Abstract
Hsp90 couples ATP hydrolysis to large conformational changes essential for activation of client proteins. The structural transitions involve dimerization of the N-terminal domains and formation of ‘closed states’ involving the N-terminal and middle domains. Here, we used Hsp90 mutants that modulate ATPase activity and biological function as probes to address the importance of conformational cycling for Hsp90 activity. We found no correlation between the speed of ATP turnover and the in vivo activity of Hsp90: some mutants with almost normal ATPase activity were lethal, and some mutants with lower or undetectable ATPase activity were viable. Our analysis showed that it is crucial for Hsp90 to attain and spend time in certain conformational states: a certain dwell time in open states is required for optimal processing of client proteins, whereas a prolonged population of closed states has negative effects. Thus, the timing of conformational transitions is crucial for Hsp90 function and not cycle speed.
Hsp90 is an ATP-dependent molecular chaperone that controls a large set of client proteins1–6. Approximately 20% of the proteome appears to directly or indirectly depend on Hsp90 (ref. 7). Hsp90 is a homodimer in which each monomer consists of an N-terminal domain (N domain, which contains the nucleotide-binding pocket8) connected via a long charged linker to the middle domain (M domain). The C-terminal domain is responsible for dimerization9, which is a prerequisite for efficient ATPase activity10–12. The last five amino acids of Hsp90 form the MEEVD motif, which serves as the binding site for a large class of cochaperones containing a peptide-binding TPR domain13.
ATP binding and hydrolysis, which are important for the function of Hsp90 in vivo14–16, are coupled to a slow conformational cycle in which the Hsp90 dimer progresses from an N-terminally open state to a closed state. The open state is V-shaped and involves dimerization via the C domains and no interaction of the N domains17. In the crystal structure of Hsp90 in the presence of the nonhydrolyzable ATP analog AMP-PNP, Hsp90 adopts a compact structure in which the N domains are also associated18. The conformational changes from the open to the N-terminally closed state are rate limiting and precede ATP hydrolysis19,20. A fluorescence resonance energy transfer (FRET)-based kinetic analysis of the conformational cycle has revealed that Hsp90 adopts specific intermediate states19 before it finally reaches a fully closed state in which ATP hydrolysis occurs. Specifically, after ATP binding, the so-called ATP lid in the Hsp90 N domain changes its position and exposes a hydrophobic N-domain patch that, together with its counterpart in the second monomer, stabilizes the N-terminally closed state18. Furthermore, this conformational change releases the contacts with the N-terminal helix, which subsequently binds to the N domain of the second subunit within the dimer. Once the N-terminal association is achieved, the Hsp90 N domains associate with the Hsp90 M domains and form the active ATPase. Some of the conformational states appear to be isoenergetic, and random fluctuations between states are possible21,22. Several cochaperones target specific conformations of Hsp90 and thus reduce structural fluctuations; in addition, they modulate specific properties of Hsp90, such as its ATPase activity. Here, we used a set of yeast Hsp90 N- and M-domain mutants with reported effects on biological activity to gain insight into the importance of the timing of conformational transitions for Hsp90 functionality and client-protein processing. Our combined in vivo and in vitro assays demonstrated that the time spent in open conformations, together with the ability to proceed to the N-terminally closed state, is important for client processing and the essential functions of Hsp90 in yeast.
Results
N- and M-domain mutations affect Hsp90’s ATPase activity
The conformational transition from the open to the closed state is rate limiting in the ATPase cycle of Hsp90. We used N- and M-domain mutations with reported effects on the ATP turnover23 and biological activity of yeast Hsp90 (refs. 12,14,15,18,24–27) (Fig. 1a and Table 1) to probe the role of specific conformational rearrangements in Hsp90. These mutants affect binding interfaces in the N and M domains that are linked to distinct functional states of Hsp90. According to their ATPase activities, the mutants fell into three categories (Fig. 1b and Supplementary Table 1). The first exhibited an increased turnover rate: A107N and Δ8-Hsp90 showed the strongest effect, and T22I showed a slight increase in ATPase activity. The second class included mutants with decreased ATPase activity, such as R346S and R380A. The third class included ATPase-deficient mutants, such as E33A and D79N. For most mutants, the Km values for ATP binding were comparable to those of the wild type (WT) (Supplementary Table 1). Interestingly, A107N and Δ8-Hsp90 exhibited a higher affinity for ATP, thus suggesting changes in the ATP-binding pocket.
Figure 1.
Localization and effects of mutations on ATPase activities. (a) Mutated residues are shown as red spheres in the crystal structure of closed yeast Hsp90 (PDB 2CG9)18. Blue, N domain; green, M domain; orange, C domain; cyan, lid structure. (b) Influence of mutations on the ATPase activity of Hsp90. Coupled enzymatic ATPase assay (gray bars). Data are shown as mean ± s.d. (n = 3 technical replicates). Inset, kcat values of the Hsp90 variants in the presence of various concentrations of ATP: black, WT; yellow, A107N; blue, Δ8; green, T22I; purple, R346S; gray, R380A; red, E33A; cyan, D79N. (c) Stimulation of Hsp90 ATPase by the cochaperone Aha1, determined by a coupled enzymatic ATPase assay. Fold increases in ATPase activity in the presence of Aha1 are shown. Inset, kcat values of the Hsp90 mutants in the presence of various Aha1 concentrations: black, WT; yellow, A107N; blue, Δ8; green, T22I; purple, R346S; gray, R380A; red, E33A; cyan, D79N. (d) Viability of the Hsp90 variants deduced by a 5-fluoroorotic acid (5-FOA) shuffling assay. Shuffling experiments were carried out at least three times and represent technical replicates started from independent single yeast colonies. (e) Western blot analysis showing expression of Hsp90 in yeast cells after 5-FOA shuffling. PGK1, loading control.
Table 1. Reported characteristics of analyzed Hsp90 variants.
| Hsp90 variant | Effects | References |
|---|---|---|
| Δ8 | Increased ATPase activity; increased N-terminal dimerization in the presence of ATP | 23 |
| T22I | Temperature-sensitive mutant of T22, which is covered by the lid in the open state; increased ATPase activity | 12,24,25 |
| E33A | Hydrolysis deficient but able to bind ATP; reported lethality in yeast | 14,15 |
| D79N | Mutant of D79, which directly contacts the adenine base of the bound ATP; loss of the ability to bind ATP; loss of viability of yeast cells | 14,15 |
| A107N | Increased ATPase rate through stabilization of the N-terminally dimerized state | 12,18 |
| R346S | Decreased ATPase activity | 26 |
| R380A | Mutant of R380, which is located in the catalytic loop of the M domain and orients the γ-phosphate for hydrolysis; decreased ATPase activity in vitro and lethality in yeast | 18,27 |
The altered ATPase activities indicated that the conformational cycle was affected. We used the cochaperone Aha1 as a conformational sensor for structural alterations. Aha1 accelerates the cycle by promoting the formation of the unfavorable closed conformation19,28,29. All variants, except E33A and D79N, were stimulated by Aha1, albeit to a different extent (Fig. 1c and Supplementary Table 1). Interestingly, the R346S variant with inherently low ATPase activity exhibited a WT-like kcat value, thus demonstrating that conformational transitions preceding the formation of closed states are compromised in the absence of Aha1. In contrast, R380A, which exhibited an ATPase activity as slow as that of R346S, was stimulated only slightly by Aha1, thus suggesting that different conformational steps are affected.
