Abstract
Reverse transduction, also known as substrate-mediated gene delivery, is a strategy in which viral vectors are first coated onto a surface that subsequently comes into contact with mammalian cells. The cells internalize the surface-attached vectors, resulting in transgene expression. We hypothesized that forcing the interaction between cells and adeno-associated virus (AAV) vectors through a reverse transduction format would increase in vitro gene delivery efficiencies of the vectors in transduction-resistant cells. We tested this hypothesis by comparing the gene delivery efficiencies of three AAV serotypes using either standard or reverse transduction approaches. Our study reveals reverse transduction of AAV7 and AAV9 can significantly improve their delivery efficiencies. In contrast, AAV2 does not perform better under the reverse transduction format. Interestingly, increased vector uptake by cells does not provide a complete explanation for the increased transduction efficiency. Our findings offer a simple and practical method for improving transduction outcomes in vitro in cell types less permissive to a particular AAV vector.
Keywords: adeno-associated virus, gene delivery, reverse transduction, substrate-mediated, viral vector
Introduction
Substrate-mediated delivery is a strategy in which viral or non-viral vectors are first immobilized onto a surface after which cells are placed into contact with the modified surface. This gene delivery approach may be valuable in tissue engineering endeavors. Vectors encoding a transgene of interest can be attached to a biomaterial scaffold, then introduced at a site of injury for spatially-regulated administration to cell populations in situ and ultimately localized tissue regeneration. Recent progress in this realm has been reviewed thoroughly by Kasputis et al (Kasputis, Farris, Guerreiro, Taylor, & Pannier, 2017).
Many substrate-mediated delivery approaches rely on non-viral vectors, often perceived as less toxic and less costly to produce than viral vectors. However, viruses possess distinct advantages over their non-viral counterparts because they have an intrinsic capacity for cellular entry and can promote sustained gene expression. The ~25 nm adeno-associated virus (AAV) is one such candidate that holds excellent therapeutic promise. Among its desirable features include a simple genome, natural ability to infect mammalian cells, non-pathogenicity in humans, and capacity to yield long-term transgene expression (Buchlis et al., 2012). These assets have promoted its widespread usage in human clinical trials.
Reverse transduction in the context of AAV has been explored previously, but by no means extensively. One aspect entails leveraging this strategy for high-throughput screening applications. A prior study evaluated transduction outcomes of several mammalian cell lines in 96-well tissue culture plates immobilized with different AAV serotypes (Dong et al., 2010). The multiplicities of infection (MOIs) – number of viral genomes added per cell – necessary to produce optimal results were reported in the range of 20,000–40,000. Another study demonstrated the feasibility of developing live-cell microarrays by depositing AAV2 onto nitrocellulose-coated slides, resulting in effective reverse transduction when HeLa cells were seeded on top of these surfaces (McConnell, Schweller, Diehl, & Suh, 2011).
AAV-based reverse transduction has also been employed in various studies relevant to tissue engineering. For example, transduction efficiency resulting from AAV2 patterned onto different substrate coatings, include extracellular matrix (ECM) components, has been evaluated (McConnell, Gomez, & Suh, 2012; McConnell, Slater, Han, West, & Suh, 2011). Understanding the interaction between AAV vectors and ECM proteins is particularly relevant because the latter are present in all tissues and organs, as well as being frequently incorporated into scaffolds for tissue regeneration. A number of other studies have also pursued a combined biomaterial and AAV approach. In one instance, AAV2 encoding for vascular endothelial growth factor (VEGF) and receptor activator of nuclear factor kappa-B ligand (RANKL) have been incorporated into bone allografts, and following implantation in mice, bone remodeling was observed (Ito et al., 2005). Bone morphogenetic protein-2 (BMP2)-expressing AAV2 has also been coated onto hydroxyapatite in a similar fashion and demonstrated to induce bone formation in a rat model (Nasu et al., 2009). More recently, a group generated poly(L-lactic acid) scaffolds incorporating AAV encoding BMP2 and evaluated the osteogenic potential in vitro and in vivo with seeded human mesenchymal stem cells (hMSCs) (Xue et al., 2017). Despite encouraging outcomes seen from the aforementioned examples, some studies have also reported low transduction efficiencies. In one instance, a self-complementary AAV2.5 vector harboring the LacZ reporter transduced less than 2% of hMSCs in vitro (at MOIs 20,000 and 40,000) (Dupont et al., 2012).
The performance of a given gene delivery vehicle exhibits large variability amongst different cell types, and AAV is certainly not exempt from this challenge. Ellis and colleagues conducted a comprehensive in vitro screen, whereby they performed standard transduction on 34 human and animal cell lines with a panel of AAV serotypes. Widely variable gene expression outcomes were observed, with a handful of cell types refractory to AAV even at a high MOI of 100,000 (Ellis et al., 2013). In order to expand the use of AAV vectors for as many applications as possible, easy methods for improving the testing of AAV transduction would be valuable, especially in cell types difficult to transduce in vitro. While AAV2 is most well characterized and frequently used historically, expanding the delivery capabilities of other serotypes would be advantageous in order to harness their full potential.
