Abstract
The naphterpins and marinones are naphthoquinone meroterpenoids with an unusual aromatic oxidation pattern that is biosynthesized from 1,3,6,8-tetrahydroxynaphthalene (THN). We propose that cryptic halogenation of THN derivatives by vanadium-dependent chloroperoxidase (VCPO) enzymes is key to this biosynthetic pathway, despite the absence of chlorine in these natural products. This speculation inspired a total synthesis to mimic the naphterpin/marinone biosynthetic pathway. In validation of this biogenetic hypothesis, two VCPOs were discovered that interconvert several of the proposed biosynthetic intermediates.
Keywords: biomimetic synthesis, biosynthesis, dearomatization, meroterpenoids, total synthesis
The chemical reactions in a cell obey the same logic of organic chemistry as those that occur in a round-bottomed flask. As such, we can often use our knowledge of organic reactivity to predict a probable biosynthetic pathway to a natural product from its relatively limited pool of biochemical reagents. The total synthesis of a whole biosynthetic pathway offers an opportunity to interrogate a biogenetic hypothesis by providing both the substrates and the expected products for experiments with enzymes that are discovered through a genome mining approach. This collaborative approach gives insight into the chemistry of life, while also adding biosynthetic enzymes to the toolkit of synthetic chemists in the quest for the fastest and most efficient ways to construct complex organic molecules.
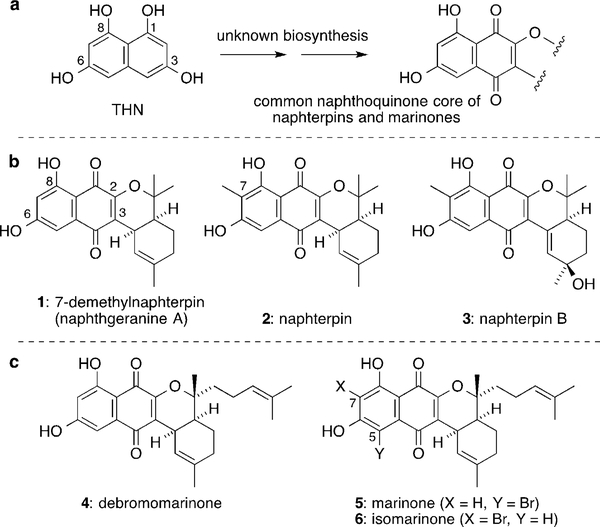
In the past 30 years, many naphthoquinone meroterpenoid antibiotics biosynthetically derived from 1,3,6,8-tetrahydroxynaphthalene (THN) have been isolated from marine and soil strains of Streptomyces bacteria, including the naphterpins,[1] marinones,[2] napyradiomycins,[3] and merochlorins.[4] 7-Demethylnaphterpin (1, also known as naphthgeranine A)[1b,c] is the simplest member of the naphterpin/marinone family (Figure 1). It possesses a naphthoquinone ring system oxygenated at C-2, C-6, and C-8 and is cis-fused to a geranyl side chain (attached by a C-C bond at C-3). Naphterpin[1a] (2) has an additional C-7 methyl substituent, and it is co-isolated with its oxidized analogues naphterpins B (3) and C (not shown).[1e]
Figure 1.
a) The mechanism of the biosynthesis of many naphthoquinone Streptomyces meroterpenoids from their biosynthetic precursor THN was previously unknown. This includes the naphterpins (b) and the marinones (c).
