Abstract
We apply integrative approaches to expression quantitative loci (eQTLs) from 44 tissues from the GTEx Project and genome-wide association study (GWAS) data. About 60% of known trait-associated loci are in linkage disequilibrium with a cis-eQTL, over half of which were not found in previous large-scale whole blood studies. Applying polygenic analyses to metabolic, cardiovascular, anthropometric, autoimmune, and neurodegenerative traits, we find that eQTLs are significantly enriched for trait associations in relevant pathogenic tissues and explain a substantial proportion of the heritability (40-80%). For most traits, tissue-shared eQTLs underlie a greater proportion of trait associations, though tissue-specific eQTLs have a greater contribution to some traits, such as blood pressure. By integrating information from biological pathways with eQTL target genes and applying a gene-based approach, we validate previously implicated causal genes and pathways, and propose new variant- and gene-associations for several complex traits, which we replicate in the UK BioBank and BioVU.
A primary goal of the Genotype-Tissue Expression (GTEx) project1 is to elucidate the biological basis of GWAS findings for a range of complex traits, by measuring eQTLs in a broad collection of normal human tissues. Several recent papers have described the GTEx v6p data, where cis-eQTLs were mapped for 44 tissues from a total of 449 individuals (70-361 samples per tissue)2 using a single-tissue method3 that detects eQTLs in each tissue separately, and a multi-tissue method4 that increases the power to detect weak effect eQTLs. Here we leverage the extensive resource of regulatory variation from multiple tissues to elucidate the causal genes for various GWAS locus and to assess their tissue specificity (Fig. 1a). We highlight the challenges of using eQTL data for the functional interpretation of GWAS findings and identification of tissue of action. Using several polygenic approaches (Table 1), we provide comprehensive analyses of the contribution of eQTLs to trait variation. Finally, by integrating eQTL with pathway analysis, and replication in DNA biobanks tied to electronic health records (UK Biobank5 and BioVU6; see URLs), we propose new trait associations and causal genes for follow-up analyses for a range of complex traits.
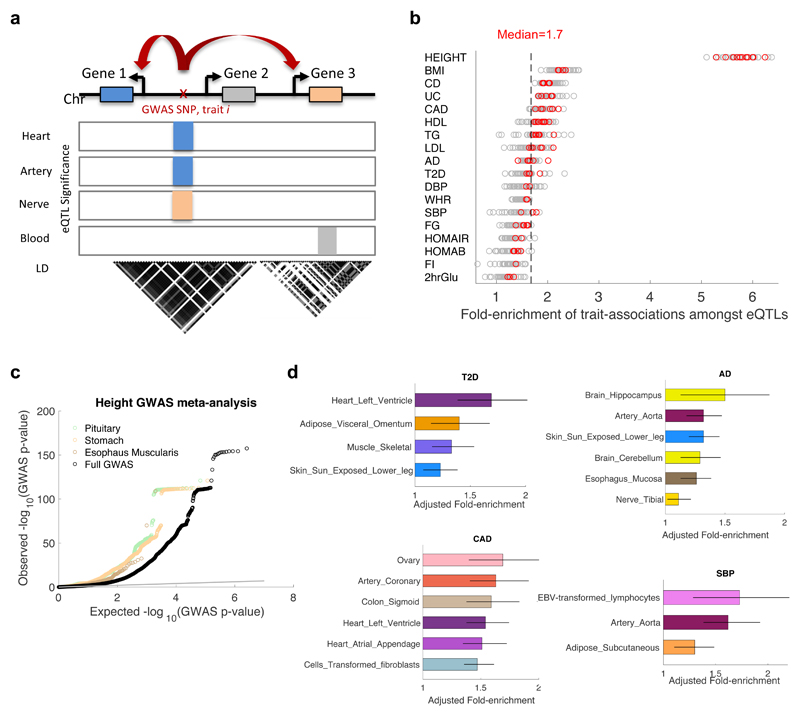
Fig. 1. Incorporating eQTLs from 44 tissues into GWAS of complex traits.
a, Schematic diagram demonstrating how eQTL annotation from various tissues can be used to propose one or more potential causal genes whose regulation is either tissue-specific (orange) or tissue-shared (blue) for a trait-associated (GWAS) variant. A gene close to the trait-associated variant (grey) may have an eQTL that is not in LD with the trait-associated variant. b, Fold-enrichment of eQTLs (FDR≤0.05) with GWAS p≤0.05 compared to a uniform null distribution of GWAS p-values, shown for 44 tissues by 18 complex traits. Red circles: tissue-trait pairs that pass Bonferroni correction (P<6.3x10-5; 89 out of 792 tissue-trait pairs tested). Dashed line: median fold-enrichment of all significant tissue-trait pairs. The ‘best eQTL per eGene’ set per tissue was used here. c, Quantile-quantile (Q-Q) plot of variant association p-values from a large GWAS meta-analysis of Height (n=253,288) for all variants tested (black), and for eQTLs in tissues most highly enriched for height associations: pituitary (green), stomach (peach), and esophaus muscularis (brown). All significant variant-gene eQTL pairs were plotted. d, Top ranked tissues based on their adjusted fold-enrichment of trait associations amongst eQTLs (compared to the best eQTL for all non-significant eGenes) that pass Bonferonni correction (P<6.3E-05) for type 2 diabetes (T2D, n=69,033), Alzheimer’s disease (AD, n=54,162), coronary artery disease (CAD, n=184,405), and systolic blood pressure (SBP, n=69,395) (Supplementary Table 2). Estimated lower and upper bound 95% confidence intervals for the adjusted fold-enrichment are shown (see Methods).
Table 1. Summary of polygenic methods used to test contribution of eQTLs to trait variation.
| Method* | Goal | Description and assumptions | Limitations | eQTL set used | GWAS data types |
|---|---|---|---|---|---|
| eQTLEnrich, Rank and permutation-based GWAS-eQTL enrichment method | Tests whether eQTLs from a given tissue are significantly enriched for trait associations more than would be expected by chance and estimates adjusted fold-enrichment. | Estimates the probability of observing a given fold-enrichment of top ranked trait associations (e.g. GWAS p<0.05) amongst eQTLs in a given tissue, relative to the fold-enrichment of nonsignificant eVariants (adjusted fold-enrichment), using a null distribution derived from multiple randomly sampled variants matched on MAF, distance to TSS, and local LD. Per GWAS tested, tissues are ranked based on their adjusted fold-enrichment. | Adjusted fold-enrichment is correlated with GWAS sample size. | Best eQTL per eGene | Variant association p-values |
| TORUS, Bayesian and MLE approach for quantifying GWAS-eQTL enrichment | Estimates an enrichment parameter that represents the relationship between the log-odds ratio of the trait associations being causal and their eQTL effect size. | Estimates the relationship between the (absolute value of) single variant eQTL z-scores and the corresponding log odds of a variant being causally associated with the complex trait of interest. A confident positive estimate of the log odds ratio indicates the increased odds of a variant being causally associated with the trait with stronger effect of eQTL association. Uses z-scores from all gene-variant pairs for a given tissue, and assumes a single causal trait association per LD block (following the assumption of fgwas). | Enrichment parameter estimation (esp. standard error) is correlated with tissue sample size of eQTLs | All variant-gene pairs tested | Variant association test statistics |
| π1 method | Estimates the fraction of eQTLs in a given tissue that are likely to be associated with a given complex trait. | Estimates the fraction of true trait associations amongst eQTLs in a given tissue, using the π1 statistic, which assumes a standard uniform distribution for the null distribution and independence between variants. | Results not robust to small variant sets. | Best eQTL per eGene | Variant association p-values |
| Summary statistics-based heritability estimation | Estimates the relative contribution of eQTLs in aggregate to the heritability of complex traits, using LD Score regression applied to publicly available GWAS summary statistics. | Estimates the per-variant effect of the trait association by an annotated eQTL vs. an unannotated variant. A larger difference indicates a higher degree of enrichment of contribution of eQTLs to trait associations. | Works optimally when the per-variant variance is not correlated with the LD score. | All significant variant-gene pairs | Variant association test statistics |
| Mixed-effects-model heritability estimation | Estimates proportion of complex trait variance explained by eQTL variants in aggregate using GWAS genotype data. | Estimates the heritability attributable to eQTL variants using the Restricted Maximum Likelihood approach. The approach assumes a normal distribution of trait effect sizes for the eQTL variants and uses a genetic similarity matrix generated from the eQTL variants. | Requires genotype data. | All significant variant-gene pairs | Individual genotype data |
See URLs for links to methods’ software.
Results
Relevance of eQTLs from 44 tissues to trait associations
We tested the extent to which cis-eQTLs (using the ‘best eQTL per eGene’ at a genome-wide FDR≤0.05 per tissue) from each of the 44 tissues2 were enriched for trait associations (GWAS p≤0.05) using eQTLEnrich (see Methods and Supplementary Fig. 1). Testing 18 complex traits (metabolic, cardiovascular, anthropometric, autoimmune and neurodegenerative, listed in Supplementary Table 1) with available GWAS summary statistics, we found significant enrichment for trait associations amongst eQTLs (Bonferroni-adjusted P<6.3x10-5) for 11% of 792 tissue-trait pairs tested, with a median fold-enrichment per trait ranging from 1.19 to 5.75 (Fig. 1b and Supplementary Table 2), and different tissues significant per trait (Supplementary Fig. 2). The enrichment results also suggest hundreds of modest-effect associations amongst eQTLs in various tissues for all traits tested (Supplementary Fig. 3 and Supplementary Table 2). While the adjusted fold-enrichment (see Methods) is unaffected by differences in number of eQTLs per tissue (Supplementary Fig. 4), increased enrichment was observed for GWAS with larger sample sizes, such as Height7 (N>250k), where there is greater detection power (Fig. 1c). Enrichment amongst eQTLs was also found for less-powered GWAS, such as HOMA-IR8 (N~37k), where no variants passed genome-wide significance (Supplementary Fig. 5). The tissues in which eQTLs were most strongly enriched for associations included relevant tissues, such as aortic artery for systolic blood pressure (SBP), coronary artery for coronary artery disease (CAD), skeletal muscle for type 2 diabetes (T2D), colon for Crohn’s disease (CD), and hippocampus for Alzheimer’s disease (AD) (Fig. 1d and Supplementary Table 2). However, the most enriched tissues per trait also included less biologically obvious tissues, suggesting either shared regulation with the actual tissues of action or new pathogenic tissues. Notably, eQTLs in (commonly studied) whole blood were enriched for associations with about half of the traits tested (P<6.3x10-5; e.g., Ulcerative Colitis (UC), and low- and high-density lipoprotein cholesterol (LDL and HDL); Supplementary Table 2), demonstrating the utility of blood for broadly studying the underlying genetic mechanisms of some associations, but also emphasizing the importance of studying gene regulation in a biologically diverse set of disease-relevant tissues.
