SUMMARY
At the cellular level, α-tubulin acetylation alters the structure of microtubules to render them mechanically resistant to compressive forces. How this biochemical property of microtubule acetylation relates to mechanosensation remains unknown, although prior studies have shown that microtubule acetylation influences touch perception. Here, we identify the major Drosophila α-tubulin acetylase (dTAT) and show that it plays key roles in several forms of mechanosensation. dTAT is highly expressed in the larval peripheral nervous system (PNS), but it is largely dispensable for neuronal morphogenesis. Mutation of the acetylase gene or the K40 acetylation site in α-tubulin impairs mechanical sensitivity in sensory neurons and behavioral responses to gentle touch, harsh touch, gravity, and vibration stimuli, but not noxious thermal stimulus. Finally, we show that dTAT is required for mechanically induced activation of NOMPC, a microtubule-associated transient receptor potential channel, and functions to maintain integrity of the microtubule cytoskeleton in response to mechanical stimulation.
Graphical Abstract
In Brief
Yan et al. identify the major microtubule acetylase in Drosophila and show that the enzyme and microtubule acetylation broadly control mechanosensation, but not other sensory modalities. Acetylation is required for mechanosensation by the TRP channel NOMPC, and possibly other channels, by virtue of its effects on microtubule mechanical stability and/or dynamics.
INTRODUCTION
Mechanosensation is a signal transduction process in which mechanical forces are converted into the neuronal signals that mediate hearing, balance, proprioception, and touch. In the peripheral nervous system (PNS), this conversion is mediated by ion channels that are gated by mechanical stimuli (Coste et al., 2010; O’Hagan et al., 2005; Walker et al., 2000). Mechanosensitive ion channels appear to have evolved multiple times and, as a result, several different channel families contribute to mechanosensation in animals, notably including transient receptor potential (TRP) channels, epithelial Na+ channel (ENaC)/degenerin family channels, and piezos (Katta et al., 2015). Mechanical force is thought to activate the channels by inducing conformational changes, and two distinct models have been proposed to explain how force gates these channels. In the force from lipids model, plasma membrane deformation generates the tension required for channel gating via direct interaction between the mechanoreceptor and lipids of the plasma membrane (Christensen and Corey, 2007; Kung, 2005). In the force from filaments model, the channel is tethered to a non-compliant structure by a gating spring, and movement of the membrane-bound channel relative to the immobile structure induces tension within the spring to open the channel (Howard and Hudspeth, 1987; Jin et al., 2017; Liang et al., 2013). In this model, the gating spring is an elastic tether that connects the channel to extracellular structures, such as the extracellular matrix, or intracellular components, such as the cytoskeleton (Jin et al., 2017).
Our understanding about how tethered mechanoreceptors interact with the cytoskeleton largely comes from recent studies of mechanosensitive TRP channels. Notably, mammalian TRPV1 and Drosophila TRPN/no mechanoreceptor potential C (NOMPC) directly interact with microtubules (Cheng et al., 2010; Prager-Khoutorsky et al., 2014). In mammalian osmosensory neurons, TRPV1 binds a dense network of subcortical microtubules via cytoplasmic tubulin-binding motifs (Prager-Khoutorsky et al., 2014). Under hypertonic conditions, cells shrink and their membranes press against microtubules to generate an elastic compression that opens the channel. NOMPC possesses an elongated N-terminal cytoplasmic domain with 29 tandem ankyrin repeat (AR) domains that bind to microtubules (Cheng et al., 2010; Zhang et al., 2015). NOMPC forms tetramers in which the AR domains are organized into a quadruple bundle of helical, spring-like structures (Jin et al., 2017). The AR domains and microtubule interactions are necessary for NOMPC touch-evoked responses, leading to the model that the AR helical elements function as a gating spring (Howard and Bechstedt, 2004; Jin et al., 2017; Liang et al., 2013; Zhang et al., 2015). Since direct interaction with the microtubule cytoskeleton is necessary for TRPV1 and NOMPC to function as mechanoreceptors, modulating mechanical properties of microtubules could be a control point for these channels.
Post-translational modifications of microtubules regulate their function during mechanosensation in several model systems. In C. elegans, the α-tubulin acetylase MEC-17 was identified in a screen for mutations with impaired touch sensitivity (Chalfie and Au, 1989). MEC-17 was the founding member of a family of α-tubulin acetylases with conserved homologs in all of the organisms that possess cilia (Akella et al., 2010). Depleting zebrafish MEC-17 produced a variety of developmental defects in embryos, including reduced startle responses to touch (Akella et al., 2010). In mice, mutants lacking the Atat1 homolog in sensory neurons exhibited reduced mechanosensitivity and were unresponsive in assays for touch and pain (Kalebic et al., 2013b; Kim et al., 2013). These studies broadly implicate microtubules as conserved elements in mechanosensation and highlight a key regulatory role for acetylation.
Microtubule acetylation was discovered >30 years ago (L’Hernault and Rosenbaum, 1985), yet understanding of its biological function was hindered until the recent identification of α-tubulin acetylases. α-Tubulin acetylation occurs on lysine 40 (K40) in the microtubule lumen and has generally been associated with populations of long-lived microtubules. Recent studies in which individual microtubules were mechanically stressed by repeated cycles of bending showed that they could be damaged, resulting in decreased microtubule stiffness and localized material fatigue (Portran et al., 2017). K40 acetylation enhanced microtubule flexibility and increased mechanical resilience by altering the lattice structure, allowing microtubules to comply with deformative forces without breaking (Xu et al., 2017) and suggesting that α-tubulin acetylation regulates the mechanical resilience of microtubules. If the primary role for K40 acetylation is to tune microtubule mechanical properties, how might this biochemical activity relate to its requirement during mechanosensation?
Here, we report the identification of Drosophila α-tubulin acetylase (dTAT) and our characterization of its role in mechanosensation. dTAT is broadly required for α-tubulin acetylation and is enriched in the PNS. Blocking α-tubulin acetylation broadly affected mechanosensation, but not several other sensory modalities, while causing minimal effects on dendrite morphogenesis in the PNS. Using calcium imaging, we found that mutation of dTat or non-acetylatable alleles of α-Tubulin84B (αTub84B) attenuated gentle touch responses of NOMPC-expressing class III dendrite arborization (c3da) neurons. We further found that dTAT is required for NOMPC-dependent mechanically induced membrane depolarization. However, dTAT does not regulate gentle touch responses via effects on NOMPC-microtubule interactions or NOMPC localization. Instead, we found that dTAT modulates mechanical stability and/or dynamics of microtubules to control gentle touch responses and other forms of mechanosensation. First, hyperacetylation or taxol-induced microtubule stabilization sensitize larvae to gentle touch. Second, taxol treatment rescues mechanosensory behavior defects of dTat and non-acetylatable αTub84B mutants. Third, dTat mutant sensory dendrites contain more microtubule plus ends during development and in response to mechanical stimulation than controls, reflecting an increase in mechanically induced microtubule breakage or dynamics. Thus, modulation of microtubule stability appears to be a critical control point for mechanosensation. We also observed that, as in mice, cells lacking dTAT exhibit greater cortical stiffness, although we do not know whether this activity contributes to mechanosensation.
RESULTS
dTAT Is the Major Microtubule Acetylase in Drosophila
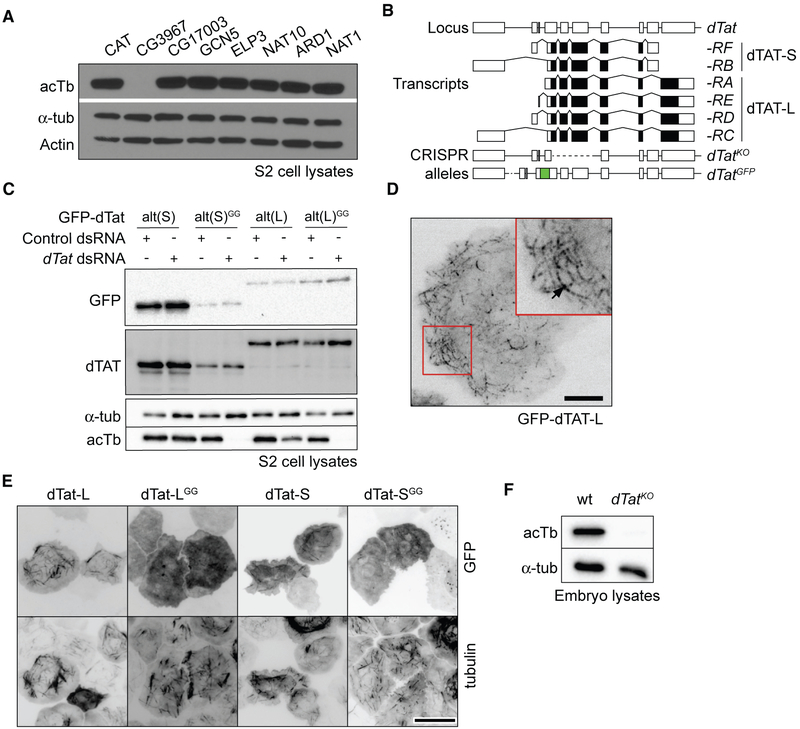
Five different acetylases are capable of modifying α-tubulin in mammalian cells or Caenorhabditis elegans, including GCN5, elongator protein 3 (ELP3), N-acetylase 10 (NAT10), the ARD1-NAT1 complex, and α-tubulin acetylase (αTAT)/MEC-17 (Akella et al., 2010; Conacci-Sorrell et al., 2010; Creppe et al., 2009; Ohkawa et al., 2008; Shida et al., 2010). We tested Drosophila homologs of these acetylases (see STAR Methods for details on candidate identification) for roles in microtubule acetylation by depleting each candidate in S2 cells with RNAi and monitoring acetylated α-tubulin (acTb) levels by immunoblot (Figure 1A). Depletion of only one candidate, CG3967, resulted in loss of acTb. We conclude that CG3967 is the major αTAT in S2 cells and hereafter refer to it as dTAT.
Figure 1. dTAT Is the Major Microtubule Acetylase in Drosophila.
(A) Western blots of lysates from S2 cells treated with dsRNA to the indicated genes (CAT [chloramphenicol acetyltransferase]; used as a negative control).
(B) Schematic of the dTat locus which encodes six documented transcripts and two polypeptides (dTAT-L and dTAT-S) with alternative 3′ coding exons. Lines depict introns, and boxes depict coding (shaded) and non-coding (empty) exons. The dTatKO allele deletes the first three coding exons shared by all dTat transcripts and dTatGFP contains GFP coding sequences fused in-frame upstream of the start codon shared by all isoforms.
(C) Western blots of lysates from S2 cells treated with dTat or CAT dsRNA and additionally expressing RNAi-resistant versions of GFP-dTAT.
(D and E) Fluorescence microscopy of S2 cells expressing GFP-dTAT fusions showing the punctate, filamentous localization of GFP-dTAT-L (D) and colocalization of GFP-dTAT with α-tubulin (E). Scale bar, 5 μm in (D), 15 μm in (E).
(F) Western blotting of wild-type and dTatKO mutant embryo lysates.
See also Figure S1.
Using the same strategy, we identified the α-tubulin deacetylase. In mammalian cells, both HDAC6 and Sirt2 deacetylate microtubules (Hubbert et al., 2002; North et al., 2003). We targeted Drosophila HDAC6 and the two Drosophila Sirt2-related genes (Sirt1 and Sirt2) with RNAi and found that acTb levels were only elevated by HDAC6 double-stranded RNA (dsRNA) (Figure S1A). We corroborated these results by immunoblotting samples from HDAC6KO, Sirt1KO, and Sirt2KO null mutants (Figure S1B). Taken together, our data indicate that Drosophila α-tubulin acetylation is regulated by the antagonistic activities of dTAT and HDAC6.
The dTat locus is predicted to generate a long (L) and a short (S) isoform encoding 461 residue (50 kDa) and 291 residue (30 kDa) proteins, respectively (Figure 1B). Both isoforms share an N-terminal 201 residue catalytic domain but diverge in their C-terminal tails, which lack predicted secondary structure. To define the roles of the two isoforms, we synthesized GFP-tagged versions of each isoform using alternate codons (dTatalt-L and -S) to make them resistant to RNAi. Both isoforms rescued microtubule acetylation in cells treated with dsRNA to deplete endogenous dTAT (Figure 1C). In contrast, catalytically inactive mutant versions of both isoforms (G133W, G135W; Topalidou et al., 2012) failed to rescue microtubule acetylation.