Yeast viability does not require Hsp90 ATP hydrolysis
To investigate the biological consequences of the alterations in the conformational cycle of Hsp90, we tested the variants for their ability to support viability in a yeast strain in which both Hsp90 isoforms were deleted, and Hsp90 was provided in a plasmid24. Control experiments confirmed that after loss of the WT-URA plasmid, the strain could not grow on medium lacking uracil (Supplementary Fig. 1a). Expression of the variants T22I, E33A, A107N and R346S supported viability similar to that of the WT protein (Fig. 1d,e). The expression of the D79N and R380A variants caused lethality, as previously reported14,15,27. Surprisingly, in contrast to the results of previous studies14–16, we found that yeast cells expressing E33A as the sole source of Hsp90 were able to grow, although we detected no ATPase activity in vitro. To further investigate this effect, we tested the ability of the Hsp90 E33A mutant under control of the endogenous promoter to support growth of different yeast strains. We additionally performed tetrad dissections (Supplementary Fig. 1b,c). All of these experiments confirmed our initial findings. In contrast, the Δ8 Hsp90 variant was not able to support yeast viability despite its increased ATPase activity. Finally, although R380A and R346S displayed similar ATPase activities, only R346S supported viability. These results showed that the ability to hydrolyze ATP is not sufficient to confer the essential function of Hsp90.
Hsp90 mutants differ in accumulation of the closed state
To obtain insight into the structural consequences of mutating specific residues, we used small-angle X-ray scattering (SAXS) to determine the shapes of the Hsp90 variants (Supplementary Fig. 2). In the absence of nucleotide, WT yeast Hsp90 adopted an open conformation characterized by a broad distribution in the interatomic distance distribution (P(r)) curve (Fig. 2a and Supplementary Table 2; radius of gyration (Rg) of 64.2 Å; maximum dimension (Dmax) of 240 Å), as previously reported30,31. Most variants tested showed a WT-like shape in the absence of nucleotide. The only exception was the lethal Δ8-Hsp90 mutant, which adopted a partly compact state, as indicated by the reduced Rg of 61.9 Å and a redistribution of the P(r) to smaller distances. This mutant appeared to be closed to some extent even in the absence of nucleotide (Fig. 2a).
Figure 2.
SAXS analysis of Hsp90 variants. (a–c) P(r) curves for WT and mutant Hsp90 in the absence of nucleotide (a) or the presence of 4 mM ATP-γS (b) or ATP (c). Bottom, mutants for which significant differences in the P(r) were observed; top, mutants for which no differences or only minor differences were observed. Black, WT; yellow, A107N; blue, Δ8; green, T22I; purple, R346S; gray, R380A; red, E33A.
In the presence of ATP-γS, a slowly hydrolyzed ATP analog, all variants displayed a narrow range of distribution in the P(r) curves, a result indicative of a more compact state (Fig. 2b). Δ8 was further compacted, and the mutant E33A also showed a compact shape. Thus, E33A was able to undergo specific conformational transitions in response to binding ATP-γS. Compared with the WT protein, the R380A and R346S mutants were less efficient at adopting the closed state.
In the presence of ATP, Δ8, T22I, A107N and the ATPase-deficient mutant E33A showed a redistribution of distances in the P(r) curves, thus indicating that they adopted a more compact conformation, whereas the WT protein and R346S and R380A variants displayed open conformations (Fig. 2c). Thus, the SAXS analysis showed that several of the mutants—particularly Δ8 and E33A, and to lesser extent T22I and A107N—exhibited scattering properties consistent with the increased presence of closed states, in contrast with the properties of the WT protein.
Structural effects of mutations in the N domain
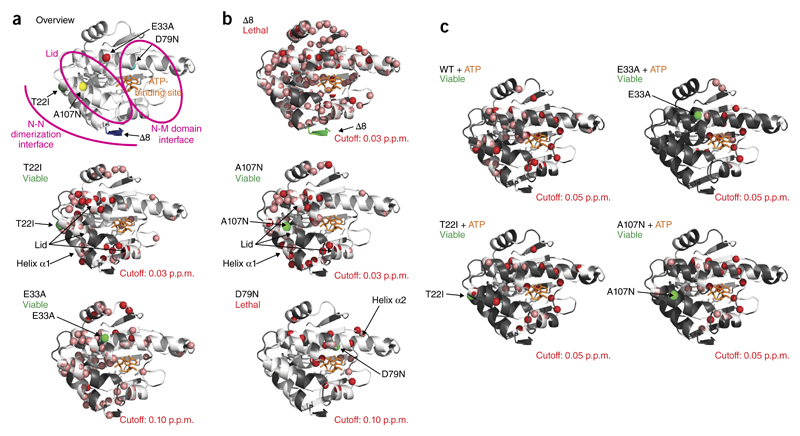
NMR 1H,15N correlation spectra of the mutations within the N domain (Supplementary Fig. 3) indicated mutation-specific conformational changes in the N domain, as evidenced by the chemical shift-changes between WT and mutants (Fig. 3a,b and Supplementary Fig. 4). The deletion of the first eight amino acids (Δ8) led to substantial chemical-shift changes for residues in various regions of the N domain (Fig. 3b). The other mutant with strongly increased ATPase activity, A107N, exhibited local changes in the lid and in helices α1 and α2. For the variant T22I, the same structural elements as in A107N were affected (Fig. 3b). For both variants, various NMR chemical shifts, even those far from the location of the mutated residue, indicated a complex allosteric network of interactions in the N domain. T22 and A107 are located in helix α1 and the lid, respectively, and are involved in N-terminal closure of the full-length Hsp90 dimer (described below).
Figure 3.
NMR analysis of N-domain mutants. (a) Overview of mutations in the N domain of Hsp90. (b) Amide chemical-shift differences larger than the indicated cutoff value observed in 1H,15N-HSQC spectra of the Hsp90 N-domain mutants compared with the WT protein, as indicated by red spheres on the crystal structure of residues 1–210 of yeast Hsp90 (PDB 1AM1)8. Residues with large chemical-shift changes in the mutant could not be unambiguously assigned and are indicated by salmon spheres; residues not assigned in the WT protein are shown in gray. Bound ADP is shown in orange to indicate the nucleotide-binding site. The position of the mutation is indicated by a green sphere. (c) Differences in chemical shifts between the free mutant and the complex with ATP are indicated, as in b. Shifting residues whose endpoints could not be assigned in the complex are indicated by salmon spheres; unassigned residues in the free mutant or overlapping residues, which could not be assigned in the mutant, are in gray.
The two mutations that lacked ATPase activity showed distinct patterns. Whereas the E33A mutation affected all α-helices, D79N had more local effects in the β-sheet and in helix α2, results consistent with direct contacts being made to ATP8. The more widespread chemical-shift changes observed for E33A indicated that this residue is important for the overall conformation and dynamics of the N domain.
In the presence of ATP, changes for A107N and T22I were in the vicinity of the nucleotide-binding site (Fig. 3c). As expected, E33A but not D79N responded to ATP (Fig. 3c). ATP affected the β-sheet, helix α2 and the lid. We observed similar changes in the presence of AMP-PNP (Supplementary Fig. 5), thus demonstrating that nucleotide binding is sufficient to induce structural changes in the E33A mutant.