In this work, we evaluated three AAV serotypes previously shown to display high (AAV2), moderate (AAV9), and low (AAV7) transduction efficiencies in HeLa cells in vitro (Ellis et al., 2013). Previous work examined standard transduction efficiencies for different AAV serotypes in various cell lines. However, to our knowledge, no comparisons between standard and reverse transduction protocols have been made directly side-by-side. In standard transduction, cells are first seeded on tissue culture plates to adhere to the surface and proliferate. Viral vectors are then added to the cells the next day (Figure 1A). Conversely, in reverse transduction viral vectors are first coated onto tissue culture plates and aspirated from the wells the following day. A suspension of cells is then added to the virus-coated wells (Figure 1B). Under the reverse transduction format, cells may be forced into contact with the surface-immobilized viral vectors while settling on the bottom of the well, whereas for standard transduction viral vectors rely on diffusion through culture media to interact with cells. Based on this difference in virus-cell contact routes, we hypothesized reverse transduction would lead to greater gene delivery efficiencies compared to standard transduction, and the transduction of less permissive cells would be improved by using the reverse transduction approach. We further investigated whether increased transduction efficiencies could be attributed to improved cellular uptake. The studies presented herein offer a simple method for more effective transduction in vitro.
Figure 1.
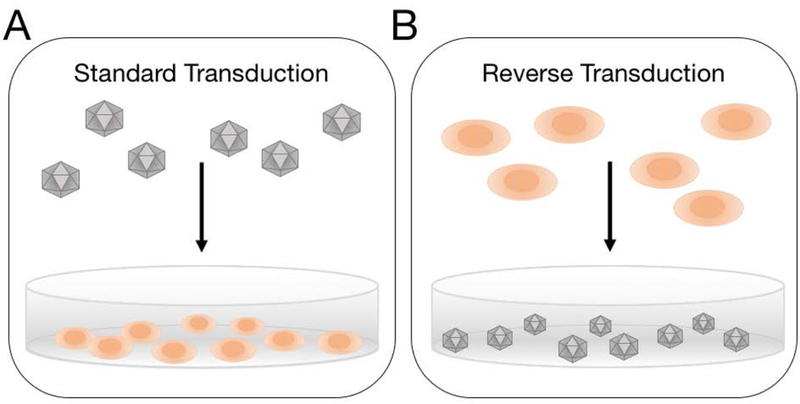
Schematic of the two transduction strategies evaluated. (A) In standard transduction, cells are seeded in wells of a tissue culture plate prior to introducing virus. (B) In reverse transduction, virus particles are pre-coated onto the wells of a tissue culture plate, and cells are subsequently seeded.
Materials and Methods
Virus production
Recombinant adeno-associated virus serotypes 2, 7, and 9 (AAV2, AAV7, and AAV9) were prepared as follows. HEK 293T cells were transfected with a mixture of polyethylenimine and three plasmids: pXX6–80 (adenovirus helper), scAAV-GFP (self-complementary vector with green fluorescent protein expressed under a constitutive CMV promoter), and either pXX2, pAAV2/7, or pAAV2/9. At 48 h post-transfection, the cells were harvested for virus extraction by iodixanol gradient ultra-centrifugation. Virus was either maintained in iodixanol solution or subjected to buffer exchange in 1X Gradient Buffer (diluted from a 10X stock solution composed of sterile water, 1M Tris at pH 7.6, 5M NaCl, and 1M MgCl2) containing 0.001% Pluronic F-68 (Thermo Fisher Scientific) using Amicon Ultra Centrifugal Filters (Millipore Sigma). The number of viral genomes in each preparation were determined through quantitative polymerase chain reaction (qPCR) with Power SYBR Green Master Mix (Applied Biosystems) and primers (Integrated DNA Technologies) generating an amplicon from within the CMV promoter upstream of the GFP transgene cassette: CMV Forward (5’-TCACGGGGATTTCCAAGTCTC-3’) and CMV Reverse (5’-AATGGGGCGGAGTTGTTACGAC-3’). Samples were run on a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA).
Standard AAV transduction
HeLa cells were seeded at 1.35 × 105 cells per well on 24-well tissue culture plates that were coated with poly(L-lysine). After 24 h, virus suspended in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco) with 10% Fetal Bovine Serum (FBS) (Gemini) and 1% Penicillin/Streptomycin (P/S) (Gibco) was added to the cells. Culture medium was replaced 24 h following transduction.