Prior studies have shown that αTAT interacts with microtubules in vitro (Akella et al., 2010; Kalebic et al., 2013a; Shida et al., 2010; Szyk et al., 2014). Likewise, GFP-tagged dTATalt-L and dTATalt-S exhibited robust co-localization with microtubules (Figures 1D and 1E). In contrast, catalytically inactive dTATalt-LGG and dTATalt-SGG mostly localized in the cytoplasm, suggesting that acetylase activity is required for microtubule localization.
To study dTat function in flies, we deleted the first four exons of the gene to produce a null mutant ((dTatKO) (Figure 1B). Immunoblotting confirmed that dTatKO homozygous mutants lacked detectable acTb (Figure 1F). We conclude that dTAT is the major αTAT in flies, as in S2 cells.
dTAT Is Highly Expressed in PNS Neurons
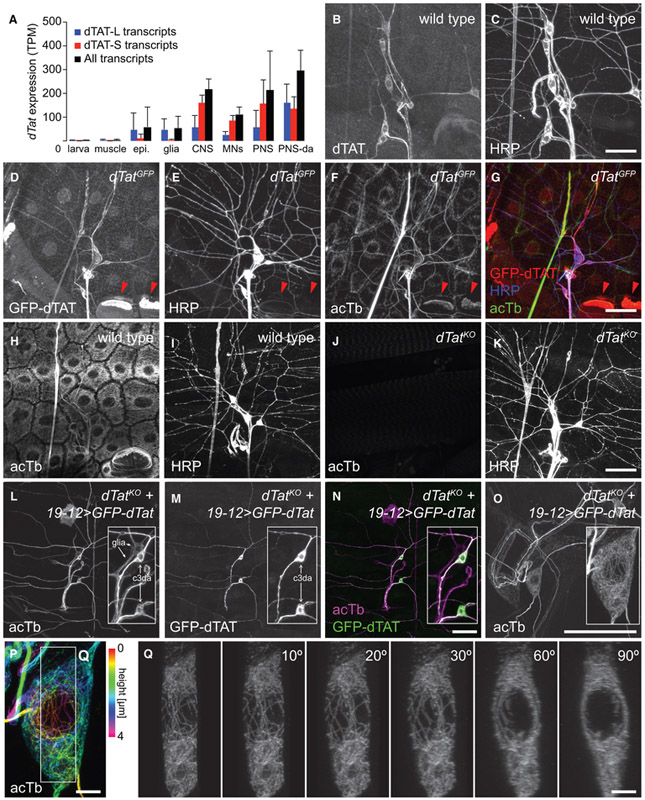
To define biological functions of dTAT, we first examined dTat expression patterns. We fluorescence-activated cell sorting (FACS) isolated different GFP-labeled cell types and subjected the cell lysates to RNA-sequencing (RNA-seq) analysis (Figure 2A). Among the larval cell types that we surveyed, including muscle, epithelia, glia, and neurons, dTat expression is the highest in neurons. PNS neurons, particularly da neurons that mediate responses to mechanical, thermal, light, and proprioceptive stimuli, highly express isoforms for both dTAT-S and dTAT-L, suggesting that the PNS is likely an important functional site for dTAT.
Figure 2. dTAT Is Enriched in the Peripheral Nervous System.
(A) RNA-seq analysis of dTat expression. Bars depict mean expression levels of dTat transcripts in the indicated cell types. TPM, transcripts per million. Error bars, SDs. N = 4+ independent samples for each condition.
(B–G) Distribution of endogenous dTAT. Maximum intensity projections of larval body walls immunostained with antibodies to dTAT (B) and horseradish peroxidase (HRP) to label sensory neurons (C).
(D–G) Maximum intensity projections of dTatGFP larval body walls stained with antibodies to GFP (D), HRP (E), and acTb (F).
(G) Overlay of dTAT-GFP, acTb, and HRP signals. We note high levels of dTAT-GFP and acTb in apodemes (red arrowheads), which are consistent with a prior characterization of acTb distribution (Jenkins et al., 2017).
(H–K) dTat is required for acTb accumulation in vivo. Maximum intensity projections are shown for wild-type third-instar larvae (H and I) stained with antibodies to acTb (H) and HRP (I) and dTatKO mutant third-instar larvae (J and K) stained with antibodies to acTb (J) and HRP (K).
(L–O) acTb distribution in sensory neurons. Maximum intensity projections are shown for dTatKO mutant third-instar larvae expressing UAS-GFP-dTat-L via 19-12-Gal4 and stained with antibodies to acTb (L) and GFP (M).
(N) Overlay of images from (L) and (M). Insets, zoomed images of cell bodies; arrows mark the cell types (c3da neurons and peripheral glia) labeled by 19-12-Gal4.
(O) Maximum intensity projection of expanded body wall tissue from dTatKO mutant third-instar larvae expressing UAS-GFP-dTat-L via 19-12-Gal4 stained with acTb antibodies.
(P) Maximum intensity projection of c3da neuron from (O), with color marking the height within the specimen.
(Q) 3D rendering of acTb staining in the boxed portion of c3da neuron from (P) viewed en face or rotated as indicated.
Scale bars, 50 μm in (B–N), 20 μm in pre-expansion dimensions (O), and 2 μm in pre-expansion dimensions (P and Q).
Next, we examined dTAT protein distribution in situ. Consistent with our RNA-seq results, anti-dTAT immunoreactivity was concentrated in neurons, particularly PNS-da neurons (Figures 2B and 2C). dTAT exhibited elevated accumulation in the soma, axon, and primary dendrites, and undetectable levels in terminal dendrites of da neurons. Likewise, when we monitored GFP-dTAT distribution in larvae homozygous for an allele (dTatGFP) that produces a GFP-dTAT fusion protein expressed from the endogenous dTat locus and supports microtubule acetylation in vivo, we found that GFP-dTAT was expressed most highly in PNS neurons (Figures 2D–2G), where it was enriched in the soma, axons, and primary dendrites but largely absent from terminal dendrites.
We next investigated patterns of acTb accumulation in wild-type and dTatKO mutant larvae. Immunostaining with a monoclonal antibody to acTb revealed a dense network of acTb immunoreactivity in all of the cells of the larval body wall, including prominent labeling of the PNS (Figures 2H and 2I). In contrast, dTatKO mutant larvae lacked detectable acTb immunoreactivity, demonstrating that dTat is required for tubulin acetylation (Figures 2J and 2K). Resupplying dTat selectively to mechanosensory c3da neurons via Gal4-mediated expression of the dTat long isoform containing an N-terminal GFP tag (UAS-GFP-dTat-L) rescued microtubule acetylation in a cell-autonomous manner, demonstrating that dTat expression in the PNS is sufficient for tubulin acetylation (Figures 2L–2N). This rescue assay allowed us to monitor acTb distribution in PNS neurons because acTb is present only in dTat-expressing cells. Similar to GFP-dTAT-L, acTb was enriched in the c3da soma, axons, and primary dendrites but largely absent from terminal dendrites. Thus, dTat is both necessary and sufficient for microtubule acetylation, and acTb distribution mirrors dTAT distribution in neurons.
In cultured mechanosensory dorsal root ganglion (DRG) neurons, acTb appears to be concentrated in a submembrane band (Morley et al., 2016). We therefore hypothesized that acTb would be similarly concentrated in mechanosensory c3da neurons. This is not what we found. Using expansion microscopy, which yields a spatial resolution of ~70 nm (Jiang et al., 2018), we observed a dense network of acTb throughout the soma that extended into axons and dendrites (Figure 2O). Although acTb immunoreactivity was restricted to a thin band around the nucleus of c3da neurons, the network of acTb coursed throughout other regions of the soma (Figures 2P and 2Q).
dTAT Is Required for Larval Mechanosensitivity
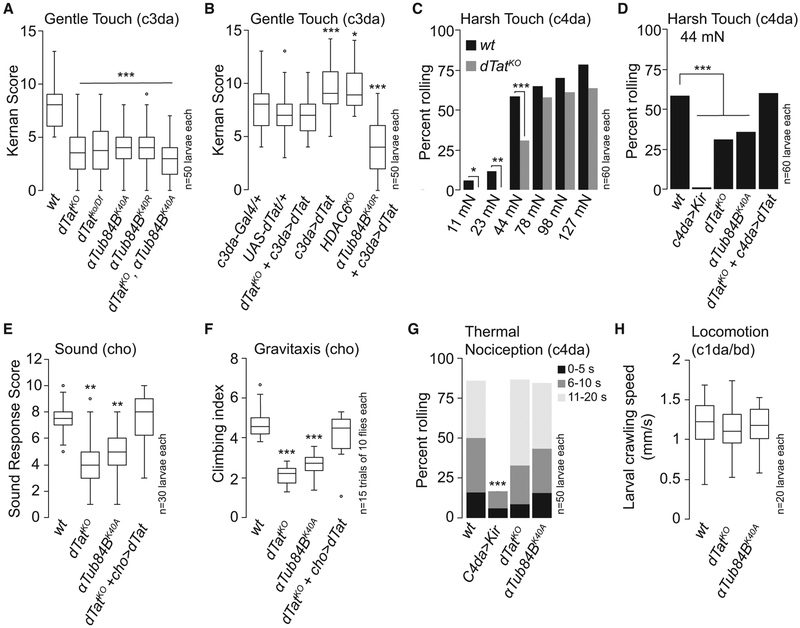
The microtubule cytoskeleton is involved in mechanosensation, particularly touch responses (Bounoutas et al., 2009; Tanner et al., 1998; Zhang et al., 2015), and mutations in mouse αTAT1 and C. elegans mec-17 and atat-2 reduce mechanosensitivity (Morley et al., 2016; Shida et al., 2010; Topalidou et al., 2012). In Drosophila, gentle touch activates the TRP channel NOMPC, which relies on microtubule interactions for gating (Zhang et al., 2015), in c3da neurons to elicit stereotyped behaviors, including backward locomotion and turning (Kernan et al., 1994). We therefore tested dTat mutants for touch sensitivity defects. dTat mutation led to a ~67% decrease in gentle touch responses, with both dTatKO homozygotes and dTatKO in combination with a deficiency spanning the dTat locus (dTatKO/Df) exhibiting similar defects (Figure 3A), suggesting that loss of dTat was the root cause of the defects. Next, we assayed gentle touch responses of mutants in which lysine 40 (K40) of the major α-tubulin isotype αTub84B, which accounts for >90% of α-tubulin expression in da neurons (Figure S2), is mutated to a non-acetylatable residue (Jenkins et al., 2017). We found that αTub84BK40A and αTub84BK40R mutants exhibited defects in gentle touch responses that were comparable to dTatKO mutants, strongly suggesting that microtubule acetylation regulates gentle touch responses (Figure 3A). Finally, (dTatKO, αTub84BK40R double mutant larvae exhibited comparable gentle touch defects to either single mutant alone, suggesting that dTat and αTub84B function in the same pathway for gentle touch responses.
Figure 3. dTat Regulates Drosophila Mechanosensation.
(A and B) dTat is necessary for gentle touch responses. Gentle touch responses are shown for dTat and non-acetylatable αTub84B mutant larvae (A) and in dTat rescue and overexpression larvae (B). Boxplots depict larval behavioral responses of the indicated genotypes to gentle touch at 96 hr after egg laying (AEL). In this and subsequent panels, boxes mark first and third quartiles, bands mark medians, whiskers mark 1.53 × interquartile range (IQR), and outliers are shown as points. *p < 0.05, ***p < 0.001 compared to wild-type (WT); Kruskal-Wallis rank sum test with Dunn’s post hoc test and Bonferroni correction for multiple comparisons.
(C and D) dTat is necessary for harsh touch responses.
(C) Bars depict the proportion of wild-type or dTat mutant larvae responding to von Frey fiber stimulation delivering the indicated amount of force. dTat mutants exhibit significant defects in response to 11, 22, and 44 mN stimulus. *p < 0.05, **p < 0.01, ***p < 0.001, compared to wild-type controls; unpaired t test with Welch’s correction.
(D) Bars depict the proportion of larvae of the indicated genotype that exhibited nociceptive rolling in response to 44 mN von Frey fiber stimulation. ***p < 0.001 compared to wild-type; chi square test.