The mutations affect distinct conformational steps
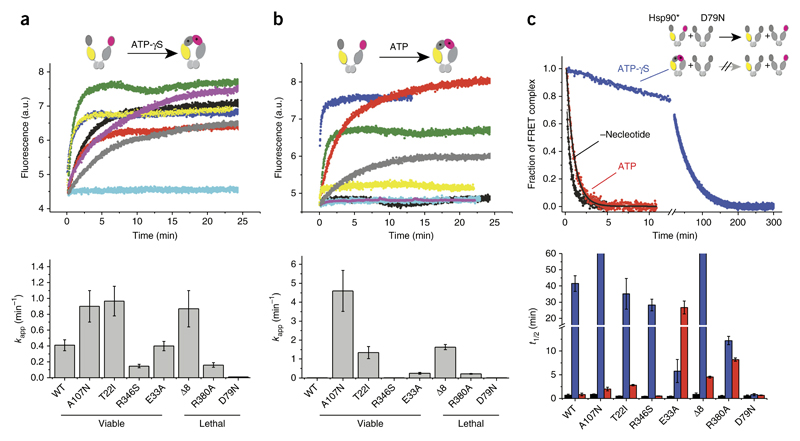
To probe whether the mutations affected the transition to the closed state, we used a FRET system in which the M and N domains in the dimer were labeled with a donor or acceptor dye, respectively19. After mixing of the components, all Hsp90 variants exchanged subunits and formed FRET complexes (Supplementary Fig. 6 and Supplementary Table 3). Addition of ATP-γS to labeled WT Hsp90 induced N-terminal closing and concomitant increases in FRET efficiency and thus the population of the closed state of Hsp90 (ref. 19) (Fig. 4a). All mutants except D79N showed changes in FRET efficiency in response to ATP-γS, thus indicating N-terminal closing (Fig. 4a and Supplementary Table 2). As compared with the kinetics of the WT, the kinetics was equally fast or was accelerated for mutants with increased ATPase activity (Δ8, T22I and A107N) and was decelerated for variants with decreased ATPase rates (R346S and R380A). These findings were in line with the conformational changes being the rate-limiting events during the ATPase cycle19,20.
Figure 4.
N-terminal closing of the different Hsp90 variants. Monitoring of nucleotide-induced closing kinetics via FRET. (a,b) Changes in acceptor fluorescence of the different Hsp90 variants in response to the binding of 2 mM ATP-γS (a) or 2 mM ATP (b) (top). Black, WT; yellow, A107N; blue, Δ8; green, T22I; purple, R346S; gray, R380A; red, E33A; cyan, D79N. The increase in signal was fitted to a monoexponential function to obtain the apparent rate constants kapp (min−1). Data are shown as mean ± s.d. (n = 3 technical replicates) (bottom). (c) N-terminal dimerization stability of the Hsp90 variants, deduced by FRET chase experiments. The chase was induced by addition of an excess of unlabeled Hsp90 D79N variant to 400 nM preformed Hsp90 FRET complexes in the absence of nucleotide (black) or the presence of 2 mM ATP-γS (blue) or 2 mM ATP (red). Top, chase experiments for WT Hsp90. Bottom, apparent half-lives (t1/2) derived from a nonlinear fit of the acceptor signal changes. Data are shown as mean ± s.d. (n = 3 technical replicates).
In the presence of ATP, for WT Hsp90, the closed state is only marginally populated19, and thus we observed no change in the FRET signal. This observation was also true for A107N, R346S and D79N. However, for the lethal mutants Δ8 and R380A and the viable variants T22I and E33A, the acceptor fluorescence increased with the addition of ATP (Fig. 4b, Supplementary Fig. 6 and Supplementary Table 3), thus suggesting that the closed state is more favored in these mutants than in the WT. Interestingly, this effect was observed for mutants with higher as well as lower ATPase activities. Thus, the specific step in the cycle affected by the mutation, not the kinetics of the overall reaction, is important for biological function.
Mutants differ in the accumulation of closed conformations
To gain further insight into the nature of the N-terminally closed state, we performed chase experiments by adding an excess of unlabeled Hsp90 D79N to preformed Hsp90 FRET complexes. Because the disruption of the FRET complexes was strongly influenced by the N-terminal dimerization properties of Hsp90 variants19, we were able to test the stability of the closed state (Fig. 4c). Without ATP, all variants showed kinetics with comparable half-lives (Fig. 4c and Supplementary Table 4), thus implying similar C-terminal dimerization. When we used ATP-γS to induce the N-terminally closed state, all mutants, except D79N, responded with dramatically increased apparent half-lives of the FRET complexes (Fig. 4c and Supplementary Table 4). In the presence of ATP, the chase with unlabeled Hsp90 resulted in a complex disruption for WT, D79N and R346S, a result similar to that observed without nucleotide. However, we observed a dramatic increase in the half-lives of the FRET complexes for the lethal variants Δ8 and R380A, as well as for the ATPase-deficient mutant E33A. The effect was less pronounced for the variants T22I and A107N compared with Hsp90 WT. Thus, several mutants capable of hydrolyzing ATP (R380A, Δ8, T22I and A107N) displayed accumulation of N-terminally dimerized intermediate states under continuous cycling conditions and ongoing hydrolysis.
To characterize the intermediate states that accumulated, we assessed the binding of the cochaperone Aha1 to the variants in the presence of ATP. Aha1 accelerates conformational changes in Hsp90, thus leading to the closed-1 state19. As expected, all variants were able to bind Aha1 (Supplementary Fig. 7a). When we preincubated the FRET complexes with ATP and Aha1, for all variants except R346S, the chase with unlabeled Hsp90 resulted in a slower complex disruption, as also observed in the absence of Aha1. However, the apparent half-lives of Hsp90 FRET complexes, except for that with D79N, differed from that of the WT in the presence of ATP and Aha1. We observed an additional increase for the variants Δ8, T22I, R380A and to a lesser extent A107N (Supplementary Fig. 7b). Together, these results support the idea that the mutations influenced the conformational changes leading to the closed-1 state.
Mutations affect formation of closed-1 and closed-2 states
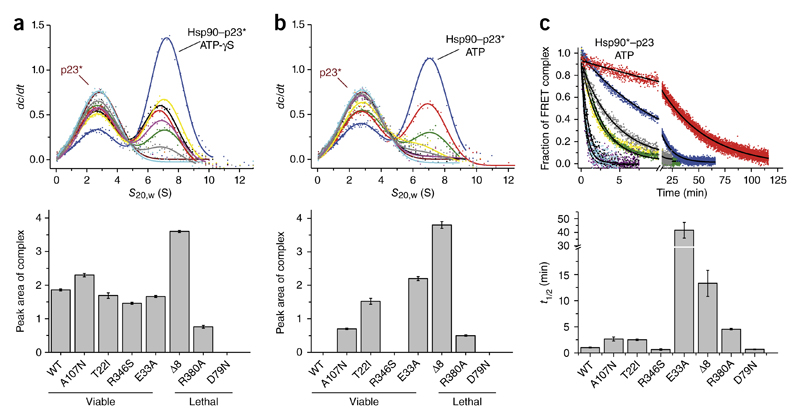
We had previously observed two different N-terminally dimerized conformations. In closed-1, the N domains are dimerized, but the N-M interaction is not yet fully formed32. In contrast, the closed-2 state exhibits the N-M interaction33. From the experiments above, we were unable to deduce which state was populated in the variants. To differentiate between the closed states, we used the cochaperone p23, whose binding is restricted to the closed-2 state18,34. We performed analytical ultracentrifugation (AUC) to monitor the interaction (Fig. 5).
