage of combining two surfactants in the same formulation.22,23 Particle size, polydispersity index (PDI), and zeta potential (ZP) of both niosomes were analyzed for comparison purposes. When the pCMS-EGFP plasmid was added at different cationic lipid/DNA ratios (w/w), the resulted nioplexes were physicochemically analyzed. In vitro experiments were performed to evaluate the behavior of both vectors in NT2 cells regarding their cellular uptake, protein expression efficiency, and cell viability. Afterwards, the formulation that showed better results was administered to primary neuronal culture, and then, into rat cerebral cortex in order to evaluate protein expression in in vivo conditions.
Methods
Elaboration of cationic niosomes
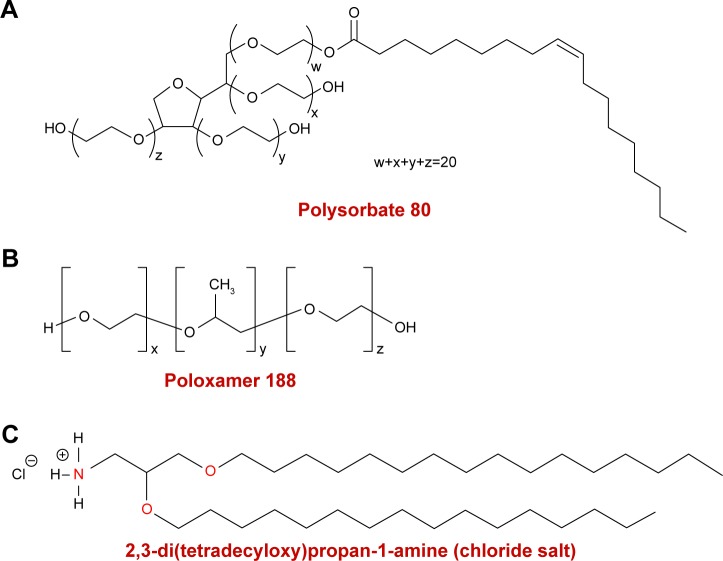
The hydrochloride salt of the cationic lipid DTPA-Cl was synthesized as indicated in previous work.24 Once the cationic lipid (D) was produced, it was used to elaborate niosomes that were elaborated by modified reverse-phase evaporation technique.25 An amount of 5 mg of D was dissolved in 1 mL of dichloromethane organic solvent, and sonicated in 5 mL of surfactant (either P80 alone or equal weights of both P and P80). The details of chemical structure of each component of the niosome formulations are represented in Figure 1. The emulsions were obtained after a sonication process (Branson Sonifier 250®; Branson Ultrasonics Corporation, Danbury, CT, USA) at 45 W for 30 seconds. When dichloromethane was evaporated, dispersions of niosome vesicles were obtained in the aqueous phase. Then, niosome formulations were named as DP80 and DPP80 both of them containing DTPA-Cl (D) cationic lipid and P80 or P+P80 as nonionic surfactants at a cationic lipid/nonionic surfactant ratio of 1/5 (Table 1).
Figure 1.
Chemical structure of: (A) polysorbate 80, (B) poloxamer 188, and (C) cationic lipid DTPA.
Abbreviation: DTPA, 2,3-di(tetradecyloxy)propan-1-amine.
Table 1.
Components (in mg) and physical characterization of niosome formulations regarding particle size (nm), PDI, and zeta potential (mV)
| Niosomes | Components (mg)
|
Characterization
|
||||
|---|---|---|---|---|---|---|
| DTPA (D) | Poloxamer 188 (P) | Polysorbate 80 (P80) | Size (nm) | PDI | Zeta potential (mV) | |
|
| ||||||
| DP80 | 5 | – | 25 | 54.02±0.94 | 0.52±0.10 | 41.9±7.10 |
| DPP80 | 5 | 12.5 | 12.5 | 90.41±0.65 | 0.42±0.01 | 44.1±4.39 |
Note: Data represent mean±SD (n=3).
Abbreviations: DTPA, 2,3-di(tetradecyloxy)propan-1-amine; PDI, polydispersity index.
Plasmid propagation and elaboration of complexes
The pCMS-EGFP plasmid (5,541 bp; PlasmidFactory, Bielefeld, Germany), was propagated, as previously described.14 Nioplexes were obtained after mixing a stock solution of pCMS-EGFP plasmids (0.5 mg/mL) with different volumes of suspensions where niosomes were dispersed (1 mg cationic lipid/mL) to obtain nioplexes at different cationic lipid/DNA ratios (w/w).
Physicochemical analysis of niosomes and complexes
The particle size, superficial charge (ZP), and morphology of lipid formulations were analyzed as described previously.24,26 To evaluate the interactions between the niosomes and the genetic material, an agarose gel electrophoresis assay was performed.24
In vitro transfection in NT2 cells
NT2 cells (ATCC®−CRL, 1973) were cultivated and processed as previously described.27 Cells were exposed to complexes (1.25 µg of pCMS-EGFP/well). After 4 hours of incubation, transfection medium was removed and refreshed with complete medium. Cells were allowed to grow for 24 hours until being analyzed by fluorescence microscopy (EclipseTE2000-S; Nikon Instruments, Melville, NY, USA) and flow cytometry (FACSCalibur; BD, San Jose, CA, USA) as described previously.14 Positive control (Lipofectamine® 2000 [L2K], Gibco®; Life Technologies S.A., Madrid, Spain) was prepared following the manufacturer’s protocol.
Cellular uptake and intracellular disposition of complexes
NT2 cells were cultured as previously mentioned in Section “In vitro transfection in NT2 cells.” After 4 hours of incubation with complexes, the transfection medium was removed and cells were washed thoroughly with PBS, trypsinized, and analyzed by FACSCalibur flow cytometer to evaluate cellular uptake. In total, 10,000 events were collected and analyzed for each sample. Each sample was analyzed in triplicate.