(E and F) Boxplots depict larval vibration responses (E) and adult gravitaxis (F) in the indicated genotypes. *p < 0.05, **p < 0.01, ***p < 0.001 compared to wild-type; Kruskal-Wallis rank sum test with Dunn’s post hoc test and Bonferroni correction for multiple comparisons.
(G) Bars depict the proportion of larvae that exhibited nociceptive rolling responses to stimulus with a 39.5°C thermal probe. Responding larvae were grouped in three bins according to response latency: 0–5 s (black), 6–10 s (gray), and 11–20 s (light gray). ***p < 0.001 compared to wild-type; chi square test.
(H) Boxplots depict the rate of larval locomotion for the indicated genotypes. dTat and αTub84B mutants exhibited comparable rates of locomotion to wild-type (Kruskal-Wallis rank sum test). The number of larvae or adults tested is shown for each condition.
See also Figure S2.
To determine the site of action for dTat in control of gentle touch responses, we next performed genetic rescue assays. Expressing UAS-GFP-dTat-L in c3da neurons rescued dTat mutant gentle touch defects, demonstrating that dTat function in c3da neurons is sufficient to support gentle touch responses (Figure 3B). Overexpressing UAS-GFP-dTat-L in c3da neurons of wild-type but not αTub84BK40A mutant larvae significantly enhanced gentle touch responses, suggesting that increasing acTb levels potentiates mechanosensitivity in c3da neurons. Consistent with this notion, HDAC6KO mutant larvae also exhibited heightened gentle touch responses (Figure 3B).
We next asked whether dTat was involved in other forms of mechanosensation, including harsh touch, vibration response, and gravitaxis. Harsh touch activates c4da nociceptive neurons to elicit stereotyped nocifensive rolling responses (Zhong et al., 2010), so we stimulated larvae with von Frey filaments and monitored touch-evoked rolling responses as a measure for dTat function in harsh touch. dTatKO mutant larvae exhibited significantly reduced nocifensive responses to stimuli ranging from 11 to 44 mN but not from 78 to 127 mN (Figure 3C), revealing that harsh stimuli can bypass the requirement for acTb in mechanosensory responses. These mechanonociceptive defects likely reflect a cell-autonomous role for acetylation in c4da neurons, as dTatKO mechanonociception defects were phenocopied by non-acetylatable αTub84BK40A mutants and rescued by c4da-specific GFP-dTat-L expression (Figure 3D).
Whereas da neurons mediate responses to gentle and harsh touch, chordotonal (cho) neurons mediate vibration and gravity responses (Kamikouchi et al., 2009; Zhang et al., 2013). To determine whether dTat was required for mechanosensory responses in cho neurons, we assayed dTat mutants for defects in larval vibration responses and adult gravitaxis. Wild-type larvae exhibited a stereotyped startle response to vibration delivered in the form of a sound stimulus (70 dB, 500 Hz tone) that was compromised in dTatKO and non-acetylatable αTub84BK40A mutants (Figure 3E). As with touch responses, these defects likely reflect a neuronal requirement for dTat as expressing UAS-GFP-dTat-L in cho neurons of dTatKO mutants rescued the vibration startle defects. Finally, adult gravitaxis behavior was impaired in dTatKO and non-acetylatable αTub84BK40A mutants, and gravitaxis defects could be rescued by neuronal expression of UAS-GFP-dTat-L in dTatKO mutants (Figure 3F). These results demonstrate that dTat and microtubule acetylation are broadly required for Drosophila mechanosensation.
These mechanosensory defects could reflect general defects in sensory transduction or a more specific effect on mechanosensation. To differentiate between these possibilities, we assayed for dTat functions in other sensory modalities. First, we found that larval responses to noxious heat, which are mediated by c4da neurons (the very same neurons that mediate harsh touch responses) (Hwang et al., 2007), were unaffected in dTatKO and non-acetylatable αTub84BK40A mutants (Figure 3G). Second, application of the TRPA1 channel agonist allyl isothiocyanate (AITC), which directly activates c4da neurons to elicit nocifensive escape responses (Kaneko et al., 2017), generated comparable responses in wild-type and dTatKO larvae (56.6% + 7.1% of control and 60.1% + 6.8% of dTatKO larvae exhibit nociceptive rolling within 30 s of AITC application; n = 6 trials of 50 larvae each). Third, we found that wild-type controls, (dTatKO, and non-acetylatable αTub84BK40A mutants exhibited comparable rates of larval locomotion (Figure 3H). These results demonstrate that dTat and microtubule acetylation do not broadly regulate sensory transduction but instead preferentially affect mechanosensory responses.
dTAT Is Largely Dispensable for Dendrite Morphogenesis
How might dTAT influence mechanotransduction? Knock down of the ARD1-NAT acetylase, which reduces microtubule acetylation, and pharmacological inhibition of the microtubule deacetylase HDAC6, which increases acetylation, compromise dendrite growth in hippocampal cultures (Ageta-Ishihara et al., 2013; Ohkawa et al., 2008). Likewise, α-Tub84B K40 mutation affects dendrite branching in Drosophila c4da neurons (Jenkins et al., 2017). While each of these studies suggests a role for acetylation in dendrite development, both ARD1-NAT and HDAC6 target additional substrates, and α-tubulin K40 may have a structural role and may be subject to additional modifications, including methylation (Park et al., 2016). Thus, the roles of dTAT and α-tubulin acetylation in dendrite development are still unclear.
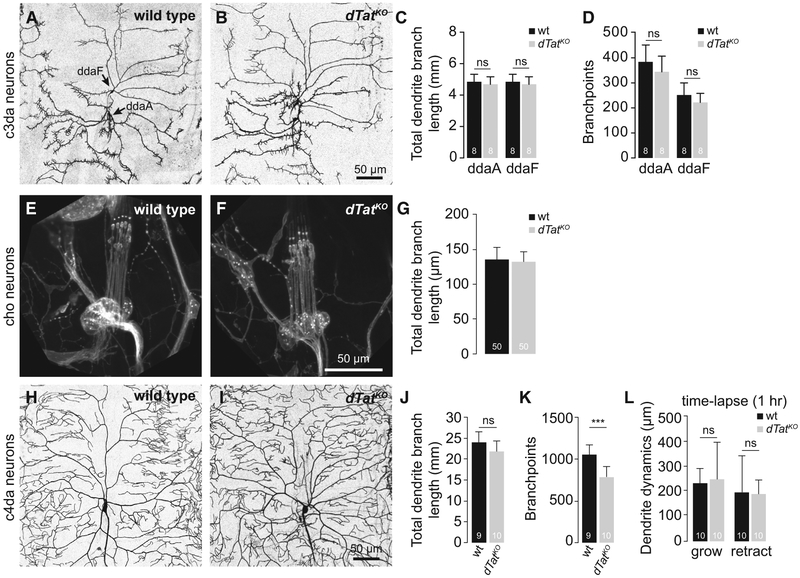
We therefore examined whether defects in mechanosensory neuron morphogenesis caused by dTat mutation may contribute to mechanosensory defects. Using nompC-Gal4 (Peterson and Stowers, 2011) to visualize c3da neurons, which mediate larval responses to gentle touch, we found that dTat mutation had no obvious effect on dendrite arborization (Figures 4A–4D). Similarly, when we used antibody staining to label cho neurons, which mediate larval response to vibration, we found that dTat mutation had no obvious effect on cho neuron morphogenesis (Figures 4E–4G). Thus, dTat is dispensable for dendrite morphogenesis in mechanosensory c3da and cho neurons.
Figure 4. dTat Is Largely Dispensable for Dendrite Morphogenesis in Mechanosensory Neurons.
(A and B) Representative images of (A) wild-type control and (B) dTatKO mutant c3da neurons labeled with nompC-Gal4, UAS-mCD4-tdGFP.
(C and D) Means and SDs are shown for (C) dendrite branch length and (D) number of branch points of the c3da neurons ddaA and ddaF.
(E and F) Representative images of cho neurons from (E) wild-type and (F) dTatKO mutant third-instar larvae labeled with anti-HRP antibodies.
(G) Bar graphs depict means and SDs for cho dendrite length of the indicated genotypes.
(H and I) Representative images of c4da neurons labeled with ppk-CD8-GFP are shown for (H) wild-type and (I) dTatKO mutant third instar larvae.
(J–L) Means and SDs are shown for (J) dendrite branch length, (K) number of branch points, and (L) dendrite dynamics measured over a 1-hr time lapse (96–97 hr AEL).
***p < 0.001; ns, not significant compared to wild-type; unpaired t test with Welch’s correction. The number of neurons analyzed for each sample is indicated.
Scale bars, 50 μm.
See also Figure S3.
Next, we monitored the effects of dTat mutation on dendrite morphogenesis in c4da neurons using ppk-CD4-tdGFP (Han et al., 2011) to selectively visualize c4da dendrite arbors. dTat mutant c4da dendrites had no overt defects in the establishment or maintenance of dendrite coverage (Figures 4H and 4I), in overall growth (Figure 4J), or in growth dynamics (Figure 4L). However, we noted a minor but consistent effect on the number and position of dendrite branch points (Figures 4K and S3). Prior studies suggested that microtubule acetylation regulates microtubule susceptibility to katanin severing (Sudo and Baas, 2010), but we found that dTat mutation had no effect on katanin-induced dendrite arbor remodeling (Figure S3). These results demonstrate that although dTat plays a minor role in c4da neuron dendrite development, it is not broadly required for mechanosensory neuron morphogenesis. Similarly, mouse Atat1 (Morley et al., 2016) and C. elegans mec-17 (Akella et al., 2010; Shida et al., 2010) are largely dispensable for sensory neuron morphogenesis, suggesting that the wide-ranging effects of dTAT on mechanosensation are likely caused by other mechanisms.
dTat Is Required for NOMPC-Dependent Mechanotransduction
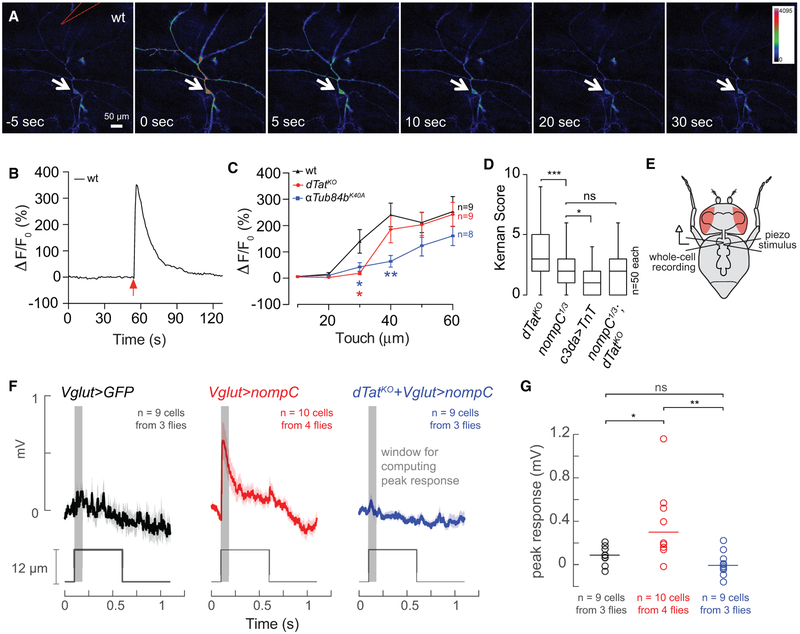
The dTat mutant mechanosensory defects could reflect defects in mechanosensation or in transmission downstream of mechanosensation. To differentiate between these possibilities, we used calcium imaging to measure mechanosensory responses of c3da neurons in wild-type and dTat mutant larvae. We immobilized semi-intact larval preparations expressing the transgenic calcium sensor GCaMP6s in c3da neurons, provided mechanical stimulus via focal body wall displacement, and monitored calcium responses in c3da neurons using a confocal microscope (Figure 5A). Consistent with previous reports (Yan et al., 2013), we found that touch stimulus induced robust calcium responses in c3da neurons (Figure 5B). In wild-type larvae, increased stimulus strength led to a progressive increase in calcium responses over a low range of stimuli (10–40 μm displacement), beyond which responses reached plateau (Figure 5C). dTatKO and αTub84BK40A mutants exhibited significant reductions in touch-induced calcium transients that were most pronounced in the low force range. For example, 30 μm displacement yielded half-maximal calcium responses in wild-type larvae and negligible responses in dTatKO and αTub84BK40A mutants. These defects were progressively attenuated with increased stimulus strength, suggesting that the mechanosensory requirement for dTat in c3da neurons can be overcome by increasing stimulus strength, similar to what we observed in c4da neurons (Figures 3C and 5C). These results further suggest that dTat functions in PNS neurons to regulate mechanosensory responses.
