Abstract
Nucleic Acid Aptamers (NAAs) are a class of synthetic DNA or RNA molecules that bind specifically to their target. We recently introduced an aptamer termed R1.2 against membrane Immunoglobulin M (mIgM) expressing B-cell neoplasms using Ligand Guided Selection (LIGS). While LIGS-generated aptamers are highly specific, their lower affinity prevents aptamers from being used for translational applications. Highly specific aptamers with higher affinity can increase targetability, boosting the application of aptamers as diagnostic and therapeutic molecules. Herein, we report that dimerization of R1.2, an aptamer generated from LIGS, leads to high affinity variants without compromising the specificity. Three dimeric aptamer analogues with variable linker lengths were designed to evaluate the effect of linker length in affinity. The optimized dimeric R1.2 against cultured B-cell neoplasms, four donor B-cell samples and mIgM-positive Waldenström’s Macroglobulinemia (WM) showed specificity. Furthermore, confocal imaging of dimeric aptamer and anti-IgM antibody in purified B-cells suggests co-localization. Binding assays against IgM knockout Burkitt’s Lymphoma cells utilizing CRISPR/Cas9 further validated specificity of dimeric R1.2. Collectively, our findings show that LIGS-generated aptamers can be re-engineered into dimeric aptamers with high specificity and affinity, demonstrating wide-range of applicability of LIGS in developing clinically practical diagnostic and therapeutic aptamers.
Keywords: Ligand Guided Selection, Aptamer, Dimerization, B-cell lymphoma, mIgM
Introduction
Hematological diseases are commonly diagnosed on the basis of abnormalities in gene and protein expression, or based on tissue morphology [1-4]. The resulting knowledge collectively leads to improved clinical diagnoses, including revision of existing classifications, and increased data for scientific study. For example, the World Health Organization (WHO) introduced a classification of neoplasms in 2001, which was later updated in 2008 and 2016 with new guidelines, introducing new variants and new entities[2-4]. Despite such new guidelines, misclassification of cases still occurs in high numbers, especially in developing countries[5]. This can be attributed to the mishandling of specimens and lack of technical improvements in clinics[5]. This calls for improved detection platforms for point-of-care testing. In this regard, nucleic acid aptamers (NAAs) are very well suited for designing molecular tools for point-of care detection. NAAs are a class of synthetic DNA or RNA molecules that bind specifically to their target, ranging from small molecules to proteins and whole cells[6, 7]. Since NAAs are chemical in nature, they generally show extended shelf life, as well as compatibility with a variety of solvents and heat, thus allowing a wide range of applications[8, 9]. Aptamers are selected using a simple iterative method called Systematic Evolution of Ligands by Exponential Enrichment (SELEX)[10, 11]. Since the inception of SELEX, a number of variants to the method have been developed, and alternative strategies to design SELEX libraries with enhanced chemical diversity have been introduced[7, 12]. The hallmark of NAAs arises from their target affinity and specificity, which are particularly important when developing diagnostic and therapeutic molecules against cell-surface receptor proteins in their native state. Many groups prominent in the aptamer research field have introduced variants of SELEX to improve the screening of NAAs. Few of these methods include, for instance, the use of whole cells as the sample, whole cells with flow cytometry for biomarker discovery, crossover-SELEX and reverse-crossover SELEX[12]. Indeed, these methods have already produced several successful molecular probes against cell-surface targets, and they have been applied in proof-of-concept studies[12-19]. We recently introduced a novel variant termed Ligand-guided Selection (LIGS)[20, 21]. LIGS is designed to identify highly specific aptamers from an enriched SELEX library against a whole cell. The core principle of LIGS is based on competition whereby aptamers against a predetermined cell-surface receptor compete against a secondary ligand for the same receptor to select aptamers with the greatest specificity for that target[7]. Using this method, we introduced an aptamer (R1) against membrane IgM (mIgM) expressed on Burkitt’s lymphoma cells, a form of mature B-cell lymphoma[20]. The post-SELEX truncation of aptamers generates favorable secondary folds while destabilizing the formation of undesirable secondary structures, leading to increase in affinity. Therefore, we later truncated aptamer R1 to generate a shorter second-generation aptamer R1.2 to improve its affinity without compromising its specificity[22]. The target of aptamer R1.2, mIgM, is expressed on the surface of B lymphocytes at multiple stages of their development in the bone marrow[23]. Mature B-cells express mIgM as a part of the B-cell receptor complex (BCR complex), which is involved in regulating B-cell survival, differentiation and development[23].
Based on the diagnostic potential of aptamer R1.2, we herein investigated the impact of dimerization of R1.2 to improve its affinity. Accordingly, we designed three dimeric aptamer analogues of R1.2, and investigated the linker-length connecting the aptamer as a function of affinity. We then analyzed the specificity of all three analogues, against mIgM-expressing cell lines using mIgM negative cultured B- and T- cell lines as controls. We further validated the specificity of dimeric aptamer utilizing mIgM knockout Burkitt’s lymphoma cell line engineered utilizing CRISPER/CAS9 technology. Furthermore, binding analysis of one of the dimeric analogues of R1.2 against mIgM-positive B-cells from healthy donor peripheral blood mononuclear cells (PBMC) showed positive binding towards mIgM positive B-cells, but not towards Ig expressing CD3 positive T-cells, indicating that LIGS-generated aptamers are able to specifically recognize its epitope expressed on primary cells. In addition, confocal imaging of dimeric aptamer and anti-IgM antibody in purified B-cells from PBMCs suggested co-localization, while co-incubation of anti-IgM antibody and dimeric aptamer resulted in rapid internalization into cultured lymphoma cells. Finally, we show that dimeric R1.2 could specifically bind to mIgM-positive WM cells, but not to non-B cells. Collectively, these findings demonstrate that dimerization did not alter specificity of LIGS-generated aptamer R1.2 while increasing dimeric aptamer’s affinity towards cultured cells and primary cells that express the same epitope, and that these aptamers can be re-engineered into molecular probes with high functional affinity without compromising specificity.
Methods
Materials and detailed protocols of formulations of buffers are available in supporting file.