Applying a Bayesian-based enrichment method that accounts for eQTL effect size and considers all significant variant-gene pairs, TORUS9,10 (Supplementary Note and Table 1), similarly showed substantial enrichment for trait associations amongst eQTLs (Supplementary Fig. 6 and Supplementary Table 3).
Since traits may be determined by tissue-specific processes, we further examined just the subset of tissue-specific eQTLs (defined as eQTLs significant in a given tissue and at most 4 other tissues, ~10% of tissues, using multi-tissue analysis; see Methods and Supplementary Fig. 7a). Using eQTLEnrich, we found significant enrichment in fewer tissue-trait pairs when restricting to tissue-specific eQTLs (Supplementary Table 4 and Supplementary Fig. 7b) than with all eQTLs (Supplementary Table 2). Among the top results were adipose-specific eQTLs for diastolic blood pressure (DBP) and aorta-specific eQTLs for SBP, proposing different tissue-specific processes that may underlie DBP and SBP.
Cis-eQTL characterization of known trait associations
Since regulatory effects are enriched for top-ranked trait associations, we asked how many of the genome-wide significant associations (P<5x10-8) from the NHGRI-EBI GWAS catalog might be acting via eQTLs, and in what tissues. We annotated 5,895 genome-wide significant associations (P<5x10-8; hereafter “trait-associated variants”), identified primarily in samples of European descent (Supplementary Table 5), with GTEx eQTLs from both single-tissue (FDR≤0.05) and multi-tissue analyses (METASOFT4, m-value≥0.9) using a linkage disequilibrium (LD) cutoff of r2>0.8 (see Methods; Supplementary Table 6). Considering all significant variant-gene eQTL pairs, we observed that 61.5% of the 5,895 trait-associated variants were in LD (r2>0.8) with at least one eQTL from any tissue (Supplementary Tables 7).
To characterize the target gene and tissue patterns of trait-associated variants in LD with an eQTL, we extracted a set of 3,718 independent trait-associated variants across all traits in unlinked loci (r2<0.1) (see Methods) and considered only protein-coding, lincRNA, and antisense genes (Supplementary Tables 8). Notably, 58.0% (2,158) of the trait-associated variants were in LD (r2>0.8) with at least one eQTL, when considering all significant variant-gene pairs, half of which (1,197) were the actual reported GWAS variant, and 27.8% (1,034) of all variants were in LD with the ‘best eQTL per eGene’ (see Methods and Supplementary Table 7). This is a ~5-fold increase over that reported in the GTEx pilot phase11 for eQTLs from nine tissues with fewer samples (27.8% versus 5.9% for ‘best eQTL per eGene’ set). A third of the increase is due to the expanded number of tissues, which resulted in 308 trait-associated variants in LD with an eQTL in only a non-pilot tissue, while the increased sample size (relative to the pilot-phase) leads to an additional ~3-fold increase. Consistent with the eQTLEnrich results, the independent set of genome-wide significant variants were significantly enriched for eQTLs in LD with them, across the 44 tissues (P<10-4 using variants matched on minor allele frequency [MAF], distance to nearest gene, and LD as the null; see Supplementary Note).
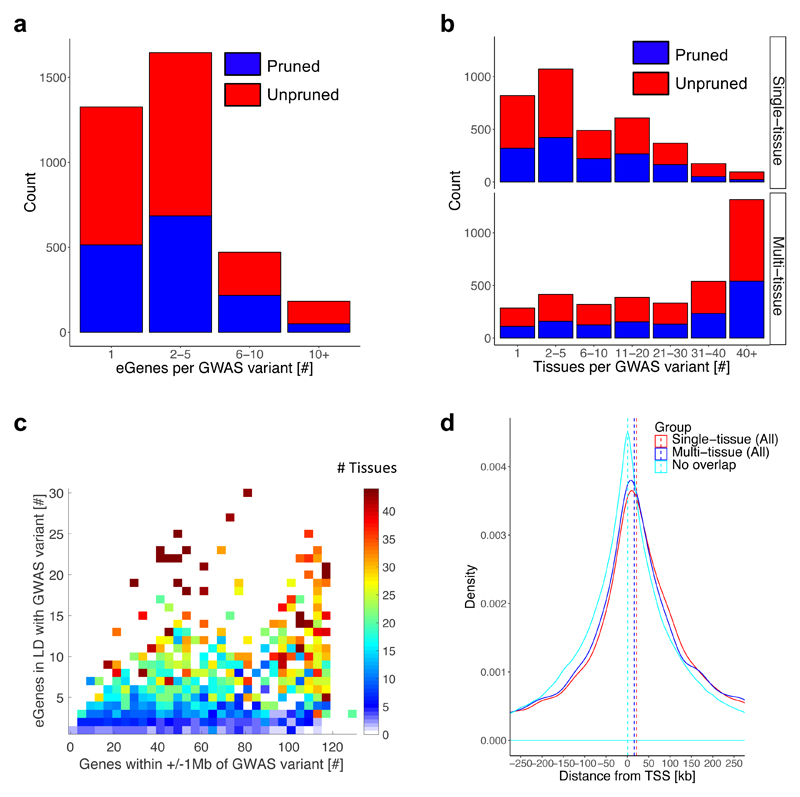
To determine whether trait-associated variants tended to have regulatory effects on multiple genes, or target the same gene in multiple tissues, we examined the distribution of the number of eQTL target genes and implicated tissues per trait-associated variant, using the independent set of 3,718 trait associations (Fig. 2a,b). Of the trait-associated variants in LD (r2>0.8) with at least one eQTL, 62% were in LD with an eQTL that targeted more than one gene (median 2.0 genes ± 3.8; using all eQTLs per eGene, Fig. 2a), and 77% were in LD with eQTLs that are significant in more than one tissue (median 5.0±11.6 tissues) (Fig. 2b). In contrast, among eQTLs in LD with trait-associated variants, those that target only a single gene were more tissue-specific than those that target multiple genes (Fig. 2c). Using eQTLs from the multi-tissue analysis (see Methods) further increased the number of tissues for eQTLs in LD with trait-associated variants (median 31.0±16.9 tissues; Fig. 2b), with a single tissue implicated by eQTLs for only 4.7% (173) of the trait-associated variants, primarily (88%) non-whole blood. Overall, for more than 50% of trait-associated variants, more than one causal gene and one tissue are implicated as potential mechanisms of action. Importantly, the use of eQTLs versus a physical window (e.g. of ±1Mb), substantially reduces the number of proposed causal genes in trait-associated loci (Fig. 2c) for follow-up analyses and inspection.
Fig. 2. eQTL annotation of variants from GWAS catalog.
a, Distribution of number of target genes for one or more eQTLs (from any of 44 tissues) with which a trait-associated variant is in LD (r2>0.8), considering only protein-coding, antisense and lincRNA genes. All significant variant-gene pairs per eGene from single-tissue analysis were used. Colors of stacked bars denote an LD-pruned threshold at r2>0.1 (blue) or unpruned (red) GWAS catalog variants with association p<5x10-8. b, Distribution of number of tissues implicated for each of the trait-associated variants in LD (r2>0.8) with at least one eQTL, using either all significant eQTLs per eGene discovered from the single-tissue (top panel) or multi-tissue (bottom panel) analysis. c, Number of eGenes implicated per trait-associated variant based on eQTLs (from 44 tissues) in LD with each trait-associated variant, is shown compared to number of genes within ±1Mb of the GWAS variant. The pruned set of GWAS catalog variants was used. Number of tissues implicated per variant, averaged in bins of 4 along the x-axis, is reflected in blue to red color gradient. d, Distribution of distance of eQTLs to TSS of their target genes in a ±250kb window, shown for eQTLs in LD (r2≥0.8) with a GWAS catalog variant based on single-tissue analysis (red; median distance to TSS: 21kb, interquartile range -66kb to 129kb) or multi-tissue analysis (blue), relative to eQTLs that are not in strong LD (r2<0.8) with any of the GWAS catalog variants (cyan; median distance to TSS: 0.7kb, interquartile range: -87kb to 91kb).
Of the three gene biotypes examined, 85% of the target genes of eQTLs in LD with one or more trait-associated variants are protein-coding genes and 15% are noncoding: 7% lincRNA and 8% antisense genes (Supplementary Table 8). Most proposed causal genes for known trait associations are protein-coding, but for ~4% (134) of trait associations, only noncoding genes are implicated, primarily lincRNAs (Supplementary Table 6). For example, the neuroblastoma-associated variant rs6939340 is in LD (r2=0.86) with an eQTL (rs9466271) acting on the neuroblastoma associated transcript 1, NBAT112, in multiple tissues, including nerve and brain.
Further, a common assumption is that the nearest gene to the trait-associated variant is the likely causal gene. However, for only 50% of trait-associated variants in LD with at least one eQTL was the target gene the nearest gene, illustrating the limitations of proximity-based assignment in identifying potentially causal genes. In addition, the distance of eQTLs in LD with trait-associated variants to the transcription start site (TSS) of their target gene was significantly greater than that of all other eQTLs (Wilcoxon rank sum P=3.0x10-59), and more likely to be downstream of the TSS (Fig. 2d and Supplementary Fig. 8).