Figure 5. dTat Is Required for NOMPC-Mediated Mechanotransduction.
(A) Calcium responses to mechanical stimulus delivered via a polished glass electrode (position marked in red in top left image) in larvae expressing UAS-GCaMP6s in c3da neurons (nompC-Gal4).
(B) Representative trace of calcium response measured from the c3da soma.
(C) Means and SDs for calcium responses [ΔF/F0 = (Fpeak–F0)/F0] from larvae of the indicated genotypes in response to different mechanical stimuli. *p < 0.05, **p < 0.01 compared to wild-type; two-way ANOVA followed by Bonferroni’s multiple comparisons test.
(D) Larval behavioral responses to gentle touch stimulus at 96 hr AEL are shown for the indicated genotypes. Bar graphs show means and SDs for Kernan scores. *p < 0.05, ***p < 0.001; ns, not significant compared to wild-type; Kruskal-Wallis rank sum test for multiple independent samples with Dunn’s post hoc test and Bonferroni correction for multiple comparisons. Brackets indicate pairs being compared.
(E) Schematic for recording preparation used in (F and G).
(F) Average traces (±SEM) from whole-cell patch-clamp recordings for wild-type MNs expressing UAS-GFP (Vglut > GFP, left), wild-type MNs expressing UAS-GFP-nompC (Vglut > nompC, middle), and MNs from a dTatKO mutant larva expressing UAS-GFP-nompC (dTatKO + Vglut > nompC, right). Red lines depict mechanical stimulus.
(G) Scatterplot depicting mean (line) and individual measurements (points) of maximum depolarization minus baseline resting potential for the indicated genotypes. *p < 0.05, **p < 0.01; ns, not significant compared to wild-type; one-way ANOVA followed by Bonferroni’s multiple comparisons test.
The number of independent samples measured for each genotype is shown in each panel.
See also Table S1.
Given that larval gentle touch responses require the NOMPC channel, whose function depends on microtubule interactions, we next conducted genetic epistasis analysis to determine whether dTAT and NOMPC function in the same pathway. While mutation in either dTat or nompC impaired larval gentle touch responses, nompC mutation had more severe effects (Figure 5D). Expressing tetanus toxin in c3da neurons to block their synaptic output resulted in an even stronger defect than nompC mutation (Figure 5D), suggesting that nompC-independent pathways contribute to gentle touch responses (Tsubouchi et al., 2012). If dTat and nompC function in independent pathways, we reasoned that nompC; dTat double mutants should have more severe defects than either single mutant alone. Instead, gentle touch defects of the double mutant and nompC mutants were indistinguishable (Figure 5D), suggesting that dTat and nompC function in the same genetic pathway for gentle touch.
We next tested whether dTat was required for NOMPC-mediated mechanotransduction. For these experiments, we first tested whether NOMPC expression could confer mechanosensitivity to neurons that were normally unresponsive to mechanical stimuli. Whereas many PNS neurons, including c1da, c3da, c4da, and cho neurons, express mechanosensitive ion channels, we found that motor neurons (MNs) do not, although MNs do exhibit high levels of dTat expression (Table S1; Figure 2A). Consistent with these expression data, when we conducted in vivo whole-cell recordings from leg MNs in the adult ventral nerve cord (Figure 5E), we found that mechanical neuropil stimulation did not produce a consistent response (Figures 5F and 5G). By contrast, when we expressed UAS-nompC-GFP in leg MNs, mechanical neuropil stimulation reliably evoked depolarizing responses. This mechanically evoked depolarization was abrogated by dTat mutation. Thus, NOMPC expression is sufficient to confer mechanosensitivity to MNs, and dTat is required for NOMPC-dependent mechanotransduction.
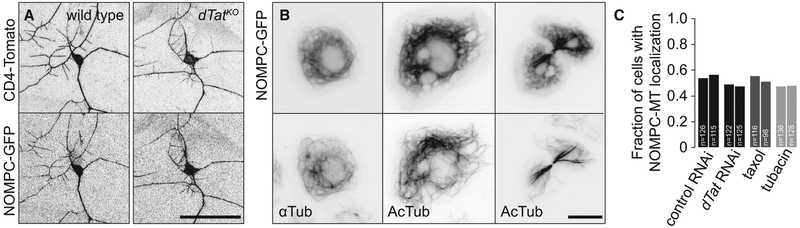
We next tested hypotheses for how dTat regulates NOMPC function. First, microtubule acetylation plays established roles in intracellular transport (Bhuwania et al., 2014; Reed et al., 2006); thus, we tested roles for dTat in NOMPC localization. We found that dTat mutant c3da neurons exhibit normal NOMPC-GFP distribution when NOMPC-GFP is ectopically expressed (Figure 6A), suggesting that dTat regulates NOMPC function instead of localization. Second, NOMPC-microtubule interactions are critical to NOMPC function (Zhang et al., 2015); thus, we investigated dTat effects on NOMPC-microtubule interactions in S2 cells. When expressed in S2 cells, NOMPC-GFP co-localizes with microtubules and is sufficient to confer mechanosensitivity to this normally non-responsive cell line (Yan et al., 2013). Although we found that NOMPC-GFP co-localized extensively with acTb (Figure 6B), neither eliminating microtubule acetylation with dTat RNAi (Figure 6C) nor treating S2 cells with the HDAC6 inhibitor tubacin or taxol to increase acTb levels significantly reduced NOMPC-GFP co-localization with microtubules, indicating that acTb is not a critical determinant of NOMPC-microtubule interactions.
Figure 6. dTat Is Dispensable for NOMPC-Microtubule Interactions.
(A) Maximum intensity projections showing GFP-NOMPC distribution in c3da neurons wild-type control (left) and dTatKO mutant (right) larvae additionally expressing the membrane marker CD4-tdTomato.
(B) S2 cells stably transfected with UAS-GFP-nompC were immunostained with antibodies to GFP and acTb or α-tubulin, as indicated. Images show cells in interphase (left, middle) and during anaphase (right).
(C) S2 cells stably transfected with UAS-GFP-nompC were treated with control RNAi, dTat RNAi, taxol, or tubacin; immunostained using GFP and tubulin antibodies; and the fraction of cells exhibiting NOMPC-microtubule co-localization was visually scored in a blind experiment. Chi-square tests revealed no differences in NOMPC-microtubule co-localization among the different treatments. The number of cells analyzed is shown for each treatment.
dTAT Regulates Microtubule Breakage and/or Dynamics to Promote NOMPC-Dependent Mechanotransduction
Recent reports have established a role for acTb in regulating microtubule dynamics and in conferring microtubules with resistance to mechanical breakage (Portran et al., 2017; Xu et al., 2017). Because NOMPC interacts with microtubules via its AR domain and this interaction is required for mechanosensation (Cheng et al., 2010; Zhang et al., 2015), we speculated that loss of dTat would perturb the mechanical properties of microtubules and/or microtubule dynamics to inhibit mechanosensation. This hypothesis was supported by three independent lines of experimentation.
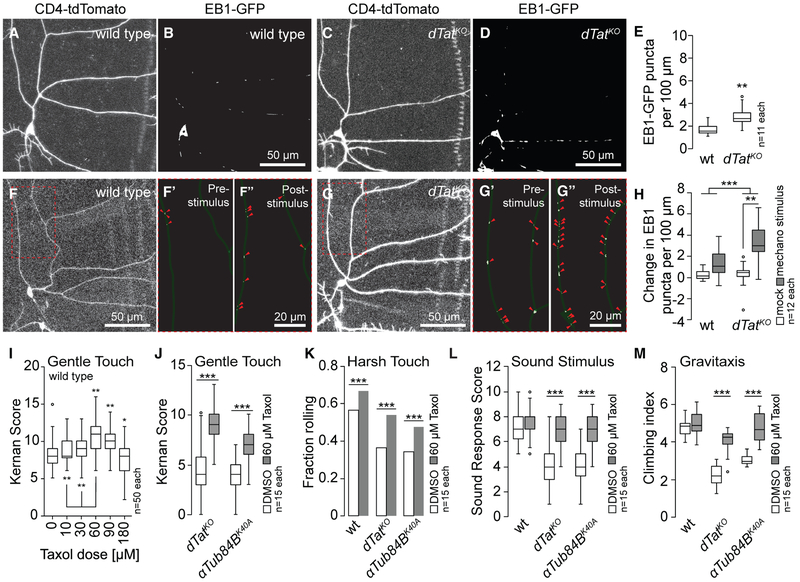
First, we examined microtubule plus end numbers and dynamics by expressing a GFP-tagged version of EB1 (UAS-EB1-GFP) in c3da neurons additionally expressing a membrane-targeted red fluorescent protein (UAS-CD4-tdTomato). In third instar larvae, dTat mutant c3da neurons exhibited a ~70% increase in EB1-GFP+ puncta (control, 1.74 ± 0.51; dTat mutant, 2.99 ± 0.90) (Figures 7A–7E and S4). This increase in the number of growing plus ends could be caused by an increased rate of new microtubule nucleation events, destabilization of existing microtubules leading to increased rescue events, and periods of polymerization, or by increased frequency of microtubule breakage leading to the production of new plus ends (Goodwin and Vale, 2010). Next, we examined whether mechanical stimulus could alter microtubule plus end numbers by visualizing EB1-GFP puncta immediately before and after mechanical stimulus. We found that mechanical stimulus induced a significant increase in EB1-positive puncta in dTat mutant but not wild-type control c3da neurons (Figures 7F–7H). These results indicate that microtubule mechanical stability and/or dynamics are altered in dTat mutants compared to wild-type controls and that mechanical stimulus can trigger changes in the number of growing plus ends in the absence of microtubule acetylation.
Figure 7. dTat Promotion of Microtubule Stability Modulates Mechanotransduction.
(A–D) Representative images of c3da neurons from (A and B) wild-type (CD4-tdTomato, A; EB1-GFP, B) and (C and D) dTatKO mutant larvae expressing CD4-tdTomato to label membranes (C) and EB1-GFP to label microtubule plus ends (D).
(E) Boxplot depicts the number of EB1+ puncta per 100 mm of dendrite length. **p < 0.01 compared to wild-type; unpaired t test with Welch’s correction.
(F–H) Microtubules of dTatKO mutants are prone to mechanically -induced breakage. Images of representative (F) wild-type and (G) dTat mutant c3da neurons that were subjected to mechanical stimulus are shown. EB1-GFP labeling is shown immediately before (F’ and G’) and after stimulus (F’’ and G’’). Arrowheads mark EB1 puncta.
(H) Boxplot depicting quantification of EB1+ puncta. Two-way ANOVA analysis revealed a significant interaction effect between mechanical treatment and genotype on EB1 puncta number. F(2,12) = 7.539, p = 0.009. Simple main effects analysis showed a significant difference in EB1 puncta number between treated and untreated dTat mutant larvae (p = 0.001), but not between treated and untreated wild-type larvae (p = 0.184). **p < 0.01, ***p < 0.001. Raw images are shown in Figure S4.
(I–M) Taxol-induced microtubule stabilization rescues mechanosensory defects of acetylation mutants.
(I) Boxplot depicting gentle touch response of larvae fed the indicated dose of taxol for 3 hr. *p < 0.05, **p < 0.01, compared to vehicle-fed controls; one-way ANOVA with a post hoc Dunnett’s test.
(J) Boxplot depicting gentle touch response of dTat and αTub84b mutant larvae fed vehicle or 60 μM taxol for 3 hr.
(K) Bars depict the proportion of larvae of the indicated genotype that exhibited a nociceptive rolling responded to 44 mN von Frey stimulus.
(L and M) Boxplots depicting sound stimulus responses (L) and gravitaxis behavior (M) of wild-type, dTat, and μTub84B mutant flies fed vehicle or 60 μM taxol for 12 hr.
***p < 0.001 compared to vehicle controls; unpaired t test with Welch’s correction for (J–M). The number of independent samples measured for each genotype is shown in each panel.
See also Figures S4, S5, S6, and S7, and Table S2.