Figure 5.
Binding of the cochaperone p23 (Sba1) to the Hsp90 variants. (a,b) Binding of fluorescein-labeled p23 to Hsp90 variants, as analyzed by analytical ultracentrifugation with fluorescence detection in the presence of 2 mM ATP-γS (a) or 2 mM ATP (b). Left, p23* alone (brown; asterisk indicates labeled protein) or p23* in complex with WT (black), A107N (yellow), Δ8 (blue), T22I (green), R346S (purple), R380A (gray), E33A (red) and D79N (cyan) are shown. Bottom, areas of the complex peaks, obtained by fitting. Error bars, standard error of the fit. s20,w, standardized sedimentation coefficient; dc/dt, concentration change over time. (c) N-terminal dimerization stability of the Hsp90 variants in the presence of ATP and p23, obtained by FRET chase experiments. Apparent half-lives (t1/2) were derived from a nonlinear fit of the acceptor signal changes. Data are shown as mean ± s.d. (n = 3 technical replicates).
Whereas all viable Hsp90 variants bound p23 in the presence of ATP-γS, similarly to the WT, the two lethal variants Δ8 and R380A bound more and less p23, respectively, and D79N did not bind p23 at all (Fig. 5a and Supplementary Fig. 8a). For R380A, this result indicated a destabilization of the closed-2 state; moreover, the lower p23 binding of this mutant also occurred in the presence of ATP, and the closed-1 state appeared to accumulate under cycling conditions (Fig. 5b and Supplementary Fig. 8b). In contrast, the Δ8-variant exhibited an increased p23 binding consistent with a population of the closed-2 state in the presence of ATP (Fig. 5b and Supplementary Fig. 8b). However, without ATP, Δ8 did not bind to p23 (Supplementary Fig. 8c). Furthermore, in the presence of ATP, p23 also bound to the variants T22I, E33A and A107N (Fig. 5b and Supplementary Fig. 8b). For E33A, the closed-2 state was the final conformation reached in the presence of nucleotide, because hydrolysis could not occur.
When we preincubated FRET complexes with ATP and p23, the chase with Hsp90 resulted in complex disruption for the D79N and R346S, a result similar to that observed for the WT (Fig. 5c and Supplementary Table 4). For the variants E33A and Δ8, we observed an increase in the half-lives of the FRET complexes (Fig. 5c and Supplementary Table 4). We found a less pronounced effect for A107N, T22I and R380A compared with Hsp90 WT (Fig. 5c and Supplementary Table 4). However, the closed states of the variants A107N, T22I and R380A were not further stabilized by p23, because the half-lives were similar for those variants without p23 (Supplementary Table 4). These findings suggested that the mutations modulated the population of the closed-1 and closed-2 states.
Population of closed states decreases Hsp90 chaperone function
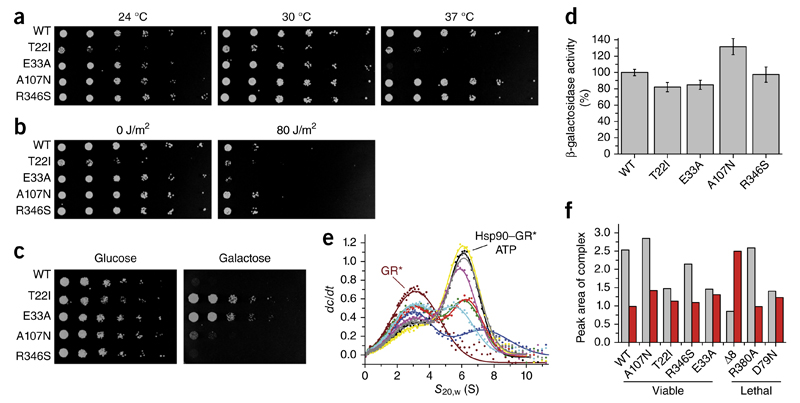
The mutations affected yeast growth to different extents. Some of the viable variants showed growth defects (Fig. 1 and Fig. 6a), such as T22I which grew slowly, thus confirming its temperature-sensitive phenotype24. We detected normal growth under physiological temperatures and sensitivity under increased temperatures for E33A. Only the R346S and A107N mutants behaved like the WT protein.
Figure 6.
Influence of Hsp90 variants in vivo. (a) Temperature sensitivity of yeast cells expressing Hsp90 WT and variants as the sole Hsp90 source at 24 °C, 30 °C or 37 °C. The assay was carried out at least three times, and data represent technical replicates from independent single yeast colonies. (b) Influence of the Hsp90 variants on DNA-repair activity in yeast cells exposed to UV light (80 J/m2) and incubated at 30 °C. As a control, one plate was not treated with UV radiation (0 J/m2). The assay was carried out at least three times, and data represent technical replicates from independent single yeast colonies. (c) Influence of Hsp90 variants on v-Src activation. Yeast cells expressing Hsp90 variants as the sole Hsp90 source and v-Src under the control of a galactose promoter were spotted on glucose (control, no v-Src expression)- and galactose-containing dropout plates (v-Src expression) and incubated at 30 °C. The assay was carried out at least three times, and data represent technical replicates started from independent single yeast colonies. (d) Influence of Hsp90 mutations on GR processing in yeast cells. GR activation, measured with a β-galactosidase-coupled assay, in yeast cells expressing different Hsp90 variants. Experiments were carried out at least three times, and data represent technical replicates (n = 3) started from independent single yeast colonies. Error bars, s.d. (e) Binding of the GR to different Hsp90 mutants, monitored by analytical ultracentrifugation with fluorescence detection and derived from dc/dt plots. GR* alone (brown; asterisk indicates labeled protein) or GR* in complex with WT (black), A107N (yellow), Δ8 (blue), T22I (green), R346S (purple), R380A (gray), E33A (red) and D79N (cyan) are shown. (f) Peak areas of the GR*–Hsp90 complex in the presence (gray bars) or absence of ATP (red bars).
To test for specific functions of Hsp90, we induced DNA damage by UV light35,36, which activates the Hsp90-dependent nucleotide excision repair machinery35,36. Yeast cells expressing the T22I or E33A variants displayed increased sensitivity to UV damage, whereas those expressing A107N and R346S behaved similarly to the WT control (Fig. 6b).
Furthermore, we determined the ability of the variants to activate the two well-established Hsp90 clients viral Src kinase (v-Src) and glucocorticoid receptor (GR) in yeast. Expression of active v-Src is toxic37,38 and inhibits yeast growth. The variants T22I and E33A showed reduced toxicity indicating impaired v-Src activation (Fig. 6c), whereas the other Hsp90 variants tested (A107N and R346S) exhibited WT-like v-Src maturation. To assess GR activation in yeast, we used a β-galactosidase-based reporter assay24. GR processing was slightly reduced in cells expressing the T22I or E33A variants (Fig. 6d). For T22I, this result was consistent with published data24. A107N and R346S supported WT-like maturation of GR. Thus, of the functional mutations, the variants E33A and T22I in particular appeared to broadly affect client protein processing.