Marinones (4–6), isolated from Streptomyces sp. CNQ-509,[2] have an extra prenyl group compared to 1 that is derived from cyclization of a farnesyl side chain. Marinone (5) and isomarinone (6) have a bromine substituent at C-5 and C-7, respectively. Several structurally similar naphthoquinone meroterpenoids have been isolated from trees of the Bignoniaceae family, such as pyranokunthone A, which was synthesized by Trauner using an intramolecular hetero-Diels–Alder reaction.[5] Presumably, a similar hetero-Diels–Alder reaction is involved at a late stage of the biosynthesis of the naphterpins and marinones. However, in the case of these Streptomyces meroterpenoids, the mechanism of the biosynthesis of the highly oxidized naphthoquinone ring system is currently unknown. Although naphterpins and marinones have been shown to be biosynthesized from THN through 13C labeling studies,[6] the oxidation pattern of THN does not obviously correlate with the oxidation pattern of these natural products. Furthermore, THN is nucleophilic at C-2 and C-4, but the naphterpins and marinones possess an isoprene substituent at the non-nucleophilic C-3 position.
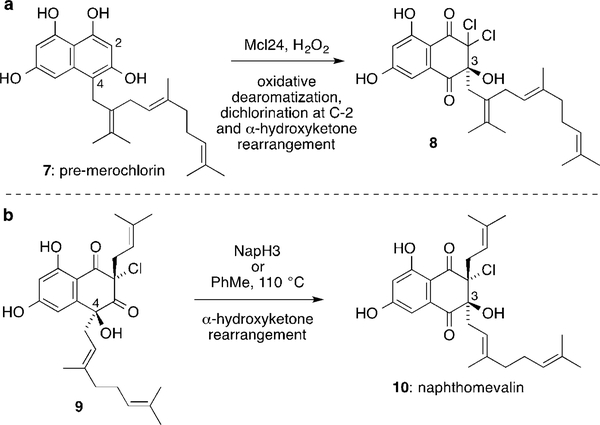
We recently reported two vanadium-dependent chloroperoxidase (VCPO)[7] enzymes, Mcl24 and NapH3,[8] that catalyze α-hydroxyketone rearrangements to shift terpene side chains[9] from C-4 to C-3 in merochlorin and napyradiomycin biosynthesis, respectively (Scheme 1). Mcl24 catalyzes oxidative dearomatization and dichlorination of the THN ring system of 7 followed by a 1,2-shift to give 8, whereas NapH3 just catalyzes the 1,2-shift of 9 to give naphthomevalin (10), the simplest member of the napyradiomycin family. The discovery of these enzymes, combined with associated biomimetic synthetic work, led us to propose a biosynthesis of the napyradiomycins.[8] Herein, we extend this biosynthetic logic to include the naphterpins and marinones.
Scheme 1.
VCPO enzymes known to catalyze α-hydroxyketone rearrangements a) Mcl24 in merochlorin biosynthesis. b) NapH3 in napyradiomycin biosynthesis.
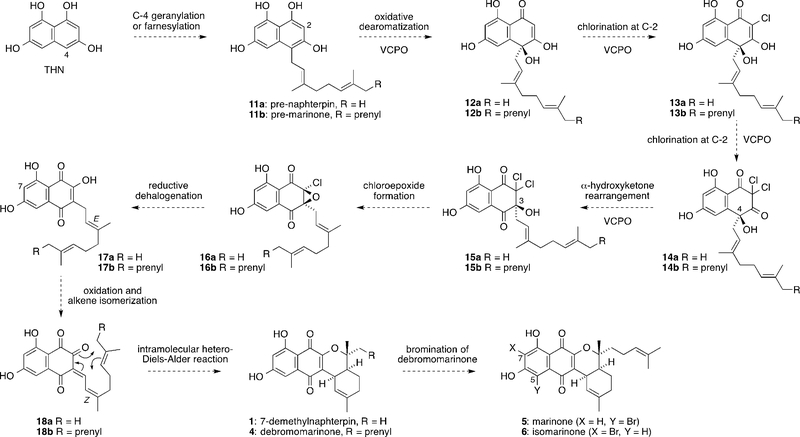
Our proposed biosynthesis of 7-demethylnaphterpin (1) and debromomarinone (4) from THN is outlined in Scheme 2. Firstly, THN is known to be biosynthesized by the condensation of five malonyl-coenzyme A units followed by aromatization of the resultant pentaketide under the control of a single type III polyketide synthase.[10] Next, we propose that THN undergoes geranylation or farnesylation at C-4 to give 11a and 11b. This reaction is putatively catalyzed in vivo by the NphB aromatic prenyltransferase in naphterpin biosynthesis[11] or by CnqP3 or CnqP4 in marinone biosynthesis.