Determination of binding affinity at 4 °C
Binding affinity of each dimeric R1.2 molecule towards target cells was evaluated by using BJAB cells. 1 μM working solution of each dimeric aptamer sequence was prepared by diluting the respective 10 μM solution using the binding buffer containing 0.2 M KCl. Following 10 min of heating at 95°C and folding on ice for 45 min, a range of aptamer concentrations was prepared by serial dilution. About 1.0 × 105 BJAB cells were suspended in 75 μL of cell suspension buffer and incubated with 75 μL of aptamer solution of different concentrations for 1 hr on ice, making the final volume 150 μL. At the end of 1 hr incubation time, 2 mL of wash buffer at 4°C were added for one wash, and the cells were then reconstituted in 250 μL of wash buffer. Aptamer binding towards BJAB cells was analyzed with a FACSCalibur Flow Cytometer (Cytek Biosciences) by counting 5000 events for each concentration. The equilibrium dissociation constant (Kd) of aptamer-cell interaction was obtained by plotting the difference in median fluorescence intensity against concentration. Specific binding was calculated using GraphPad Prism 5 (La Jolla, CA, USA). Binding affinity for dimeric R1.2 molecules at room temperature (RT) was tested using conditions similar to those at 4°C, except 1.5×105 BJAB cells were used, and incubation was performed in a 25°C incubator.
CRISPR/cas9 Experimental Design
sgRNA Plasmid Construction
The sgRNA was cloned into a 3rd generation lentiCRISPRv2 (Addgene Plasmid #52961) plasmid containing BsmBI sites downstream from the human U6 promoter with a cas9 expression vector[24]. Both forward and reverse sgRNA oligonucleotides were annealed without phosphorylation at a concentration of 100 μM in 10 μL water. The mixture was incubated in a thermocycler for 5 min at 95°C. Then, the heat was gradually reduced until the oligonucleotides reached RT (25°C) at 5°C/minute. The annealed sgRNA oligos were inserted into the lentiSRISPRv2 following golden gate CRISPR assembly as previously described [25, 26]. In a 20 μL reaction, 1μg of lentiCRISPRv2 was combined with 1 μL BsmBI (ThermoScientific ER0451), 2 μL 10X FastDigest buffer, 1 μL annealed oligos, 1.5 μL T4 DNA ligase buffer (NEB M0202S), and 2 μL 10X T4 ligase buffer. The ligation mixture was then incubated in a thermocycler at 37°C for 12–15 hours. For transformation of the plasmid, 2 μL of the ligation mixture was added into 50 μL of sub cloning efficiency stabl3 cells (Invitrogen C737303) using appropriate protocol described by Invitrogen. Then, 75 μL of the transformation was spread on a pre-warmed ampicillin selected plates (100 μg/mL ampicillin) and incubated overnight at 37°C. The colonies from the plate were grown in LB media, and the DNA plasmid were extracted with QIAprep Spin Miniprep Kit at high concentration. The plasmid constructs were then sequence verified using EtonBioscience sequencing service with the U6 promoter (LKO.1 5’) as the primer.
Lenti-viral Particle Productions
HEK-293T (ATCC CRL-3216) cells were used for packaging of plasmids and production of viral particles. The cells were maintained in DMEM media (ATCC 30–2002) supplemented with 10% fetal bovine serum (FBS), and 100 units/mL penicillin-streptomycin (P/S). In addition to the lentiSRISPRv2 plasmids containing the sgRNA insert and cas9 expression vector, two other packaging plasmids were used as envelope expressing plasmid; pMD2.G (Addgene Plasmid #12259) and psPAX2 (Addgene Plasmid #12260). To generate lentiviral particle 1.5 millions HEK-293T cells were plated in tissue culture treated T-25 flask the night before and incubated in 5% CO2 tissue culture incubator at 37°C. The following day, cells were transfected using lipofectamine 2000 (Invitrogen 11668027) transfection method. At first, 5 μg of lentiCRISPRv2, and 1.25 μg of the packaging plasmids were mixed in a tube together with 0.5 mL of Opti-MEM I serum medium (Gibco 31985070) incubated for 5 min. In a separate tube, 10 μL of lipofectamine 2000 was mixed with 0.5 mL of Opti-MEM I serum medium and the plasmid mixture from the first tube was added, then, the mixture was incubated together for ~20 min at room temperature. The media of HEK293T cells were removed and replaced with 2 mL of Opti-MEM I serum medium. Then, the cells were transfected with the lipofectamine mixture and combined very gently. About 8–10 hours post-transfection, the lipofectamine media was aspirated from the cells and replaced with complete DMEM media. The viral supernatant was collected twice at 24 hours and 48 hours post-transfection, clarified at 3000 RPM for 10 min, passed through a 0.45 μm filter, pooled and used either fresh or snap frozen in small aliquots for later use.
Delivery of CRISPR/cas9 into target cells
The CRISPR/cas9 system can be easily delivered to human cells by transfection with a plasmid that encodes the cas9 protein and sgRNA, however cell lines can vary widely in their transfection efficiency[27]. Therefore, in order to increase the possibilities of knockout we used SKLY-16 Burkitt’s lymphoma cell line, which was a generous gift from David Scheinberg lab at the Memorial Sloan Kettering Cancer Center. Prior to transduction with viral particle, SKLY-16 cells were assessed for the expression of appropriate IgM markers to verify the cell line authenticity for the knockout. The cells were maintained in RPMI 1640 medium supplemented with 10% dialyzed fetal bovine serum (FBS), and 100 units/mL penicillin-streptomycin (P/S). For transduction, we utilized spinoculation method by adding polybrene (Santa Cruz Biotechnology NC9840454) to enhance the efficiency of the retroviral infection to the mammalian cells[28, 29]. Cells were cultured to high confluency prior to the infection and maintained in appropriate culture media without P/S to increase the puromycin selection after the knockout. We utilized three infection conditions with different viral particle volume as followed, 0 μL, 500 μL (IgHM), 750 μL (IgHM). For transduction, 1.0×106 cells were placed in 15 mL sterile conical tubes with a total volume of 5 mL and a final polybrene concentration of 10 μg/mL. Then, appropriate volume of viral particles was added to each tube and incubated at RT under a tissue culture hood for ~60 min. For spinoculation, the cells in conical tube were spun at 800g/2000RPM in a pre-warmed 32°C centrifuge for 30 min. The virus containing media was aspirated from the cells and re-suspended in 3 mL of fresh media, placed in 6 well plates, and incubated in 5% CO2 at 37°C in tissue culture incubator.