The intracellular distributions of the complexes were analyzed by confocal laser scanning microscopy (CLSM, Olympus Fluoview 500). Specific fluorescence-labeled commercial reagents were used to evaluate co-localization with Fluorescein isothiocyanate (FITC)-pCMS-EGFP plasmid by Mander’s co-localization coefficient (M) as previously reported.27
Primary cortical neuron culture
Experimental procedures for in vitro isolation and culture of primary cortex cells were carried out according to the directive 2010/63/EU of the European Parliament and of the Council, and the RD 53/2013 Spanish regulation on the protection of animals use for scientific purposes. Additionally, all procedures were approved by the Miguel Hernandez University Committee for Animal Use in Laboratory. Dissociated cultures of primary cortical neurons were obtained from E17–E18 rat embryos (Sprague–Dawley) and preserved in Hanks’ Balanced Salt Solution during extraction. Then, trypsin was added to the medium and incubated at 37°C for chemical dissociation. Subsequently, the tissue was dissociated in neurobasal medium/FBS and cell density was determined using a hemocytometer. Cells were seeded on glass coverslips following the same transfection protocol previously described for cellular uptake and trafficking studies. Then, the samples were incubated overnight at 4°C with the primary antibodies and rabbit anti-glial fibrillary acidic protein (GFAP) (1:300), as a marker of glial cells and mouse anti-GFP (1:100) diluted in PBS containing TritonX100 (0.5%). Later, coverslips were washed in PBS and incubated for 1 hour with specific Alexa Fluor 555 and Alexa Fluor 488 conjugated secondary antibodies (Thermo Fisher Scientific, Madrid, Spain). Hoechst 33342 was used to label the nuclei. Finally, coverslips were mounted for imaging and analyzed with a Leica TCS SPE fluorescence microscope.
In vivo studies in rat brain
Adult male Sprague–Dawley rats were used for in vivo experiments. All procedures were carried out in accordance with the Spanish and European Union regulations for the use of animals in research and supervised by the Miguel Hernandez University Standing Committee for Animal Use in Laboratory. Pretreatment of rats and injection of the complexes were performed as previously reported.18
Analysis of protein expression in brain
About 72 hours after surgery, animals were sacrificed and perfusion with PBS followed by paraformaldehyde (4%) was performed for fixation. Then, rat brains were preserved in paraformaldehyde (4%) and cryoprotected in sucrose solution (30%) with PBS before slicing. A cryostat (HM 550; Microm International GmbH, Walldorf, Germany) was used to obtain slices of 20 µm from coronal frozen sections close to the area of injection. Next, slices were mounted for immunohistochemistry analysis with rabbit anti-NeuN (1:100) to label neurons and with mouse anti-GFP (1:100) diluted in PBS containing TritonX100 (0.5%) and incubated at 4°C overnight. Thereafter, slices were washed in PBS and incubated for 1 hour in dark with specific Alexa Fluor 555 and Alexa Fluor 488 conjugated secondary antibodies for visualization. Nuclei were counterstained with Hoechst 33342.18
Statistical analysis
Statistical differences between groups at significance levels of >95% were calculated by ANOVA and Student’s t-test. In all cases, P-values <0.05 were regarded as significant. Normal distribution of samples was assessed by the Kolmogorov–Smirnov test and the homogeneity of the variance by the Levene test. Numerical data were presented as mean±SD.
Results
Physicochemical characterization of niosomes/complexes
Table 1 shows the characterization of both DP80 and DPP80 niosome formulations. The niosomes prepared only with P80 (DP80) had sizes around 54 nm. Interestingly, when equal weights of both tensioactives, P and P80, were incorporated, the size obtained was significantly larger (90.4 nm) with a slight decrease in PDI (0.42, instead of 0.52). Regarding the surface charge, all niosomes were in the positive territory ranging from 42 to 44 mV.
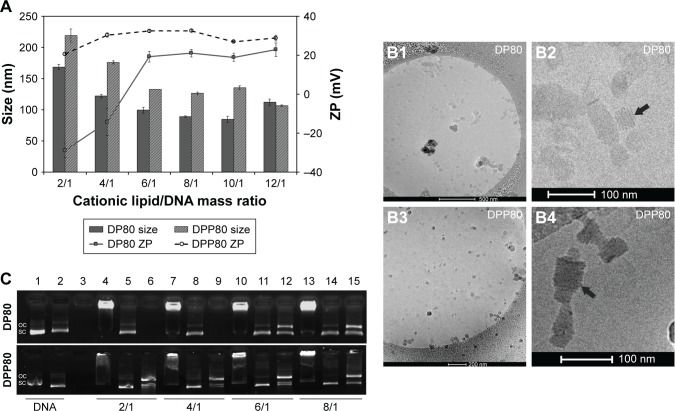
Figure 2 summarizes some parameters of DP80 and DPP80 complexes in terms of size, superficial charge (ZP), morphology, and plasmid-binding capacity. DPP80 complexes depicted sizes larger than DP80 (P<0.05), except for 12/1 ratio (where no true difference was noticed, P>0.05). For both formulations, complexes gradually tended to decrease in size while ratio increased. In general, values of size/charge were significantly affected by the DNA incorporated into these complexes. Starting with the mass ratio of 2/1, the sizes of DP80 and DPP80 complexes were around 168 and 219 nm, respectively. A gradual reduction was discerned to become almost 106 and 111 nm at 12/1 ratio. The PDI values of both nioplexes were around 0.2 (as depicted in Table S1). Regarding ZP of DP80 complexes (Figure 2A, lines), it started with negative values at 2/1 and 4/1 ratios (−29 and −14 mV, respectively) which gradually increased towards the positive territory to remain around +20 mV. The behavior of DPP80 complexes was different. In this case, all ZP values were positive. In addition, ZP values increased from 20 mV at 2/1 ratio to 32.5 mV at 8/1 ratio, then decreased in the last two ratios (10/1 and 12/1) (Figure 2A). When assessed by cryo-TEM (Figure 2B), both DP80 and DPP80 complexes at 6/1 (w/w) ratio appeared as electron-dense particles with a discrete distribution. Interestingly, DPP80 (Figure 2B3) depicted less tendency to form aggregates compared with DP80 (Figure 2B1). At a higher magnification (Figure 2B2 and B4), complexes were mostly elongated with a lamellar morphology (arrows). Agarose gel electrophoresis assays (Figure 2C) revealed the capacity of DP80 niosomes to bind efficiently to DNA (lanes 4, 7, 10, and 13 that corresponded to 2/1, 4/1, 6/1, and 8/1 ratios, respectively). On the other hand, the ability of DPP80 niosomes to capture DNA increased proportionally with the increase in the ratio (Figure 2C). Upon addition of sodium dodecyl sulphate (SDS) the DNA bound to the niosomes was released from all studied complexes. The absence of white bands in wells 5, 8, 11, and 14 (2/1, 4/1, 6/1, and 8/1 ratios) confirmed this fact. Interestingly, the supercoiled (SC) bands observed in lanes 6, 9, 12, and 15 (Figure 2C, lower panel) confirmed that the plasmid DNA released from the DPP80 niosome formulation was protected from degradation at all ratios tested. However, DP80-released DNA was not protected at ratios of 2/1 and 4/1 (Figure 2C, upper panel).