[12] We then propose that 11a/11b are subjected to VCPO-catalyzed oxidative dearomatization to initially give 12a/12b, followed by VCPO-catalyzed chlorination at C-2 to give monochlorides 13a/13b and dichlorides 14a/14b. A VCPO-catalyzed α-hydroxyketone rearrangement (which shifts the geranyl substituent from C-4 to C-3) would then give 15a/15b. Computational studies have shown that this rearrangement is thermodynamically favorable, and the proposed multi-tasking VCPO chemistry is analogous to the known reactivity of the VCPO Mcl24 in merochlorin biosynthesis (Scheme 1a).[8] Exposure of 15a/15b to mildly basic conditions should induce cyclization to give a-chloroepoxides 16a/16b. A similar cyclization has been proposed to occur in the biosynthesis of A80915G from naphthomevalin.[13] A handful of marine a-chloroepoxide natural products have been previously reported,[14] but none have yet been proposed as intermediates in biosynthetic pathways. Next, we propose reductive dehalogenation of α-chloroepoxides 16a/16b to give hydroxynaphthoquinones 17a/17b. Related reductions of naphthoquinone epoxides to give naphthoquinones have been proposed in the biosynthesis and used in the biomimetic synthesis of zeylanone[15] and the juglocombins.[16] Importantly, the C-7 methylated analogue of 17a, which we suggest is the direct biosynthetic precursor of naphterpin (2), has been isolated as a Streptomyces metabolite.[17]
Scheme 2.
Proposed biosynthesis of 7-demethylnaphterpin and debromomarinone from THN.
Next, oxidation and facile E-to-Z double-bond isomerization of 17a/17b would give the reactive enones 18a/18b, which could be converted to 7-demethylnaphterpin (1) and debromomarinone (4) through intramolecular hetero-Diels–Alder reactions. This oxidative cyclization is similar to the biosynthesis of Δ1-tetrahydrocannabinolic acid from cannabigerolic acid catalyzed by THCA synthase.[18] The proposed oxidation of 17a/17b also has precedent in the biosynthesis of chlorizidine A in a marine strain of Streptomyces.[19] The naphterpins and marinones are isolated as enantiopure compounds, so the oxidative cyclization of achiral 17a/17b must be under enzymatic stereocontrol. Finally, late-stage, vanadium-dependent bromoperoxidase-catalyzed bromination of 4 at C-5 or C-7 would give marinone (5) or isomarinone (6), respectively. Despite the absence of chlorine in the naphterpin and marinone natural products, we propose that this rather elaborate biosynthetic pathway involves cryptic chlorination[20] to selectively oxidize the THN ring and promote the a-hydroxyketone rearrangement. Genomic analyses of marinone-producing Streptomyces sp. CNQ-509 supports this biosynthetic hypothesis through the clustering of VCPO and aromatic prenyltransferase homologues with a THN synthase.[12] In the case of naphterpin biosynthesis, only three genes have yet been reported, none of which are VCPOs.[11a]