To determine the reasons for these observations, we analyzed the interaction with GR by AUC. GR preferentially interacts with ATP-bound Hsp90 (ref. 31). Accordingly, D79N showed the weakest affinity for GR, as expected from its deficiency in binding nucleotide (Fig. 6e, f). Surprisingly, the Δ8 variant also exhibited strongly reduced GR binding. Furthermore, for E33A, and to a lesser extent for T22I, GR binding was compromised in the presence of ATP, thus suggesting that mutants for which the closed-2 state accumulates in the presence of ATP show weaker client interactions. Without nucleotide, all mutants interacted similarly with GR (Fig. 6f and Supplementary Fig. 8d). Thus, the binding site was not affected by the mutations. Only Δ8-Hsp90 bound strongly to the client protein, thereby indicating that the GR-interacting state had already accumulated.
Discussion
In this study, we used a set of Hsp90 mutants exhibiting changes in ATP hydrolysis, cochaperone interaction, biological function and client binding26,39–41 to probe the conformational cycle of Hsp90. These mutants allowed us to elucidate the mechanism of this molecular machine, which is characterized by large and slow conformational transitions (Fig. 7a). In this respect, the association of the N-terminal domains to form the closed-1 state and the subsequent interaction of the N and M domains to form the closed-2 state is important, because these processes are the rate-limiting steps of the ATPase cycle. The mutations are located in the N-terminal loop (which closes over the ATP-binding site), the N-terminal helix α1 and the N-M interface (which includes the catalytic loop (Pro375–Ile388)). Helix α1 is part of the N-terminal dimerization interface18. In the open state, helix α1 and the lid are in spatial proximity8. Closure of the lid and N-terminal dimerization are both coupled to ATP binding12.
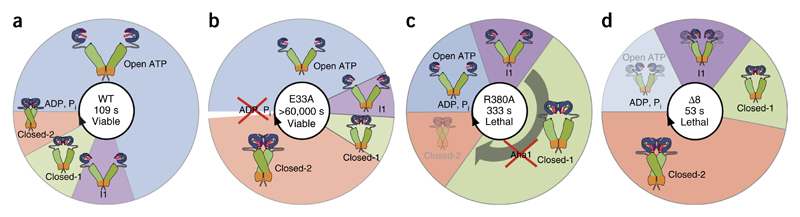
Figure 7.
Importance of cycle timing in the function of Hsp90. The scheme illustrates the differences in dwell times in different states (blue, open; violet, I1; green, closed-1; orange, closed-2) for WT Hsp90 (ref. 19) and mutants analyzed in this study. The sizes of the sectors indicate the relative time spent by the mutated protein in the different states during one round of hydrolysis. The average cycle time is indicated by the numbers in the centers of the circles. Mutants showing less intense effects, such as R364A, A107N and T22I, or those that did not bind nucleotide, such as D79N, are not depicted. (a) Hsp90 WT. (b) E33A. The white sector in the scheme for E33A indicates that hydrolysis did not detectably occur, and therefore the mutant does not appear to pass beyond this state. Release of ATP appears to rest the cycle. The variant shifts primarily between the open and the closed-2 state. (c) R380A. The conformational cycle of this mutant is dominated by closed states, especially closed-1. The defect cannot be corrected by Aha1. (d) Δ8. This mutant adopts a compact state even in the absence of nucleotide. It hydrolyzes ATP efficiently but populates primarily the closed-2 state.
Analysis of Hsp90 mutants has suggested that for viability of yeast cells, ATP hydrolysis is essential but that a range of ATPase activities of Hsp90 is tolerated26,40,42. Our study showed that, in agreement with previous reports, ATP hydrolysis is not required for viability, and even mutants with high ATPase rates23 can be lethal42,43. The most striking examples were Δ8 and E33A: Δ8 Hsp90 exhibited a two-fold increase in ATP turnover rate relative to that of the WT protein and was therefore expected to be a good substitute for WT Hsp90. However, this mutant was completely defective in vivo. In contrast, E33A bound ATP but did not hydrolyze it and therefore was expected to be catalytically dead. However, we found that yeast cells expressing only E33A Hsp90 were viable. These results cannot be explained on the basis of the current understanding of the Hsp90 cycle. Our mechanistic analysis revealed that the timing of the steps of the Hsp90 cycle is important. In this context, FRET experiments, SAXS and NMR analysis together with the effect of conformation-sensitive cochaperones provided direct insight into the presence of specific conformational states.
Δ8 Hsp90 was especially interesting in this context, because this mutant did not populate the open conformation as effectively as the WT protein. Instead, it exhibited a compact, N-terminally associated form. Thus, even in the absence of nucleotides, Δ8 appeared to be in the closed-1 state, whereas the addition of ATP induced the closed-2 state. This model is consistent with the differences between the mutant Δ8 and the WT protein seen in NMR chemical shifts that spread over the entire N-terminal domain in the absence of ATP, thus suggesting a different conformation that allows N-terminal dimerization (Fig. 3b).
In contrast, the E33A variant unexpectedly did not hydrolyze ATP at detectable rates but supported growth. Our analysis showed that this mutant still responded to ATP binding with defined conformational changes. Ultimately, ATP binding led to the formation of the closed-2 state, because p23 bound in vitro and in vivo39. The D79N variant, in contrast, was not able to bind ATP (Supplementary Fig. 4), could not undergo conformational changes and therefore was not able to support viability.
These findings suggest that the speed of the cycle plays a subordinate role in the function of Hsp90. Instead the conformations that are adopted along the way and, importantly, the time spent by Hsp90 in a specific conformational state are critical. Thus, the cycling between conformational states has tremendous consequences for the biological effects of Hsp90 (Fig. 7a).
Together, our results revealed several characteristics of the conformational cycle: (i) Dramatic changes in ATP hydrolysis are tolerated if they allow conformational changes to occur, as observed for E33A and T22I (Fig. 7b). These mutations apparently do not affect functionality to an extent that precludes viability, but they render yeast sensitive to stress. (ii) The deceleration of the transition between the closed-1 and closed-2 states, as is the case of R380A, and the accumulation of the closed-1 state under cycling conditions, affects Hsp90 function (Fig. 7c). (iii) Mutations that alter initial conformational changes of the chaper-one can still support growth (A107N and R346S). (iv) The accumulation of closed states at the expense of open states is deleterious, and the respective mutants are dysfunctional even if they show ATPase activity, as is the case for Δ8 (Fig. 7d).
We found that mutations affecting the timing of the conformational transitions strongly impaired the biological function of Hsp90 regardless of their position. In this context, several mutants capable of hydrolyzing ATP (R380A, Δ8, T22I and A107N) exhibited accumulation of N-terminally dimerized intermediate states under continuous cycling conditions and ongoing hydrolysis. This finding was in line with the results of our SAXS experiments, in which Δ8, E33A, T22I and A107N, in contrast to WT Hsp90, tended to populate closed states in the presence of ATP. These results were consistent with NMR data for T22I and A107N, which showed similar chemical shift-changes that affected the N-terminal dimerization interface of Hsp90. The FRET experiments and cochaperone binding further demonstrated that these mutants exhibited accumulation of the closed-2 state, in contrast to the normal predominance of the open conformation in the presence of ATP19,22. E33A, T22I and Δ8 bound less GR in the presence of ATP, thus suggesting that this conformation is not competent for efficient client binding.