Puromycin Selection and Cell Sorting
After post-transfection, the cells were selected with puromycin (MP Biomedicals 0219453925). Initially, after 48 hours of post-transduction, cells were treated with 1.0 μg/mL puromycin and transferred into T25 flask with a total volume of 10 mL RPMI media without P/S. This initial treatment was done to ensure and only select the cells with a lentiCRISPR construct, which contain puromycin resistance were selected. At this stage, the cells were routinely assessed with propidium iodide staining of dead cells with flow cytometry for negative screening. After initial puromycin screening cells with negative IgHM expression and a rate of knockout were transduced with 750 μL of lentiviral particles. Finally, these mIgM (–) populations of SKLY-16 cells were further isolated by FACS (fluorescence-activated cell sorting) analysis.
Specificity Assay with CRISPR-Cas9 knockout SKLY-16 cells at 4°C
Specificity assays were conducted for DR1.2_3S, DR1.2_5S and DR1.2_7S with wild-type and CRISPR-Cas9 knockout SKLY-16 cells. These assays were performed by incubating 75 μL of 1 μM working solution of each dimeric aptamer or random control with 1.0×105 cells in 75 μL of cell suspension buffer on ice for 45 min. The incubation period was followed by washing twice with 1.5 mL wash buffer each time. Cells were reconstituted in 200 μL wash buffer. Binding was analyzed using flow cytometry by counting 5000 events. Percent specific binding was determined using the equation and quantified using GraphPad Prism software. Reagents used for this experiment were kept at 4oC.
Specificity Assay with Cultured Cells at 4°C
Specificity assays were conducted for all three dimeric R1.2 aptamers separately with six different cell lines, including the B-cell lines, BJAB, Ramos, SKLY-16, CA46 and Toledo, and the T-cell line, MOLT-3. These assays were performed by incubating 75 μL of 1 μM working solution of each dimeric aptamer or random control with 1.0 ×105 cells in 75 μL of cell suspension buffer on ice for 1 hour, followed by washing twice with 1.5 mL wash buffer each time. Cells were reconstituted in 250 μL wash buffer. Finally, binding was analyzed using flow cytometry by counting 5000 events for each cell line. Expression of mIgM on all five cell lines was also analyzed by incubating 1.0×105 cells in 75 μL volume using a final concentration of 0.5 μg/mL anti-IgM monoclonal antibody (mAb) (Novus Biologicals), followed by flow cytometric analysis. Percent specific binding was determined using the equation and quantified using GraphPad Prism software. Specificity assays at RT (25°C) were also performed in a manner similar to those at 4 °C, except that incubation was performed in a 25°C incubator in a final volume of 150 μL. Reagents used for this experiment were kept at room temperature.
Specificity Assay with Primary Cells at 25°C
Peripheral blood mononuclear cells (PBMCs) were isolated from the whole blood of 4 different healthy donors using Ficoll-Paque PLUS (GE Healthcare). B-cells were separated from PBMCs by using human CD19 microbeads, according to the manufacturer’s manual (Miltenyi Biotec). Specificity assays were conducted at 25°C in a manner similar to that described above, except that the primary cells were reconstituted in cell suspension buffer containing anti-human CD3-Percp-Cy5.5 and anti-human CD19-PE-Cy7 (BD Pharmingen, 1:100 dilution), in order to differentiate between B-cells and T-cells, respectively, during flow cytometric analysis.Expression of mIgM on the primary B-cells was analyzed by incubating the cells suspended in 75 μL of cell suspension buffer with 2.5 μL of anti-IgM mAb (Novus Biologicals), or isotype control using 1:50 dilution, followed by flow cytometric analysis. Cells were reconstituted in 250 μL wash buffer containing DAPI (4’,6-diamidino-2-phenylindole) (Sigma Aldrich) in 1:3000 dilution for the staining of live cells.
WM bone marrow mononuclear cells were obtained through ficoll gradient centrifugation from bone marrow aspirates of three WM patients with CD20+IgM+kappa+ clonal B-cells. The samples were stained with anti-human CD3-Percp-Cy5.5, anti-human CD19-PE-C, anti-human CD20-APC and anti-human kappa light chain-Alexa700(BD Pharmingen, 1:100 dilution), and the specificity assay was conducted at 25°C in a manner similar to that described above.
Microscopy Imaging
Primary B-cells separated from the extracted PBMCs were obtained from healthy donors’ blood samples as described above. Cells were reconstituted in cell suspension buffer and incubated with 75 μL of 2 μM DR1.2_7S or random control and 2.5 uL of 1:100 dilution of anti-IgM mAb (Novus Biologicals) for 45 mins at RT. The cells were washed with 2 mL of wash buffer, followed by reconstitution in 50 μL of wash buffer containing Hoechst 33342 Fluorescent Stain (10 mg/mL) using a 1:3000 dilution for live cell fluorescent staining of DNA and nuclei. The solution was then transferred to a glass slide and covered with a cover slip for imaging using an upright confocal microscope.
Results
Dimerization of aptamer R1.2
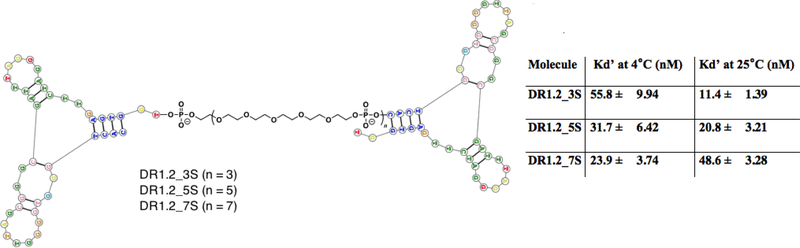
The original aptamer R1 was 79 bases in length but showed low affinity of 315 ± 44 nM [20]. Later, we introduced a truncated second-generation 42-mer aptamer R1.2 with improved affinity constants of 35.5 ± 8.94 nM at 4°C [22]. While significant improvement to the affinity was achieved in the truncated version, we aimed to further improve the affinity by systematic linear dimerization of R1.2, as described before[30]. Accordingly, dimeric aptamers were engineered by linking two R1.2 aptamers with spacer molecules comprised of Poly Ethylene Glycol (PEG) units. It has been previously shown that dimerization of an aptamer can effectively enhance its affinity [30]. Investigation of the length of linker connecting the two aptamers was done by designing three different versions of dimeric aptamers utilizing 3, 5 and 7 spacer molecules (Fig. 1). Following solid-state synthesis, each dimeric construct was analyzed for affinity. Affinity at 4°C significantly improved for all dimeric versions, despite the length of the linker (Fig. 1 and supplementary figure 1). We previously reported that R1.2 binds to circulating soluble IgM, which prevents using R1.2 as a therapeutic molecule. Therefore, we are currently focusing on evaluating diagnostic applicability of aptamer R1.2 scaffolds. Thus, affinities of dimeric scaffold were evaluated only at 4°C and 25°C. Interestingly, all three dimeric aptamer constructs showed significantly improved affinity at 25°C (Fig. 1 and Supplementary Fig. 2).