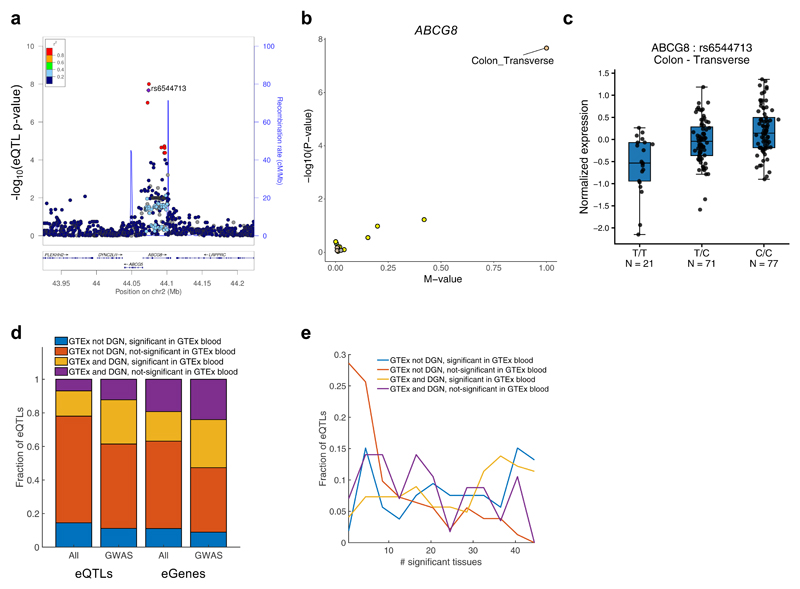
Since eQTLs are ubiquitous in the genome2, LD between an eQTL and trait-associated variant can occur by chance. Hence, we applied two colocalization methods, Regulatory Trait Concordance (RTC)13,14 and eCAVIAR15 (Supplementary Note), to three traits: SBP, DBP and CAD. Out of 21 (SBP), 19 (DBP), and 37 (CAD) associated variants (P<5x10-8), that are in LD with an eQTL, there is colocalization support for 67%, 58% and 32% of the loci, respectively, by at least one of the methods (Supplementary Table 9 and Supplementary Fig. 9). Some high-confidence genes suggested by high-LD and supported by both co-localization methods include rs1412444-LIPA and rs6544713-ABCG8 for CAD, rs1173771-NPR3 and rs17477177 with CCDC71L and CTB-30L5.1 (a lincRNA) for SBP, and rs2521501-MAN2A2 for both SBP and DBP (results and significant tissues in Supplementary Table 9). For CAD, the lead variant (rs6544713)16, located in the intron of ABCG8, is in almost complete LD with the best eQTL for ABCG8 (rs4245791; r2=0.99), which is specific to transverse colon (Fig. 3a,b) and has a 2.45-fold effect on expression17 (ALT vs. REF allele). ABCG8 plays a critical role in cholesterol metabolism by limiting intestinal dietary sterol uptake and by secreting sterol into bile. Recessive mutations in ABCG8 cause sitosterolaemia, a disorder characterized by premature atherosclerosis and abnormal sterol accumulation18. The minor T-allele at rs6544713 is associated with lower expression of ABCG8 in transverse colon (Fig. 3c), and increased CAD risk and higher low-density lipoprotein cholesterol (LDL) levels19. The three top eQTLs for ABCG8, which are in strong LD with the CAD-associated variant rs6544713 (r2>0.95), overlap gastrointestinal (GI) and liver enhancers based on Roadmap Epigenomics Project20 data.
Fig. 3. Proposing causal genes in inaccessible tissues.
a, LocusZoom48 plot showing that the lead variant at ABCG5/8 locus for CAD (n=184,405) and LDL cholesterol (n=95,454) (rs6544713; purple diamond) is in LD (r2=0.99), and colocalizes, with an eQTL signal for ABCG8 in transverse colon, using eCAVIAR and RTC. No other gene in the locus was implicated based on LD and colocalization. b, Forest PM-plot49 of single-tissue eQTL – log10(P-value) against the METASOFT posterior probability, m-value (indicating multitissue support), demonstrating rs6544713-ABCG8 eQTL is specific to transverse colon. c, Box plot showing correlation between rs6544713 and normalized ABCG8 expression in transverse colon, corrected for covariates used in cis-eQTL analysis. Box edges depict interquartile range, whiskers 1.5x the interquartile range, and center lines the median. Minor T-allele, associated with lower expression, is associated with increased CAD risk and higher LDL19. d, Fraction of best eQTL per eGenes (‘eQTLs’) or ‘eGenes’ significant in at least one GTEx tissue identified (yellow and purple) or not identified (blue and red) in DGN blood study at FDR≤0.05, further stratified by being significant (FDR≤0.05) (blue and yellow) or non-significant (red and purple) in GTEx blood. We compared all (21,643) eQTLs in GTEx (‘All’) to the subset of eQTLs in LD (r2≥0.8) with a GWAS variant (‘GWAS’; 471 independent trait-associated variants from GWAS catalog). e, Distribution of number of significant tissues per ‘best eQTL per eGene’ (FDR<0.05) sets in LD with GWAS variants, stratified by discovery in DGN (n=922) and being a GTEx blood eQTL (n=338; color-code as in d).
Breadth vs. depth of tissues: eQTL analysis of GWAS loci
Most eQTL analyses have been limited to a few readily accessible tissues (primarily blood), though with large sample sizes (900-5000). A specific goal of the GTEx study, in contrast, was to survey a wide range of (often inaccessible) tissues from the body, though with necessarily smaller sample sizes. To assess the relative value of breadth in sample type versus depth of sample size in the functional characterization of trait associations, we compared cis-eQTLs found in at least one of the 44 tissues to those discovered in two large cis-eQTL studies of whole blood (DGN21,22 n=922; Westra et al.23 n=5,311). We found that 80% of all ‘best eQTL per eGene’ variants and 63% of all eGenes found in ≥1 tissue in GTEx were not found in DGN, an RNA-seq based study (FDR<0.05; see Methods and Fig. 3d). Of just the subset of eQTLs in LD (r2>0.8) with 467 independent trait-associated variants from the GWAS catalog, 62% were not found in DGN, and of these, 82% were not significant in GTEx whole blood (Fisher’s Exact P=3.3x10-27; Fig. 3d). Due to differences in analytic methods, we also inspected the overlap at the eGene level. Importantly, 47% of all eGenes identified in GTEx across the 44 tissues were not found in DGN, of which 81% were identified only in non-blood tissues in GTEx (Fisher’s Exact test P=1.1x10-15; Fig. 3d). In contrast, only 3% of DGN eGenes were not detected in GTEx in any of the 44 tissues, even though DGN detected 1.3-fold more eGenes than GTEx whole blood. Notably, the GTEx eQTLs not found in DGN, in particular non-blood eQTLs, tended to be more tissue-specific than GTEx eQTLs that were also found in the larger DGN blood study (Wilcoxon rank sum P=1.0x10-16; Fig. 3e). Similar patterns were observed with the much larger, microarray-based study by Westra et al. (see Methods and Supplementary Figures 10 and 11). Hence, while larger studies provide better discovery power for a specific tissue of interest, there is great value to the diversity of tissues in proposing new biological hypotheses, especially tissue-specific ones, for a considerable number of trait associations (see examples listed in Supplementary Tables 10 and 11).
Trait heritability attributable to cis-eQTLs
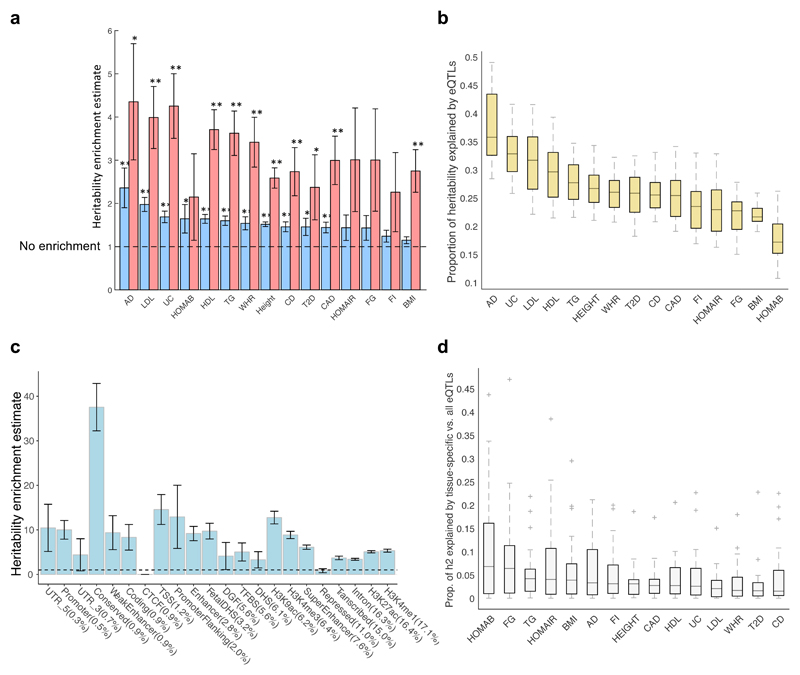
To quantify the proportion of genetic contribution to trait variation (heritability) that may be attributed to regulatory variation from across the 44 tissues, we applied (summary-statistics-based) LD score regression (LDSR)24 to 15 of the 18 traits tested for enrichment above, with available GWAS meta-analysis effect sizes (Supplementary Table 1 and Methods). Using all significant (single-tissue) eQTL variant-gene pairs from the 44 tissues, we found that while the eQTLs comprise on average 33% of the variants tested in all GWAS meta-analyses, they explained 52.1% of the variant-based heritability, showing a 1.6-fold concentration of heritability (see Methods; Fig. 4a and Supplementary Table 12). The combined set of eQTLs explains from 38.0 ± 2.7% (for BMI) to 78.2+15.2% (for AD) of the traits’ heritability (Supplementary Table 12), of which 10% to 16% are tissue-specific eQTLs (see Methods and Supplementary Table 13). By restricting our analysis to the top 10 eQTLs per eGene, which are likely to be enriched for causal variants25, proportionately, we found an even greater contribution of eQTLs to the variant-based heritability (3.2-fold concentration of heritability; Fig. 4a and Supplementary Table 14). Considering the contribution of eQTLs from each tissue separately, we found that the proportion of heritability explained by eQTLs for the different tissue-by-trait pairs tested ranged from a median of 5.9% to 9.9% per trait (ranging from 0% to 32.7 ± 7.7%), based on single-tissue eQTL analysis (Supplementary Table 15), and a median of 18.4% to 35.8% per trait (ranging from 10.8 ± 2.1% to 49 ± 9.5%), based on eQTLs from the multi-tissue eQTL analysis (Fig. 4b and Supplementary Table 16). By partitioning the heritability from the full set of significant eQTL variant-gene pairs by different structural/functional genomic features26 (see Methods), we found the highest concentration of heritability was for conserved genomic regions, and the lowest for repressor regions (Fig. 4c).
Fig. 4. Heritability estimates explained by eQTLs in 44 tissues.
a, Heritability enrichment estimates for 15 traits, defined as the proportion of heritability explained by all eQTLs (blue bars) or top 10 significant eQTL variants per eGene (red bars) aggregated across the 44 tissues divided by the fraction of GWAS variants that are eQTLs, using LD score regression analysis (Supplementary Tables 12 and 14). ** Heritability enrichment p-value passes Bonferroni correction, p<0.0017; * Heritability enrichment p<0.05. b, Distribution of proportion of heritability of 15 traits explained by eQTLs in 44 tissues, computed by multi-tissue (METASOFT) analysis (Supplementary Table 16). c, Heritability enrichment estimate computed for subsets of eQTLs that fall in different genomic features taken from 26, sorted in ascending order by percentage of eQTLs in each functional category shown in brackets. eQTLs from all 44 GTEx tissues based on single-tissue analysis were used. TFBS, transcription factor binding site. DGF, digital genomic footprint. d, Distribution of proportion of heritability explained by eQTLs acting on tissue-specific genes (Methods and Supplementary Table 17) divided by the proportion of heritability explained by all eQTLs (Supplementary Table 16) in each of the 44 tissues, computed by multi-tissue (METASOFT) analysis. All significant variant-gene pairs per eGene were used in all panels. 4a,c show the standard error from the LD score regression method. In 4b,d the boxes depict the interquartile range, whiskers depict 1.5x the interquartile range, center lines show the median, and ‘+’ represent the outliers.