Cochaperones can be used as conformational sensors to pinpoint defects in cycling. In the case of Aha1, we observed effects exclusively for mutants with defects in the transition from the open to the N-terminally closed state; these results were consistent with the function of Aha1 in promoting N-terminal closure28. Comparison of the mutants R380A and R346S provided further clues about important features of the conformational cycle. R380 is located in the so-called catalytic loop of the M domain, a structural element that contacts the ATP-binding site in the N domain, and R346 is part of the N-M interface formed after nucleotide binding. Both mutants led to an almost identical decrease in ATPase activity, to approximately one-third that of the WT. If ATP turnover were the critical determinant of Hsp90 function, then these mutants should behave similarly. However, the mutants showed opposite in vivo effects. Whereas the R380A substitution was lethal, R346S supported viability. What is the critical difference between the two mutants? Strikingly, Aha1 doubled the ATP hydrolysis rate of only R380A. In contrast, R346S was stimulated almost 80-fold, reaching a maximum velocity similar to that of the WT protein. Thus, R346S had difficulties in the transition from open to closed-1. In contrast, for R380A, this step was unaffected, but the subsequent formation of the closed-2 state was compromised and consequently led to the accumulation of the closed-1 state. Thus, although both N-M-interface mutants exhibited a slowed closing reaction, the precise point at which the defect occurred played a decisive role in the reaction.
The picture that emerges is that Hsp90 must populate the open and closed-1 conformations for a certain time for efficient client binding. This model is in accordance with results obtained for the interaction of the GR ligand-binding domain (LBD) with Hsp90, in which the use of nonhydrolyzable nucleotides has been found to reduce GR binding31. Thus, even if ATP turnover is possible, the functionality of Hsp90 can be compromised to an extent that excludes in vivo function, depending on how much time Hsp90 spends in certain conformations under cycling conditions. It is conceivable that some post-translational modifications may regulate Hsp90 by affecting this balance44,45. In conclusion, the timing of the cycle, i.e., the time that Hsp90 spends in a certain conformation, determines the interaction with client proteins and thus Hsp90’s in vivo function, regardless of the overall cycle speed.
Online Methods
Cloning and protein purification
All Hsp90 point mutants were generated with QuikChange (Stratagene) site-directed mutagenesis with pET28a containing WT yeast Hsp82 as the template vector. The proteins were expressed in the Escherichia coli strain BL21(DE3) RIL (Stratagene) and were purified as described previously44 and stored in 40 mM HEPES, pH 7.5, 150 mM KCl, 5 mM MgCl2 (standard buffer) at −80 °C until usage. Hsp90 cochaperones and the GR LBD mutant were purified as described previously31,46.
Protein labeling
Labeling of the Hsp90 cysteine variants was performed with ATTO-488 maleimide and ATTO-550 maleimide (ATTO-TEC), as previously described19. p23 was labeled via an engineered cysteine at position 2 with 5-iodacetamidofluorescein (Invitrogen) according to the manufacturer’s protocol. Aha1 was labeled with 5(6)-FAM via a reactive succimidyl ester group bound to lysine (Invitrogen), according to the manufacturer’s protocol. The GR LBD mutant was randomly labeled with ATTO-488 malemide (ATTO-TEC) on cysteine residues. Free label was removed via Superdex 75 FPLC (Pharmacia Biotech) as previously described32.
ATPase activity
ATPase assays were performed as previously described, with an ATP-regenerating system47. Assays were measured with an Hsp90 concentration of 3 μM in standard buffer supplemented with 2 mM ATP (Roche) at 30 °C. The Hsp90-specific ATPase activity was inhibited by addition of 50 μM of the inhibitor radicicol (Sigma). The remaining activity was subtracted as background. The assays were evaluated with Origin software (OriginLab). The Km values for ATP of each Hsp90 variant were derived from the ATPase activities with following equation:
The stimulation of ATPase activity was measured under comparable conditions, except with 1 μM of Hsp90 in the presence of varying concentrations of Aha1 in 40 mM HEPES, pH 7.5, 50 mM KCl and 5 mM MgCl2. The stimulation factor was determined according to the following equation, with Vmax in the presence of Aha1 and kcat in the absence of Aha1:
NMR spectroscopy
All spectra were recorded on a Bruker AVIII 600 MHz spectrometer with a cryogenic triple-resonance gradient probe. A construct comprising the first 210 residues of yeast Hsp90 was used to record the 1H,15N-HSQC spectra of the WT protein and the mutants Δ8, T22I, E33A, D79N and A107N. Protein concentrations ranged from 110 to 465 μM. In the case of the E33A and D79N mutants, 15N-edited NOESY spectra were recorded to confirm assignments of the mutants. All spectra were acquired in a 20 mM sodium phosphate buffer, pH 6.5, containing 100 mM sodium chloride, 2 mM EDTA, 1 mM DTT and 5% D2O. For the Hsp90 WT N domain and the variants Δ8, T22I, E33A and A107N, spectra with a protein concentration of 300 μM in the presence of an excess of ATP (4 mM) and of 6 mM magnesium sulfate were measured. The mutants E33A and D79N were also measured in complex with a 2:1 excess of AMP-PNP. All spectra were processed with NMRPipe/Draw48 and analyzed with CcpNmr analysis49. The chemical-shift perturbations (CSPs) were calculated according to the following formula, where δ is the proton (H) or nitrogen (N) chemical shift of the WT or mutant (mut):
Small-angle X-ray scattering
SAXS data for solutions of the nucleotide-free and ATP- and ATP-γS-bound forms of WT yeast Hsp90 and Hsp90 mutants were recorded on an in-house SAXS instrument (SAXSess mc2, Anton Paar) equipped with a Kratky camera, a sealed X-ray tube source and a two-dimensional Princeton Instruments PI-SCX:4300 (Roper Scientific) CCD detector. The scattering patterns were measured with a 60-min exposure time (360 frames, each 10 s) for several solute concentrations ranging from 0.8 to 3.3 mg/ml (Fig. 4). Radiation damage was excluded on the basis of a comparison of individual frames of the 60-min exposures, wherein no changes were detected. A range of momentum transfer of 0.012 < s < 0.63 Å−1 was covered
where 2θ is the scattering angle, and λ is the X-ray wavelength, in this case 1.5 Å.
All SAXS data were analyzed with the ATSAS package (version 2.5). The data were processed with SAXSQuant (version 3.9) and desmeared with GNOM50. The forward scattering (I(0)), the radius of gyration, (Rg), the maximum dimension (Dmax) and the interatomic distance distribution function ((P(r)) were computed with GNOM50. The masses of the solutes were evaluated by comparison of the forward scattering intensity with that of a human serum albumin reference solution (molecular mass 69 kDa).
Fluorescence measurements
Fluorescence spectra and subunit exchange were carried out as previously described19.
Nucleotide binding kinetics
To analyze the conformational rearrangements after nucleotide binding, Hsp90 heterodimers (400 nM) were formed by mixture of an equal amount of donor-labeled and acceptor-labeled Hsp90 in standard buffer. The experiment was started by addition of 2 mM nucleotide (ATP, ATP-γS, all from Roche), and the increase in fluorescence intensity was recorded with a Fluoromax 3 or Fluoromax 2 fluorescence spectrophotometer (Horiba Jobin Yvon) at 575 nm after excitement at 490 nm at 30 °C. The apparent rate constants of the conformational changes were determined by fitting the data to a monoexponential function with Origin software (OriginLab).