Figure 1.
Design of dimeric assemblies of aptamer R1.2 and their affinity. Dimeric aptamer R1.2 linked by spacer molecule comprised of Polyethylene glycol (PEG). Three analogues were designed with 3, 5 or 7 spacer molecules. All constructs were synthesized using standard solid-state phosphoramid ite chemistry, and affinity was evaluated against mIgM positive BJAB cells (a Burkitt’s Lymphoma cell line). Monomeric R1.2 showed an affinity of 35.5 ± 8.94 nM at 4°C.
Evaluation of Specificity
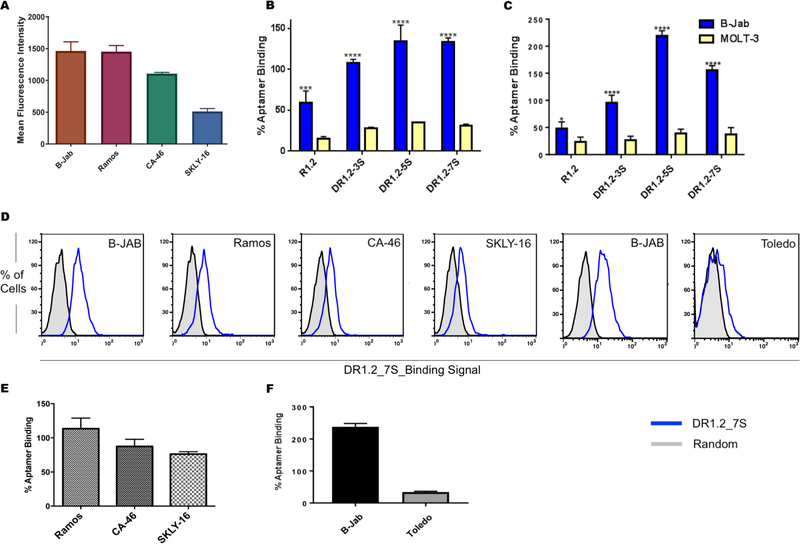
The specificity of all three dimeric constructs was evaluated against cultured cell lines known to express membrane bound Immunoglobulin M (mIgM). One cell line that highly expresses mIgM is BJAB, a Burkitt’s lymphoma cell line, which was used as a positive control (Fig. 2A). A T-cell lymphoma cell MOLT-3, negative for mIgM, was utilized as a negative control. Specificity was first investigated at 4°C (Fig. 2B and Supplementary Fig. 3) and then at 25°C (Fig. 2C, and Supplementary Fig. 3). Substantial higher dimeric aptamer binding was observed compared to monomeric aptamer R1.2 (Fig. 2B and C), suggesting the enhancement of affinity without compromising the specificity of dimeric aptamer constructs. This observation demonstrates that dimerization is an effective strategy to improve aptamer binding to its epitope. In order to investigate whether improved affinity also leads to improved signal, three more cell lines, Ramos, CA-46 and SKLY-16, which express low level of surface IgM, were compared and used for binding analysis. DR1.2_7S could specifically recognize these two cell lines with higher signal-to-noise ratio, suggesting that improvements to affinity do, indeed, lead to higher signal (Fig. 2D and 2E). Furthermore, DR1.2_7S did not bind to the diffuse large B-cell lymphoma cell line Toledo, which lacks chromosomal translocation that expresses mIgM, further demonstrating the specificity of the dimeric R1.2 aptamer construct (Fig. 2F). We previously reported that the original aptamer R1 and its truncated version R1.2 did not show cross-reactivity with Ig-α and -β positive cells Jurkat.E6 cells. As expected the dimeric variants show no binding to Jurkat.E6 cells further suggesting that dimerization did not alter specificity (Supplementary Fig. 4).
Figure 2.
Anti-IgM antibody binding (A) and conclusion from three independent specificity analyses of dimeric aptamers towards BJAB and MOLT3 cells at 4°C (B) and at 25°C (C). Binding of DR1.2_7S against Ramos, CA-46, SKLY-16, BJAB and Toledo cells (D). Conclusion from three independent analyses of specificity of DR1.2_7S with Ramos, CA46, SKLY16 (E). Overall conclusion from three independent specificity analyses against mIgM negative Toledo cells (F). ( 2way ANOVA test **** : P ≤ 0.0001).