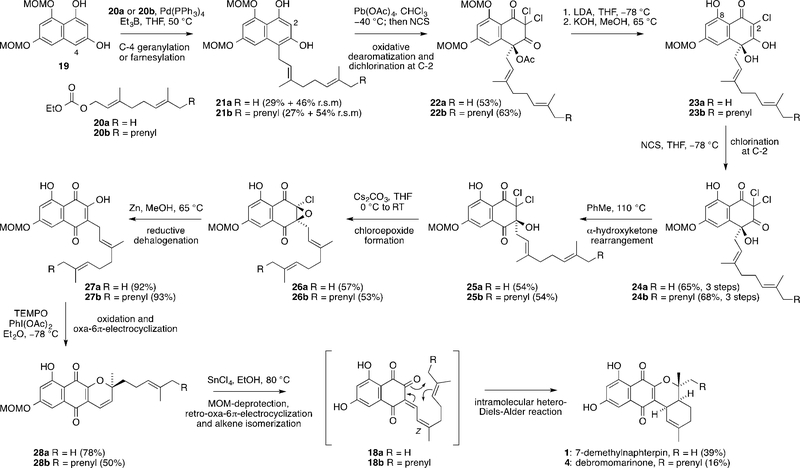
We completed a total synthesis of the whole biosynthetic pathway leading to 7-demethylnaphterpin (1) and debromomarinone (4) from a protected THN derivative using a sequence of key reactions that occur in the same order as the proposed biosynthesis. The synthesis was therefore designed to give access to as many proposed biosynthetic intermediates as possible, for use in later biosynthetic studies. Firstly, di-MOM-protected THN derivative 19 was geranylated[8] at C-4 with 20a or farnesylated with 20b to give 21a and 21b, respectively, which were then dearomatized with Pb(OAc)4 and dichlorinated at C-2 with NCS in a one-pot process to give 22a/22b (Scheme 3). LDA-mediated dechlorination and basic hydrolysis of 22a/22b gave 23a/23b, which were re-chlorinated using NCS to give 24a/24b in good yield over 3 steps. Acetate hydrolysis of 22a/22b to give 24a/24b directly was impossible due to competing fragmentation through a haloform reaction. MOM removal at the C-8 phenol also occurred during the KOH-mediated acetate hydrolysis step. Heating 24a/24b in PhMe at 110°C induced a thermal α-hydroxyketone rearrangement to give 25a/25b. The α-hydroxyketone rearrangement of 24a/24b was not found to be catalyzed by protic or Lewis acids, and attempts to catalyze the reaction with base led to fragmentation. Treatment of 25a/25b with Cs2CO3 in THF formed α-chloroepoxides 26a/26b, which were reduced with Zn in MeOH to give 27a/27b in high yield.[21] Oxidation of 27a/27b with TEMPO/PhI(OAc)2[22] in Et2O then gave tricycles 28a/28b through oxa-6π-electrocyclization. As has been observed previously, 6π-electrocyclization is usually kinetically favored over an intramolecular hetero-Diels–Alder reaction in such systems.[5] However, heating 28a/28b with SnCl4 in EtOH caused isomerization (and MOM removal) to give 1 in 39% yield (after recrystallization from EtOAc as an orange solid) or 4 in 16% yield through a cascade of retro-oxa-6π-electrocyclization, alkene isomerization, and a final intramolecular hetero-Diels–Alder reaction of the presumed intermediates 18a/18b. The yield of 4 is lower than the yield of 1, probably due to the instability of the exposed prenyl group of 4 in the presence of SnCl4.
Scheme 3.
Biomimetic total synthesis of the naphterpin/marinone biosynthetic pathway. MOM=methoxymethyl, THF=tetrahydrofuran, NCS=N-chlorosuccinimide, LDA=lithium diisopropylamide, TEMPO=(2,2,6,6-tetramethylpiperidin-1-yl)oxyl.