To conduct tissue-specific assessment of the eQTL contribution to heritability, we evaluated the proportion of heritability attributed to those eQTLs that target ‘tissue-specific genes’ (i.e., genes showing higher expression in a given tissue than in all other tissues - see Methods), using LDSR, and found it to be a limited fraction of the heritability attributed to all eQTLs (Fig. 4d and Supplementary Table 17). Biologically plausible patterns of tissue-specific heritability concentration were observed across the different traits analyzed (Supplementary Fig. 12 and Supplementary Note).
Since the estimated proportion of heritability is modestly correlated with GWAS sample size (which explains R2=2.3-13.7% of variance in LDSR-derived heritability; Supplementary Fig. 13c,f), we investigated the pattern of heritability attributed to eQTLs across tissues for several WTCCC traits27, where GWAS sample size is identical for all traits and genotype data are available, and also found biologically plausible (tissue- and trait-dependent) patterns of heritability (Supplementary Fig. 14, Supplementary Table 18 and Supplementary Note).
Using eQTLs to discover new trait associations and genes
Estimating the true positive rate
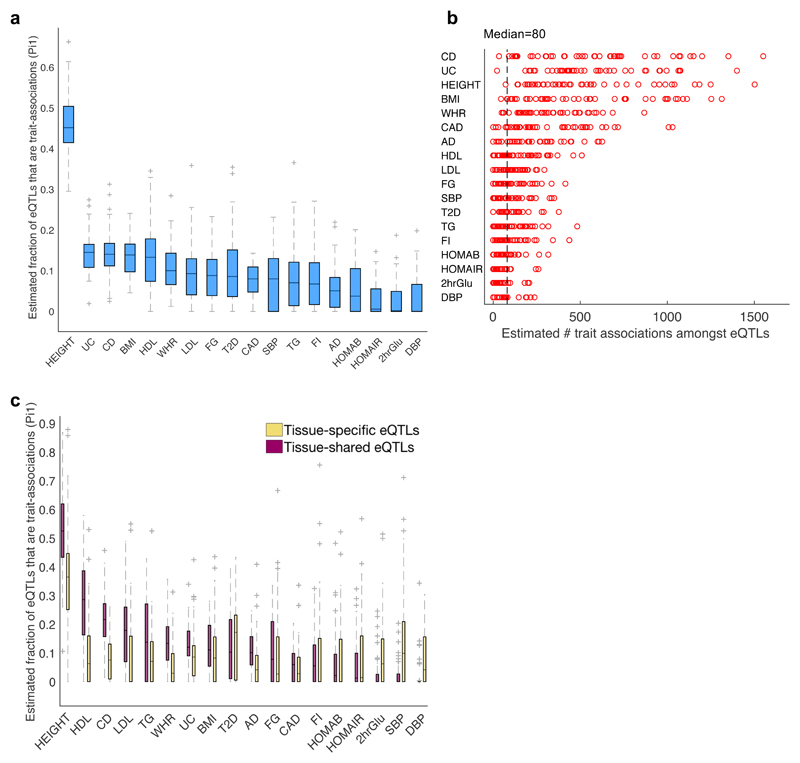
Since many more associations are likely to underlie trait variation than those currently passing genome-wide significance28 (e.g., Fig. 1b and Supplementary Fig. 3), we tested whether we could use eQTLs to identify novel associations, and to propose causal genes and potential tissues of action for these associations. We estimated the true positive rate (π1 statistic)29 of trait associations amongst eQTLs (using the ‘best eQTL per eGene’ sets) in the 44 tissues for the 18 traits tested above (see Methods). The average π1 across the 44 tissues per trait ranged from 2.9% to 45.5% for the 18 traits (Fig. 5a and Supplementary Table 19), suggesting that hundreds of trait associations (known and new) are acting via eQTLs in different tissues for all traits (Fig. 5b; lower bound estimates: median of 80 trait associations, and up to 1551 trait associations across all tissue-trait pairs tested). Consistent with the eQTLEnrich results, the anthropometric (height and BMI) and autoimmune (CD and UC) traits showed high π1 in most tissues, while other traits showed high π1 in only a subset of tissues (Supplementary Fig. 15). Clustering traits based on π1 across tissues (see Methods), we found that CD and UC clustered together (Pearson’s r=0.39, p=0.008), suggesting that eQTLs may contribute substantially to the known genetic correlation between these traits; waist-hip-ratio (WHR) clustered with T2D, more strongly than with BMI (Pearson’s r=0.37, p=0.01 versus Pearson’s r=0.12, p=0.44), consistent with reports that WHR is a better predictor of T2D30,31; and CAD clustered with SBP, a known CAD risk factor32 (Supplementary Fig. 15).
Fig. 5. Estimated true positive rate of trait associations amongst eQTLs in 44 tissues.
a, Distribution of estimated true positive rate (π1 statistic29) of trait associations (considering the full spectrum of GWAS p-values) amongst eQTLs across 44 tissues shown for 18 complex traits. b, Estimated number of true trait associations that are eQTLs in each of the 44 tissues, computed for 18 complex traits by multiplying π1 by the number of eQTLs analyzed per GWAS. The median number per trait ranges from 0 to 554, with a median of 80 trait associations per tissue-trait pair (dashed line) and a maximum of 1551 for CD. These are lower bound estimates due to incomplete overlap of variants between the GTEx and GWAS studies (see Methods). c, Distribution of estimated true positive rate (π1 statistic) of trait associations amongst tissue-specific eQTLs (yellow; significant in ≤~10% of tissues including the given tissue, based on METASOFT) versus tissue-shared eQTLs (pink; significant in ≥90% of tissues and the given tissue, based on METASOFT) was computed for 44 tissues by 18 traits. The ‘best eQTL per eGene’ set per tissue were used for all π1 analyses (Supplementary Table 19). In 5a,c the boxes depict the interquartile range, whiskers depict 1.5x the interquartile range, center lines show the median, and ‘+’ represent the outliers.
Similar to the eQTLEnrich analysis, the tissues with highest estimated π1 contained relevant pathogenic tissues, such as hippocampus for AD and skeletal muscle for T2D, but also less obvious tissues, such as the reproductive tissues. We therefore examined the relative contribution of tissue-specific eQTLs (significant in at most 10% of tissues) versus tissue-shared eQTLs (significant in over 90% of tissues) to trait associations (see Methods). Most traits showed, on average, higher absolute numbers and higher rates of trait associations (π1) among tissue-shared eQTLs (median π1=9.3%, range: 0-88%) relative to tissue-specific eQTLs (median π1=5.6%, range: 0-87%) (Fig. 5c, Supplementary Fig. 17a and Supplementary Table 19). Thus, at least some of the less obvious tissues with high π1 are capturing some component of shared regulation with the actual pathogenic tissues. On the other hand, 2-hour glucose tolerance levels (2hrGlu), SBP, and DBP showed on average a larger number of tissue-specific versus tissue-shared eQTLs amongst their trait associations (see Methods; Supplementary Fig. 17b and Supplementary Table 19). This result persisted after normalizing for differences in number of tissue-specific and tissue-shared eQTLs in each tissue (Supplementary Fig. 17c) and was not dependent on GWAS sample size (Supplementary Fig. 18).
Pathway-guided discovery and replication in DNA biobanks
To identify the true positive trait associations that contribute to the observed enrichment, we searched for target genes of eQTLs with top-ranked GWAS p-values (P≤0.05) that are enriched in biological pathways or functionally related gene sets, such as genes that share mouse knock-out phenotypes. We applied eGeneEnrich (see Methods) to several tissue-trait pairs (Supplementary Table 20) for a number of traits (AD, CAD, LDL, SBP, and T2D) that showed significant enrichment based on eQTLEnrich or π1 estimates, both of which are not affected by tissue sample size (Supplementary Figures 4 and 16, and Supplementary Tables 2 and 19). Multiple gene sets were nominally enriched (eGeneEnrich adjusted P<0.05) for each tissue-trait pair tested (Supplementary Table 20). The proposed causal genes and corresponding best eQTLs were then tested for replication in large-scale biobanks (see below).
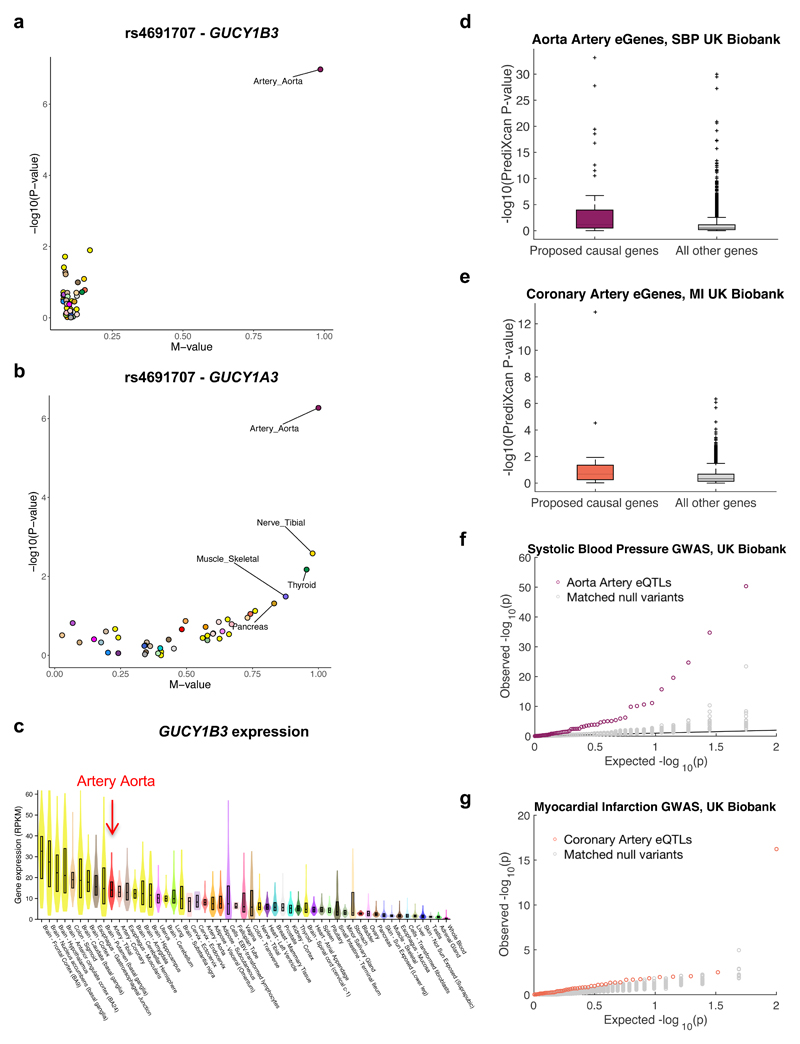
To identify tissue-specific processes, we also applied eGeneEnrich to target genes of tissue-specific eQTLs. We analyzed the target genes of aorta-specific eQTLs with SBP P<0.05 (that showed one of the strongest tissue-specific eQTL-GWAS enrichment; Supplementary Table 4), using a GWAS meta-analysis of 69k individuals33, and found significant enrichment in gene sets related to body weight and the cardiovascular system. These gene sets suggested, for example, an aorta-specific eQTL acting on two protein-coding genes, GUCY1A3 and GUCY1B3, and a non-coding gene, RP11-588K22.2, as a novel association with SBP (Fig. 6a,b). Notably, the best aorta eQTL for GUCY1B3 (rs4691707) recently reached genome-wide significance in a 5-fold larger GWAS meta-analysis of ~342,000 individuals34, but aorta would have not been prioritized as a tissue of action, based solely on the expression of GUCY1B3 or GUCY1A3 across tissues (Fig. 6c and Supplementary Fig. 19).