CRISPR/CAS9 knockout of mIgM and Specificity Analysis
Designing of sgRNA
The target gene of deletion in this study was human immunoglobulin gene (IgM). We designed single guided RNA (sgRNA) to edit immunoglobulin heavy constant mu (IgHM)[31]. The 20 nucleotide sgRNAs with a preceding 5’-NGG protospacer adjacent motif (PAM) region was manually designed using available online design tools, chopchop and synthego[32, 33]. The target site for the sgRNA selection on the specific gene was designed as previously described by Zhang et. al [34]. After designing the sgRNA, the off target sequence effects were confirmed by using NCBI blast [35]. The IgHM 20 nt genomic target sgRNA backbone was designed with sticky overhang in the forward sequence. Additionally, the complementary annealing oligos for the sgRNA reverse were also designed as:
IgHM sgRNA-fwd 5’-TCAGGCCCCTGTGATCCACG-3’ sgRNA-rev 5’-CGTGGATCACAGGGGCCTGA-3’
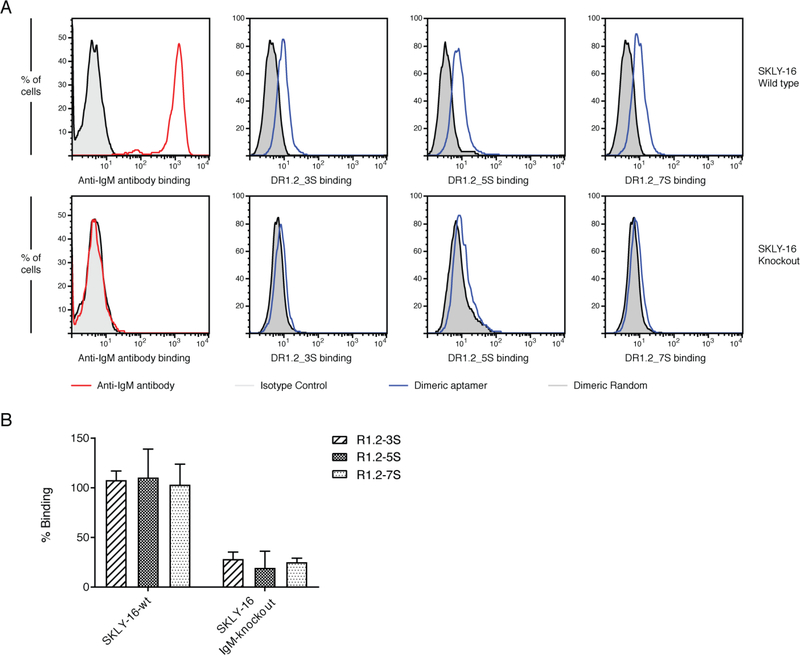
Following sgRNA plasmid construction and lenti-viral packaging as described in methods, SKLY-16 cells were transfected and selected via puromycin and FACS cells sorting (Supplementary Fig. 5). Flow cytometric analysis utilizing anti-IgM antibody of expression of mIgM on the CRISPR treated and FACS sorted cells showed that anti-IgM no longer binds to knockout cells indicating successful knockout of the target gene (Fig. 3A panel 1). We then analyzed binding of all three dimeric constructs with wild type SKLY-16 and mIgM knockout SKLY-16 cells to validate specificity. As shown in figure 3 A-B, mIgM negative SKLY-16 cells do not bind to the three dimeric R1.2, further validating specificity of R1.2 constructs.
Figure 3.
Analysis of specificity of dimeric R1.2 constructs against mIgM negative SKLY16 knockout cells. Evaluation of negative mIgM in CRISPR/CAS9 SKLY-16 knockout cells against SKLY16 wild type and analysis of binding of dimeric aptamer constructs to SKLY-16 knockout and wild-type (A). Overall conclusion from three independent binding studies against knockout SKLY-16 cells (B).
Evaluation of dimeric aptamer specificity against B-cells from healthy donors and WM samples
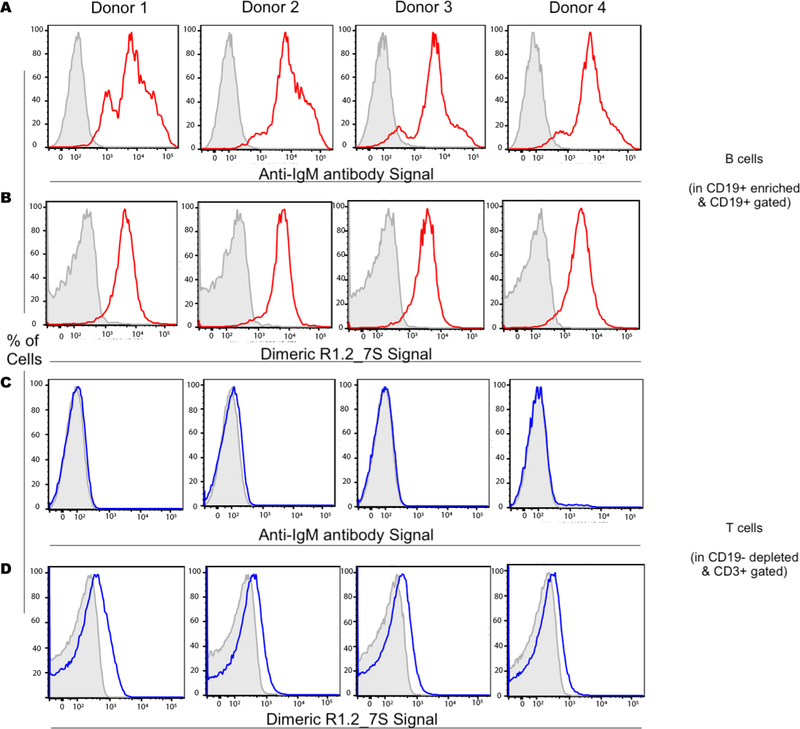
Next, the specific binding of dimeric aptamer DR1.2_7S was analyzed against B-cells from healthy donor mononuclear cells. B-cells were magnetically enriched (MACS sorting) using CD19 beads. The positively selected, B cell enriched fraction of the sample was used for the analysis of IgM staining in B-cells (gated on CD19+ cells), while the B-cells depleted flow through was used for the analysis of IgM staining in T cells (gated on CD3+ cells). As expected CD19+ B-cells showed mIgM positivity through anti-IgM antibody staining. In a similar fashion, CD19+ B-cells were positive for DR1.2_7S binding (Fig. 4A and B). Furthermore, CD3+ T-cells (in CD19 depleted samples) tested negative for both dimeric aptamer and anti-IgM antibody (Fig. 4C and D). As observed in flow cytometric analyses in Fig. 4B and 4D, T-cells, which are known to express Immunoglobulin on their surface, show no binding to dimeric aptamer R1.2, suggesting that dimerization of R1.2 does not alter its specificity towards mIgM expressed on B-cells. These collective observations suggest that aptamers generated from LIGS can successfully recognize membrane markers expressed in both cultured and primary B-cells.
Figure 4.
Analysis of DR1.2_7S binding to mIgM-positive B-cells against mIgM-negative Tcells from peripheral blood mononuclear cells (PBMCs). Anti-IgM antibody was used as a positive control. Gray, tinted histograms depict an isotype control for anti-IgM antibody stained samples or a random sequence control for anti-IgM aptamer control stained samples. Colored lines represent staining with either binding of anti-IgM antibody or binding of DR1.2_7S.