FRET chase experiments
After heterodimer formation, the subunit exchange was recorded by addition of a ten-fold excess of unlabeled Hsp90 D79N, and the decay of fluorescence intensity was recorded with a Fluoromax 3 or Fluoromax 2 fluorescence spectrophotometer (Horiba Jobin Yvon) at 575 nm after excitement at 490 nm at 30 °C. For experiments in the presence of different nucleotides (ATP and ATP-γS, all from Roche) and/or cochaperones (4 μM Aha1 or 4 μM p23), Hsp90 variants were preequilibrated 30 min in the presence of 2 mM of the respective nucleotide to allow the formation of the closed state. The apparent half-life of the reaction was determined by fitting the data with the function for exponential decay with Origin software (OriginLab).
Analytical ultracentrifugation
To analyze the binding of fluorescein-Sba1, 5(6)-FAM-Aha1 and ATTO 488-GR to Hsp90, AUC was performed in a Beckman ProteomeLab XL-A (Beckman) equipped with a fluorescence detection system (Aviv Biomedica). Sedimentation-velocity experiments were performed with labeled protein supplemented with various combinations of unlabeled proteins in standard buffer at 42,000 r.p.m. A Ti-50 rotor (Beckman) was used at 20 °C. To determine the sizes of complexes, the raw data were converted to dc/dt profiles by subtraction of nearby scans and conversion of the difference into dc/dt plots, as previously described51. The plots generally correlated with those from the SEDVIEW dc/dt program52. dc/dt profiles were analyzed to determine the s values and the areas of the corresponding peaks.
In vivo function of Hsp90 variants
The in vivo function of the different Hsp90 variants was tested with a plasmid-shuffling approach based on a system described previously24. We used the ΔPCLDα Saccharomyces cerevisiae strain (a derivative of W303) obtained from S. Lindquist’s laboratory24. This yeast strain is deficient in genomic hsp82 and hsc82 and contains a plasmid encoding hsp82 to rescue lethality. This plasmid, pKAT6, carries a URA selection marker, which enables selection for cells that had lost the WT Hsp82 plasmid in the medium supplemented with 5-FOA. Hsp90 WT and the mutant variants were constitutively expressed from a 2ì high-copy-number plasmid under the control of a constitutive glyceraldehyde-3-phosphate dehydrogenase gene (GPD) promoter (p423GPD vector). The cells surviving the shuffling were tested for loss of the URA plasmid by plating on selection medium lacking URA. To confirm the growth support by the Hsp90 E33A variant, several yeast shuffling strains were used14,15,24,39.
Tetrad dissection analysis
A diploid strain (Mat a/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; lys2Δ0/LYS2; MET15/met15Δ0; ura3Δ0/ura3Δ0; HSP82::kanMX4/HSP82; HSC82::kanMX4/HSC82); was constructed by mating of BY4741 HSC82:: kanMX4 and BY4742 HSP82::kanMX4 and was transformed with p415-GPD-HSP82E33A or the empty vector as control. After sporulation, tetrad dissection was performed, and cells were grown on leucine-depleted medium for 3 d. Finally, cells were replica-plated on YPD medium containing 200 μg/ml G418 to test for the distribution of the kanMX4-cassette.
Yeast cell lysis and western blotting
After 5-FOA shuffling, single clones were grown at 30 °C in minimal medium (5 ml) to stationary phase. After an OD600 of 5 was reached, cells were harvested (4,500 r.p.m., 5 min). The cell pellets were washed with H2O, and the OD600 was measured again. Cells were spun down (4,500 r.p.m., 5 min) and resuspended in 200 μl H2O; then 200 μl of 0.2 M NaOH was added, and cells were incubated for 3 min at RT. Cells were harvested again (13,000 r.p.m., 1 min), and supernatant was discarded. The pellet was resuspended in 100 μl (OD600 of 5) sample buffer and boiled for 5 min at 95 °C. Proteins were separated by SDS–PAGE and blotted on a PVDF membrane for 1 h.
The following primary antibodies were used to probe the membrane for 1 h at RT: anti-yeast Hsp90 (Pineda Antibody Service)33, polyclonal, produced in rabbit (1:40,000 dilution in PBS 0.1% Tween 1% milk) and anti-PGK1 (Invitrogen, cat. no. A6457), monoclonal, produced in mouse (1:20,000 in PBS 0.1% Tween 1% milk). The secondary antibodies were anti-mouse and anti-rabbit IgG-peroxidase antibodies (Sigma-Aldrich, cat. nos. A9044 and A0545, respectively), diluted 1:20,000 in PBS 0.1% Tween 1% milk, were used to detect the respective first antibody for 45 min at RT. For all antibodies, validation is provided on the manufacturers’ websites. Detection was performed with Western Bright ECL Spray (Advansta) and ImageQuant LAS4000 (GE Healthcare). Uncropped blots are shown in Supplementary Data Set 1.
Growth assays: temperature sensitivity and nucleotide excision repair assay
Drop dilution assays were performed to study the influences of various Hsp90 mutants on the growth of S. cerevisiae cells. After 5-FOA, several colonies were plated onto histidine and leucine (–His/–Leu)-depleted medium. After a 2-d incubation time at 30 °C, liquid –His/–Leu medium was inoculated and incubated again (30 °C; 18 h). The OD600 of every culture was measured and set to 0.5 by dilution with –His/–Leu medium. The first dilution sample had a total volume of 1 mL. On the basis of this first dilution sample, serial dilution consisting of five more 1:5 dilutions with a sample volume of 200 μL each was carried out. The serial dilution was finally plated on –His/–Leu medium, with 5 μL of sample per drop. Dilution steps 1–6 for each mutant were plated in studies of growth at different temperatures (24 °C, 30 °C and 37 °C). To study nucleotide excision repair, the plates were exposed (without the lid of the petri dish) to UV-light radiation (80 × 100 μJ/cm2, 10 s) with a CL-100 ultraviolet cross-linker (UVP). As control, one plate was not exposed to UV light. The growth of each Hsp90 mutant was always compared to WT Hsp90, which was present on every plate. Yeast cell growth was documented after 48 h.
v-Src and GR maturation assay
The influence on client maturation was investigated as previously described44.
Supplementary Material
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Acknowledgmentsts
We acknowledge C. Göbl and C. Hartlmüller for help with the SAXS measurements, B. Tremmel for help with protein expression and purification, J. Soroka and J. Reinstein for inspiring discussions and comments on the manuscript and S. Lindquist (Whitehead Institute) for providing reagents. F.T. acknowledges a scholarship from the Studienstiftung des deutschen Volkes. This work was supported by the Bavarian Ministry of Sciences, Research and the Arts (Bavarian Molecular Biosystems Research Network, to T.M.), the Austrian Academy of Sciences (APART-fellowship to T.M.), the Austrian Science Fund (grant no. FWF: P28854 to T.M.), the Deutsche Forschungsgemeinschaft (grant no. SFB1035 to J.B. and M.S.) and the Emmy Noether program (grant no. MA 5703/1-1 to T.M.).