Reverse AAV transduction
Amicon-purified virus was diluted in 1X Gradient Buffer containing 0.001% Pluronic F-68 based on desired MOIs and dispensed onto tissue culture plates. The plates were then stored at 4°C overnight. Prior to cell seeding, the virus solution was aspirated from the wells. HeLa cells were seeded at 2.7 × 105 cells per well onto virus-coated 24-well tissue culture plates. Culture medium was replaced 24 h post-transduction.
Fluorescence imaging to visualize transduced cells
At 48 h post-transduction, HeLa cells in the tissue culture plates were imaged on a Nikon Eclipse TE300 inverted fluorescence microscope (Nikon, Tokyo, Japan) using a 10X objective. Brightfield images were taken with an exposure time of 4 ms. A FITC filter and fluorescence lamp set to 25% output were used to capture GFP-positive cells, employing a 500 ms exposure time. The Bio-Formats package in MATLAB was used for image post-processing to improve brightness and contrast. Look-up table (LUT) values were restricted between 0.005 and 0.02 and applied uniformly across all images.
Flow cytometry to quantify gene delivery efficiency
HeLa cells from both standard and reverse transduction assays were detached from their respective tissue culture plates using trypsin followed by serum media neutralization of the protease. All cell suspensions were run on a BD FACScan system (Becton Dickinson, San Jose, CA) with modified optical configuration, as previously described (Olson, Hartsough, Landry, Shroff, & Tabor, 2014). 20,000 cells were collected for each sample. The percentage of GFP-positive cells as well as geometric mean fluorescence intensity (gMFI) normalized to SPHERO™ Rainbow Calibration Particles (Spherotech) were determined using FlowCal open-source software (Castillo-Hair et al., 2016). Transduction index (TI) was calculated as GFP-positive cells multiplied by gMFI for each experimental sample (Judd et al., 2014). TI is used as the measure for gene delivery efficiency because it increases linearly with MOI (Judd et al., 2014).
Virus internalization assay
For standard transduction, HeLa cells were seeded at 5.4 × 105 cells per well on 6-well tissue culture plates pre-coated with poly(L-lysine). After 24 h, virus suspended in DMEM with 10% FBS and 1% P/S was added to the cells at MOI 5000. For reverse transduction, 1.08 × 106 cells per well were added to virus-coated 6-well tissue culture plates. 24 h following standard or reverse transduction, cells were harvested and pelleted. An EZNA Tissue DNA Kit (Omega Bio-tek) was used per manufacturer’s instructions to isolate total DNA from the cells. The number of viral genomes comprising each DNA sample was quantified via qPCR, and results were normalized to total DNA content as measured on a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific).
Statistical analysis
All results are reported as mean ± standard error of the mean (SEM). An unpaired Student’s t-test was used to determine if there was any significant difference at p < 0.05 between standard and reverse transduction methods for a specific AAV serotype at a given MOI. Statistical analyses were run in R using the t.test( ) function with inputs of all the TI values comprising the groups being compared.
Results and Discussion
Reverse transduction improves efficiency for some AAV serotypes
We performed a direct comparison between two transduction formats – standard and reverse. In standard transduction, HeLa cells are first seeded in a tissue culture plate and allowed to adhere to the surface and proliferate overnight. Virus mixed with culture medium is then introduced to the cells. In reverse transduction, virus is first incubated overnight in the tissue culture plate. After aspirating this solution, cells resuspended in culture medium are seeded onto the wells. For both formats, the culture medium is replenished at 24 h post-transduction, and cells are harvested for analysis at the 48 h timepoint. The same amount of virus is used in both transduction configurations. However, due to differences in the timing of transduction, different numbers of cells are added to the system for the two methods. In the standard format, we must account for cell growth and division between the time they are seeded on plates and when they are transduced with AAV. Therefore, a lower cell seeding density is employed, such that the desired cell confluency will be reached 24 h later. In the reverse format, cells are transduced at the time of seeding, with no time provided for cell growth and division. Therefore, twice as many cells are added to wells for the reverse transduction method compared to the standard. In both transduction formats, the cell confluency at time of exposure to AAV was 90%. Additionally, assays with AAV7 and AAV9 were performed with higher MOIs than with AAV2 because HeLa cells are less permissive to these serotypes.
When comparing the two transduction formats, greater numbers of GFP-positive cells are obtained for standard transduction with AAV2 and for reverse transduction with AAV7 and AAV9 (Figure 2). The brightfield images demonstrate similar numbers of cells are present in the two formats by the time of transgene expression detection. In order to corroborate the microscopy images and quantify transgene expression, flow cytometry was used. As expected, increasing the MOI of AAV2 yields greater transduction efficiency in HeLa cells for both formats (Figure 3). Surprisingly, however, reverse transduction does not improve the performance of AAV2 relative to standard transduction. In fact, standard transduction seems to yield greater gene expression for this AAV serotype.
Figure 2.