Fig. 6. Discovery and replication of novel associations and genes.
a, PM-plot49 of best eQTL for GUCY1B3 in artery aorta (n=197) (rs4691707) showing –log10(P-value) from single-tissue eQTL analysis versus the multi-tissue, m-value. b, rs4691707 is also an eQTL for GUCY1A3, though less specific to artery aorta, being significant (m-value>0.9) also in nerve tibial (n=256) and thyroid (n=278). c, Violin plots of GUCY1B3 expression across 44 tissues. Overlaid boxes indicate interquartile ranges and center-lines the median. Artery aorta is not the top ranked tissue for GUCY1B3 based on expression alone. d-e, Box plots of PrediXcan p-values (-log10) with UK Biobank GWAS for SBP and aorta artery genes (d) and MI and coronary artery genes (e), comparing eGeneEnrich-proposed genes to remaining genes expressed in the corresponding tissues. For both traits, proposed genes show significantly lower p-values, as assessed by Wilcoxon Rank-Sum one-tailed test (P=1.5x10-7 for d, P=5.8x10-5 for e). The boxes indicate interquartile ranges, whiskers 1.5x interquartile range, center-lines median values, and ‘+’ represent the outliers. f, Q-Q plot of replication association p-values from UK Biobank GWAS of SBP for artery aorta eQTLs (purple), enriched for SBP associations in a discovery GWAS33, compared to 100 null variant sets (gray; empirical P<0.01). g, Q-Q plot of replication association p-values from a UK Biobank GWAS of MI for coronary artery eQTLs (orange), enriched for CAD associations in a discovery GWAS50, compared to 100 null variant sets (gray; empirical P<0.01). In 6f,g the eQTLs and null variants have association p<0.05 in the corresponding discovery GWAS.
We tested for independent support for the proposed causal target genes from the discovery gene set analysis (eGeneEnrich adjusted P<0.05) in two large-scale repositories – UK Biobank5, a prospective study with extensive phenotypic data, and BioVU6, an electronic health records-linked DNA biobank (see Methods). First, using the gene-level association method, PrediXcan35,36, we evaluated the contribution of the genetic component of gene expression to trait variance in the UK Biobank for two traits with sufficient sample size: SBP and myocardial infarction (MI), as a proxy for CAD (see Methods). The eGeneEnrich-proposed causal genes for SBP in aorta or MI in coronary artery (Table 2 and Supplementary Table 20) each had significantly lower replication p-values than the remaining genes analyzed by PrediXcan in the specific tissue (Wilcoxon Rank-Sum one-tailed test P=1.5x10-7 for SBP and P=5.8x10-5 for MI; Fig. 6d,e and Supplementary Table 21). At FDR<0.05, 33 (58%) of the proposed causal genes replicated for SBP, some of which have been previously implicated, such as FURIN (P=6.94x10-34), a gene important for the renin-angiotensin system and sodium-electrolyte balance37,38, ARHGAP42 (P=1.66x10-28), shown to contribute to variation in blood pressure by modulating vascular resistance39, and GUCY1B3 (P=2.65x10-19), implicated in the development of hypertension in mice, and 15 (28%) proposed genes replicated for CAD (Supplementary Table 21). The significant association of the expression of HLA-C (P=2.96x10-5) with MI lends further support to an important role for a chronic inflammatory process in the development of atherosclerosis40,41.
Table 2. Complex trait causal genes proposed by gene set enrichment and PrediXcan analyses of top ranked eQTL target genes.
| Trait | eQTL Tissue | eGene | # significant gene sets** |
PrediXcan UK Biobank q-value |
|---|---|---|---|---|
| SBP | Aorta artery* | FURIN | 22 | 1.16E-32 |
| SBP | Aorta artery* | ARHGAP42 | 1 | 1.39E-27 |
| SBP | Aorta artery* | GUCY1A3 | 23 | 2.05E-19 |
| SBP | Aorta artery* | GUCY1B3 | 31 | 1.11E-18 |
| SBP | Aorta artery* | PRKAR2B | 33 | 5.71E-17 |
| SBP | Aorta artery* | CSK | 25 | 7.27E-13 |
| SBP | Aorta artery | ACADVL | 6 | 7.35E-12 |
| SBP | Aorta artery* | PRDM6 | 2 | 6.23E-11 |
| SBP | Aorta artery | SLC4A7 | 12 | 3.46E-07 |
| SBP | Aorta artery | MED8 | 1 | 1.54E-06 |
| SBP | Aorta artery | ARVCF | 1 | 1.68E-06 |
| SBP | Aorta artery | MED19 | 1 | 3.81E-05 |
| SBP | Aorta artery | ATF1 | 1 | 1.30E-04 |
| SBP | Aorta artery | HFE | 2 | 1.40E-04 |
| SBP | Aorta artery | PCDHA4 | 1 | 1.40E-04 |
| SBP | Aorta artery | FBLN7 | 1 | 1.86E-04 |
| SBP | Aorta artery* | GTF2IRD1 | 35 | 2.40E-04 |
| SBP | Aorta artery* | MRAS | 5 | 5.74E-04 |
| SBP | Aorta artery | RTN4 | 1 | 4.72E-03 |
| SBP | Aorta artery | GRID1 | 9 | 5.85E-03 |
| SBP | Aorta artery | FSCN2 | 12 | 7.20E-03 |
| SBP | Aorta artery | TCF4 | 1 | 1.40E-02 |
| SBP | Aorta artery | JPH2 | 1 | 1.64E-02 |
| SBP | Aorta artery | TMEM8B | 1 | 2.57E-02 |
| SBP | Aorta artery | DCHS1 | 9 | 2.98E-02 |
| SBP | Aorta artery | ULK2 | 1 | 3.71E-02 |
| CAD | Coronary Artery | PHACTR1 | 1 | 2.00E-12 |
| CAD | Coronary Artery | HLA-C | 4 | 2.24E-04 |
| CAD | Coronary Artery | ANAPC13 | 1 | 3.31E-02 |
| CAD | Coronary Artery | CDC25A | 4 | 3.31E-02 |
| CAD | Coronary Artery | CEP63 | 2 | 3.31E-02 |
| CAD | Coronary Artery | CTSK | 6 | 3.31E-02 |
| CAD | Coronary Artery | HLA-DOB | 4 | 3.31E-02 |
| CAD | Coronary Artery | GSTT2 | 2 | 3.95E-02 |
| CAD | Coronary Artery | NME1 | 4 | 3.95E-02 |
| CAD | Coronary Artery | SRD5A3 | 1 | 3.95E-02 |
| CAD | Coronary Artery | NPHP3 | 1 | 4.04E-02 |
| CAD | Coronary Artery | BAG6 | 4 | 4.81E-02 |
| CAD | Coronary Artery | DDT | 1 | 4.81E-02 |
| CAD | Coronary Artery | DDTL | 1 | 4.81E-02 |
| CAD | Coronary Artery | RPS28 | 2 | 4.81E-02 |
denotes aorta-specific eQTLs (significant in at most 4 tissues other than aorta).
The list of gene sets, from four different databases, in which the eQTL target genes were enriched, based on eGeneEnrich (adjusted P<0.05; see Methods), along with additional results, can be found in Supplementary Table 21. See Methods (“Replication framework using large-scale biobanks”) for description of the statistical approach (PrediXcan) used for the replication analysis. SBP, systolic blood pressure; CAD, coronary artery disease.
Second, we tested for replication of association of the best eQTL variants for the proposed causal genes (eGenes) (Supplementary Table 20) in the UK Biobank. The proposed aorta eQTLs were more likely to be replicated for SBP than matched null variants with GWAS p<0.05 (Fig. 6f; fold-enrichment=11.9, empirical P<0.01; see Methods), and similarly for coronary artery eQTLs and MI (Fig. 6g; fold-enrichment = 4.9, empirical P<0.01 for MI; see Methods), implicating robust novel variant-level associations for SBP and CAD (list of eQTLs with replication p<0.05 and those that pass Bonferroni correction in Supplementary Tables 22 and 23).
Finally, we found substantial replication (17%) of the eGeneEnrich-proposed genes in the specific tissue for the remaining GWAS traits (AD, LDL, T2D, as well as SBP and CAD), by applying PrediXcan to related clinical phenotypes in BioVU (Supplementary Table 20 and Supplementary Note), most of which are new associations (Supplementary Table 6).
Taken together, these results demonstrate a new and robust framework for identifying true positive associations, at both the gene and variant levels, for complex traits.
Discussion
Characterizing the biological mechanisms underlying genetic variants associated with disease predisposition and other complex traits has proven to be an enormous, but critical challenge. Here we conducted integrative analyses of eQTL and GWAS data for a broad spectrum of complex traits. Using a diverse set of tissues, we assessed the contribution of regulatory variants to trait variation through several approaches, including enrichment analysis, heritability analysis, and true positive rate estimation, and investigated the relative contribution of tissue-specific eQTLs. Our analyses demonstrate a substantial polygenic contribution from eQTLs, including tissue-shared and tissue-specific ones, to a range of complex traits. A broader sampling of cell types with larger sample sizes promises greater resolution on the impact of regulatory variants on disease risk and trait variation.