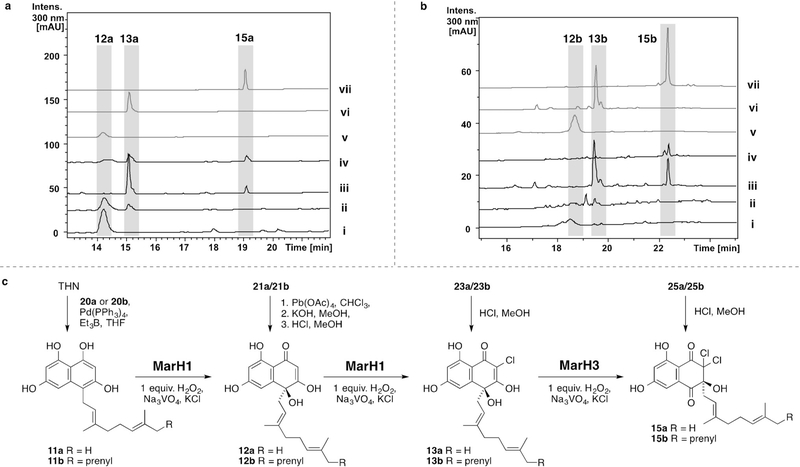
Our total syntheses of 7-demethylnaphterpin (1) and debromomarinone (4) are the first for members of the naphterpin/marinone family, but more efficient strategies for constructing these natural products can be envisaged. Indeed, a previous non-biomimetic synthesis of a protected form of 1 is shorter.[23] However, the value of our synthesis lies not in the final destination, but in the journey, since each step of the synthesis represents a potential biosynthetic intermediate (after facile MOM removal). The dearomatized biosynthetic intermediates 12a/b, 13a/b and 15a/b were prepared through this route (Scheme 4c) for use as standards in biosynthetic studies. To interrogate the individual biosynthetic steps, three VCPO homologues: marH1, marH2, and marH3, were individually cloned from the putative Streptomyces sp. CNQ-509 marinone biosynthetic gene cluster and heterologously expressed in Escherichia coli. The recombinant proteins were purified using Ni2+ affinity chromatography and initially interrogated using the monochlorodimedone (MCD) assay (see Figures S4–S6 in the Supporting Information). Two VCPO homologues (MarH1, MarH3) showed halogenation activity and were further examined using synthetic substrates.
Scheme 4.
Reversed-phase HPLC chromatograms (300 nm) of MarH enzyme assays (i–iv) and comparison to synthetic standards (v–vii) with naphterpin (a) and marinone (b) substrates. i) 11a/11b, no enzyme, 2 equiv. H2O2; ii) 11a/11b, MarH1, 2 equiv. H2O2; iii) 13a/13b, MarH3, 1 equiv. H2O2; iv) 11a/11b, MarH1, and MarH3, 3 equiv. H2O2; v) 12a/12b; vi) 13a/13b; vii) 15a/15b. c) Reaction conditions for the chemical syntheses (vertical arrows) and MarH enzymatic conversions (horizontal arrows) of biosynthetic intermediates 11a/11b, 12a/12b, 13a/13b, and 15a/15b. Additional control experiments can be found in Figures S7–S26.
Thorough in vitro characterization of the reactions between pre-naphterpin (11a) and pre-marinone (11b)[24] and the recombinant MarH enzymes was performed using reversed-phase HPLC to monitor the formation of biosynthetic intermediates (Schemes 4a,b and Figures S7–S26).[25] In the presence of sodium vanadate and two stoichiometric equivalents of hydrogen peroxide, MarH1 catalyzed the oxidative dearomatization of 11a/11b at C-4 and subsequent monochlorination at C-2, yielding 12a/12b and 13a/13b, respectively. Additional chlorination of 13a/13b by MarH1 was not observed following incubation with excess hydrogen peroxide, so this substrate was then interrogated with MarH3. In vitro incubation of 13a/13b with MarH3 subsequently catalyzed additional chlorination at C-2 and an α-hydroxyketone rearrangement, moving the terpene sidechain from C-4 to C-3 to produce 15a/15b. Furthermore, a one-pot reaction of 11a/11b with MarH1, MarH3, and sodium vanadate yielded the dichlorinated 1,2-shifted 15a/15b as the major product; progression along the biosynthetic pathway was directly correlated to the number of molar equivalents of hydrogen peroxide added (Figures S15,S24). MarH2 failed to show any catalytic halogenating or α-hydroxyketone rearrangement activities with meroterpenoid substrates, either individually or in combination with other MarH enzymes. We rationalize its inactivity despite high sequence similarity with other VCPO enzymes as being due to substitution of the key vanadate-coordinating histidine residue with an asparagine[26] (Figure S2).