Footnotes
Author Contributions
B.K.Z. and F.T. performed experiments and data analysis. M.R. recorded and analyzed NMR data. T.M. performed and analyzed SAXS measurements. F.H.S. generated Hsp90-knockout strains and performed yeast tetrad analysis. K.R. performed the AUC experiments and analyzed data, J.B. designed experiments. B.K.Z., K.R., M.S. and J.B. wrote the manuscript. D.A.R. provided ATTO488-labeled GR and performed AUC runs.
Competing Financial Interests
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Zhao R, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Young JC, Moarefi I, Hartl FU. Hsp90: a specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClellan AJ, et al. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 5.Röhl A, Rohrberg J, Buchner J. The chaperone Hsp90: changing partners for demanding clients. Trends Biochem Sci. 2013;38:253–262. doi: 10.1016/j.tibs.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Mayer MP, Le Breton L. Hsp90: breaking the symmetry. Mol Cell. 2015;58:8–20. doi: 10.1016/j.molcel.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 7.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 8.Prodromou C, et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 9.Nemoto T, Ohara-Nemoto Y, Ota M, Takagi T, Yokoyama K. Mechanism of dimer formation of the 90-kDa heat-shock protein. Eur J Biochem. 1995;233:1–8. doi: 10.1111/j.1432-1033.1995.001_1.x. [DOI] [PubMed] [Google Scholar]
- 10.Bose S, Weikl T, Bügl H, Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. doi: 10.1126/science.274.5293.1715. [DOI] [PubMed] [Google Scholar]
- 11.Wegele H, Muschler P, Bunck M, Reinstein J, Buchner J. Dissection of the contribution of individual domains to the ATPase mechanism of Hsp90. J Biol Chem. 2003;278:39303–39310. doi: 10.1074/jbc.M305751200. [DOI] [PubMed] [Google Scholar]
- 12.Prodromou C, et al. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones. 2004;9:109–121. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Panaretou B, et al. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mishra P, Bolon DN. Designed Hsp90 heterodimers reveal an asymmetric ATPase-driven mechanism in vivo. Mol Cell. 2014;53:344–350. doi: 10.1016/j.molcel.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiau AK, Harris SF, Southworth DR, Agard DA. Structural analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 18.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 20.Weikl T, et al. C-terminal regions of Hsp90 are important for trapping the nucleotide during the ATPase cycle. J Mol Biol. 2000;303:583–592. doi: 10.1006/jmbi.2000.4157. [DOI] [PubMed] [Google Scholar]
- 21.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 22.Krukenberg KA, Förster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richter K, Reinstein J, Buchner J. N-terminal residues regulate the catalytic efficiency of the Hsp90 ATPase cycle. J Biol Chem. 2002;277:44905–44910. doi: 10.1074/jbc.M208457200. [DOI] [PubMed] [Google Scholar]
- 24.Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15:3917–3925. doi: 10.1128/mcb.15.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaughan CK, Piper PW, Pearl LH, Prodromou C. A common conformationally coupled ATPase mechanism for yeast and human cytoplasmic HSP90s. FEBS J. 2009;276:199–209. doi: 10.1111/j.1742-4658.2008.06773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubert DA, He Y, McNulty BC, Tornero P, Dangl JL. Specific Arabidopsis HSP90.2 alleles recapitulate RAR1 cochaperone function in plant NB-LRR disease resistance protein regulation. Proc Natl Acad Sci USA. 2009;106:9556–9563. doi: 10.1073/pnas.0904877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer P, et al. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 28.Retzlaff M, et al. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Koulov AV, et al. Biological and structural basis for Aha1 regulation of Hsp90 ATPase activity in maintaining proteostasis in the human disease cystic fibrosis. Mol Biol Cell. 2010;21:871–884. doi: 10.1091/mbc.E09-12-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cunningham CN, Southworth DR, Krukenberg KA, Agard DA. The conserved arginine 380 of Hsp90 is not a catalytic residue, but stabilizes the closed conformation required for ATP hydrolysis. Protein Sci. 2012;21:1162–1171. doi: 10.1002/pro.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz OR, et al. Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol Cell. 2014;53:941–953. doi: 10.1016/j.molcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Richter K, Reinstein J, Buchner J. Integration of the accelerator Aha1 in the Hsp90 co-chaperone cycle. Nat Struct Mol Biol. 2013;20:326–331. doi: 10.1038/nsmb.2502. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Richter K, Buchner J. Mixed Hsp90–cochaperone complexes are important for the progression of the reaction cycle. Nat Struct Mol Biol. 2011;18:61–66. doi: 10.1038/nsmb.1965. [DOI] [PubMed] [Google Scholar]
- 34.Richter K, Walter S, Buchner J. The Co-chaperone Sba1 connects the ATPase reaction of Hsp90 to the progression of the chaperone cycle. J Mol Biol. 2004;342:1403–1413. doi: 10.1016/j.jmb.2004.07.064. [DOI] [PubMed] [Google Scholar]
- 35.Toogun OA, Dezwaan DC, Freeman BC. The hsp90 molecular chaperone modulates multiple telomerase activities. Mol Cell Biol. 2008;28:457–467. doi: 10.1128/MCB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Echtenkamp FJ, et al. Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol Cell. 2011;43:229–241. doi: 10.1016/j.molcel.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mimnaugh EG, Worland PJ, Whitesell L, Neckers LM. Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the hsp90 stress protein and the pp60v-src tyrosine kinase. J Biol Chem. 1995;270:28654–28659. doi: 10.1074/jbc.270.48.28654. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, Lindquist S. Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci USA. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JL, Halas A, Flom G. Nucleotide-dependent interaction of Saccharomyces cerevisiae Hsp90 with the cochaperone proteins Sti1, Cpr6, and Sba1. Mol Cell Biol. 2007;27:768–776. doi: 10.1128/MCB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawle P, et al. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol Cell Biol. 2006;26:8385–8395. doi: 10.1128/MCB.02188-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter K, et al. Intrinsic inhibition of the Hsp90 ATPase activity. J Biol Chem. 2006;281:11301–11311. doi: 10.1074/jbc.M510142200. [DOI] [PubMed] [Google Scholar]
- 42.Zurawska A, et al. Mutations that increase both Hsp90 ATPase activity in vitro and Hsp90 drug resistance in vivo. Biochim Biophys Acta. 2010;1803:575–583. doi: 10.1016/j.bbamcr.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Tsutsumi S, et al. Charged linker sequence modulates eukaryotic heat shock protein 90 (Hsp90) chaperone activity. Proc Natl Acad Sci USA. 2012;109:2937–2942. doi: 10.1073/pnas.1114414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soroka J, et al. Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 45.Mollapour M, Neckers L. Post-translational modifications of Hsp90 and their contributions to chaperone regulation. Biochim Biophys Acta- 2012;1823:648–655. doi: 10.1016/j.bbamcr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buchner J, Weikl T, Bügl H, Pirkl F, Bose S. Purification of Hsp90 partner proteins Hop/p60, p23, and FKBP52. Methods Enzymol. 1998;290:418–429. doi: 10.1016/s0076-6879(98)90035-0. [DOI] [PubMed] [Google Scholar]
- 47.Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- 48.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 49.Vranken WF, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 50.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 51.Stafford WF., III Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 52.Hayes DB, Stafford WF. SEDVIEW, real-time sedimentation analysis. Macromol Biosci. 2010;10:731–735. doi: 10.1002/mabi.201000075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.