Figure 2.
Characterization of DP80 and DPP80 complexes.
Notes: (A) Particle size (nm) and ZP (mV). The data represent mean±SD (n=3). (B1–B4) Cryo-TEM pictures of nioplexes. Arrows in B2 and B4 point to the multi-lamellar pattern. (C) Agarose gel electrophoresis assay at different cationic lipid/DNA ratios of DP80 and DPP80 complexes (upper and lower panels, respectively), based on both formulations. Lanes are shown as: 1–3, uncomplexed DNA; 4–6, mass ratio 2/1; 7–9, mass ratio 4/1; 10–12, mass ratio 6/1; and 13–15, mass ratio 8/1. Lanes 2, 5, 8, 11, and 14 depict niocomplexes treated with SDS, while lanes 3, 6, 9, 12, and 15 were treated with DNase I+SDS. OC and SC (open circular and supercoiled forms, respectively).
Abbreviations: SDS, sodium dodecyl sulphate; ZP, zeta potential.
Protein expression and viability studies
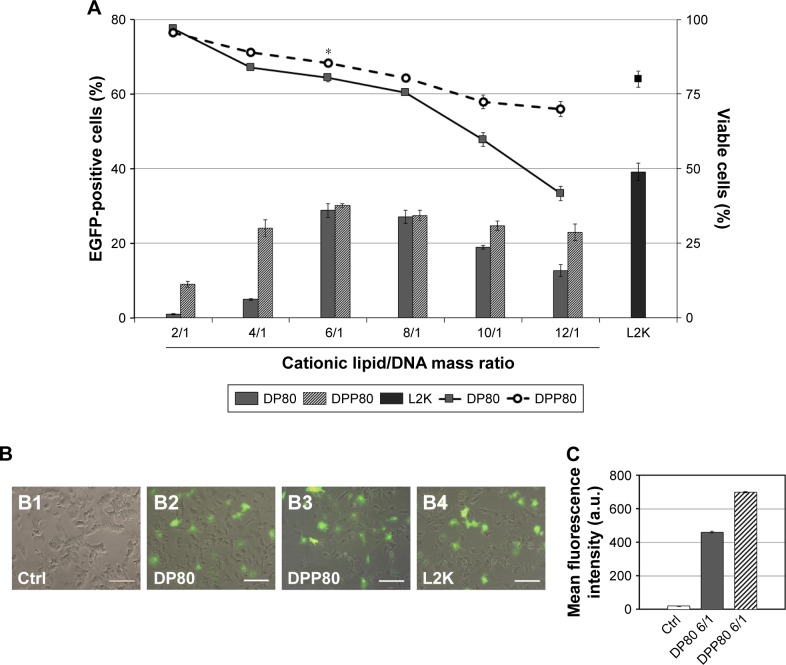
Figure 3A shows a peak of 28.8% of transfected cells when DP80 was used at 6/1 mass ratio. Regarding DPP80 formulation, the best transfection percentage (30.1%) was observed again at 6/1 mass ratio, followed by a gradual decline. No protein expression was observed on cells when free “uncomplexed” DNA was analyzed (data not shown). The viability of transfected cells (Figure 3A, lines) depicted a clear negative relationship with the cationic lipid/DNA mass ratio. DPP80 (dashed line) showed higher cell viability results compared with DP80 formulation (continuous line) at all ratios studied, unless at 2/1 ratio, where cell viability was around 100% in both formulations. At 6/1 ratio, cell viability value was 85.4%, significantly higher (P<0.05) than that of DP80 at the same 6/1 ratio and Lipofectamine 2000 positive control (80.4% and 80%, respectively). Figure 3B2–B4 revealed fluorescence overlay and phase-contrast micrographs of NT2 cells at 24 hours post-transfection of both formulations at 6/1 ratios as well as L2K positive control. In all cases, NT2 cells retained their normal morphology compared with the untransfected (control [Ctrl]) cells (Figure 3B1). Mean fluorescence intensity (MFI) of cells transfected by complexes based on DPP80 niosomes at a ratio of 6/1 had mark edly higher values (P<0.05) compared with those obtained by complexes based on DP80 niosomes (Figure 3C).
Figure 3.
Transfection studies in NT2 culture cells.
Notes: (A) Flow cytometry analysis of percentage of cells that express EGFP (bars) and percentage of live cells (lines) at different cationic lipid/DNA ratios (w/w). Values represent mean±SD (n=3). *P<0.05 compared with DP80 at 6/1 and L2K at 2/1. Overlay of fluorescence and DIC pictures of control cells (B1), DP80 at 6/1 (B2), DPP80 at 6/1 (B3), and L2K at 2/1 mass ratios (B4). Scale bars=100 µm. (C) Mean fluorescence intensity (a.u.).
Abbreviations: a.u., arbitrary unit; Ctrl, control; DIC, digital image correlation; L2K, Lipofectamine® 2000.
Trafficking studies
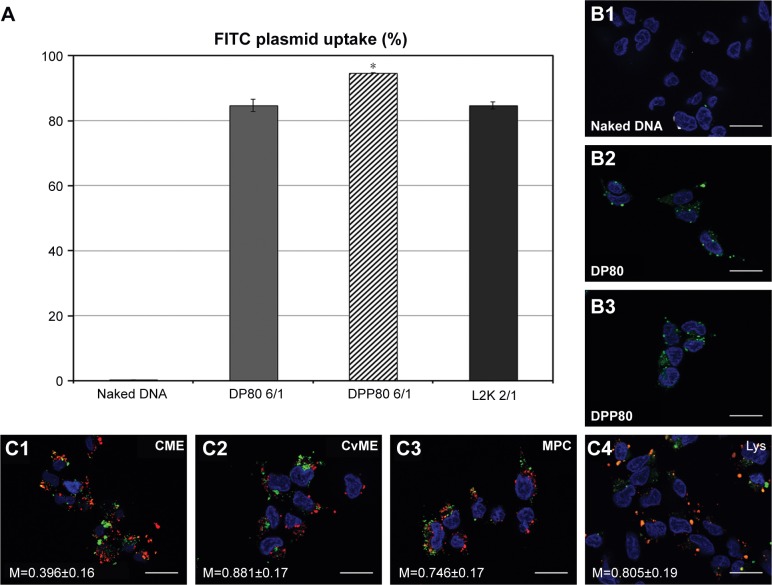
Both FITC-labeled complexes were applied at the cationic lipid/DNA (w/w) mass ratio that depicted the best transfection percentage (6/1; as shown in Figure 3A). With an uptake percentage of about 95%, DPP80 complexes exceeded the other two formulations (DP80 and L2K of 84% and 85%, respectively). Control cells transfected with DNA alone did not show any uptake (Figure 4A and B1). However, when FITC-labeled DP80 and DPP80 complexes were used, some fluorescence signal within the cytoplasm of most of the cells in the field was observed, as shown in Figure 4B2 and B3. The internalization studies of FITC-labeled DPP80 complexes at 6/1 ratio are represented in Figure 4. At 4 hours after incubation of FITC-labeled complexes with fluorescence markers of CME ([clathrin mediated endocytosis]; AlexaFluor 546-Transferrin; Figure 4C1), CvME ([caveolae mediated endocytosis]; AlexaFluor 555-Cholera Toxin; Figure 4C2), MPC ([macropinocytosis]; AlexaFluor 594-Dextran; Figure 4C3), and late endosome/lysosome (LysoTracker Red DND-99; Figure 4C4), Mander’s M1 co-localization coefficient was 0.39, 0.88, 0.75, and 0.81, respectively.