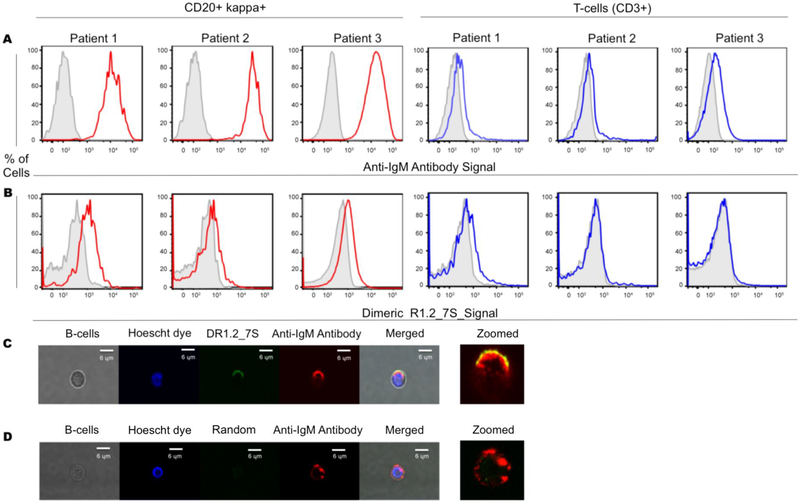
Next, we analyzed the aptamer construct DR1.2_7S binding to B-cells and T-cells from WM samples. All three WM samples showed specific binding with DR1.2_7S. However, we did not observe significant signal/noise ratio for WM cells suggesting that epitope of the DR1.2_7S could have been altered leading to lower affinity towards the epitope of the aptamer and resulting in lower signal/noise ratio (Fig. 5A and B). Importantly, we did not observe any non-specific DR1.2_7S binding to CD3+ T-cells, which known to express Ig-α and Ig-β on their cell surface indicating that while positive signal is lower for WM cells compared to purified B-cells from PBMCs, the binding observed for B-cell population of WM cells is specific towards mIgM expressed on WM cells.
Figure 5.
Specificity of DR1.2_7S towards B-cells gated from WM samples. Anti-IgM and DR1.2_7S binding to B-cells and T-cells (A and B). Confocal microscopic analysis of DR1.2_7S and anti-IgM binding to purified B-cells from PBMCs (C) and dimeric random sequence and anti-IgM binding to purified B-cells from PBMCs (D).
Confocal Microscopy Imaging
Confocal microscopic analysis of dimeric aptamer DR1.2_7S and anti-IgM antibody on the cellular membrane of B-cells isolated from PBMCs suggests co-localization of dimeric aptamer and anti-IgM antibody on B-cells (Fig. 5C and D). Interestingly, confocal fluorescence microscopic analysis of DR1.2_7S with anti-IgM antibody against Burkitt’s lymphoma cell line BJAB gave evidence of internalization and co-localization of the dimeric aptamer and anti-IgM antibody in the same intracellular compartment (Supplementary Fig. 6). Further investigation of internalization kinetics of dimeric analogues of R1.2 in cultured lymphoma cells is currently underway.
Discussion
Multivalency plays a ubiquitous role in cell-surface receptor interaction with ligands. One of nature’s most sophisticated designs is that of multivalent analogues of antibodies with dimeric, tetrameric and decameric assemblies[36]. Such molecular architecture contributes to the exquisite specificity and functional affinity, or avidity, of these assemblies, leading to a decrease in dissociation rate from the cell surface[36]. While antibodies have been successful in a variety of applications, synthetic analogues able to mimic antibody activities are highly desirable. In this regard, aptamers, unlike small-molecule drugs, show great potential owing to their flexibility towards multimerization. A number of approaches to design multimeric aptamer scaffolds have been introduced. For example, a dimeric thrombin aptamer was designed to enhance inhibitory activity[37], dimeric aptamer complexes were designed using anti-PSMA aptamer with rigid linkers composed of nucleic acids, and multimeric aptamers were designed against heat-shock factor to enhance functional affinity[38, 39]. Thus, multivalency is a widely accepted molecular engineering concept in generating high-affinity ligands utilizing low- affinity ones. Here we took advantage of linear assembly of aptamers utilizing polyethylene glycol phosphoramidite on a DNA synthesizer to generate dimeric R1.2 analogues. Interestingly, at 25°C, the affinity against BJAB cells as a function of linker length is not statistically distinct. However, at 4°C, affinities appear to show a correlation with the linker lengths of the dimeric design. It is often challenging to define binding events of a dimeric aptamer construct due to variable receptor density on cell surfaces. In addition, at higher temperatures, it has been reported that lateral angles of mIgM receptor can change due to its mobility, which can contribute to variable binding patterns.[30] Thus, the mobility of mIgM receptors as a function of temperature can challenge exact predictability of binding pattern of dimeric aptamers at higher temperatures.
Recently, our group introduced a variant of complex target SELEX, termed LIGS[20]. LIGS enables the discovery of specific aptameric ligands enriched in a SELEX library against a predetermined receptor protein utilizing competitive elution. Aptamer interaction is specific towards one target; in particular, aptamers generated through LIGS technology, are expected to possess high specificity towards one cell surface receptor. Accordingly, we used LIGS to select an aptamer, termed R1, against mIgM expressed in Burkitt’s lymphoma cell lines. One of the goals in this report is to demonstrate the application of molecular engineering to design dimeric aptamers to enhance affinity without compromising specificity. Here, we demonstrate in this report the re-engineering of R1.2 into dimeric analogues using variable linkers to facilitate specific recognition of mIgM expressed in B-cells. Typically B- and T- cells mediate immune responses via recognizing antigens mediated by immunoglobulins expressed on their membranes. Typically, T-cells express immunoglobulin α and β chains, which are comprised of T cell receptor.[40] Lack of binding of dimeric R1.2 with cultured T-cells and the primary Tcells obtained from PBMCs suggest that dimeric R1.2 can distinguish IgM expressed on B-cell membrane from other Igs expressed on non-B cells. Moreover, the specificity of dimeric aptamers R1.2 constructs is further confirmed by knocking out mIgM expression in Burkitt’s lymphoma cell line SKLY-16.
