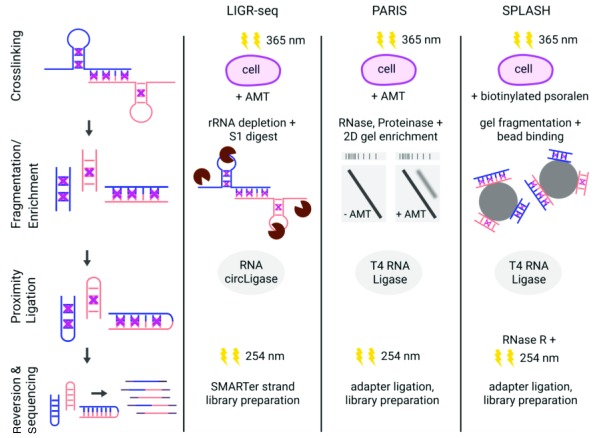
Figure 1. Schematic comparison of Direct Duplex Detection methods.
Overview of the protocols for LIGR-seq 21, PARIS 22, and SPLASH 23. The first column shows the principal steps of the three experimental procedures: crosslinking (violet crosses) was conducted in vivo by 365 nm irradiation and 4′-aminomethyltrioxsalen (AMT) or biotinylated psoralen treatment. Fragmentation was performed enzymatically (LIGR-seq and PARIS) or chemically (SPLASH). Crosslinked RNAs were additionally enriched either by size separation using two-dimensional (2D) gel electrophoresis (crosslinked RNAs above the main diagonal were eluted; PARIS) or by biotin-streptavidin binding to magnetic beads (SPLASH). Proximity ligation was carried out using different ligases. Crosslinks were reverted by 254 nm irradiation. For sequencing, different library preparation strategies were performed. Colors of RNA strands (blue and orange) indicate different RNA molecules. LIGR-seq, ligation of interacting RNA followed by high-throughput sequencing; PARIS, psoralen analysis of RNA interactions and structures; SPLASH, sequencing of psoralen-crosslinked, ligated, and selected hybrids.