Second, we examined the localization of the MAP1B-like protein, futsch. Futsch is required during Drosophila development for microtubule organization during axonal growth and synaptogenesis (Hummel et al., 2000; Roos et al., 2000). As a microtubule-associated protein, futsch has been used as a marker for microtubule polymer and an indicator of microtubule stability (Jenkins et al., 2017; Ruiz-Canada et al., 2004). Consistent with a role for acTb in regulating microtubule dynamics or stability, we observed a significant reduction in futsch immunoreactivity in c3da neurons of dTatKO or αTub84B mutants compared to wild-type controls (Figure S5); non-acetylatable αTub84B mutants similarly reduce futsch immunoreactivity in other da neurons (Jenkins et al., 2017). These results confirm that loss of acTb alters composition of the microtubule cytoskeleton in c3da neurons and potentially its stability.
Third, we examined whether taxol-mediated microtubule stabilization could potentiate mechanosensory responses. We found that acute taxol feeding led to dose-dependent increases in gentle-touch sensitivity in third-instar larvae over a range of 0–60 μM taxol (Figure 7I) with no obvious effects on neuron morphology (c3da neuron dendrite length, mean ± SD: 4.84 ± 0.76 mm, DMSO fed; 4.69 ± 0.88 mm, 60 μM taxol fed; n = 8 neurons each). Mutation of nompC rendered taxol-fed larvae insensitive to gentle touch as did c3da neuron-specific expression of tetanus toxin (Figure S6), indicating that the increased responses to gentle touch require nompC and reflect enhanced activity of c3da neurons. To test whether mechanosensory defects of dTatKO and non-acetylatable αTub84BK40A mutants reflect a decrease in microtubule stability, we treated larvae with vehicle or 60 μM taxol and measured behavioral responses to gentle touch (Figure 7J). Remarkably, taxol feeding significantly enhanced gentle touch responses of dTatKO and <xTub84BK40A mutants. Likewise, taxol feeding potentiated responses of wild-type larvae to harsh touch and significantly enhanced harsh touch responses of dTatKO and αTub84BK40A mutants (Figure 7K). Finally, although taxol feeding did not enhance vibration responses or gravitaxis in wild-type controls, it did restore these behaviors to wild-type levels in dTatKO and αTub84BK40A mutants (Figures 7L and7M).
dTAT Regulates the Rigidity of Cultured S2 Cells
Our studies thus far support a model in which microtubule acetylation by dTat broadly regulates mechanosensation via effects on microtubule stability. We next examined whether dTAT regulates cellular rigidity, which could contribute to dTat mutant mechanosensation defects. Loss of the microtubule acetylase α-tubulin N-acetyltransferase 1 (ATAT1) results in increased cellular rigidity of cultured mouse DRG neurons (Morley et al., 2016). To test whether this biophysical function was conserved in Drosophila, we used atomic force microscopy (AFM) to examine the elastic moduli of dTAT-depleted S2 cells and compared them to control-treated cells. We found that dTat RNAi-treated cells exhibited a statistically significant increase in cortical stiffness, with dTat RNAi cells exhibiting a 22% increase in stiffness (Figure S7; Table S2). These results indicate that the role of dTAT/ATAT1 in regulating the mechanical properties of cells is conserved; whether this activity contributes to its role in mechanosensation remains to be determined.
DISCUSSION
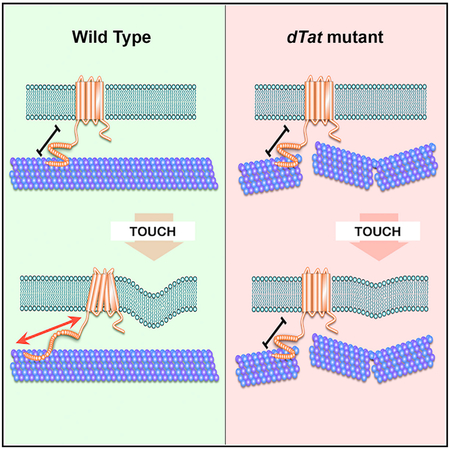
Microtubule acetylation is required for touch sensation in several model systems (Akella et al., 2010; Morley et al., 2016); however, the molecular mechanisms that underlie its role in mechanosensation are poorly understood. In this study, we identified the major αTAT in Drosophila, dTAT, and found that it is broadly required for mechanosensation. In response to gentle touch, dTat functions in the same pathway as the TRP channel nompC and is required for NOMPC-dependent touch-evoked neuronal responses. dTat mutation causes an increase in dendritic microtubule plus ends in mechanosensory neurons, and taxol stabilization of the microtubule cytoskeleton rescues mechanoreceptivity, suggesting that microtubule breakage and/or alterations in microtubule dynamics underlie the loss of touch sensitivity. We also observed that dTAT depletion in S2 cells altered their mechanical properties to make them more rigid, an effect that could alter NOMPC activation in neurons by increasing its threshold for activation. Collectively, our results suggest that K40 acetylation functions to stabilize the microtubule cytoskeleton and may tune cellular mechanics to promote NOMPC activation through its cytoplasmic microtubule-associated tension gate. Our results demonstrating defects in vibration perception and gravitaxis in dTat mutants reveal that microtubule acetylation plays a broader role in mechanosensation than was previously recognized. We speculate that microtubule acetylation likewise promotes the activation of other mechanosensory channels by facilitating microtubule-mediated mechanotransduction.
The results of our genetic, electrophysiological, and cell biological experiments indicate that dTat is required for mechanosensation by the TRP channel NOMPC. This requirement is not related to NOMPC trafficking and unlikely due to interactions with microtubules. Instead, we observed an increase in EB1-labeled microtubule plus ends in dTat mutant sensory dendrites and showed that stabilizing the cytoskeleton by feeding mutant animals taxol rescued touch sensitivity. In light of recent findings that K40 acetylation weakened inter-protofilament interactions, allowing microtubules to comply with deformative forces without breaking (Portran et al., 2017; Xu et al., 2017), our results suggest that in the absence of K40 acetylation, microtubules in c3da neurons are mechanically damaged, thereby decreasing NOMPC-microtubule interactions and attenuating the channel’s ability to transduce mechanical stimuli. We cannot exclude the possibility that regulation of microtubule dynamics is a major functional role for acetylation in PNS neurons; further investigation into the influence of microtubule structure and dynamics on NOMPC-microtubule interactions should help resolve whether acetylation controls mechanosensation primarily via the regulation of microtubule mechanical resilience or dynamics.
Loss of dTat does not phenocopy nompC null mutants in all respects. NOMPC functions in proprioceptive c1da neurons to control larval locomotion (Cheng et al., 2010), but dTat mutant larvae exhibit grossly normal locomotion. Although the absence of obvious defects in our assay does not preclude the possibility that dTat exerts a subtle influence on larval locomotion, the effects of dTat mutation are significantly less pronounced than the ~50% decrease in larval crawling speed observed in nompC mutants (Cheng et al., 2010). We speculate that this may be due to neuron class-specific susceptibility to the loss of acTb. For example, the microtubule network in c1da neurons may be more resistant to loss of dTat than in c4da neurons. We also discovered that adult gravitaxis is perturbed in dTat mutants. While NOMPC is dispensable for gravitaxis, TRPA and TRPV family channels are required for gravity sensing (Kamikouchi et al., 2009; Sun et al., 2009). Likewise, dTat mutants have defects in harsh touch responses, which involve TRP, piezo, and Ppk/ENaC channels but not NOMPC, as well as defects in larval hearing, which involve several TRP channels in addition to NOMPC. These results raise the possibility that interactions between acTb and other channels may play important roles in mechanosensation.
Prior studies of microtubule acetylation in touch perception have arrived at molecular mechanisms that differ from our model but are not mutually exclusive. In C. elegans touch receptor neurons, sensory dendrites are packed with a cross-linked bundle of long, specialized 15-protofilament microtubules specific to this cell type (Chalfie and Sulston, 1981). Mutation of the αTATs mec-17 and atat-2 resulted in the loss of these unique microtubules and insensitivity to touch (Bounoutas et al., 2009). Thus, K40 acetylation is required for the assembly of a specialized population of microtubules involved in C. elegans mechanotransduction. Although there is no evidence for specialized microtubules in Drosophila da neuron dendrites, the results from worm touch receptors mirror our results in that microtubule acetylation is required to maintain an intact microtubule network. Further electron microscopy (EM) study of the cytoskeleton in da neurons will be necessary to definitively determine whether they possess specialized microtubule arrays.
In mouse peripheral sensory neurons, acetylated microtubules are enriched in a submembranous band in the soma that is distinct from the cytoplasmic microtubule network (Morley et al., 2016). This submembranous band appears to confer rigidity to the plasma membrane, and cultured DRG neurons from Atat1 knockout mice exhibited increased membrane stiffness (Morley et al., 2016). The authors proposed that in this system, microtubule acetylation tunes the mechanical properties of the membrane and, in its absence, cells are less elastic and require more force to trigger mechanosensitive channels. Our results in S2 cells indicate that regulation of cellular stiffness is a conserved function for microtubule acetylases. However, the molecular mechanism linking microtubule acetylation to cellular stiffness and the extent to which this alteration of cellular stiffness contributes to mechanosensation are unknown. To address these important outstanding issues, it will be necessary to identify the molecular components that link microtubules to the mechanical properties of the cell cortex and examine their contribution to mechanosensation independently of microtubule function. Although we have found no evidence for a specialized submembranous network of acetylated microtubules in Drosophila PNS neurons, it will be interesting to determine whether dTat regulates cellular elasticity in these cells as well. It is therefore possible that microtubule acetylation regulates mechanosensitivity through multiple mechanisms: by regulating microtubule structure and by tuning the mechanical properties of neurons.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jay Parrish (jzp2@uw.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly husbandry
Flies were maintained on standard cornmeal-molasses-agar media and reared at 25°C under 12 h alternating light-dark cycles.
Fly stocks
Alleles used in this study are listed in the Key Resources Table below and a complete list of experimental genotypes is available in Table S3.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Tubulin (clone DM1A) | Sigma-Aldrich | Cat# T9026; RRID: AB_477593 |
| Mouse monoclonal anti-Tubulin (clone 6-11B-1) | Sigma-Aldrich | Cat#T7451, RRID: AB_609894 |
| Mouse monoclonal anti-Tubulin (clone DM1A); FITC conjugate | Sigma-Aldrich | Cat# F2168, RRID: AB_476967 |
| Mouse monoclonal anti-Myc (clone 9E10) | Developmental Studies Hybridoma Bank (DSHB) | Cat# 9E 10, RRID: AB_2266850 |
| Mouse monoclonal anti-Futsch (clone 22C10) | DSHB | Cat# 22c10, RRID: AB_528403 |
| Rabbit polyclonal anti-GFP antibody | Fisher | Cat# A-11122, RRID: AB_22159 |
| Rabbit polyclonal anti-dsRed antibody | Clontech | Cat# 632496, RRID: AB_10013483 |
| Rabbit polyclonal anti-dTat antibody | This study | N/A |
|
Chemicals, Peptides, and Recombinant Proteins | ||
| Paclitaxel | Sigma-Aldrich | T7402 |
| Tubacin | Sigma-Aldrich | SML0065 |
|
Deposited Data | ||
| D. melanogaster larval PNS RNA-Seq data | Williams et al., 2016 | NCBI-SRA GSE72884 |
| D. melanogaster whole larva RNA-Seq data | Boiko et al., 2017 | NCBI-SRA GSE99711 |
| D. melanogaster larval cell type RNA-Seq data | This study | NCBI-SRA GSE120305 |
|
Experimental Models: Cell Lines | ||
| D. melanogaster: Cell line S2 | Derosophila Genomics Resource Center | S2-DGRC |
|
Experimental Models: Organisms/Strains | ||
| w[1118] ; dTat[KO] | This study | NA |
| w[1118] ; dTat[GFP] | This study | NA |
| w[1118] ; UAS-GFP-dTat-L | This study | NA |
| y[1] M{vas-Cas9.RFP-}ZH-2A w[1118]/FM7a, P{w[+mC] = Tb[1]}FM7-A | Bloomington Drosophila Stock Center (BDSC) | 55821 |
| y[1] w[67c23] P{y[+mDint2] = Crey}1b; sna[Sco]/CyO; Dr[1]/TM3, Sb[1] | BDSC | 34516 |
| w[1118] | BDSC | 6326 |
| w[1118] ; ppk-Gal4 | BDSC | 32079 |
| y[1] w[*] HDAC6[KO] | BDSC | 51182 |
| nompC[1] cn[1] bw[1]/CyO | BDSC | 42268 |
| nompC[3] cn[1] bw[1]/CyO | BDSC | 42258 |
| w[1118]; P{w[+mC] = AyGAL4}17b | BDSC | 4413 |
| y[1] w[*]; PBac{y[+mDint2] w[+mC] = nompC-GAL4.P}VK00014; Df(3L)Ly, sens[Ly-1]/TM6C, Sb[1] Tb[1] | BDSC | 36361 |
| w[*]; P{w[+mC] = UAS-TeTxLC.tnt}R3 | BDSC | 28997 |
| w*; P{UAS-Hsap\KCNJ2.EGFP}7 | BDSC | 6595 |
| w[*]; M{w[+mC] = UAS-Kat60.M}ZH-51D/CyO | BDSC | 64117 |
| w[1118]; P{w[+mC] = UAS-Eb1.EGFP.H}G | BDSC | 36861 |
| y[1] w[*]; P{w[+mC] = UAS-CD4-tdTom}7M1 | BDSC | 35841 |
| w[1118]; P{w[+mC] = ppk-CD4-tdTom}10a/TM6B, Tb[1] | BDSC | 35845 |
| w[1118]; Df(3L)BSC113/TM6B, Tb[1] | BDSC | 8970 |
| w[1118], ppk-mCD8-GFP | Jiang et al., 2014 | NA |
| w[1118] ; αTub84B[K40A] | Jenkins et al., 2017 | NA |
| w[1118]; aTub84B[K40R] | Jenkins et al., 2017 | NA |
| w[1118] ; αTub84B[Δ] | Jenkins et al., 2017 | NA |
| w[1118]; P{w[+mW.hs] = GawB}21-7 | Song et al., 2007 | NA |
| w[1118]; P{w[+mW.hs] = GawB}19-12 | Xiang et al., 2010 | NA |
|
Oligonucleotides | ||
| Oligonucleotide sequences used in this study are listed in Table S4. | This study | NA |
|
Recombinant DNA | ||
| pBID-UAS-GFP-dTat-L | This study | NA |
| pET28a dTat (1-196) | This study | NA |
| pMT B V5/His dTat(alt)-S | This study | NA |
| pMT B V5/His dTat(alt)-S GG (G133Y, G135Y) | This study | NA |
| pMT B V5/His dTat(alt)-S | This study | NA |
| pMT B V5/His dTat(alt)-L GG (G133Y, G135Y) | This study | NA |
Cell Culture
Culture and RNAi of Drosophila S2 cells were performed as previously described (Rogers and Rogers, 2008). S2 cells (Drosophila Genomics Resource Center, Bloomington, IN) were cultured in SF900II medium supplemented with 100x antibiotic-antimycotic (Invitrogen, Carlsbad, CA).