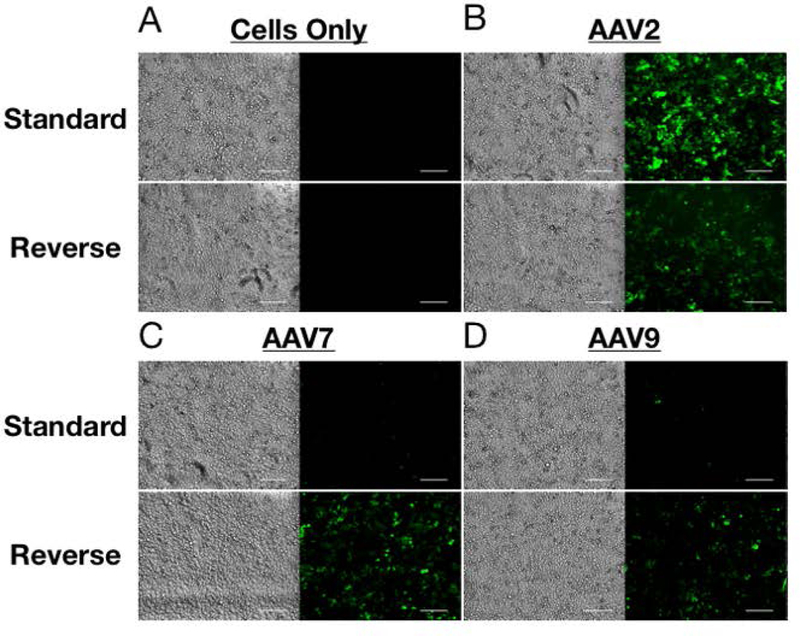
Representative brightfield and fluorescence images comparing standard and reverse transductions approaches for (A) cells only, (B) AAV2 at MOI 1000, (C) AAV7 at MOI 10000, and (D) AAV9 at MOI 10000. Similar numbers of cells between the two methods were observed at time of imaging. Scale bar is 200µm.
Figure 3.
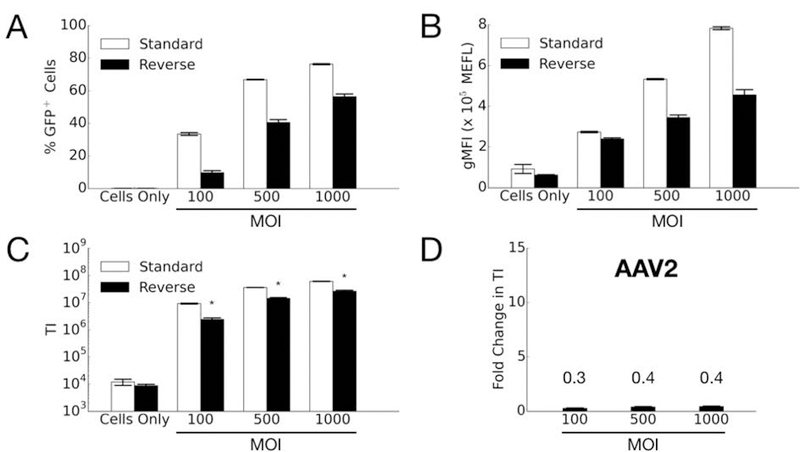
Comparison of AAV2 transduction efficiency in HeLa cells by standard and reverse transduction methods at various MOIs. (A) Percentage of GFP-positive cells; (B) Geometric mean fluorescence intensity (gMFI) in units of Molecules of Equivalent Fluorophore (MEFL); and (C) Transduction index (TI), calculated as the product of (A) and (B). Error bars denote standard error of the mean (SEM) from two independent experiments conducted in duplicate (standard transduction) or triplicate (reverse transduction). Asterisk indicates significant difference (p<0.05) between standard and reverse transduction methods. (D) Fold change in transduction efficiency is determined by dividing the averaged TI from reverse transduction by the averaged TI of the standard approach.
We then tested the two transduction formats for AAV7 and AAV9. In contrast to the results for AAV2, the transduction efficiency for both AAV7 (Figure 4) and AAV9 (Figure 5) markedly increases with reverse transduction compared to standard. One interesting observation for AAV7 is the fold-change increase in efficiency due to using the reverse format increases with higher MOI (Figure 4D). In other words, for AAV7 the fold-improvement gained by using reverse transduction is better when more viruses are used. It is possible there may be some synergistic effects when using higher MOIs (e.g. viruses can escape endosomes more easily when there are more viruses in the vesicle (Lagache, Danos, & Holcman, 2012)), but further studies are necessary to explain this result. For AAV9, on the other hand, the reverse format improves the transduction efficiency at a similar level across all MOIs tested (Figure 5D).
Figure 4.