In conclusion, we discovered that Streptomyces bacteria use VCPO enzymes to oxidize THN derivatives via cryptic chlorination in the biosynthesis of the naphterpin and marinone families of meroterpenoid natural products. The biosynthetic sequence is initiated by VCPO-mediated oxidative dearomatization and dichlorination, followed by an α-hydroxyketone rearrangement. Subsequent loss of chloride anion leaving groups allows the formation of the highly oxidized naphthoquinone core. We mimicked the entire biosynthetic pathway in biomimetic syntheses of 7-demethylnaphterpin and debromomarinone that use simple reaction conditions to exploit the predisposed reactivity of the intermediates in a logical manner. Several proposed biosynthetic intermediates were also synthesized and used as substrates and standards to help elucidate the function of two VCPO enzymes, MarH1 and MarH3, which initiate marinone biosynthesis. Further work to discover and characterize enzymes that control the final steps of naphterpin/marinone biosynthesis is underway.
Supplementary Material
Acknowledgements
This work was supported by an Australian Research Council Future Fellowship (FT170100437) awarded to J.H.G., a grant from the US National Institutes of Health (AI047818) to B.S.M., and an NSERC postdoctoral fellowship to S.M.K.M.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.201804351.
Contributor Information
Lauren A. M. Murray, Department of Chemistry, University of Adelaide Adelaide, SA 5005 (Australia)
Dr. Shaun M. K. McKinnie, Center for Marine Biotechnology and Biomedicine Scripps Institution of Oceanograph, University of California, San Diego, La Jolla, CA 92093 (USA) and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA 92093 (USA)
Dr. Henry P. Pepper, Department of Chemistry, University of Adelaide Adelaide, SA 5005 (Australia)
Reto Erni, Center for Marine Biotechnology and Biomedicine Scripps Institution of Oceanograph, University of California, San Diego, La Jolla, CA 92093 (USA) and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA 92093 (USA).
Dr. Zachary D. Miles, Center for Marine Biotechnology and Biomedicine Scripps Institution of Oceanograph, University of California, San Diego, La Jolla, CA 92093 (USA) and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA 92093 (USA)
Michelle C. Cruickshank, Department of Chemistry, University of Adelaide Adelaide, SA 5005 (Australia)
Dr. Borja López-Pérez, Department of Chemistry, University of Adelaide Adelaide, SA 5005 (Australia)
Prof. Bradley S. Moore, Center for Marine Biotechnology and Biomedicine Scripps Institution of Oceanograph, University of California, San Diego, La Jolla, CA 92093 (USA) and Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, La Jolla, CA 92093 (USA).
Dr. Jonathan H. George, Department of Chemistry, University of Adelaide Adelaide, SA 5005 (Australia).