METHOD DETAILS
Generation of dTat alleles
The dTatKO and dTatGFP alleles were engineered using CRISPR/Cas9 mediated gene editing (Gratz et al., 2014). Target sites were selected upstream of the first coding exon and in the second intron of dTat using the CRISPR Optimal Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/). chiRNA plasmids were generated by annealing sense and antisense gRNA oligos, digesting with BbsI, and ligating into the pU6-BbsI-chiRNA expression vector. Donor vectors were generated by cloning homology arms into pHD-DsRed. For (dTatKO, homology arms were designed to delete an 869 base pair fragment spanning the first three coding exons, beginning 16 base pairs upstream of the start codon. The donor vector for dTatGFP was generated as follows: pHD-dsRed was digested with EcoRI and the 5′ homology arm corresponding to sequences immediately upstreat of the dTat start codon, GFPand dTat PCR fragments were cloned into the backbone by Gibson assembly (NEB Gibson Assembly Kit) with GFP fused in-frame with the N terminus of dTat and the LoxP-flanked 3×P3-dsRed marker within the second intron. The newly assembled plasmid was then digested with XhoI and the 3′ homology arm was inserted using Gibson assembly. All primer sequences are available in Table S4.
chiRNA and pHD-DsRed plasmids were co-injected into embryos expressing Cas9 in the germline (BL55821: y[1] M{vas-Cas9.RFP-}ZH-2A w[1118]/FM7a, P{w[+mC] = Tb[1]}FM7-A), stocks were established from RFP-positive founder males, and 3×P3-RFP markers were removed using Cre-mediated reduction (BL34516: y[1] w[67c23] P{y[+mDint2] = Crey}1b; sna[Sco]/CyO; Dr[1]/TM3, Sb[1]) as previously described (Gratz et al., 2014).
RNA-Seq of larval cells
Four to seven samples of one hundred cells each were isolated and RNA-Seq libraries were prepared as described previously (Boika et al., 2017). Briefly, third instar larvae expressing UAS-Red Stinger in the target cell type (epithelia, a58-Gal4; muscles, mef-Gal4; peripheral neurons, elav-Gal4; central neurons, elav-Gal4; motor neurons, ok371-Gal4; central glia, repo-Gal4) were dissected to isolate the tissue of interest. Bodywalls or brains were dissociated and Red-Stinger-labeled cells were isolated by flow cytometry into RNAqueous lysis buffer. Samples were sequenced as 51 base single end reads on a HiSeq 2500 running in high-output mode at the UCSF Center for Advanced Technology, with read depths ranging from 1.5 to 18.4 million reads. Reads were demultiplexed with CASAVA (Illumina) and read quality was assessed with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the D. melanogaster transcriptome, FlyBase genome release 6.10, using STAR version 2.5.2b (Dobin et al., 2013) with the option ‘–quantMode TranscriptomeSAM’. Transcript expression was modeled from these STAR alignments using Salmon (Patro et al., 2017) in alignment-based mode. The raw sequencing reads and gene expression estimates are available in the NCBI Sequence Read Archive (SRA) and in the Gene Expression Omnibus (GEO) under accession number GSE120305. The da neuron and whole larvae datasets were previously described and published and are available from SRA and GEO under accession numbers GSE72884 and GSE99711, respectively.
Plasmid Constructs
Expression constructs for dTAT were generated using PCR from a synthetic template (dTATalt) in which the wobble position of each codon was conservatively replaced, making them refractory to RNAi (Blue Heron Biotech). The GFP-dTATalt fusions were cloned by 5′ overlapping extension PCR using primers listed in Table S3. GFP-dTATalt inserts were cloned into the KpnI/ApaI sites of pMT B V5/His (Invitrogen) for copper-inducible expression in S2 cells or into the EcoRI/NotI sites in pBID-UASC (Addgene) for generating transgenic fly lines. The catalytically inactive version of dTAT was generated by mutating two glycine residues (G133 and G135) to tryptophan to disrupt acetyl CoA binding (Topalidou et al., 2012).
Identification of candidate Drosophila acetylases
Drosophila homologs of GCN5, ELP3, ARD1, and NAT1 have been studied in other contexts, but their roles in microtubule acetylation have not been characterized. We additionally identified the GNAT domain-containing lethal(1)G0020/CG1994 as the likely NAT10 homolog (53.8% identity) and CG3967 and CG17003 as potential aTAT/MEC-17 homologs (CG3967, 38.8% identity; CG17003, 34.7% identity)
S2 cell culture, RNAi, and immunofluorescence
Culture and RNAi of Drosophila S2 cells were performed as previously described (Rogers and Rogers, 2008). S2 cells (Drosophila Genomics Resource Center, Bloomington, IN) were cultured in SF900II medium supplemented with 100x antibiotic-antimycotic (Invitrogen, Carlsbad, CA). DNA templates for dsRNA synthesis were obtained by PCR amplification of the pFastBacHT-CAT expression plasmid (Invitrogen), BDGP cDNA clones, or S2 cell genomic DNA using the gene-specific primer sequences (Table S4). As a negative control, a sequence from chloramphenicol acetyltransferase (CAT) was amplified and transcribed into dsRNA. In vitro transcription reactions were performed with T7 RNA polymerase purified in house. Cells were transfected using Fugene HD (Promega) according to the manufacturer’s instructions. Stable cell lines were selected by supplementing culture medium with 10 μg/mL blasticidin or 500 ug/mL hygromycin (Invitrogen). Immunofluorescence was performed by plating cells into fabricated 35 mm glass bottom culture dishes pre-coated with concanavalin A in serum-free Schneider’s medium (Sigma). After cells had attached and spread for 1 hour, they were fixed with 10% formaldehyde (EM Sciences) in HL3 buffer (70 mM sodium chloride; 5 mM potassium chloride; 20 mM magnesium chloride hexahydrate; 10 mM sodium bicarbonate; 5 mM trehalose; 115 mM sucrose; 5 mM HEPES; pH 7.2) for 10 minutes. Cells were permeabilized and blocked with 5% bovine serum albumin in TBST (TBS + 0.1% Triton X-100) before staining with primary and secondary antibodies. Cells were imaged on an Eclipse Ti-E microscope with a 100x oil NA-1.45 objective, driven by NIS Elements software. Images were captured with a cooled charge-coupled device camera (CoolSNAP HQ, Roper Scientific).
Antibodies
The following antibodies were used in this study: anti-acTb (6-B11-1, Sigma), anti-α-tubulin (DM1α, Sigma), FITC-labeled DM1α monoclonal antibodies (Sigma), anti-GFP (A-11122, Fisher), anti-DsRed (632496, Clontech), Anti-Myc (9E11, DSHB), Anti-Futsch (22C10, DSHB), Cy5-conjugated anti-HRP (Jackson immunoresearch), and Alexa Fluor conjugated secondary antibodies (Fisher). In order to generate antibodies against dTAT, we used PCR to amplify the catalytic domain (residues 1 to 196) and cloned this fragment into the NheI/XbaI sites of pET28a or the BamHI/NotI sites of pGEX6P2. Recombinant dTAT 1-196 was expressed in E. coli and purifiied on NiNTA resin (QIAGEN) and glutathione-Sepharose, respectively. Purified 6xhis-dTAT1-196 protein was used to generate polyclonal antibodies in rabbits (Pocono Rabbit Farm) and the antibodies were further affinity-purified on GST-dTAT 1-196 bound to amino-link resin (Thermo Fisher Scientific). Secondary antibodies for immunofluorescence were purchased from Jackson Immunoresearch. HRP-conjugated secondary antibodies for immunoblots were purchased from Sigma.
Imaging of larval samples
Live imaging
Larvae were mounted in 90% glycerol under No. 1 coverslips and imaged using a Leica SP5 microscope with a 40 × 1.2 NA lens. For time-lapse analysis, larvae were imaged at the indicated time, recovered to yeasted agar plates with vented lids, aged at 25°C, and imaged again.
EB1 assays
Larvae were carefully mounted and imaged, then immediately placed in a small plastic Petri dish and stimulated with forceps pinches to segments A2, A3, and A4 and re-imaged under the same conditions. Maximum projections of confocal z stacks (1 μm z step size) were set to identical threshold levels to eliminate non-punctate signals and quantified in Fiji (Schindelin et al., 2012).
Immunostaining
Larval immunostaining was performed as described (Jiang et al., 2018) with the exception of fixation in freezing methanol for 15 min for AcTb staining. Antibody dilutions were as follows: acTb, 1:1000; GFP, 1:100; 22c10 (1:200); HRP-Cy5 (1:100), secondary antibodies (1:200).
Expansion microscopy
Following immunostaining with Mouse anti-GFP, clone 3E6 (Invitrogen #A11120, 1:100) and Goat anti-Mouse Alexa488 (Thermofisher A31561, 1:100) as previously described (Jiang et al., 2018), samples were mounted on lysine-coated #1.5 cover glass in polydimethylsiloxane wells and incubated in monomer solution (2 M NaCl, 8.625% Sodium Acrylate, 2.5% Acrylamide, 0.15% Bisacrylamide in PBS) for 1 h at 4°C prior to gelation. A stock of 4-hydroxy-2,2,6,6-tetramenthylpiperidin-1-oxyl (4-hydroxy-TEMPO) at 1% (wt/wt) in water was added to the incubation solution and diluted to concentration of 0.01%. Concentrated stocks of tetramethyle-thylenediamine (TEMED) and ammonium persulfate (APS) at 10% (wt/wt) in water were added sequentially to the incubation solution and diluted to concentrations of 0.2% (wt/wt). The tissues were then incubated at 37°C for 3-4 h. After gelation, the gels were cut and placed in a small 12-well chamber and 1mg/ml of Chitinase in PBS (pH 6.0) was used to digest the cuticles for ~4 d at 37°C. Chitinase-treated samples were incubated with 1000 units/ml collagenase solution (prepared with buffer 1x HBSS lacking calcium, magnesium, and phenol red) with 0.01 M CaCl2 and 0.01 M MgCl2 overnight in a 37°C shaking incubation chamber. Samples were then rinsed with PBS twice for 5 min and digested in 8 units/ml proteinase K solution in digestion buffer (40 mM Tris pH 8.0, 1 mM EDTA, 0.5% Triton, 0.8 M Guanidine HCl) for 1 h at 37°C. Subsequently, samples were removed from the digestion solution and were allowed to expand in excess water overnight. After expansion, the expanded gel was trimmed to fit onto the coverglass, excess water was removed, and the gel was mounted on a lysine-coated cover glass for imaging. Confocal microscopy was performed on a Leica SP5 inverted confocal scanning microscope using a 63 × 1.2 NA water lens.