We observed a five-fold increase in the number of known trait-associated variants in LD with at least one best eQTL per eGene in the 44 tissues compared to the GTEx pilot phase with 9 tissues. Notably, for over half of these trait-associated variants, more than one target gene, in one or more tissues, was suggested by the linked eQTLs, raising the possibility that more than one causal gene, and possibly tissue, might underlie many of the associations. This pattern was also observed from colocalization analysis (also shown for v6p in2). Measuring eQTLs in individual cell types might increase resolution and narrow down the list of candidate genes and cell types. Furthermore, gene- and causal inference-based methods (such as PrediXcan35 or a Mendelian Randomization approach42) and additional functional validation (such as with CRISPR-mediated genome editing43,44) will be important in determining the causal genes at trait-associated loci. The proposed causal gene for trait-associated variants based on the strongest eQTL-derived target gene was, notably, often discordant (~50%) with proximity-based assignment, reinforcing the importance of eQTL analysis for prioritizing causal genes.
Our study implicates non-coding target genes, in particular lincRNAs and antisense genes that are polyadenylated, for about 15% of trait associations. This is of particular interest as many non-coding RNAs have regulatory functions (e.g., associated with chromatin-modifying complexes45), and participate in regulatory networks46. This suggests that among the trait-associated variants acting via non-coding RNA targets, some may be trans-eQTLs.
For the complex traits tested, eQTLs explain a substantial proportion of the genetic contribution to trait variation (10-50% per tissue), only a small fraction of which is due to eQTLs acting on tissue-specific genes. The proportion of heritability explained by all eQTLs (40-80%) is likely to increase with greater tissue sample size, which will lead to improved detection of eQTLs with weaker regulatory effects and additional independent eQTL signals per gene. The observation that tissue-shared eQTLs comprise a larger fraction of the trait associations than tissue-specific eQTLs for many of the tissue-trait pairs tested poses challenges in distinguishing pathogenic tissues from shared regulation among tissues. Alternatively, it also suggests that the underpinnings of many noncoding trait associations may be decipherable even if the actual pathogenic tissue is not available. Integrating additional layers of information, such as the tissue-specificity of eQTLs14,47, expression of transcriptional regulators, or broader cellular network effects on the locus in different cell types, may assist in detecting relevant tissue(s) of action.
While tissue-shared regulation appears to underlie an appreciable proportion of the genetic component of complex traits, we find multiple examples for which the trait associations are tissue-specific eQTLs, that were not found in previous, much larger whole blood eQTL studies. Our polygenic analyses also demonstrate the importance of a broad sampling of tissues; for some traits, enrichment for trait associations amongst eQTLs is most prominent only in a subset of difficult-to-acquire tissues.
By integrating prior biological knowledge (of pathways and mouse phenotype ontologies) with top-ranked trait-associated eQTLs in relevant tissues, followed by additional analysis for independent support in large-scale DNA biobanks, we were able to propose and replicate potentially causal genes and novel trait associations. Our work suggests that gene-based approaches that test the contribution of the genetically determined expression to trait variation35, coupled with better understanding of biological networks in a diverse set of tissues, promise to greatly enhance the functional interpretation of GWAS findings and identification of disease-relevant genes.
Online Methods
All statistical tests based on theoretical distributions were two-sided, unless noted otherwise.
Genotype Tissue Expression (GTEx) Project
All eQTLs used in the paper were computed from 44 tissues in GTEx release v6p2. Complete descriptions of the donor enrollment and consent process, and the biospecimen procurement methods, sample fixation, and histopathological review procedures were previously described51. Description of single-tissue and multi-tissue eQTL analyses can be found in Supplementary Note.
eQTL analyses of trait-associated variants
eQTL annotations of genome-wide significant associations with complex traits
To assess the utility of GTEx eQTLs (release v6p) for providing functional insights into trait-associated variants, we used all genome-wide significant associations (p-value ≤5x10-8) from the NHGRI-EBI GWAS Catalog version 1.0.1, release 2016-07-10 (see URLs), which contains significant associations from published GWAS studies for 659 distinct complex diseases and traits (referred to as “trait-associated variants”) and 563 unique phenotype ontologies (Experimental Factor Ontology), supplemented with 25 genome-wide significant variants for coronary artery disease16,50. In total, these data represented 11,010 entries corresponding to 7,076 unique dbSNP identifiers (Supplementary Table 5). For our analyses, we excluded entries that did not have a single dbSNP identifier for the association (n=179 entries), as well as all entries without mention of the use of European samples in either the discovery or replication sample set (n=1,885 entries; n=1,181 unique dbSNP identifiers).
Using PLINK 1.9052 (see URLs) on all non-Finnish northern European samples from the 1000 Genomes Phase 3 release53, all variants in strong LD (r2>0.8) with the remaining 5,895 unique GWAS index variants were identified. These index variants were then annotated with four categories of GTEx eQTLs, based on overlap of the GWAS index or their LD-proxy variants with: (1) the most significant eQTL for an eGene within ±1MB window around the transcription start site (TSS) (“best eQTL per eGene”; FDR≤0.05) in ≥1 tissue; (2) all significant variant-gene pairs for an eGene in ≥1 tissue (FDR≤0.05); (3) the most significant variant for an eGene in ≥1 tissue based on the multi-tissue method, METASOFT4 (see Supplementary Note) with significant evidence for an eQTL (m-value≥0.9); (4) all significant variant-gene pairs for an eGene showing significant evidence for an eQTL (m-value≥0.9) in ≥1 tissue based on METASOFT4.
The GWAS catalog was annotated with all analyzed GTEx genes, but for downstream analyses only the “protein_coding”, “lincRNA” and “antisense” biotypes were considered. Since entries in the complete GWAS catalog could comprise multiple index variants at the same locus for a single or different traits, LD-pruning was performed to provide a list of independent GWAS variants for downstream analyses. Variants associated with more than one trait were considered only once. Starting with the variants with the greatest number of eQTL annotations, pruning was performed according to three LD thresholds (r2>0.8, 0.5 and 0.1), as recorded in Supplementary Table 6. For analyses presented in this paper r2>0.1 was used unless mentioned otherwise. The eQTL annotated GWAS catalog is in Supplementary Table 6 and posted on the GTEx Portal (see URLs).
Comparison of GTEx eQTLs to previous large whole blood eQTL studies
We compared the eQTLs and eGenes discovered in any of the 44 tissues in GTEx to those cis-eQTLs discovered in two previous whole blood eQTL studies of substantially larger sample sizes: (1) a microarray-based study of 5,311 samples imputed to HapMap2 by Westra et al23, and (2) an RNA-Seq study of 922 samples from the Depression Genes and Networks (DGN)21,22, imputed to 1000 Genomes Project Phase 1. For the comparison with the Westra et al. study, we considered only protein-coding eGenes and eQTLs within +/- 250kb of the TSS of the target gene (14,303 eGenes). For the comparison with the DGN study, we considered protein-coding, lincRNA, and antisense gene types (23,219 eGenes) and eQTL variants within +/-1Mb of the TSS of the target gene, which were also tested in DGN (21,643 ‘best eQTL per eGenes’). For eQTL comparison, a single best eQTL variant was chosen per eGene across tissues - the variant with the largest number of significant tissues, determined by m-value≥0.9 in METASOFT and/or FDR≤0.05 in the single-tissue analysis.
We computed the proportion of eGenes and ‘best eQTL per eGenes’ discovered in ≥1 tissue in GTEx, but not found in DGN, and compared them (and their tissue specificity) to that of GTEx eQTLs found in DGN (Fig. 3d,e). For comparison with Westra et al. we considered only eGenes, due to impartial overlap of variants tested between GTEx and Westra et al. (Supplementary Figures 10 and 11). Furthermore, we determined the proportion of independent trait-associated variants (from the GWAS catalog) that are in LD (r2>0.8) with ≥1 eQTL, none of which was found in DGN or Westra et al. (Supplementary Tables 10 and 11). In cases where multiple eQTLs were in LD with a given GWAS variant, the eQTLs were grouped into one count; being significant in the non-GTEx blood study took precedence over not being identified in the study, and being significant in whole blood in GTEx took precedence over not being significant in blood.
Polygenic analyses of top ranked trait associations using eQTLs
GWAS meta-analysis data
Polygenic analysis is an approach aimed at relating phenotypic variation to multiple genetic variants simultaneously. It differs from conventional single-variant tests of association by allowing large numbers of loci (potentially in the thousands) to be tested for their contribution to the genetic architecture of phenotype. We analyzed 18 complex traits with available GWAS summary statistics, as well as several extensively studied WTCCC phenotypes27, for which genotype and phenotype data are available. These phenotypes span a wide range of complex traits, including metabolic, cardiovascular, anthropometric, autoimmune, and neurodegenerative phenotypes (Supplementary Table 1), allowing us to conduct comprehensive polygenic analyses (Table 1) of their genetic basis, using the eQTLs from the single-tissue and multi-tissue analyses.
Tissue-specific and tissue-shared eQTLs
For the GWAS-eQTL fold-enrichment and π1analyses in the paper, tissue-specific eQTLs were defined as eQTLs with m-value≥0.9 in METASOFT and/or FDR≤0.05 in the single-tissue analysis in 1-5 tissues (up to ~10% of tissues; the most highly similar tissues, except brain, are in sets of 2-3), including the tissue of interest, and tissue-shared eQTLs were defined as eQTLs with m-value≥0.9 in METASOFT and/or FDR≤0.05 in the single-tissue analysis in 40-44 tissues (over 90% of tissues), including the tissue of interest (Supplementary Fig. 7a).