Currently, all hematological diseases are diagnosed by examining their CD expression profiles utilizing anti-CD antibodies[1, 2, 4, 5, 41]. Recently, differential expression of mIgM has been used to describe and distinguish early and late onset of pediatric Burkitt’s lymphoma[42]. Based on their synthetic nature, nucleic acid aptamers, or NAAs, are sufficiently versatile for the development of diagnostic tools. Thus, with improved affinity and specificity, dimeric R1.2 could recognize mIgM expressed on the cell surface of isolated B-cells obtained from PBMCs. These data demonstrate that LIGS-generated aptamers are specific toward both cultured and primary cells expressing the same receptor. It has been shown that aptamers targeting receptor molecules could internalize into cellular compartments. Accordingly, our lab is currently studying the internalization kinetics of dimeric aptamer R1.2 and downstream signaling of BCR crosslinking induced by dimeric aptamer binding in cultured B-cell lymphoma cells.
WM accounts for 1 to 2% of hematologic cancers[41]. Moreover, higher levels of mIgM expression in WM define a subset of patients with high BCR signaling profiles and more aggressive clinical course[43]. Dimeric R1.2 binding to WM cells and observed low signal/noise is intriguing given the high expression levels of mIgM in WM, when assessed by polyclonal anti-IgM antibodies. Given that the BCR complex is often pre-clustered in B cell malignancies, we cannot exclude the possibility that the epitope recognized by DR1.2_7S is masked or high expression levels of mIgM leads to steric crowding, hindering the dimeric aptamer binding to its epitope[44]. Another observation discussed greatly in previous reports is the lower signal output from aptamers compared to antibodies. Typically, antibodies are conjugated to approximately 4–5 fluorophores, which leads to higher signal output from an antibody, while aptamers are conjugated to one fluorophore leading to lower signal output. Enhancement of signal amplification of aptamers is typically addressed by a coupling of enzymatic reactions, incorporating isothermal amplifications and coupling of multi-dye incorporated nanoparticles[45]. However, analysis of clinical specimen calls for simple approaches that could complement existing establish methods. In that regard, we are investigating the ability of appending multi-dye dendrimer constructs to enhance the signal in detecting the mIgM expression in the clinical B cell lymphomas, which will address the observed low signal output by dimeric aptamers.
In conclusion, we have introduced a dimeric re-engineering of aptamer R1.2, which was initially identified using LIGS. We demonstrated that multimerization of R1.2 could further improve the functional affinity of the aptamer, while its specificity remained unchanged. Microscopic analysis utilizing purified B-cells and cultured cells implied co-localization with anti-IgM antibody, and in cultured cells, it appears that the aptamer and the anti-IgM antibody rapidly internalized. Specificity analysis against mIgM-positive B-cells obtained from healthy donors and WM patient samples showed specific recognition, demonstrating that LIGS-generated aptamers could be developed into clinically practical diagnostic tools and therapeutic agents.
Supplementary Material
Highlights.
Dimerization of an aptamer selected by Ligand Guided Selection significantly improves affinity
Dimerization of an aptamer does not alter specificity
LIGS generated aptamer recognize same epitope expressed on primary samples
Acknowledgments
Authors are grateful for funding for this work by NIGMS grant SC1 GM122648–02, Junior Faculty Research Awards in Science and Engineering and NSF CBET award 1605904. Authors acknowledge Lia Palomba MD, Medical Oncologist, Lymphoma Service, MSKCC for generous assistance in providing healthy donor samples and WM samples.
Footnotes
Disclosures/Conflict of Interest
Authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Borowitz MJ, Bousvaros A, Brynes RK, Cousar JB, Crissman JD, Whitcomb CC, Kerns BJ, Byrne GE Jr., Monoclonal antibody phenotyping of B-cell non-Hodgkin’s lymphomas. The Southeastern Cancer Study Group experience, Am J Pathol, 121 (1985) 514–521. [PMC free article] [PubMed] [Google Scholar]
- [2].Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES, The 2016 revision of the World Health Organization classification of lymphoid neoplasms, Blood, 127 (2016) 2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Harris NL, Jaffe ES, Stein H, Banks PM, Chan JK, Cleary ML, Delsol G, De Wolf-Peeters C, Falini B, Gatter KC, et al. , A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group, Blood, 84 (1994) 1361–1392. [PubMed] [Google Scholar]
- [4].Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES, The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications, Blood, 117 (2011) 5019–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Perry AM, Diebold J, Nathwani BN, MacLennan KA, Muller-Hermelink HK, Bast M, Boilesen E, Armitage JO, Weisenburger DD, Non-Hodgkin lymphoma in the developing world: review of 4539 cases from the International Non-Hodgkin Lymphoma Classification Project, Haematologica, 101 (2016) 1244–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Batool S, Bhandari S, George S, Okeoma P, Van N, Zumrut HE, Mallikaratchy P, Engineered Aptamers to Probe Molecular Interactions on the Cell Surface, Biomedicines, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mallikaratchy P, Evolution of Complex Target SELEX to Identify Aptamers against Mammalian Cell-Surface Antigens, Molecules, 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Keefe AD, Pai S, Ellington A, Aptamers as therapeutics, Nat Rev Drug Discov, 9 (2010) 537–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].White RR, Sullenger BA, Rusconi CP, Developing aptamers into therapeutics, Journal of Clinical Investigation, 106 (2000) 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ellington AD, Szostak JW, In vitro selection of RNA molecules that bind specific ligands, Nature, 346 (1990) 818–822. [DOI] [PubMed] [Google Scholar]
- [11].Tuerk C, Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 249 (1990) 505–510. [DOI] [PubMed] [Google Scholar]
- [12].Takahashi M, Aptamers targeting cell surface proteins, Biochimie, 145 (2018) 63–72. [DOI] [PubMed] [Google Scholar]