Calcium imaging
Third-instar larvae were dissected in calcium imaging solution (310mOsm, pH7.2) containing (in mM): NaCl 120, MgCl2 4, KCl 3, NaHCO3 10, Glucose 10, Sucrose 10, Threalose 10, TES 5, HEPES 10. Larvae were pinned ventral side up on Sylgard® 184 silicone elastomer plates. After opening the larval body from the ventral side, internal organs were removed, and the muscle covering the dorsal c3 da neuron (ddaF) was also gently removed to facilitate imaging of ddaF in segments A4 to A6.
Stimulation electrodes (sealed/polished with a diameter of ~10 μm) were mounted in contact with the internal side of the larval body wall within the dendritic filed of the target c3 da neuron. The tip position was set to avoid direct contact with dendritic tips for potential damage. Following 20ms transient vertical stimulations, each with increasing displacements (10, 20, 30, 40, 50 and 60 μm; applied with a Sutter MP-285 micromanipulator), the electrode was returned to the starting position. GCaMP6s fluorescence was excited with a 488nm solid-state laser and GCaMP fluorescence was imaged at a 0.97 Hz frame acquisition rate using a W Plan-APOCHROMAT 20 × /1.0 objective lens and a Zeiss LSM 700 confocal microscope. Changes in calcium levels in the cell body were measured using the following formula:
where F0 is the average fluorescence in 30 s right before vertical mechanical stimulation. Fpeak is defined as the maximum fluorescence upon stimulation.
Behavior assays
Gentle touch
Each larva was stimulated with an eyelash stroke on thoracic segments while in a bout of linear locomotion. The stimulus was applied and scored four times per larva, with responses scored as previously described (Kernan et.al 1994). Tests were performed with the experimenter blind to genotype in this and all other behavior assays.
Harsh Touch
Larvae were placed in a plastic Petri dish with enough water, so larvae remained moist, but not floating in the dish. Von frey filaments made from fishing line and affixed to glass capillaries were applied to the dorsal side of the larvae between segments A3-A6 until the filament buckled, exhibiting a pre-determined force. Forces of 44mN, 78mN and 98mN were used in this study. A positive response was scored if one complete nocifensive roll occurred after the first mechanical stimulus.
Sound/vibration
Wandering third instar larvae were picked from a vial and washed with PBS. 10 larvae were placed on a 1% agar plate on top of a speaker and stimulated as previously described (Zhang et al., 2013). A 1 s 70dB, 500Hz pure tone was played 10 times with 4 s of silence in between. Video recordings captured larval behavior, with the number of times out of 10 each larva exhibited sound startle behavior as its individual score. 3 separate trials were performed for each genotype. Videos were recorded with AmScope MU300 Microscope digital camera. Larval startle behavior was scored as responsive with the following behaviors: mouth-hook retraction, pausing, excessive turning, and/or backward locomotion.
Gravitaxis
A RING apparatus was assembled as described (Kamikouchi et al., 2009) using 2.3 cm diameter, 9.5 cm polypropylene Drosophila vials. 25 flies were collected, aged until all the flies were 5-8 days old, and transferred to a gravitaxis vial sealed with parafilm. The apparatus was rapped on a table five times in rapid succession to initiate the gravitaxis response. Videos of the flies were captured and position of each fly in the tube was determined 4 s after the response initiation. Flies were allowed to rest 1 minute, and this was repeated for 5 total trials, for n = 1. Assays were repeated for each genotype for total n = 3
Thermal Nociception
Local heat probe assays were performed as previously described (Chattopadhyay et al., 2012). Washed larvae were placed on a piece of vinyl and stimulated on their dorsal midline at segment A4 with a thermal probe maintained at 38c°C for a maximum of 20 s or until completion of a rolling nocifensive response.
Larval locomotion
Larvae were washed and placed on an agar plate with a paint brush, allowed to habituate for 1 minute, and subsequently 10 s videos of individual crawling larvae were recorded in LAS as uncompressed avi files. Files were converted to flymovieformat with any2ufmf and analyzed in Ctrax (Branson et al., 2009). Videos were recorded on Leica DFC310 FX camera on an AmScope FMA050 mount.
Taxol Feeding
Third instar larvae were transferred to 35 mM dishes of cornmeal-molasses food containing DMSO (vehicle) or 60uM taxol (unless otherwise indicated) for 3 hours and then subject to behavior analysis. Adult flies were starved overnight (16 hours) and then fed a 5% sucrose solution containing DMSO or 60uM taxol for 5 hours prior to behavioral analysis.
Electrophysiology
We adapted the methods of (Gong et al., 2013) to record NOMPC-mediated mechanically evoked responses from central neurons in the Drosophila ventral nerve cord (VNC). Adult physiology preparations were as previously described with some modification (Tuthill and Wilson, 2016). Flies were cold-anesthetized and fixed with their ventral side facing up to the underside of a custom-milled steel platform using UV-cured glue (KOA 300, KEMXERT). The ventral head and anterior thorax were partly inserted through a hole in the platform. The top side of the platform, and thus also the exposed parts of the head and thorax, were continually perfused with oxygenated saline. All six legs were glued to the holder with UV-cured glue. A small hole was manually dissected in the cuticle of the ventral thorax to expose the prothoracic neuromeres, and the perineural sheath was gently removed with fine forceps to expose neuronal cell bodies.
The preparation was perfused at ~2-3 ml/min with saline (103 mM NaCl, 3 mM KCl, 5 mM TES, 8 mM trehalose, 10 mM glucose, 26 mM NaHCO3, 1 mM NaH2PO4, 1.5 mM CaCl2, and 4 mM MgCl2; pH 7.1, osmolality adjusted to 270-275 mOsm) bubbled with 95% O2/5% CO2. Recordings were performed at room temperature. Cell bodies were visualized using an infrared LED (Smartvision) and a 40 × water-immersion objective on an upright compound microscope equipped with a fluorescence attachment (Sutter SOM). Whole-cell patch-clamp recordings were targeted to GFP-labeled cell bodies in the prothoracic region of the VNC. The internal patch pipette solution contained (in mM): 140 potassium aspartate, 10 HEPES, 1 EGTA, 4 MgATP, 0.5 Na3GTP, 1 KCl, and 13 biocytin hydrazide (pH 7.2, osmolarity adjusted to ~265 mOsm). We distinguished MNs from other glutamatergic neurons by the characteristic and reliable positions of their cell bodies (Baek and Mann, 2009; Brierley et al., 2012), as well as their intrinsic properties (input resistance, resting membrane potential, and spike waveform).
All recordings were made in current-clamp mode using an Axopatch 700A amplifier. Data were low-pass filtered at 5 kHz before they were digitized at 10 kHz by a 16 bit A/D converter (National Instruments, USB-6343), and acquired in MATLAB. Stable recordings were typically maintained for ~1 hour. A small hyperpolarizing current (approximately −5 to −10 pA) was injected to compensate for the depolarizing seal conductance (Gouwens and Wilson, 2009). Analysis of electrophysiology data was performed with custom scripts written in MATLAB and Python.
Motor neurons were mechanically stimulated with a closed-loop piezoelectric actuator (Physik Instrumente P-841.60, 90 μm travel range, with E-509.S1 sensor/piezo servo-control module). Mechanical stimuli were generated in MATLAB and sent to the amplifier at 5 kHz using an analog output DAQ (National Instruments 9263). Mechanical stimuli were generated in MATLAB and sent to the amplifier at 5 kHz using an analog output DAQ (National Instruments 9263). The stimulating pipette was positioned next to the ipsilateral VNC neuromere under visual control. A 12 μm square wave was used to indent the neuropil. Because the MN cell bodies are segregated from the VNC neuropil, the mechanical stimuli had no visible effect on the cell body and patch pipette. Stimulation of the contralateral neuromere failed to evoke a response.
Atomic force microscope measurements
The elastic moduli of control- and dTat RNAi-treated cells were measured using an MFP-3D Bio AFM (Asylum Research, Santa Barbra, CA). The AFM head was mounted on an Olympus IX71 inverted optical microscope to aid in accurately positioning the cantilever probe directly above a single cell. Cells were probed using a Novascan silicon nitride AFM cantilever of nominal spring constant 0.03N/m with a 4.5um polystyrene bead attached. Before force measurements were made on each sample, the cantilever spring constant was calibrated using the built-in thermal tune method in the IGOR/Asylum Software. These calibrations ranged from 0.039-0.042N/m.
S2 cell RNAi was performed for seven days as described above; we then treated with dsRNA every other day to maintain protein depletion. On the day of an experiment, confluent cells from each population (control or dTat RNAi) were plated on cleaned glass poly L-lysine-coated coverslips such that adherent cells were spaced about 2-3 cell diameters apart once attached. Stiffness measurements were acquired 45 min to 120 min after plating at room temperature. Parallel samples of control or dTat RNAi cells were plated and an approximately equivalent number of data points were acquired during each trial. The order of data acquisition was varied for each experiment.
For force versus indentation measurements the cantilever was moved at a velocity of 5 μm/s toward the cell until a force of 3 nN was reached. The cantilever was then retracted at the same rate. For most cells, this was equivalent to an indentation of 3-4 μm. For each cell, two force curves were collected and one was chosen, based on clearly defined points of contact and flat baseline approaches. The first curve was picked in approximately 80% of all cells. Force measurements were acquired for ~300 cells of each type over a period of three days of experiments.
Calibrated cantilever deflection and piezo displacement data collected were converted to produce force versus indentation curves (Figure S7). Using a custom MATLAB code, force-indentation data were fit to the Hertzian contact mechanics model to determine elastic moduli (stiffness) for each cell (Beicker et al., 2018; Cribb et al., 2016). The force versus indentation data were fit up to indentations of 750nm which corresponded to less than 10% of the cell height. The entire dataset was screened for relative RMS fitting errors greater than two sigma above the mean. Then, elastic moduli values that were above or below two sigma of the mean were excluded within each trial (Grubbs, 1950).
QUANTIFICATION AND STATISTICAL ANALYSIS
Datasets were tested for normality using Shapiro-Wilks goodness of fit tests. Details on statistical tests are provided in figure legends and the corresponding methods sections.