Figure 4.
Trafficking studies in NT2 culture cells.
Notes: (A) Flow cytometry measurement of the percentage of cells that captured plasmid labeled with FITC. Error bars represent SD (n=3). *P<0.05 compared with other groups. (B1–B3) Fluorescence micrographs with FITC-labeled naked plasmids, and DP80 and DPP80 niocomplexes (at 6/1 mass ratios), respectively (scale bar=25 µm). (C) Confocal micrographs showing intracellular distribution of DPP80 complexes, where plasmid is labeled in green color, and AlexaFluor 546-Transferrin (C1), AlexaFluor 555-Cholera Toxin (C2), AlexaFluor 594-Dextran (C3), and LysoTracker Red DND-99 markers (C4), all in red. Nuclei of cells were stained with DAPI-fluoromount G (blue) (scale bar=25 µm).
Abbreviations: CME, clathrin mediated endocytosis; CvME, caveolae mediated endocytosis; FITC, fluorescein isothiocyanate; M, Mander’s co-localization coefficient; MPC, macropinocytosis.
Protein expression in primary cortical neuron and rat brain
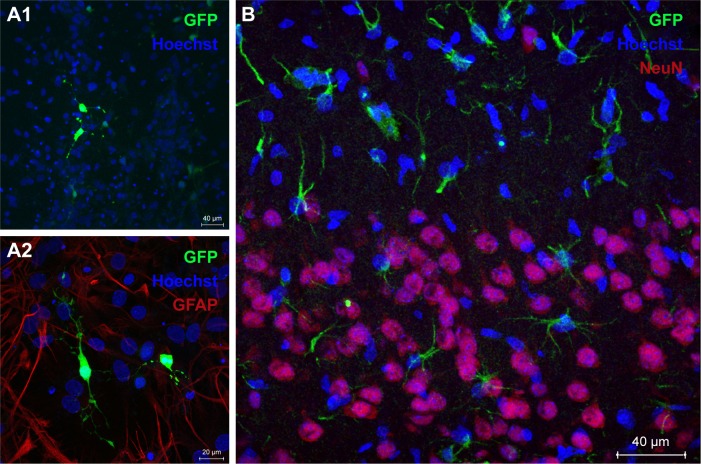
Protein expression was observed in primary cortical cultures of cells transfected with DPP80 complexes at 6/1 ratio (w/w), and is shown in Figure 5A1. In any case, none of those cells were GFAP+ve (Figure 5A2). Interestingly, 72 hours after intracortical administration of complexes, NeuN−ve (nonneuronal) positive cells with glial morphology were observed expressing EGFP in their dendrites (Figure 5B). Other pictures of brain experimented with different injections and the corresponding contralateral sections are shown in Figure S1.
Figure 5.
EGFP gene expression in primary neuronal cell cultures (A1 and A2). GFAP+ve neuroglia (red) and nuclei were stained in blue color with Hoechst 33342 (scale bars=40 and 20 µm, respectively). (B) In vivo expression green protein (scale bar=40 µm).
Note: Nuclei are shown in blue color (Hoechst), while neurons in red (NeuN+ve).
Discussion
Considering the recently shown promising results of cationic niosomes to transfect stem cells17 and neurons in both brain and retina,18,28 and the reports on literature that highlights the benefits of combining two surfactants in the same formulation,22,23 we explored in the present manuscript, the impact of combining P and P80 nonionic surfactants in the same niosome gene-delivery system based on the cationic lipid DTPA.
Our data showed that a particle was affected depending on the type of surfactant used (Table 1). The difference in size could be due to the different hydrophilic–lipophilic balance (HLB) value of surfactant used. P80 (lower HLB value) gave rise to nanovesicles of smaller sizes.29 However, the interaction between surfactant(s) and cationic lipid in niosomes should not be excluded as another influential factor that could also affect the vesicle size.30
Prior to performing transfection experiments, we characterized complexes at different mass ratios of cationic lipid and DNA content (Figure 2). There is no general rule in the scientific community about the optimal size of nonviral vector particles for gene-delivery applications. In any case, it is clear that this parameter affects their transfection efficiency,31 and in the case of niosome vesicles, it strongly depends on the mass ratio between both the cationic lipid and the DNA. As can be observed in Figure 2A, when DNA was bound to obtain both DP80 and DPP80 nioplexes at 2/1 ratio, the size exceeded 150 and 200 nm, respectively. In any case, when the ratio increased, the size of nioplexes decreased, probably due to the condensation of DNA plasmid on the surface of niosome vesicles in a more efficient multi-lamellar pattern by interaction between opposite charges, as discerned in the cryo-TEM examination (Figure 2B2 and B4, arrows). The shape of complexes has a marked effect on their final performance as a gene-delivery carrier.32 Interestingly, the lamellar pattern discerned in several complexes is believed to occur during the process of complex formation. The complete topological transformation of both lipid and DNA into complex particles would form string-like aggregates, in which complexes have a multi-lamellar pattern. The discrete morphology of complexes at 6/1 mass ratio (Figure 2B) could be attributed to the moderately positive surface charge (>20 mV).16 DPP80 complexes had a higher ZP (>30 mV), and thus depicted less aggregates as they tend to repel each other.33 Another crucial parameter that influences the transfection process is the electrostatic interactions between the negatively charged phosphate groups of DNA and the positively charged amine groups of the cationic niosomes.34 Such parameter was analyzed in agarose gel electrophoresis assays (Figure 2C). For DPP80 niosomes, all ratios analyzed released and protected plasmid DNA against degradation. However, complexes based on DP80 niosomes (Figure 2C, upper panel) failed clearly to protect the DNA at low cationic lipid/DNA rates. Despite the fact that DP80 niosomes condensed and released properly the DNA after the addition of SDS at all analyzed mass ratios, the protective capacity of DNA against enzymatic digestion is a mandatory issue for efficient gene-delivery vectors. We hypothesize that electrostatic interactions between the DP80 niosomes and DNA at low cationic lipid/DNA ratios (2/1 and 4/1) were efficient to condense the DNA, but were not strong enough to protect the DNA,24 that might be attributed to the net negative surface charge of the complexes at the aforementioned ratios.