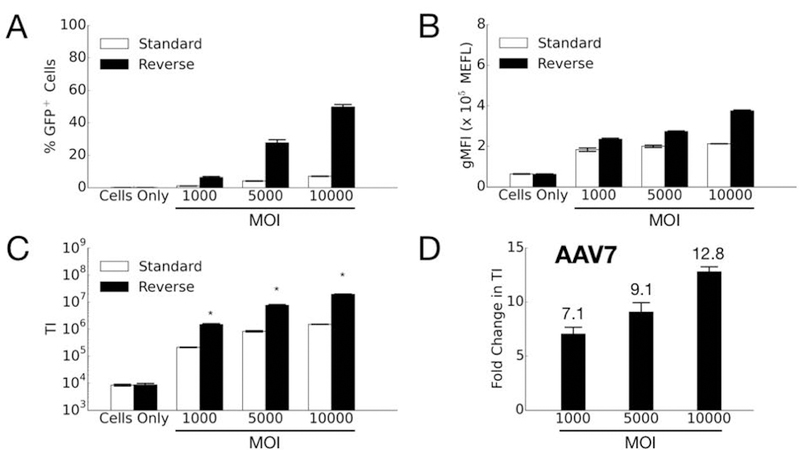
Comparison of AAV7 transduction efficiency in HeLa cells by standard and reverse transduction methods. (A) Percentage of GFP-positive cells; (B) Geometric mean fluorescence intensity (gMFI) in units of Molecules of Equivalent Fluorophore (MEFL); (C) Transduction index (TI), calculated as the product of (A) and (B). Error bars denote standard error of the mean (SEM) from two independent experiments conducted in duplicate (standard transduction) or triplicate (reverse transduction). Asterisk indicates significant difference (p<0.05) between standard and reverse transduction methods. (D) Fold change in transduction efficiency is determined by dividing the averaged TI from reverse transduction by the averaged TI of the standard approach.
Figure 5.
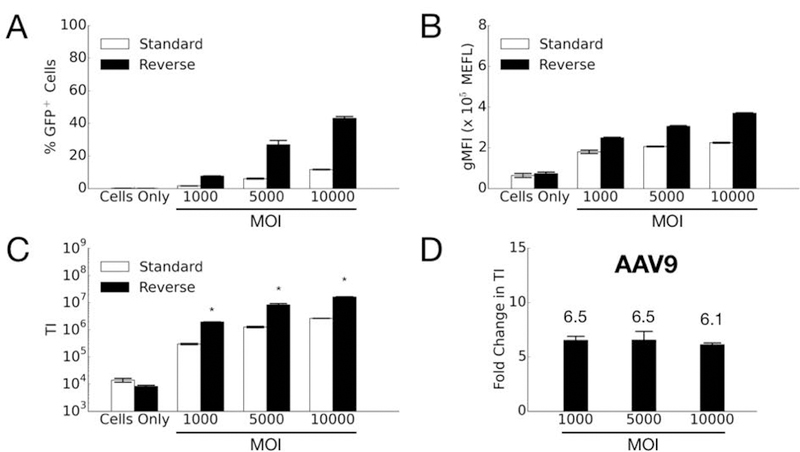
Comparison of AAV9 transduction efficiency in HeLa cells by standard and reverse transduction methods. (A) Percentage of GFP-positive cells; (B) Geometric mean fluorescence intensity (gMFI) in units of Molecules of Equivalent Fluorophore (MEFL); (C) Transduction index (TI), calculated as the product of (A) and (B). Error bars denote standard error of the mean (SEM) from two independent experiments conducted in duplicate. Asterisk indicates significant difference (p<0.05) between standard and reverse transduction methods. (D) Fold change in transduction efficiency is determined by dividing the averaged TI from reverse transduction by the averaged TI of the standard approach.
These cumulative findings suggest the effectiveness of reverse transduction is influenced by the AAV serotype and the permissiveness of a given cell line to it. HeLa cells are highly permissive to AAV2 transduction, moderately permissive to AAV9, and lowly permissive to AAV7 in vitro. Under standard transduction conditions, AAV2 works much better than AAV7 and AAV9 in HeLa cells. Therefore, reverse transduction might be particularly beneficial when using serotypes to which a given cell type is less permissive in vitro. In the case of serotypes to which a particular cell line is already highly permissive, the reverse format may not yield improvements in efficiency.
To provide further context, we compare our results to that of Ellis et al., who also packaged a self-complementary AAV expressing GFP under a constitutive CMV promoter. They reported 80%, 20%, and 38% GFP-positive cells for AAV2, AAV7, and AAV9, respectively, at MOI 100,000 in HeLa cells by standard transduction. In our study, we observed 76%, 7%, and 12% GFP-positive cells for AAV2 (MOI 1000), AAV7 (MOI 10,000), and AAV9 (MOI 10,000), respectively, for standard transduction. Additionally, we achieve 56%, 50%, and 43% GFP-positive cells for AAV2 (MOI 1000), AAV7 (MOI 10,000), and AAV9 (MOI 10,000), respectively, for reverse transduction.