- [13].Boltz A, Piater B, Toleikis L, Guenther R, Kolmar H, Hock B, Bi-specific aptamers mediating tumor cell lysis, J Biol Chem, 286 (2011) 21896–21905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chu TC, Twu KY, Ellington AD, Levy M, Aptamer mediated siRNA delivery, Nucleic Acids Res, 34 (2006) e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pastor F, Soldevilla MM, Villanueva H, Kolonias D, Inoges S, de Cerio AL, Kandzia R, Klimyuk V, Gleba Y, Gilboa E, Bendandi M, CD28 aptamers as powerful immune response modulators, Mol Ther Nucleic Acids, 2 (2013) e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Renders M, Miller E, Lam CH, Perrin DM, Whole cell-SELEX of aptamers with a tyrosine-like side chain against live bacteria, Org Biomol Chem, 15 (2017) 1980–1989. [DOI] [PubMed] [Google Scholar]
- [17].Wilner SE, Wengerter B, Maier K, de M Lourdes Borba Magalhaes, D.S. Del Amo, S. Pai, F. Opazo, S.O. Rizzoli, A. Yan, M. Levy, An RNA alternative to human transferrin: a new tool for targeting human cells, Mol Ther Nucleic Acids, 1 (2012) e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhao W, Schafer S, Choi J, Yamanaka YJ, Lombardi ML, Bose S, Carlson AL, Phillips JA, Teo W, Droujinine IA, Cui CH, Jain RK, Lammerding J, Love JC, Lin CP, Sarkar D, Karnik R, Karp JM, Cell-surface sensors for real-time probing of cellular environments, Nat Nanotechnol, 6 (2011) 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou J, Satheesan S, Li H, Weinberg MS, Morris KV, Burnett JC, Rossi JJ, Cell-specific RNA aptamer against human CCR5 specifically targets HIV-1 susceptible cells and inhibits HIV-1 infectivity, Chem Biol, 22 (2015) 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zumrut HE, Ara MN, Fraile M, Maio G, Mallikaratchy P, Ligand-Guided Selection of Target-Specific Aptamers: A Screening Technology for Identifying Specific Aptamers Against Cell-Surface Proteins, Nucleic Acid Ther, 26 (2016) 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zumrut HE, Ara MN, Maio GE, Van NA, Batool S, Mallikaratchy PR, Ligand-guided selection of aptamers against T-cell Receptor-cluster of differentiation 3 (TCR-CD3) expressed on Jurkat.E6 cells, Anal Biochem, 512 (2016) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zumrut HE, Batool S, Van N, George S, Bhandari S, Mallikaratchy P, Structural optimization of an aptamer generated from Ligand-Guided Selection (LIGS) resulted in high affinity variant toward mIgM expressed on Burkitt’s lymphoma cell lines, Biochim Biophys Acta, 1861 (2017) 1825–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Geisberger R, Lamers M, Achatz G, The riddle of the dual expression of IgM and IgD, Immunology, 118 (2006) 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening, Nat Methods, 11 (2014) 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Trevino AE, Zhang F, Genome editing using Cas9 nickases, Methods Enzymol, 546 (2014) 161–174. [DOI] [PubMed] [Google Scholar]
- [26].Vad-Nielsen J, Lin L, Bolund L, Nielsen AL, Luo Y, Golden Gate Assembly of CRISPR gRNA expression array for simultaneously targeting multiple genes, Cell Mol Life Sci, 73 (2016) 4315–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, Multiplex genome engineering using CRISPR/Cas systems, Science, 339 (2013) 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, Karn J, Hu W, Khalili K, Elimination of HIV-1 Genomes from Human T-lymphoid Cells by CRISPR/Cas9 Gene Editing, Sci Rep, 6 (2016) 22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang T, Lander ES, Sabatini DM, Viral Packaging and Cell Culture for CRISPR-Based Screens, Cold Spring Harb Protoc, 2016 (2016) pdb prot090811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ, Scheinberg DA, A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia, Nucleic Acids Res, 39 (2011) 2458–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cheong TC, Compagno M, Chiarle R, Editing of mouse and human immunoglobulin genes by CRISPR-Cas9 system, Nat Commun, 7 (2016) 10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E, CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering, Nucleic Acids Res, 44 (2016) W272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E, CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing, Nucleic Acids Res, 42 (2014) W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, Genome engineering using the CRISPR-Cas9 system, Nat Protoc, 8 (2013) 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool, J Mol Biol, 215 (1990) 403–410. [DOI] [PubMed] [Google Scholar]
- [36].Rudnick SI, Adams GP, Affinity and avidity in antibody-based tumor targeting, Cancer Biother Radiopharm, 24 (2009) 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Muller J, Wulffen B, Potzsch B, Mayer G, Multidomain targeting generates a high-affinity thrombin-inhibiting bivalent aptamer, Chembiochem, 8 (2007) 2223–2226. [DOI] [PubMed] [Google Scholar]
- [38].Boyacioglu O, Stuart CH, Kulik G, Gmeiner WH, Dimeric DNA Aptamer Complexes for High-capacity-targeted Drug Delivery Using pH-sensitive Covalent Linkages, Mol Ther Nucleic Acids, 2 (2013) e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhao X, Lis JT, Shi H, A systematic study of the features critical for designing a high avidity multivalent aptamer, Nucleic Acid Ther, 23 (2013) 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hood L, Kronenberg M, Hunkapiller T, T cell antigen receptors and the immunoglobulin supergene family, Cell, 40 (1985) 225–229. [DOI] [PubMed] [Google Scholar]
- [41].Mazzucchelli M, Frustaci AM, Deodato M, Cairoli R, Tedeschi A, Waldenstrom’s Macroglobulinemia: An Update, Mediterr J Hematol Infect Dis, 10 (2018) e2018004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Eason AB, Sin SH, Lin C, Damania B, Park S, Fedoriw Y, Bacchi CE, Dittmer DP, Differential IgM expression distinguishes two types of pediatric Burkitt lymphoma in mouse and human, Oncotarget, 7 (2016) 63504–63513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Argyropoulos KV, Vogel R, Ziegler C, Altan-Bonnet G, Velardi E, Calafiore M, Dogan A, Arcila M, Patel M, Knapp K, Mallek C, Hunter ZR, Treon SP, van den Brink MR, Palomba ML, Clonal B cells in Waldenstrom’s macroglobulinemia exhibit functional features of chronic active B-cell receptor signaling, Leukemia, 30 (2016) 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, Xu W, Shaffer AL, Wright G, Xiao W, Powell J, Jiang JK, Thomas CJ, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Johnson NA, Rimsza LM, Campo E, Jaffe ES, Wilson WH, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pierce SK, Staudt LM, Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma, Nature, 463 (2010) 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Li F, Zhang H, Wang Z, Newbigging AM, Reid MS, Li XF, Le XC, Aptamers facilitating amplified detection of biomolecules, Anal Chem, 87 (2015) 274–292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.