Afterward, both complexes were evaluated in in vitro conditions to analyze protein expression and cytotoxic effect of formulations in NT2 culture cells. This cell line has been reported to be a promising human cell source for in vivo therapeutic application in many CNS disorders.35,36 Their therapeutic importance lies in the fact that, following retinoic acid treatment, NT2 cells undergo an irreversible commitment to terminally differentiate into stable post-mitotic neurons, even long time after transplantation in animal brain.37 Moreover, NT2-mediated functional recovery in preclinical stroke models has endorsed their application in clinical trials.38 NT2 cells are stable, easy to cultivate, amenable to scale-up for cell production, and represent a CNS-like delivery vehicle for glioblastoma therapy.38–40 NT2 cells can be differentiated into both neuronal and glial cells.41 Therefore, these cells represent an appealing model to evaluate transfection efficiency by nonviral vectors based on cationic niosomes.
The data of transfection efficiency (Figure 3A) revealed the impact of the surfactant included in the formulation. When DP80 was used, the values of transfection started to increase at the 6/1 mass. Complexes at the lower ratios (2/1 and 4/1) had previously shown an inability to protect DNA against enzymatic digestion (Figure 2C), which could justify low transfection efficiencies at these ratios. However, the incorporation of P in DPP80 formulation markedly enhanced transfection at the lower ratios (2/1 and 4/1) and maintained sufficient transfection at the higher ratios. The addition of P in DPP80 complexes shifted the ZP (Figure 2A) from negative to positive values at small mass ratios (2/1 and 4/1), which would enhance uptake, and thus transfection (Figure 3A).42 Interestingly, DPP80 depicted higher MFI compared with DP80 at the cationic lipid/DNA mass ratio of 6/1 (maximum transfection). Actually, one of the most relevant concerns of gene therapy is to attain enough expression of the protein to reach physiological levels. Thus, the amount of protein expression by the transfected cells needs to be also considered along with the percentage of transfected cells when designing nonviral vectors for gene-delivery purposes.26
Regarding cell viability, DPP80 niosomes showed less toxic effect on NT2 cells than DP80 counterpart at all mass ratios analyzed, being cell viability at 6/1 mass ratio significantly superior to that reported by liposome-based L2K transfection reagent. These promising data encouraged us to perform further in vivo studies, where not only gene-delivery efficiency but also cytotoxicity needs to be considered. The higher cationic lipid/DNA mass ratios resulted in a clear toxic effect on cells which were more accused in the case of DP80 niosomes, without the increase of transfection efficiency.
The fact that both the type of surfactant and the amount of such surfactant influence the biological function of nanovesicles43 could explain the low percentage of transfected cells and viability results observed when only P (Figure S2) or P80 (Figure 3A) surfactant was used in niosome formulations (DP and DP80, respectively). Additionally, when liposome vesicles were elaborated in the absence of any kind of surfactant (referred to as D; Figure S2), both transfection efficiency and cell viability values were also lower than the values obtained with DPP80 niosomes. To analyze more deeply the complex transfection mechanism of DPP80 complexes in NT2 culture cells, we performed some trafficking studies to investigate both the cellular uptake of complexes and the posterior cytoplasmic disposition inside the cells, since it has been previously reported on the literature that those parameters could markedly influence the gene-delivery properties of vectors.14 Cellular uptake values were compared with DP80 niosomes as well as L2K transfection reagent. As can be observed in Figure 4A, the uptake of DPP80 complexes exceeded that of their DP80 counterparts that could be due to their higher surface charge (Figure 2A) or to more specific interactions of such nioplexes with some lipid components present on the cell membrane.44 Additionally, it has been reported that variations in the length of surfactant carbon chains and/or in the number of unsaturated bonds (inside their carbon chains) can affect the permeability of nanovesicles across membranes.29 However, the great difference observed on DPP80 complexes between cellular uptake (95%; Figure 4A) and percentage of transfected cells (30%; Figure 3A) suggests that other biological processes involved on the transfection process should also be evaluated.45 Therefore, we analyzed as well the endocytosis mechanism and the co-localization with the late endosome. Despite the absence of consensus on the most efficient endocytosis pathway, we analyzed the endocytosis processes mediated by clathrin, caveolae, and macropinocytosis, which could affect the fate of internalized complexes.46
The observed results in Figure 4C suggested that internalization of DPP80 complexes was mediated, mainly by caveolae and macropinocytosis. Such endocytosis pathways, normally, are connected with the lysosomes. In such compartments, the acid pH value, normally, degrades the genetic material, which in turn could reduce the percentage of transfected cells.45 Therefore, the co-localization of complexes with the late endosome/lysosome (Figure 4C4) could explain why out of 95% of NT2 that captured DPP80 complexes (Figure 4A), only around 30% of them were successful to eventually express EGFP (Figure 3).47
Next, and before proceeding for in vivo studies in rat brain, we analyzed the transfection efficiency of DPP80 complexes in primary cortical cultures of rat embryos, where different kinds of neurons and glial cells are interconnected to set up complex neuronal–glial networks. Results showed some positive EGFP cells (Figure 5A1). Next, the lack of GFAP+ve reactivity in immunohistochemical assays confirmed that such EGFP-positive cells could be neurons (Figure 5A2). After observing these promising results obtained in neurons, we proceeded to evaluate in rat brain the protein expression of DPP80 complexes after intracranial injection. We did not observe any sign of toxicity in the cortical tissue after injection (data not shown). Unlike the primary culture observations, the NeuN−ve cells (neuroglia) were the only transfected cells (Figure 5B). The discrepancies between in vitro and in vivo assays are widely recognized and could be probably due to the fundamental difference in the mechanism of gene delivery in both biological contexts.48
The preference of DPP80 complexes to transfect mainly glial cells instead of neurons could be explained by the fact that glial cells have a higher mitotic and phagocytic activity.49 Therefore, transfection process could be enhanced in such circumstances. By contrast, the lack of protein expression observed in neurons of rat cortex exposed to DPP80 complexes could be explained by the particular biological processes of these kind of cells where both endocytosis and posterior intracellular disposition are severely hampered.50 Considering these encouraging in vivo results, DPP80 complexes could be used as nonviral vectors to deliver genetic material into glial cells in the CNS, in order to tackle neurological disorders that affect these kind of cells.51 Alterations of glial cells are commonly found in many devastating neurological disorders such as Alzheimer and Parkinson, to name just the most relevant ones.51,52
Conclusion
Our results showed that physicochemically characterized DPP80 niosomes elaborated with a mixture of both P and P80 nonionic surfactants were able to transfect efficiently NT2 cells without compromising their viability after cellular uptake mainly mediated by CME and MPC. Such novel biocompatible nanocarriers were able to induce in vivo transfection into glial cells of rats after intracortical administration. Interestingly, DPP80 gene carriers not only represent a promising tool for direct in vivo transfection, but also for cell-based gene-delivery applications, since transfected NT2 cells or their derived neurons could be used as a delivery tool of therapeutic genes for different neurologic disorders.53,54
Supplementary materials
(A) EGFP expression in rat cortex. Nuclei are shown in blue (Hoechst); scale bar=40 µm. (B) Contralateral picture. Nuclei are shown in blue (Hoechst), and neurons in red (NeuN+ve); scale bar=40 µm.