References
- [1].a) Shin-ya K, Imai S, Furihata K, Hayakawa Y, Kato Y, Vanduyne GD, Clardy J, Seto H, J. Antibiot 1990, 43, 444; [DOI] [PubMed] [Google Scholar]; b) Wessels P, Göhrt A, Zeeck A, Drautz H, Zahner H, J. Antibiot 1991, 44, 1013; [DOI] [PubMed] [Google Scholar]; c) Shin-ya K, Shimazu A, Hayakawa Y, Seto H, J. Antibiot 1992, 45, 124; [DOI] [PubMed] [Google Scholar]; d) Volkmann C, Hartjen U, Zeeck A, Fiedler H-P, J. Antibiot 1995, 48, 522; [DOI] [PubMed] [Google Scholar]; e) Takagi H, Motohashi K, Miyamoto T, Shin-ya K, Furihata K, Seto H, J. Antibiot 2005, 58, 275; [DOI] [PubMed] [Google Scholar]; f) Lu C, Yang C, Xu Z, Rec. Nat. Prod 2016, 10, 430; [Google Scholar]; g) Park J-S, Kwon HC, Mar. Drugs 2018, 16, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Pathirana C, Jensen PR, Fenical W, Tetrahedron Lett. 1992, 33, 7663; [Google Scholar]; b) Hardt IH, Jensen PR, Fenical W, Tetrahedron Lett. 2000, 41, 2073. [Google Scholar]
- [3].a) Takemura S, Hirayama A, Tokunaga J, Kawamura F, Inagaki K, Hashimoto K, Nakata M, Tetrahedron Lett. 1999, 40, 7501; [Google Scholar]; b) Tatsuta K, Tanaka Y, Kojima M, Ikegami H, Chem. Lett 2002, 31, 14; [Google Scholar]; c) Snyder SA, Tang Z-Y, Gupta R, J. Am. Chem. Soc 2009, 131, 5744; [DOI] [PubMed] [Google Scholar]; d) Bernhardt P, Okino T, Winter JM, Miyanaga A, Moore BS, J. Am. Chem. Soc 2011, 133, 4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Kaysser L, Bernhardt P, Nam SJ, Loesgen S, Ruby JG, Skewes-Cox P, Jensen PR, Fenical W, Moore BS, J. Am. Chem. Soc 2012, 134, 11988; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pepper HP, George JH, Angew. Chem. Int. Ed 2013, 52, 12170; Angew. Chem 2013, 125, 12392; [DOI] [PubMed] [Google Scholar]; c) Meier R, Strych S, Trauner D, Org. Lett 2014, 16, 2634; [DOI] [PubMed] [Google Scholar]; d) Teufel R, Kaysser L, Villaume MT, Diethelm S, Carbullido MK, Baran PS, Moore BS, Angew. Chem. Int. Ed 2014, 53, 11019; Angew. Chem 2014, 126, 11199; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Diethelm S, Teufel R, Kaysser L, Moore BS, Angew. Chem. Int. Ed 2014, 53, 11023; Angew. Chem 2014, 126, 11203; [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Pepper HP, George JH, Synlett 2015, 26, 2485; [Google Scholar]; g) Yang H, Liu X, Li Q, Li L, Zhang J-R, Tang Y, Org. Biomol. Chem 2016, 14, 198; [DOI] [PubMed] [Google Scholar]; h) López-Pérez B, Pepper HP, Ma R, Fawcett BJ, Pehere AD, Wei Q, Ji Z, Polyak SW, Dai H, Song F, Abell AD, Zhang L, George JH, ChemMedChem 2017, 12, 1969. [DOI] [PubMed] [Google Scholar]
- [5].Malerich JP, Maimone TJ, Elliot GI, Trauner D, J. Am. Chem. Soc 2005, 127, 6276. [DOI] [PubMed] [Google Scholar]
- [6].a) Shin-ya K, Furihata K, Hayakawa Y, Seto H, Tetrahedron Lett. 1990, 31, 6025 [Google Scholar]; b) Kalaitzis JA, Hamano Y, Moore BS, Org. Lett 2003, 5, 4449. [DOI] [PubMed] [Google Scholar]