Increased cellular uptake does not fully explain improved transduction efficiency
We next sought to determine if increased cellular uptake of the viral vectors leads to increased gene delivery efficiencies under the standard or reverse transduction formats. We posited reverse transduction may facilitate greater virus uptake compared to standard transduction (at least for AAV7 and AAV9), as the former strategy forces cells to come into contact with viruses as they settle and attach to the bottom of the plate where the viruses are immobilized.
Interestingly, results from the cellular internalization assay neither fully support nor dispel this hypothesis (Figure 6). For AAV7, the 6.3-fold increase in cellular uptake of vectors noted for reverse transduction may possibly explain the 7.1 to 12.8-fold increases in transduction efficiency using this format. For the other two serotypes, cellular uptake results do not explain the transduction results. For AAV2, reverse transduction results in 0.3 to 0.4-fold TI compared to the standard transduction format (i.e. 2.5 to 3.3-fold higher transduction by the standard approach compared to reverse). The number of internalized viral genomes under the reverse transduction format, however, is 0.7-fold of the standard format (i.e. 1.4-fold higher by standard transduction compared to reverse), and this difference is not statistically significant. Thus the increased transduction efficiency under the standard approach is not due to a significant increase in cellular uptake of the AAV2 vectors. For AAV9, vector internalization is only 2-fold greater in reverse transduction, which is not a significant difference. Consequently, this outcome does not fully explain the ~6-fold higher gene delivery efficiency observed for the reverse format. These discrepancies suggest that AAV vector internalization into cells does not completely account for higher transduction efficiencies. Therefore, other intracellular trafficking mechanisms may also exert influence on the overall gene delivery efficiency.
Figure 6.
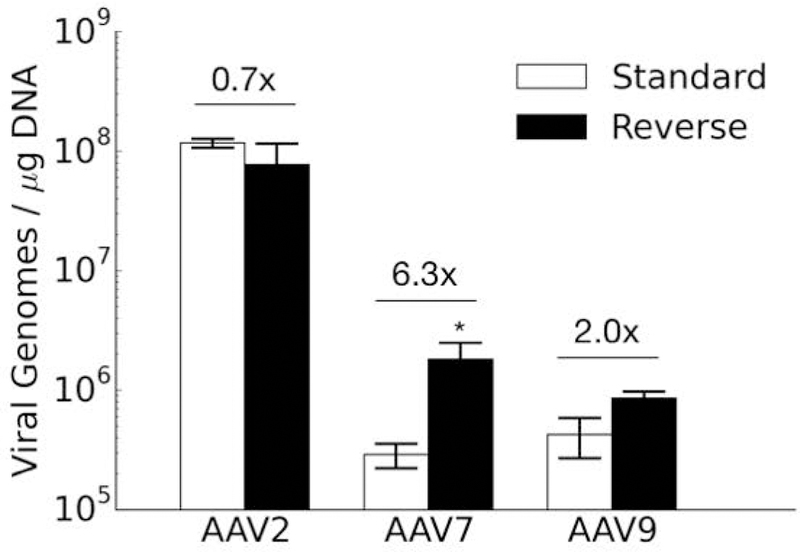
Internalization of AAV2, AAV7, and AAV9 in standard and reverse transduction formats. Viral genomes in cells detected via qPCR were normalized to the total DNA content for each sample. The fold-differences of reverse divided by standard formats are indicated. Results are from two independent experiments conducted in triplicate. Data is represented as mean ± standard error of the mean (SEM). Asterisk indicates significant difference (p<0.05) between standard and reverse transduction methods.
Capsid protein sequence alignment of the three AAV serotypes in our study reveals they share ~82% homology. The first stage of viral entry depends on AAV vectors binding to cell surface receptors. AAV2 has been shown to primarily attach to heparan sulfate proteoglycans (Summerford & Samulski, 1998) and secondarily to fibroblast growth factor, laminin, and hepatocyte growth factor (Akache et al., 2006; Kashiwakura et al., 2005; Qing et al., 1999). Five amino acids (R484, R487, R585, R588, and K532) on the AAV2 capsid have been identified as the heparan sulfate binding pocket, and they are nestled within the protrusions facing the capsid’s three-fold axes of symmetry (Kern et al., 2003; Opie, Warrington, Agbandje-McKenna, Zolotukhin, & Muzyczka, 2003). The binding receptors of AAV7 remain unidentified. AAV9’s primary receptor is N-terminal galactose (Shen, Bryant, Brown, Randell, & Asokan, 2011), and its co-receptor is laminin (Akache et al., 2006). The galactose binding pocket of AAV9 is comprised of five amino acids (N470, D271, N272, Y446, W503) situated at the base of protrusions surrounding the capsid’s three-fold axes of symmetry (Bell, Gurda, Van Vliet, Agbandje-McKenna, & Wilson, 2012). Moreover, AAVR is a recently identified receptor shown to be necessary for infection by multiple variants, including AAV2 and AAV9 (S. Pillay et al., 2016; Sirika Pillay et al., 2017). It is plausible that AAV vectors may use different routes of cellular entry and subsequent intracellular trafficking depending on whether they are subjected to standard or reverse transduction formats, and follow-up studies are necessary to parse out the differences.