(A) Transfection efficiency of D liposomes and DP niosomes in NT2 cells. Percentages of cells that express green protein (bars) and live cells (lines). Values represent mean±SD (n=3). (B1 and B2) Overlay of fluorescence and DIC pictures of NT2 cells with D liposomes (6/1) and DP niosomes (2/1), respectively (scale bar=100 µm).
Abbreviations: DIC, digital image correlation; T, transfection; V, viability.
Table S1.
Polydispersity index (PDI) of DP80 and DPP80 niocomplexes at different cationic lipid/DNA mass ratios
| Nioplexes | 2/1 | 4/1 | 6/1 | 8/1 | 10/1 | 12/1 |
|---|---|---|---|---|---|---|
|
| ||||||
| DP80 | 0.296±0.016 | 0.222±0.014 | 0.211±0.013 | 0.193±0.020 | 0.229±0.009 | 0.234±0.012 |
| DPP80 | 0.208±0.031 | 0.211±0.017 | 0.230±0.014 | 0.217±0.003 | 0.258±0.032 | 0.202±0.005 |
Note: Data represent mean±SD (n=3).
Acknowledgments
This project was supported by the Basque Country Government (CGIC10/172), Spanish Ministry of Education (Grant CTQ2017-84415-R, MAT2015-69967-C3-1R), the Generalitat de Catalunya (2014/SGR/624), and the Instituto de Salud Carlos III (CB06_01_0019, CB06_01_1028). The authors also wish to thank the intellectual and technical assistance from the ICTS “NANBIOSIS”, more specifically by the Drug Formulation Unit (U10) of the CIBER in Bioengineering, Biomaterials, and Nanomedicine (CIBER-BBN) at the University of Basque Country (UPV/EHU). Technical and human support provided by SGIker (UPV/EHU) is gratefully acknowledged. Jose Luis Pedraz and Gustavo Puras are corresponding authors for this study.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Tardieu M, Zérah M, Gougeon ML, et al. Intracerebral gene therapy in children with mucopolysaccharidosis type IIIB syndrome: an uncontrolled phase 1/2 clinical trial. Lancet Neurol. 2017;16(9):712–720. doi: 10.1016/S1474-4422(17)30169-2. [DOI] [PubMed] [Google Scholar]
- 2.Kaplitt MG, Feigin A, Tang C, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson’s disease: an open label, phase I trial. Lancet. 2007;369(9579):2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- 3.Tuszynski MH, Thal L, Pay M, et al. A phase 1 clinical trial of nerve growth factor gene therapy for Alzheimer disease. Nat Med. 2005;11(5):551–555. doi: 10.1038/nm1239. [DOI] [PubMed] [Google Scholar]
- 4.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. On-demand optogenetic control of spontaneous seizures in temporal lobe epilepsy. Nat Commun. 2013;4:1376. doi: 10.1038/ncomms2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jayant RD, Sosa D, Kaushik A, et al. Current status of non-viral gene therapy for CNS disorders. Expert Opin Drug Deliv. 2016;13(10):1433–1445. doi: 10.1080/17425247.2016.1188802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palfi S, Gurruchaga JM, Ralph GS, et al. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson’s disease: a dose escalation, open-label, phase 1/2 trial. Lancet. 2014;383(9923):1138–1146. doi: 10.1016/S0140-6736(13)61939-X. [DOI] [PubMed] [Google Scholar]
- 7.Rafii MS, Baumann TL, Bakay RA, et al. A phase1 study of stereotactic gene delivery of AAV2-NGF for Alzheimer’s disease. Alzheimers Dement. 2014;10(5):571–581. doi: 10.1016/j.jalz.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Pluvinage JV, Wyss-Coray T. Microglial Barriers to Viral Gene Delivery. Neuron. 2017;93(3):468–470. doi: 10.1016/j.neuron.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Liang H, Liu J, Wang Z. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int J Pharm. 2018;546(1–2):215–225. doi: 10.1016/j.ijpharm.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Li J, Liu N, et al. In vitro studies of phospholipid-modified PAMAM-siMDR1 complexes for the reversal of multidrug resistance in human breast cancer cells. Int J Pharm. 2017;530(1–2):291–299. doi: 10.1016/j.ijpharm.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Vanderwall AG, Noor S, Sun MS, et al. Effects of spinal non-viral interleukin-10 gene therapy formulated with d-mannose in neuropathic interleukin-10 deficient mice: Behavioral characterization, mRNA and protein analysis in pain relevant tissues. Brain Behav Immun. 2018;69:91–112. doi: 10.1016/j.bbi.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torchilin VP. Nanoparticulates as Drug Carriers. London: Imperial College Press; 2006. [Google Scholar]
- 13.Kazi KM, Mandal AS, Biswas N, et al. Niosome: A future of targeted drug delivery systems. J Adv Pharm Technol Res. 2010;1(4):374–380. doi: 10.4103/0110-5558.76435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mashal M, Attia N, Puras G, Martínez-Navarrete G, Fernández E, Pedraz JL. Retinal gene delivery enhancement by lycopene incorporation into cationic niosomes based on DOTMA and polysorbate 60. J Control Release. 2017;254:55–64. doi: 10.1016/j.jconrel.2017.03.386. [DOI] [PubMed] [Google Scholar]
- 15.Allam A, Fetih G. Sublingual fast dissolving niosomal films for enhanced bioavailability and prolonged effect of metoprolol tartrate. Drug Des Devel Ther. 2016;10:2421–2433. doi: 10.2147/DDDT.S113775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puras G, Mashal M, Zárate J, et al. A novel cationic niosome formulation for gene delivery to the retina. J Control Release. 2014;174:27–36. doi: 10.1016/j.jconrel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Puras G, Martínez-Navarrete G, Mashal M, et al. Protamine/DNA/Niosome Ternary Nonviral Vectors for Gene Delivery to the Retina: The Role of Protamine. Mol Pharm. 2015;12(10):3658–3671. doi: 10.1021/acs.molpharmaceut.5b00422. [DOI] [PubMed] [Google Scholar]