DATA AND SOFTWARE AVAILABILITY
D. melanogaster larval cell type RNA-Seq data are available from SRA and GEO under the accession number GEO: GSE120305.
Supplementary Material
Highlights.
dTAT acts as the major α-tubulin acetylase in Drosophila
Microtubule acetylation broadly and specifically regulates mechanosensation
dTAT is required for mechanosensation by the TRP channel NOMPC
Acetylation tunes microtubule stability to control mechanosensation
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH/National Institute of Neurological Disorders and Stroke (NINDS) (R01NS076614), a University of Washington Research Innovation award, and startup funds from the University of Washington to J.Z.P.; support from Mark Peifer and a University of North Carolina Biology/iBGS Pilot Award to S.L.R.; grants from the NIH/NINDS (R01NS089787 and R21NS107924) to Y.X.; a grant from the NIH/NINDS (R21NS101553) to J.W.; a Klingenstein-Simons Fellowship to J.C.T.; a grant from the NIH/National Institute of Biomedical Imaging and Bioengineering (NIBIB) (5-P41-EB002025-33) to R.S.; and a grant from the NIH/National Institute of Mental Health (NIMH) (R01MH115767) to J.C.V. Fly stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) and antibodies from the Developmental Studies Hybridoma Bank, created by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the NIH and maintained at The University of Iowa, were used in this study. We thank Mark Peifer and Peter Soba for critical reading of this manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.09.075.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ageta-Ishihara N, Miyata T, Ohshima C, Watanabe M, Sato Y, Hamamura Y, Higashiyama T, Mazitschek R, Bito H, and Kinoshita M (2013). Septins promote dendrite and axon development by negatively regulating microtubule stability via HDAC6-mediated deacetylation. Nat. Commun 4, 2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, and Gaertig J (2010). MEC-17 is an alpha-tubulin acetyltransferase. Nature 467, 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, and Mann RS (2009). Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. J. Neurosci. 29, 6904–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beicker K, O’Brien ET 3rd, Falvo MR, and Superfine R (2018). Vertical light sheet enhanced side-view imaging for AFM cell mechanics studies. Sci. Rep 8, 1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuwania R, Castro-Castro A, and Linder S (2014). Microtubule acetylation regulates dynamics of KIF1C-powered vesicles and contact of microtubule plus ends with podosomes. Eur. J. Cell Biol 93, 424–437. [DOI] [PubMed] [Google Scholar]
- Boiko N, Medrano G, Montano E, Jiang N, Williams CR, Madungwe NB, Bopassa JC, Kim CC, Parrish JZ, Hargreaves KM, et al. (2017). TrpA1 activation in peripheral sensory neurons underlies the ionic basis of pain hypersensitivity in response to vinca alkaloids. PLoS One 12, e0186888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounoutas A, O’Hagan R, and Chalfie M (2009). The multipurpose 15-protofilament microtubules in C. elegans have specific roles in mechanosensation. Curr. Biol 19, 1362–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branson K, Robie A, Bender J, Perona P, and Dickinson M (2009). High-throughput ethomics in large groups of Drosophila. Nat. Methods 6, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley DJ, Rathore K, VijayRaghavan K, and Williams DW (2012). Developmental origins and architecture of Drosophila leg motoneurons. J. Comp. Neurol 520, 1629–1649. [DOI] [PubMed] [Google Scholar]
- Chalfie M, and Au M (1989). Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 243, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Chalfie M, and Sulston J (1981). Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol 82, 358–370. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A, Gilstrap AV, and Galko MJ (2012). Local and global methods of assessing thermal nociception in Drosophila larvae. J. Vis. Exp 63, e3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LE, Song W, Looger LL, Jan LY, and Jan YN (2010). The role of the TRP channel NompC in Drosophila larval and adult locomotion. Neuron 67, 373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AP, and Corey DP (2007). TRP channels in mechanosensation: direct or indirect activation? Nat. Rev. Neurosci 8, 510–521. [DOI] [PubMed] [Google Scholar]
- Conacci-Sorrell M, Ngouenet C, and Eisenman RN (2010). Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 142, 480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, and Patapoutian A (2010). Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert M-L, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, et al. (2009). Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136, 551–564. [DOI] [PubMed] [Google Scholar]
- Cribb JA, Osborne LD, Beicker K, Psioda M, Chen J, O’Brien ET, Taylor li RM, Vicci L, Hsiao JP-L, Shao C, et al. (2016). An automated high-throughput array microscope for cancer cell mechanics. Sci. Rep 6, 27371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Wang Q, and Wang Z (2013). NOMPC is likely a key component of Drosophila mechanotransduction channels. Eur. J. Neurosci 38, 2057–2064. [DOI] [PubMed] [Google Scholar]
- Goodwin SS, and Vale RD (2010). Patronin regulates the microtubule network by protecting microtubule minus ends. Cell 143, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouwens NW, and Wilson RI (2009). Signal propagation in Drosophila central neurons. J. Neurosci 29, 6239–6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and O’Connor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs FE (1950). Sample criteria for testing outlying observations. Ann. Math. Stat 21, 27–58. [Google Scholar]
- Han C, Jan LY, and Jan Y-N (2011). Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl. Acad. Sci. USA 108, 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, and Bechstedt S (2004). Hypothesis: a helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors. Curr. Biol 14, R224–R226. [DOI] [PubMed] [Google Scholar]
- Howard J, and Hudspeth AJ (1987). Mechanical relaxation of the hair bundle mediates adaptation in mechanoelectrical transduction by the bull-frog’s saccular hair cell. Proc. Natl. Acad. Sci. USA 84, 3064–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang X-F, and Yao T-P (2002). HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458. [DOI] [PubMed] [Google Scholar]
- Hummel T, Krukkert K, Roos J, Davis G, and Klämbt C (2000). Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26, 357–370. [DOI] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, and Tracey WD (2007). Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr. Biol 17, 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BV, Saunders HAJ, Record HL, Johnson-Schlitz DM, and Wildonger J (2017). Effects of mutating α-tubulin lysine 40 on sensory dendrite development. J. Cell Sci 130, 4120–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Kim H-J, Chozinski TJ, Azpurua JE, Eaton BA, Vaughan JC, and Parrish JZ (2018). Superresolution imaging of Drosophila tissues using expansion microscopy. Mol. Biol. Cell 29, 1413–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Soba P, Parker E, Kim CC, and Parrish JZ (2014). The micro-RNA bantam regulates a developmental transition in epithelial cells that restricts sensory dendrite growth. Dev. Camb. Engl 141, 2657–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Bulkley D, Guo Y, Zhang W, Guo Z, Huynh W, Wu S, Meltzer S, Cheng T, Jan LY, et al. (2017). Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N, Martinez C, Perlas E, Hublitz P, Bilbao-Cortes D, Fiedorczuk K, Andolfo A, and Heppenstall PA (2013a). Tubulin acetyltransferase aTAT1 destabilizes microtubules independently of its acetylation activity. Mol. Cell. Biol 33, 1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N, Sorrentino S, Perlas E, Bolasco G, Martinez C, and Heppenstall PA (2013b). αTAT1 isthe major α-tubulin acetyltransferase in mice. Nat. Commun 4, 1962. [DOI] [PubMed] [Google Scholar]
- Kamikouchi A, Inagaki HK, Effertz T, Hendrich O, Fiala A, Gópfert MC, and Ito K (2009). The neural basis of Drosophila gravity-sensing and hearing. Nature 458, 165–171. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Macara AM, Li R, Hu Y, Iwasaki K, Dunnings Z, Firestone E, Horvatic S, Guntur A, Shafer OT, et al. (2017). Serotonergic modulation enables pathway-specific plasticity in a developing sensory circuit in Drosophila. Neuron 95, 623–638.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta S, Krieg M, and Goodman MB (2015). Feeling force: physical and physiological principles enabling sensory mechanotransduction. Annu. Rev. Cell Dev. Biol 31,347–371. [DOI] [PubMed] [Google Scholar]
- Kernan M, Cowan D, and Zuker C (1994). Genetic dissection of mechanosensory transduction: mechanoreception-defective mutations of Drosophila. Neuron 12,1195–1206. [DOI] [PubMed] [Google Scholar]
- Kim G-W, Li L, Ghorbani M, You L, and Yang X-J (2013). Mice lacking α-tubulin acetyltransferase 1 are viable but display α-tubulin acetylation deficiency and dentate gyrus distortion. J. Biol. Chem 288, 20334–20350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C (2005). A possible unifying principle for mechanosensation. Nature 436, 647–654. [DOI] [PubMed] [Google Scholar]
- L’Hernault SW, and Rosenbaum JL (1985). Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry 24, 473–478. [DOI] [PubMed] [Google Scholar]
- Liang X, Madrid J, Gärtner R, Verbavatz J-M, Schiklenk C, Wilsch-Bräuninger M, Bogdanova A, Stenger F, Voigt A, and Howard J (2013). A NOMPC-dependent membrane-microtubule connector is a candidate for the gating spring in fly mechanoreceptors. Curr. Biol 23, 755–763. [DOI] [PubMed] [Google Scholar]
- Morley SJ, Qi Y, Iovino L, Andolfi L, Guo D, Kalebic N, Castaldi L, Tischer C, Portulano C, Bolasco G, et al. (2016). Acetylated tubulin is essential for touch sensation in mice. eLife 5, e20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, and Verdin E (2003). The human Sir2 ortholog, SIRT2, is an NAD+−dependent tubulin deacetylase. Mol. Cell 11, 437–444. [DOI] [PubMed] [Google Scholar]
- O’Hagan R, Chalfie M, and Goodman MB (2005). The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat. Neurosci 8, 43–50. [DOI] [PubMed] [Google Scholar]
- Ohkawa N, Sugisaki S, Tokunaga E, Fujitani K, Hayasaka T, Setou M, and Inokuchi K (2008). N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cells 13, 1171–1183. [DOI] [PubMed] [Google Scholar]
- Park IY, Powell RT, Tripathi DN, Dere R, Ho TH, Blasius TL, Chiang Y-C, Davis IJ, Fahey CC, Hacker KE, et al. (2016). Dual chromatin and cytoskeletal remodeling by SETD2. Cell 166, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen LK, and Stowers RS (2011). A Gateway MultiSite recombination cloning toolkit. PLoS One 6, e24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Schaedel L, Xu Z, Théry M, and Nachury MV (2017). Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat. Cell Biol 19, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager-Khoutorsky M, Khoutorsky A, and Bourque CW (2014). Unique interweaved microtubule scaffold mediates osmosensory transduction via physical interaction with TRPV1. Neuron 83, 866–878. [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, and Verhey KJ (2006). Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol 16, 2166–2172. [DOI] [PubMed] [Google Scholar]
- Rogers SL, and Rogers GC (2008). Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nat. Protoc 3, 606–611. [DOI] [PubMed] [Google Scholar]
- Roos J, Hummel T, Ng N, Klämbt C, and Davis GW (2000). Drosophila Futsch regulates synaptic microtubule organization and is necessary for synaptic growth. Neuron 26, 371–382. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Ashley J, Moeckel-Cole S, Drier E, Yin J, and Budnik V (2004). New synaptic bouton formation is disrupted by misregulation of microtubule stability in aPKC mutants. Neuron 42, 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, and Nachury MV (2010). The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. USA 107,21517–21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Onishi M, Jan LY, and Jan YN (2007). Peripheral multidendritic sensory neurons are necessary for rhythmic locomotion behavior in Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A 104, 5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo H, and Baas PW (2010). Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J. Neurosci 30, 7215–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu L, Ben-Shahar Y, Jacobs JS, Eberl DF, and Welsh MJ (2009). TRPA channels distinguish gravity sensing from hearing in Johnston’s organ. Proc. Natl. Acad. Sci. USA 106, 13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Spector J, Goodman B, Valenstein ML, Ziolkowska NE, Kormendi V, Grigorieff N, and Roll-Mecak A (2014). Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KD, Levine JD, and Topp KS (1998). Microtubule disorientation and axonal swelling in unmyelinated sensory axons during vincristine-induced painful neuropathy in rat. J. Comp. Neurol 395, 481–492. [PubMed] [Google Scholar]
- Topalidou I, Keller C, Kalebic N, Nguyen KCQ, Somhegyi H, Politi KA, Heppenstall P, Hall DH, and Chalfie M (2012). Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr. Biol 22, 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi A, Caldwell JC, and Tracey WD (2012). Dendritic filopodia, Ripped Pocket, NOMPC, and NMDARs contribute to the sense of touch in Drosophila larvae. Curr. Biol 22, 2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuthill JC, and Wilson RI (2016). Parallel transformation of tactile signals in central circuits of Drosophila. Cell 164, 1046–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RG, Willingham AT, and Zuker CS (2000).A Drosophila mechanosensory transduction channel. Science 287, 2229–2234. [DOI] [PubMed] [Google Scholar]
- Williams CR, Baccarella A, Parrish JZ, and Kim CC (2016). Trimming of sequence reads alters RNA-Seq gene expression estimates. BMC Bioinformatics 17, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, and Jan YN (2010). Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468, 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, and Nachury MV (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Zhang W, He Y, Gorczyca D, Xiang Y, Cheng LE, Meltzer S, Jan LY, and Jan YN (2013). Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature 493, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Yan Z, Jan LY, and Jan YN (2013). Sound response mediated by the TRP channels NOMPC, NANCHUNG, and INACTIVE in chordotonal organs of Drosophila larvae. Proc. Natl. Acad. Sci. USA 110, 13612–13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Cheng LE, Kittelmann M, Li J, Petkovic M, Cheng T, Jin P, Guo Z, Gópfert MC, Jan LY, and Jan YN (2015). Ankyrin repeats convey force to gate the NOMPC mechanotransduction channel. Cell 162, 1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Hwang RY, and Tracey WD (2010). Pickpocket is a DEG/ENaC protein required for mechanical nociception in Drosophila larvae. Curr. Biol 20, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.