Rank and permutation-based GWAS-eQTL fold-enrichment analysis
To test whether a set of eQTLs in a given tissue are enriched for sub-threshold (e.g., 5x10-8<P<0.05) to genome-wide significant (p≤5x10-8) common variant associations with a given complex disease or trait, more than would be expected by chance, we developed the following rank and permutation-based method, called eQTLEnrich. Specifically, for a given GWAS and for each of the 44 tissues with eQTLs, the most significant (best) cis-eQTL per eGene was retrieved (to control for linkage disequilibrium between the multiple variants tested per gene), and the GWAS variant association p-values for each set of eQTLs were extracted (eQTLs affecting more than one gene are considered only once). The distribution of GWAS p-values for each set of eQTLs is then tested for enrichment of highly ranked trait associations compared to an empirical null distribution sampled from non-significant variant-gene expression associations (FDR>0.05), also called null-eVariants, as follows: (i) A fold-enrichment is computed for each GWAS-tissue pair as the fraction of eQTLs with GWAS variant p-value<0.05 compared to expectation (5% of eQTLs; assuming a uniform distribution of GWAS p-values, if eQTLs contain no GWAS signal); (ii) Similar fold-enrichment values are computed for 100 to 100,000 randomly sampled sets (with replacement) of null-eVariants of equal size to the eQTL set, matching on potential confounding factors (using 10 quantile bins): distance of eQTL to TSS of the target gene, MAF, and number of proxy variants (at r2≥0.5), representing local LD (see Supplementary Fig. 1); (iii) An enrichment p-value is then computed as the fraction of permutations with similar or higher fold-enrichment than the observed value; (iv) An adjusted fold-enrichment (column H in Supplementary Table 2) is computed by dividing the fold-enrichment for a specific GWAS-tissue pair by the fold-enrichment of all null-eVariants with GWAS p<0.05 for the tissue-trait pair. The adjusted fold-enrichment is used as the enrichment test-statistic for ranking tissues per trait, because it is not dependent on tissue sample size (variance in adjusted fold-enrichment explained by tissue sample size is R2=0.04%), while the enrichment p-value is weakly correlated with tissue sample size (variance in the p-value explained by tissue sample size is R2=0.64%; Supplementary Fig. 4). Lower and upper bound 95% confidence intervals were estimated using bootstrapping of randomly sampled sets of null eVariants with replacement, matching on the three potential confounding factors above. We note that our definition of null-eVariants (FDR>0.05) for this method should yield a conservative estimate of the adjusted fold-enrichment.
eQTLEnrich was applied to 18 GWAS meta-analyses (Supplementary Table 1) using eQTLs from the single-tissue analysis at FDR≤0.05 (Supplementary Table 2) and tissue-specific eQTLs (defined above; Supplementary Table 4). Significant GWAS-tissue pairs were assessed using Bonferroni correction, correcting for total number of GWAS-tissue pairs tested (P<6.3x10-5). The adjusted fold-enrichment of the tissue-specific eQTLs is also not dependent on tissue samples size, number of eQTLs analyzed, or GWAS sample size (Supplementary Fig. 20).
eGeneEnrich: Gene set enrichment analysis of top ranked eQTL target genes
When enrichment for trait associations (subthreshold to genome-wide significant) is found amongst a set of eQTLs, gene-set enrichment analysis (GSEA) can help detect the true associations over noise amongst the top ranked eQTLs. This is based on the assumption that causal genes affecting a given trait will tend to cluster in a limited number of biological processes. To this end, we developed a gene-set enrichment analysis approach, called eGeneEnrich that tests whether the top ranked target genes of eQTLs with GWAS p-values below a given cutoff (P<0.05 used here) for a given trait-tissue pair are enriched for genes in predefined gene sets, compared to a null distribution that includes only genes expressed in the given tissue, as defined below (based on method described in54,55). For each gene set gs and a set of eQTLs, l (FDR<0.05), we computed the probability (hypergeometric) of observing at least k target genes of eQTLs l with GWAS P<0.05 out of a total of m eGenes with GWAS P<0.05 that belong to gene-set gs, given that n out of N target genes of all (eQTLs and null-eVariants) ‘best-eQTL per gene’ eQTLs belong to the gene-set gs:
To account for potential bias that may arise from the subset of genes expressed in a given tissue, we computed an eGeneEnrich adjusted p-value, i.e., an empirical GSEA p-value, that is the fraction of 1,000 to 10,000 randomly sampled target genes from a null set of variants, r (null-eVariants and eQTLs with GWAS p>0.05) of equal size to the eQTL set l, that have the same or more significant probability, Pgs,r than the observed probability, Pgs,l(X ≥ k).
We tested a range of sets of functionally related genes with ≥10 genes expressed in the given tissue, including metabolic and signaling pathways, gene ontology and mouse phenotype ontology, starting with: 674 gene-sets from REACTOME (downloaded from MSigDB v5.1), 186 gene-sets from KEGG (downloaded from KEGG in 2010), 1,942 gene ontologies (GO; see URLs) and 3,792 mouse phenotype ontologies (downloaded from Mouse Genome Informatics, MGI in 2013; see URLs). Bonferroni correction was applied per resource, correcting for number of gene-sets tested that contained ≥1 target gene of a best eQTL per eGene with GWAS p<0.05. The method was applied to GWAS meta-analyses for SBP, T2D, LDL, CAD, and AD, and a number of tissues chosen based on significant eQTL enrichment for trait associations or high π1 statistic and their relevance to the trait.
Replication framework using large-scale biobanks
To evaluate the role a gene may play in the etiology of a trait, we used PrediXcan35. Evaluating the genetically determined component of gene expression in an independent dataset for contribution to trait variance may facilitate replication of proposed causal genes. Specifically, from the weights derived from the gene expression model35 and the number of effect alleles Xij at the variant j, the genetically determined component of gene expression was estimated as follows:
An observed association between the estimated genetic component of gene expression and a trait proposes a causal direction of effect, as with eQTLs.
To test for independent support for the proposed causal genes for given trait-tissue pairs from the eGeneEnrich analysis, we utilized GWAS data from two large-scale biobanks. For replication analysis of proposed genes using the 500k UK Biobank5, we performed (variant-level) GWAS of SBP (phenotype code=4080, Systolic blood pressure, automated reading; n=473,460) and myocardial infarction (MI; phenotype code=20002_1075, Non-cancer illness code, self-reported: heart attack / myocardial infarction; number of cases=10,866, number of controls=428,004), using the mixed model association method, BOLT-LMM56, and applied PrediXcan using summary statistics36. The two phenotypes were chosen for their available large sample size. Replication of a gene was tested in the same discovery tissue (aorta artery for SBP and coronary artery for CAD), and significance was assessed using the q-value approach (FDR<0.05) applied to all genes tested in the given tissue for each trait. To test for higher replication rate for the proposed genes (in the given tissue context), we compared the distribution of replication p-values for the proposed genes to that of the remaining genes with gene expression imputation models (Wilcoxon Rank Sum one-tailed test).
We also sought variant-level replication of the associations of the best eQTLs for the eGeneEnrich-proposed genes using the BOLT-LMM results for SBP and MI in the UK Biobank. To determine whether our framework for finding true positive associations yields significantly improved replication rates, we generated an empirical distribution from 100 sets of null variants of equal size to the input set, matching on distance of the eQTL to the TSS of the proposed gene, MAF, and number of LD-proxy variants (at r2≥0.5). In addition, the null variants were chosen from the best eQTLs for non-significant eGenes (FDR>0.05) and were required to show a nominal GWAS association p-value<0.05.
We sought to replicate the proposed gene-tissue pairs for all remaining traits (AD, LDL, T2D), as well as SBP and CAD, from the eGeneEnrich analysis using BioVU6. For each gene-tissue pair, we estimated the genetic component of gene expression in the implicated tissue in 18,620 BioVU samples using PrediXcan35, enabling testing of gene association with the trait despite the lack of directly measured gene expression on the samples.
Estimation of true positive trait associations amongst eQTLs using π1 statistic
We calculated the proportion (π1) of true positive trait associations amongst the set of ‘best eQTL per eGene’ (FDR≤0.05) for each of the 44 tissues (computed with the single-tissue analysis) for 18 complex traits (Supplementary Table 1), by applying Storey’s method29 (qvalue R package 2.4.2, default options) to the GWAS association p-values for each tissue-trait pair (Supplementary Table 19). The π1 statistic considers the full distribution of GWAS p-values (from 0 to 1). We used the ‘best eQTL per eGene’ to control for potential confounding effects due to LD between the multiple variants tested per eGene. The π1 statistic was not correlated with number of ‘best eQTL per eGenes’ analyzed per tissue-trait pair (r=-0.03, p=0.35; Supplementary Fig. 16b). Furthermore, the tissue sample size explained only a small percentage of the variability (R2=1%) in the π1 statistic (Supplementary Fig. 16a). The π1 statistic was not correlated with GWAS sample size after excluding the Height GWAS meta-analysis, which is an outlier with respect to its much larger sample size compared to the other meta-analyses (Pearson’s r=0.06, p=0.1; Supplementary Fig. 16c,d). We performed hierarchical clustering of the traits based on the π1 values using Euclidean distance between pairs of traits.
The estimated number of eQTLs in a given tissue that are true positive trait associations was computed as π1 × NeQTL, where NeQTL is the number of ‘best eQTL per eGene’ variants that have available summary statistics in the given GWAS meta-analysis (Fig. 5b). Note these are lower bound estimates, as the overlap of the GTEx eQTL variants, imputed using 1000 Genomes Project Phase 1 vs3 (March 2012), with publicly available GWAS data variants, imputed using HapMap2 or earlier versions of 1000 Genomes Project, was partial (~26% of ‘best eQTL per eGenes’ for HapMap2 and 73-82% for 2010 and 2011 releases of 1000 Genomes Project Phase 1; see Supplementary Table 1).
For each tissue t, we also estimated the π1 statistic for tissue-specific eQTLs and tissue-shared eQTLs, anchored to the tissue t (as defined above) (Supplementary Figures 7a and 17a). The π1 of small eQTL sets (with ≤30 eQTLs) was set to ’NA’. We calculated a tissue-specificity measure per tissue-trait pair TSt,t.s., defined as the estimated number of tissue-specific eQTLs that are true positive trait associations based on π1,tissue−specific divided by the estimated number of tissue-shared eQTLs that are true positive trait associations based on π1,tissue−shared, for tissue t:
π1,tissue−shared below 0.01 were set to 0.01. The statistic provides a measure of eQTL tissue-specificity per tissue and controls for the effect of GWAS sample size and number of eQTLs tested per tissue (Supplementary Figures 17b and 18). Normalizing by the total of number of tissue-specific and tissue-shared eQTLs per tissue: π1,tissue−specific / π1,tissue−shared gave similar results with respect to the extent that tissue-specific eQTLs versus tissue-shared eQTLs underlie trait associations for the 18 complex traits tested (see Supplementary Fig. 17c compared to Supplementary Fig. 17b).
LD Score regression and summary statistics-based heritability methods
We performed LD score regression (LDSR)24 using the ldsc software package (see URLs) following the recommended steps in the web tutorial to estimate the relative contribution of eQTLs to the heritability of complex traits. To estimate the overall contribution to heritability from the eQTLs to 15 complex traits with available GWAS meta-analysis variant effect sizes, LDSR was applied to three different sets of eQTLs aggregated across all 44 tissues: (i) all significant variant-gene pair eQTLs (FDR≤0.05) from the single-tissue analysis, (ii) all tissue-specific eQTLs based on multi-tissue analysis (defined above), and (iii) a more stringent set of just the top 10 eQTLs per eGene in each of the tissues (Supplementary Tables 12-14). We also assessed the heritability attributed to eQTLs in each tissue separately, using either the single-tissue analysis (Supplementary Table 15) or the multi-tissue, METASOFT analysis (Supplementary Table 16). To carry out tissue-specific assessment (Supplementary Table 17), we ran the group analysis module in ldsc using the METASOFT4 derived eQTLs (m-value≥0.9) that were associated with tissue-selective genes in each GTEx tissue. For each tissue, tissue-selective (specific) genes were defined using a weighted tissue selectivity score (ts_score>3), that identifies genes with higher expression levels in a given tissue compared to all other tissues57.