- 18.Ojeda E, Puras G, Agirre M, et al. The influence of the polar head-group of synthetic cationic lipids on the transfection efficiency mediated by niosomes in rat retina and brain. Biomaterials. 2016;77:267–279. doi: 10.1016/j.biomaterials.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 19.Gu JH, Ge JB, Li M, Xu HD, Wu F, Qin ZH. Poloxamer 188 protects neurons against ischemia/reperfusion injury through preserving integrity of cell membranes and blood brain barrier. PLoS One. 2013;8(4):e61641. doi: 10.1371/journal.pone.0061641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan H, Zhang W, du YZ, Hu FQ, Yz D, Fq H. Ternary nanoparticles of anionic lipid nanoparticles/protamine/DNA for gene delivery. Int J Pharm. 2010;392(1–2):224–231. doi: 10.1016/j.ijpharm.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Law SL, Chuang TC, Kao MC, Lin YS, Huang KJ. Gene transfer mediated by sphingosine/dioleoylphosphatidylethanolamine liposomes in the presence of poloxamer 188. Drug Deliv. 2006;13(1):61–67. doi: 10.1080/10717540500309024. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Li X, Wang L, Xu Y, Cheng X, Wei P. Formulation and pharmacokinetic evaluation of a paclitaxel nanosuspension for intravenous delivery. Int J Nanomedicine. 2011;6:1497–1507. doi: 10.2147/IJN.S21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muzzalupo R, Tavano L, Lai F, Picci N. Niosomes containing hydroxyl additives as percutaneous penetration enhancers: effect on the transdermal delivery of sulfadiazine sodium salt. Colloids Surf B Biointerfaces. 2014;123:207–212. doi: 10.1016/j.colsurfb.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 24.Ojeda E, Puras G, Agirre M, et al. Niosomes based on synthetic cationic lipids for gene delivery: the influence of polar head-groups on the transfection efficiency in HEK-293, ARPE-19 and MSC-D1 cells. Org Biomol Chem. 2015;13(4):1068–1081. doi: 10.1039/c4ob02087a. [DOI] [PubMed] [Google Scholar]
- 25.Ojeda E, Agirre M, Villate-Beitia I, et al. Elaboration and Physicochemical Characterization of Niosome-Based Nioplexes for Gene Delivery Purposes. Methods Mol Biol. 2016;1445:63–75. doi: 10.1007/978-1-4939-3718-9_5. [DOI] [PubMed] [Google Scholar]
- 26.Attia N, Mashal M, Grijalvo S, et al. Stem cell-based gene delivery mediated by cationic niosomes for bone regeneration. Nanomedicine. 2018;14(2):521–531. doi: 10.1016/j.nano.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Mashal M, Attia N, Soto-Sánchez C, et al. Non-viral vectors based on cationic niosomes as efficient gene delivery vehicles to central nervous system cells into the brain. Int J Pharm. 2018;552(1–2):48–55. doi: 10.1016/j.ijpharm.2018.09.038. [DOI] [PubMed] [Google Scholar]
- 28.Ochoa GP, Sesma JZ, Díez MA, et al. A novel formulation based on 2,3-di(tetradecyloxy)propan-1-amine cationic lipid combined with polysorbate 80 for efficient gene delivery to the retina. Pharm Res. 2014;31(7):1665–1675. doi: 10.1007/s11095-013-1271-5. [DOI] [PubMed] [Google Scholar]
- 29.Basha M, Abd El-Alim SH, Shamma RN, Awad GE. Design and optimization of surfactant-based nanovesicles for ocular delivery of Clotrimazole. J Liposome Res. 2013;23(3):203–210. doi: 10.3109/08982104.2013.788025. [DOI] [PubMed] [Google Scholar]
- 30.Bnyan R, Khan I, Ehtezazi T, et al. Surfactant Effects on Lipid-Based Vesicles Properties. J Pharm Sci. 2018;107(5):1237–1246. doi: 10.1016/j.xphs.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Pezzoli D, Giupponi E, Mantovani D, Candiani G. Size matters for in vitro gene delivery: investigating the relationships among complexation protocol, transfection medium, size and sedimentation. Sci Rep. 2017;7:44134. doi: 10.1038/srep44134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Bihan O, Chèvre R, Mornet S, Garnier B, Pitard B, Lambert O. Probing the in vitro mechanism of action of cationic lipid/DNA lipoplexes at a nanometric scale. Nucleic Acids Res. 2011;39(4):1595–1609. doi: 10.1093/nar/gkq921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mady MM, Darwish MM, Khalil S, Khalil WM. Biophysical studies on chitosan-coated liposomes. Eur Biophys J. 2009;38(8):1127–1133. doi: 10.1007/s00249-009-0524-z. [DOI] [PubMed] [Google Scholar]
- 34.Eastman SJ, Siegel C, Tousignant J, Smith AE, Cheng SH, Scheule RK. Biophysical characterization of cationic lipid: DNA complexes. Biochim Biophys Acta. 1997;1325(1):41–62. doi: 10.1016/s0005-2736(96)00242-8. [DOI] [PubMed] [Google Scholar]
- 35.Jain KK. Cell therapy for CNS trauma. Mol Biotechnol. 2009;42(3):367–376. doi: 10.1007/s12033-009-9166-8. [DOI] [PubMed] [Google Scholar]
- 36.Cacciotti I, Ceci C, Bianco A, Pistritto G. Neuro-differentiated Ntera2 cancer stem cells encapsulated in alginate beads: First evidence of biological functionality. Mater Sci Eng C Mater Biol Appl. 2017;81:32–38. doi: 10.1016/j.msec.2017.07.033. [DOI] [PubMed] [Google Scholar]