- [7].a) Agarwal V, Miles ZD, Winter JM, Eustaquio AS, El Gamal AA, Moore BS, Chem. Rev 2017, 117, 5619; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Moore BS, Synlett 2018, 29, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Miles ZD, Diethelm S, Pepper HP, Huang DM, George JH, Moore BS, Nat. Chem 2017, 9, 1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].A related 1,2-shift of a terpene side chain: Katsuyama Y, Li X-W, Mgller R, Nay B, ChemBioChem 2014, 15, 2349. [DOI] [PubMed] [Google Scholar]
- [10].a) Funa N, Ohnishi Y, Fujii I, Shibuya M, Ebizuka Y, Horinouchi S, Nature 1999, 400, 897; [DOI] [PubMed] [Google Scholar]; b) Austin MA, Izumikawa M, Bowman ME, Udwary DW, Ferrer JL, Moore BS, Noel JP, J. Biol. Chem 2004, 279, 45162. [DOI] [PubMed] [Google Scholar]
- [11].a) Kuzuyama T, Noel JP, Richard SB, Nature 2005, 435, 983; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) T. Kuzuyama, J. Antibiot 2017, 70, 811.28196976 [Google Scholar]
- [12].Leipoldt F, Zeyhle P, Kulik A, Kalinowski J, Heide L, Kaysser L, PLoS One 2015, 10, e0143237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Fukuda DS, Myderse JS, Baker PJ, Berry DM, Boeck LD, Yao RC, Mertz FP, Nakatsukasa WM, Mabe J, Ott J, Counter FT, Ensminger PW, Allen NE, Albourn WE Jr., Hobbs JN Jr., J. Antibiot 1990, 43, 623; [DOI] [PubMed] [Google Scholar]; b) Henkel T, Zeeck A, J. Antibiot 1991, 44, 665. [DOI] [PubMed] [Google Scholar]
- [14].a) Stabler M, Anke H, J. Antibiot 1993, 46, 968; [DOI] [PubMed] [Google Scholar]; b) Watanabe K, Sekine M, Iguchi K, J. Nat. Prod 2003, 66, 1434. [DOI] [PubMed] [Google Scholar]
- [15].Maruo S, Nishio K, Sasamori T, Tokitoh N, Kuramochi K, Tsubaki K, Org. Lett 2013, 15, 1556. [DOI] [PubMed] [Google Scholar]
- [16].Kamo S, Yoshioka K, Kuramochi K, Tsubaki K, Angew. Chem. Int. Ed 2016, 55, 10317; Angew. Chem 2016, 128, 10473. [DOI] [PubMed] [Google Scholar]
- [17].Shitakawa H, Nakajima S, Hirayama M, Kondo H, Ojiri K, Suda H (Banyu Pharmaceutical Co., Ltd., Japan: ), JP 11349522, 1999. [Google Scholar]
- [18].a) Taura F, Morimoto S, Shoyama Y, J. Am. Chem. Soc 1995, 117, 9766; [Google Scholar]; b) Shoyama Y, Tamada T, Kurihara K, Takeuchi A, Taura F, Arai S, Blaber M, Shoyama Y, Morimoto S, Kuroki R, J. Mol. Biol 2012, 423, 96. [DOI] [PubMed] [Google Scholar]
- [19].Mantovani SM, Moore BS, J. Am. Chem. Soc 2013, 135, 18032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT, Nature 2005, 436, 1191. [DOI] [PubMed] [Google Scholar]
- [21].Templeton JF, Kumar VPS, El-Sheikh AAM, Zeglam TH, Marat K, J. Chem. Soc. Perkin Trans 1 1988, 1961. [Google Scholar]
- [22].Lam HC, Spence JTJ, George JH, Angew. Chem. Int. Ed 2016, 55, 10368; Angew. Chem 2016, 128, 10524. [DOI] [PubMed] [Google Scholar]
- [23].Tapia RA, Alefria L, Valderrama JA, Cortes M, Pautet F, Fillion H, Tetrahedron Lett. 2001, 42, 887. [Google Scholar]
- [24]. See the Supporting information for the synthesis of 11a/b, according to the general procedure of Ref. [4e].
- [25].McKinnie SMK, Miles ZD, Moore BS, Methods Enzymol. 2018, 604, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Winter JM, Moore BS, J. Biol. Chem 2009, 284, 18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.