Furthermore, the potential influence of other downstream processes should be considered. After cellular uptake, the vectors escape the vesicular compartment by externalizing a phospholipase domain and translocate into the host nucleus through the use of nuclear localization signals (Girod et al., 2002; Sonntag, Bleker, Leuchs, Fischer, & Kleinschmidt, 2006). Once inside the nucleus, virus un-coating leads to the release of packaged genomes to be processed for gene expression (Sipo et al.). Prior work has also demonstrated that proteasomes may contribute to post-translational processing of AAV vectors (Denby, Nicklin, & Baker, 2005; Yan et al., 2002). Further studies addressing the differences at each of these intracellular transduction steps for standard and reverse transduction formats could help explain the results obtained here.
Conclusion
In this study, we directly compared the transduction outcomes of reverse and standard formats with three AAV serotypes (AAV2, AAV7, AAV9) that have been previously shown in the literature to transduce HeLa cells with varying efficiencies. The substrate-mediated approach leads to significant improvements in transduction efficiency for both AAV7 and AAV9 at various MOIs. However, standard transduction still yields better outcomes for AAV2. Future endeavors should extend these side-by-side comparisons of standard and reverse transduction protocols to other cell lines, especially for combinations of serotypes and difficult-to-transduce cell lines with clinical relevance. Moreover, further studies should focus on unraveling intracellular trafficking mechanisms to explain why the reverse transduction approach improves transduction efficiency for some AAV serotypes. The cellular internalization assay results suggest the level of virus uptake by cells may play a role, but that it is insufficient on its own to explain the transduction outcome. In summary, reverse transduction may offer a facile approach to effectively use AAV serotypes in cells that are normally resistant to transduction in vitro. These findings can be expanded upon for substrate-mediated gene delivery applications in tissue engineering.
Acknowledgments
This work was supported by the National Science Foundation (DMR1611044 to J.S.) and the National Institutes of Health (R01CA207497 and R01HL138126 to J.S., R21AR067527 to A.G.M. and J.J.T.), and a National Science Foundation Graduate Research Fellowship to E.J.L. We acknowledge the University of North Carolina at Chapel Hill Gene Therapy Center Vector Core for providing pXX6–80 and pXX2 and the University of Pennsylvania for providing pAAV2/7 and pAAV2/9. We thank Jeron Chen for his assistance with post-acquisition processing of microscopy images.
References
- Akache B, Grimm D, Pandey K, Yant SR, Xu H, & Kay MA (2006). The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. Journal of virology, 80(19), 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CL, Gurda BL, Van Vliet K, Agbandje-McKenna M, & Wilson JM (2012). Identification of the Galactose Binding Domain of the Adeno-Associated Virus Serotype 9 Capsid. J Virol, 86(13), 7326–7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchlis G, Podsakoff GM, Radu A, Hawk SM, Flake AW, Mingozzi F, & High KA (2012). Factor IX expression in skeletal muscle of a severe hemophilia B patient 10 years after AAV-mediated gene transfer. Blood, 119(13), 3038–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Hair SM, Sexton JT, Landry BP, Olson EJ, Igoshin OA, & Tabor JJ (2016). FlowCal: A user-friendly, open source software tool for automatically converting flow cytometry data from arbitrary to calibrated units. ACS synthetic biology, 5(7), 774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denby L, Nicklin SA, & Baker AH (2005). Adeno-associated virus (AAV)-7 and-8 poorly transduce vascular endothelial cells and are sensitive to proteasomal degradation. Gene therapy, 12(20), 1534. [DOI] [PubMed] [Google Scholar]
- Dong X, Tian W, Wang G, Dong Z, Shen W, Zheng G, … Chen J (2010). Establishment of an AAV reverse infection-based array. PLoS One, 5(10), e13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont KM, Boerckel JD, Stevens HY, Diab T, Kolambkar YM, Takahata M, … Guldberg RE (2012). Synthetic scaffold coating with adeno-associated virus encoding BMP2 to promote endogenous bone repair. Cell and tissue research, 347(3), 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis BL, Hirsch ML, Barker JC, Connelly JP, Steininger RJ, & Porteus MH (2013). A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1–9) and one engineered adeno-associated virus serotype. Virology journal, 10(1), 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod A, Wobus CE, Zádori Z, Ried M, Leike K, Tijssen P, … Hallek M. (2002). The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. Journal of General Virology, 83(5), 973–978. [DOI] [PubMed] [Google Scholar]