- 37.Hara K, Yasuhara T, Maki M, et al. Neural progenitor NT2N cell lines from teratocarcinoma for transplantation therapy in stroke. Prog Neurobiol. 2008;85(3):318–334. doi: 10.1016/j.pneurobio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Hao S, Wang B. Editorial: Review on Intracerebral Haemorrhage: Multidisciplinary Approaches to the Injury Mechanism Analysis and Therapeutic Strategies. Curr Pharm Des. 2017;23(15):2159–2160. doi: 10.2174/1381612823999170307150448. [DOI] [PubMed] [Google Scholar]
- 39.Frosina G. Stem cell-mediated delivery of therapies in the treatment of glioma. Mini Rev Med Chem. 2011;11(7):591–598. doi: 10.2174/138955711795906897. [DOI] [PubMed] [Google Scholar]
- 40.Ahmed AU, Alexiades NG, Lesniak MS. The use of neural stem cells in cancer gene therapy: predicting the path to the clinic. Curr Opin Mol Ther. 2010;12(5):546–552. [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrari A, Ehler E, Nitsch RM, Götz J. Immature human NT2 cells grafted into mouse brain differentiate into neuronal and glial cell types. FEBS Lett. 2000;486(2):121–125. doi: 10.1016/s0014-5793(00)02251-1. [DOI] [PubMed] [Google Scholar]
- 42.Midoux P, Monsigny M. Efficient gene transfer by histidylated polylysine/pDNA complexes. Bioconjug Chem. 1999;10(3):406–411. doi: 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- 43.Taymouri S, Varshosaz J. Effect of different types of surfactants on the physical properties and stability of carvedilol nano-niosomes. Adv Biomed Res. 2016;5:48. doi: 10.4103/2277-9175.178781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma B, Zhang S, Jiang H, Zhao B, Lv H. Lipoplex morphologies and their influences on transfection efficiency in gene delivery. J Control Release. 2007;123(3):184–194. doi: 10.1016/j.jconrel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 45.Goldshtein M, Forti E, Ruvinov E, Cohen S. Mechanisms of cellular uptake and endosomal escape of calcium-siRNA nanocomplexes. Int J Pharm. 2016;515(1–2):46–56. doi: 10.1016/j.ijpharm.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Zhao F, Zhao Y, Liu Y, Chang X, Chen C, Zhao Y. Cellular uptake, intracellular trafficking, and cytotoxicity of nanomaterials. Small. 2011;7(10):1322–1337. doi: 10.1002/smll.201100001. [DOI] [PubMed] [Google Scholar]
- 47.Ruiz de Garibay AP, Solinís Aspiazu MÁ, Rodríguez Gascón A, Ganjian H, Fuchs R. Role of endocytic uptake in transfection efficiency of solid lipid nanoparticles-based nonviral vectors. J Gene Med. 2013;15(11–12):427–440. doi: 10.1002/jgm.2749. [DOI] [PubMed] [Google Scholar]
- 48.Puras G, Zarate J, Díaz-Tahoces A, Avilés-Trigueros M, Fernández E, Pedraz JL. Oligochitosan polyplexes as carriers for retinal gene delivery. Eur J Pharm Sci. 2013;48(1–2):323–331. doi: 10.1016/j.ejps.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Schafer DP, Stevens B. Phagocytic glial cells: sculpting synaptic circuits in the developing nervous system. Curr Opin Neurobiol. 2013;23(6):1034–1040. doi: 10.1016/j.conb.2013.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergen JM, Park IK, Horner PJ, Pun SH. Nonviral approaches for neuronal delivery of nucleic acids. Pharm Res. 2008;25(5):983–998. doi: 10.1007/s11095-007-9439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barres BA. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60(3):430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Milligan ED, Watkins LR. Pathological and protective roles of glia in chronic pain. Nat Rev Neurosci. 2009;10(1):23–36. doi: 10.1038/nrn2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson DJ, Longhi L, Lee EB, et al. Genetically modified NT2N human neuronal cells mediate long-term gene expression as CNS grafts in vivo and improve functional cognitive outcome following experimental traumatic brain injury. J Neuropathol Exp Neurol. 2003;62(4):368–380. doi: 10.1093/jnen/62.4.368. [DOI] [PubMed] [Google Scholar]
- 54.Longhi L, Watson DJ, Saatman KE, et al. Ex vivo gene therapy using targeted engraftment of NGF-expressing human NT2N neurons attenuates cognitive deficits following traumatic brain injury in mice. J Neurotrauma. 2004;21(12):1723–1736. doi: 10.1089/neu.2004.21.1723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) EGFP expression in rat cortex. Nuclei are shown in blue (Hoechst); scale bar=40 µm. (B) Contralateral picture. Nuclei are shown in blue (Hoechst), and neurons in red (NeuN+ve); scale bar=40 µm.
(A) Transfection efficiency of D liposomes and DP niosomes in NT2 cells. Percentages of cells that express green protein (bars) and live cells (lines). Values represent mean±SD (n=3). (B1 and B2) Overlay of fluorescence and DIC pictures of NT2 cells with D liposomes (6/1) and DP niosomes (2/1), respectively (scale bar=100 µm).
Abbreviations: DIC, digital image correlation; T, transfection; V, viability.
Table S1.
Polydispersity index (PDI) of DP80 and DPP80 niocomplexes at different cationic lipid/DNA mass ratios
| Nioplexes | 2/1 | 4/1 | 6/1 | 8/1 | 10/1 | 12/1 |
|---|---|---|---|---|---|---|
|
| ||||||
| DP80 | 0.296±0.016 | 0.222±0.014 | 0.211±0.013 | 0.193±0.020 | 0.229±0.009 | 0.234±0.012 |
| DPP80 | 0.208±0.031 | 0.211±0.017 | 0.230±0.014 | 0.217±0.003 | 0.258±0.032 | 0.202±0.005 |
Note: Data represent mean±SD (n=3).