For each of the eQTL classes, we calculated the proportion of heritability explained by eQTLs, Pr(h2g), and a “heritability enrichment” score (or “concentration of heritability”), defined as the proportion of the heritability explained by the eQTL variants, divided by the proportion of all variants represented by these eQTLs in the given GWAS: Pr(h2g)/Pr(SNPs). We note that the smaller variant set size for eQTLs acting on tissue specific genes may affect precision of LDSR heritability estimates. The 2hrGlu GWAS meta-analysis (Supplementary Table 1) was found to be unsuited for LD score regression, as the mean chi-square value obtained with LDSR was 1.02, suggesting very little polygenic signal (chi-square below 1.02 was reported as not suitable for LDSR24). All other traits tested had higher chi-square values. The heritability results for 2hrGlu were not included in the summary of all LDSR analyses (Fig. 4).
To assess the heritability of human disease risk and trait variation from eQTLs within different genome features, we computed the heritability enrichment score with ldsc, defined as the proportion of heritability explained by eQTL variants in each functional category taken from26, divided by the proportion of all variants represented by these eQTLs in the GWAS. All significant variant-gene pair eQTLs from all 44 GTEx tissues based on the single-tissue analysis were used for this analysis. The functional categories analyzed are displayed in Fig. 4c.
Supplementary Material
Acknowledgements
We thank the DIAGRAM, MAGIC, GIANT, GLGC, CARDIoGRAM, ICBP, IGAP and IIBDGC consortia for making their GWAS meta-analysis summary statistics publicly available. This work was conducted using the UK Biobank Resource (application number 25331). E.R.G. acknowledges support from R01 MH101820, R01 MH090937, R01 MH113362, and R01 CA157823 and benefited immensely from a Fellowship at Clare Hall, University of Cambridge. A.V.S., F.A., M.K., G.G., and K.G.A acknowledge support from the NIH contract HHSN268201000029C to The Broad Institute, Inc. M.v.d.B. acknowledges support by a Novo Nordisk postdoctoral fellowship run in partnership with the University of Oxford. F.H. and E.E. are supported by NIH grants R01-MH101782 and R01-ES022282. X.W. acknowledges support from NIH grants R01 HG007022 and R01 AR042742. M.I.McC is a Wellcome Senior Investigator supported by Wellcome (098381, 090532, 106130, 203141) and NIH (U01-DK105535, R01-MH101814). E.T.D. acknowledges support from the Swiss National Science Foundation, European Research Council, NIH-NIMH, and Louis Jeantet Foundation. N.J.C. is supported by R01 MH113362, R01 MH101820, and R01 MH090937. The datasets used for part of the replication analysis were obtained from Vanderbilt University Medical Center’s BioVU, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH funded Shared Instrumentation Grant S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vanderbilt.edu/pub/biovu/.
Footnotes
URLs
PLINK 1.90: https://www.cog-genomics.org/plink2
eCAVIAR: https://github.com/fhormoz/caviar
Regulatory Trait Concordance (RTC): https://qtltools.github.io/qtltools/
TORUS: https://github.com/xqwen/torus
PrediXcan: https://github.com/hakyim/PrediXcan
Storey’s qvalue R package: https://github.com/StoreyLab/qvalue
LD score regression (LDSR): https://github.com/bulik/ldsc
GCTA: http://cnsgenomics.com/software/gcta/#Download
eGeneEnrich: https://segrelab.meei.harvard.edu/software/
eQTLEnrich: https://segrelab.meei.harvard.edu/software/
GTEx Portal: http://www.gtexportal.org/
Gene Ontology: http://geneontology.org/
UK Biobank: http://www.ukbiobank.ac.uk/
BioVU: https://victr.vanderbilt.edu/pub/biovu/?sid=194
NHGRI-EBI GWAS Catalog: http://www.ebi.ac.uk/gwas
Mouse Genome Informatics: http://www.informatics.jax.org/downloads/reports/index.html
Author Contributions
E.R.G and A.V.S. jointly designed the study and led the analysis. E.R.G., A.V.S., M.v.d.B., and K.G.A. wrote the manuscript. E.R.G., A.V.S., M.v.d.B., X.W., H.S.X., F.H., H.O., A.K., E.M.D., F.A., and J.Q. performed statistical analysis. E.R.G., A.V.S., M.v.d.B., X.W., H.S.X., F.H., E.M.D., D.L.N., E.E., M.K., G.G., M.I.McC, E.T.D., N.J.C., and K.G.A. interpreted the results of the analysis. All authors contributed to the critical review of the manuscript.
Competing Interests
M.I.McC serves on advisory panels for Pfizer and NovoNordisk. He has received honoraria from Pfizer, NovoNordisk, Sanofi-Aventis and Eli-Lilly, and research funding from Pfizer, Eli-Lilly, Merck, Takeda, Sanofi Aventis, Astra Zeneca, NovoNordisk, Servier, Janssen, Boehringer Ingelheim and Roche. M.v.d.B is an employee of Novo Nordisk. H.S.X. and J.Q. are employees of Pfizer.
Code Availability
Code for methods applied in the paper can be downloaded from the URLs above.
Data availability statement
The protected data for the GTEx project (e.g., genotype and RNA-sequence data) are available via access request to dbGaP accession number phs000424.v6.p1. Processed GTEx data (e.g., gene expression and eQTLs) are available on the GTEx portal: https://gtexportal.org. The NHGRI-EBI GWAS Catalog version 1.0.1, release 2016-07-10 was downloaded from www.ebi.ac.uk/gwas. The URL of the summary statistics datasets of all the GWAS meta-analyses analyzed in the paper can be found in Supplementary Table 1.
References
- 1.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GTEx Consortium. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–85. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet. 2011;88:586–98. doi: 10.1016/j.ajhg.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen X. Molecular QTL Discovery Incorporating Genomic Annotations using Bayesian False Discovery Rate Control. Annals of Applied Statistics. 2016 [Google Scholar]
- 10.Wen X, Lee Y, Luca F, Pique-Regi R. Efficient Integrative Multi-SNP Association Analysis via Deterministic Approximation of Posteriors. Am J Hum Genet. 2016;98:1114–29. doi: 10.1016/j.ajhg.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey GK, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–37. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ongen H, et al. Estimating the causal tissues for complex traits and diseases. Nat Genet. 2017;49:1676–1683. doi: 10.1038/ng.3981. [DOI] [PubMed] [Google Scholar]
- 15.Hormozdiari F, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CARDIoGRAMplusC4D Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammadi P, Castel SE, Brown AA, Lappalainen T. Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res. 2017;27:1872–1884. doi: 10.1101/gr.216747.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berge KE, et al. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–5. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 19.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–81. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kukurba KR, et al. Impact of the X Chromosome and sex on regulatory variation. Genome Res. 2016;26:768–77. doi: 10.1101/gr.197897.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown AA, et al. Predicting causal variants affecting expression by using whole-genome sequencing and RNA-seq from multiple human tissues. Nat Genet. 2017;49:1747–1751. doi: 10.1038/ng.3979. [DOI] [PubMed] [Google Scholar]
- 26.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wellcome Trust Case Control, C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eichler EE, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao Q, Nyamdorj R. Is the association of type II diabetes with waist circumference or waist-to-hip ratio stronger than that with body mass index? Eur J Clin Nutr. 2010;64:30–4. doi: 10.1038/ejcn.2009.93. [DOI] [PubMed] [Google Scholar]
- 31.Cheng CH, et al. Waist-to-hip ratio is a better anthropometric index than body mass index for predicting the risk of type 2 diabetes in Taiwanese population. Nutr Res. 2010;30:585–93. doi: 10.1016/j.nutres.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Emerging Risk Factors, C et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.International Consortium for Blood Pressure Genome-Wide Association, S et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ehret GB, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–84. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–8. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbeira Alvaro, S KP, Torres Jason M, Wheeler Heather E, Torstenson Eric S, Edwards Todd, Garcia Tzintzuni, Bell Graeme I, Nicolae Dan, Cox Nancy J, Im Hae Kyung. MetaXcan: Summary Statistics Based Gene-Level Association Method Infers Accurate PrediXcan Results. bioRxiv. 2017 [Google Scholar]
- 37.Ganesh SK, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–78. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li N, et al. Associations between genetic variations in the FURIN gene and hypertension. BMC Med Genet. 2010;11:124. doi: 10.1186/1471-2350-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rippe C, et al. Hypertension reduces soluble guanylyl cyclase expression in the mouse aorta via the Notch signaling pathway. Sci Rep. 2017;7 doi: 10.1038/s41598-017-01392-1. 1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies RW, et al. A genome-wide association study for coronary artery disease identifies a novel susceptibility locus in the major histocompatibility complex. Circ Cardiovasc Genet. 2012;5:217–25. doi: 10.1161/CIRCGENETICS.111.961243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lahoute C, Herbin O, Mallat Z, Tedgui A. Adaptive immunity in atherosclerosis: mechanisms and future therapeutic targets. Nat Rev Cardiol. 2011;8:348–58. doi: 10.1038/nrcardio.2011.62. [DOI] [PubMed] [Google Scholar]
- 42.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–7. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 43.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol. 2016;34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 45.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai Y, Dai X, Harrison AP, Chen M. RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Brief Funct Genomics. 2015;14:91–101. doi: 10.1093/bfgp/elu017. [DOI] [PubMed] [Google Scholar]
- 47.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang EY, et al. ForestPMPlot: A Flexible Tool for Visualizing Heterogeneity Between Studies in Meta-analysis. G3 (Bethesda) 2016;6:1793–8. doi: 10.1534/g3.116.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carithers LJ, et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv Biobank. 2015;13:311–9. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Segre AV, et al. Pathways targeted by antidiabetes drugs are enriched for multiple genes associated with type 2 diabetes risk. Diabetes. 2015;64:1470–83. doi: 10.2337/db14-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–90. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang RY, Quan J, Sodae R, Aguet F, Segrè AV, Allen JA, Lanz TA, Reinhart V, Crawford M, Hasson S, GTEx Consortium et al. A systematic survey of human tissue-specific gene expression and splicing reveals new opportunities for therapeutic target identification and evaluation. bioRxiv. 2018 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.