- Ito H, Koefoed M, Tiyapatanaputi P, Gromov K, Goater JJ, Carmouche J, … Samulski RJ (2005). Remodeling of cortical bone allografts mediated by adherent rAAV-RANKL and VEGF gene therapy. Nature medicine, 11(3), 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd J, Ho ML, Tiwari A, Gomez EJ, Dempsey C, Van Vliet K, … Suh J. (2014). Tunable protease-activatable virus nanonodes. ACS nano, 8(5), 4740–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwakura Y, Tamayose K, Iwabuchi K, Hirai Y, Shimada T, Matsumoto K, … Daida H (2005). Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. Journal of virology, 79(1), 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasputis T, Farris E, Guerreiro G, Taylor J, & Pannier AK (2017). Substrate-Mediated Gene Delivery THE WORLD SCIENTIFIC ENCYCLOPEDIA OF NANOMEDICINE AND BIOENGINEERING I: Volume 3: Nanotechnology in Gene Delivery (pp. 219–260): World Scientific. [Google Scholar]
- Kern A, Schmidt K, Leder C, Muller OJ, Wobus CE, Bettinger K, … Kleinschmidt JA (2003). Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J Virol, 77(20), 11072–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagache T, Danos O, & Holcman D (2012). Modeling the step of endosomal escape during cell infection by a nonenveloped virus. Biophys J, 102(5), 980–989. 10.1016/j.bpj.2011.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell KI, Gomez EJ, & Suh J (2012). The identity of the cell adhesive protein substrate affects the efficiency of adeno-associated virus reverse transduction. Acta biomaterialia, 8(11), 4073–4079. [DOI] [PubMed] [Google Scholar]
- McConnell KI, Schweller RM, Diehl MR, & Suh J (2011). Live-cell microarray surface coatings supporting reverse transduction by adeno-associates viruses. BioTechniques, 51(4), 255. [DOI] [PubMed] [Google Scholar]
- McConnell KI, Slater JH, Han A, West JL, & Suh J (2011). Microcontact printing for co-patterning cells and viruses for spatially controlled substrate-mediated gene delivery. Soft Matter, 7(10), 4993–5001. [Google Scholar]
- Nasu T, Ito H, Tsutsumi R, Kitaori T, Takemoto M, Schwarz EM, & Nakamura T (2009). Biological activation of bone‐related biomaterials by recombinant adeno‐associated virus vector. Journal of Orthopaedic Research, 27(9), 1162–1168. [DOI] [PubMed] [Google Scholar]
- Olson EJ, Hartsough LA, Landry BP, Shroff R, & Tabor JJ (2014). Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat Methods, 11(4), 449–455. [DOI] [PubMed] [Google Scholar]
- Opie SR, Warrington KH Jr., Agbandje-McKenna M, Zolotukhin S, & Muzyczka N (2003). Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J Virol, 77(12), 6995–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y. a., … Chapman MS (2016). An essential receptor for adeno-associated virus infection. Nature, 530(7588), 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay S, Zou W, Cheng F, Puschnik AS, Meyer NL, Ganaie SS, … Yan Z. (2017). Adeno-associated virus (AAV) serotypes have distinctive interactions with domains of the cellular AAV receptor. Journal of virology, 91(18), e00391–00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing K, Mah C, Hansen J, Zhou S, Dwarki V, & Srivastava A (1999). Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nature medicine, 5(1), 71. [DOI] [PubMed] [Google Scholar]
- Shen S, Bryant KD, Brown SM, Randell SH, & Asokan A (2011). Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. Journal of Biological Chemistry, 286(15), 13532–13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipo I, Fechner H, Pinkert S, Suckau L, Wang X, Weger S, & Poller W (2007). Differential internalization and nuclear uncoating of self-complementary adeno-associated virus pseudotype vectors as determinants of cardiac cell transduction. Gene Ther, 14(18), 1319–1329. [DOI] [PubMed] [Google Scholar]
- Sonntag F, Bleker S, Leuchs B, Fischer R, & Kleinschmidt JA (2006). Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. Journal of virology, 80(22), 11040–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerford C, & Samulski RJ (1998). Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. Journal of virology, 72(2), 1438–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Lin H, Bean A, Tang Y, Tan J, Tuan RS, & Wang B (2017). One-Step Fabrication of Bone Morphogenetic Protein-2 Gene-Activated Porous Poly-L-Lactide Scaffold for Bone Induction. Molecular Therapy-Methods & Clinical Development, 7, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Zak R, Luxton GWG, Ritchie TC, Bantel-Schaal U, & Engelhardt JF (2002). Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. Journal of virology, 76(5), 2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]


