Abstract
Inflammation is highly associated with colon carcinogenesis. Epigenetic mechanisms could play an important role in the initiation and progression of colon cancer. Curcumin, a dietary phytochemical, shows promising effects in suppressing colitis-associated colon cancer in azoxymethane-dextran sulfate sodium (AOM-DSS) mice. However, the potential epigenetic mechanisms of curcumin in colon cancer remain unknown. In this study, the anticancer effect of curcumin in suppressing colon cancer in an 18-week AOM-DSS colon cancer mouse model was confirmed. We identified lists of differentially expressed and differentially methylated genes in pairwise comparisons and several pathways involved in the potential anticancer effect of curcumin. These pathways include LPS/IL-1-mediated inhibition of RXR function, Nrf2-mediated oxidative stress response, production of NO and ROS in macrophages and IL-6 signaling. Among these genes, Tnf stood out with decreased DNA CpG methylation of Tnf in the AOM-DSS group and reversal of the AOM-DSS induced Tnf demethylation by curcumin. These observations in Tnf methylation correlated with increased and decreased Tnf expression in RNA-seq. The functional role of DNA methylation of Tnf was further confirmed by in vitro luciferase transcriptional activity assay. In addition, the DNA methylation level in a group of inflammatory genes was decreased in the AOM+DSS group but restored by curcumin and was validated by pyrosequencing. This study shows for the first time epigenomic changes in DNA CpG methylation in the inflammatory response from colitis-associated colon cancer and the reversal of their CpG methylation changes by curcumin. Future clinical epigenetic studies with curcumin in inflammation-associated colon cancer would be warranted.
Introduction
Inflammation has been linked to the pathogenesis of multiple cancers. Colorectal cancer, the third most commonly diagnosed cancer worldwide and one of the most widely studied cancers, is strongly associated with inflammation. Epidemiology studies have shown that patients with long-standing inflammatory bowel disease, including ulcerative colitis and Crohn’s disease, have a 2- to 3-fold higher risk of developing colorectal cancer than patients without ulcerative colitis or Crohn’s disease (1). Azoxymethane (AOM) is a classic carcinogen that induces aberrant crypt foci (2), and dextran sulfate sodium (DSS) is an inflammatory agent that damages the epithelial lining of the colon (3). Thus, short-term DSS administration is commonly used to induce acute colitis, and AOM injection followed by DSS insult is a classic animal model for simulating the pathogenesis of colitis-accelerated colon cancer (CAC).
Accumulating evidence has revealed that epigenetic mechanisms, heritable alterations that are not caused by changes to the DNA sequence, play an important role in colon carcinogenesis (4). DNA methylation, the addition of a methyl group to 5-cytosine in the CpG dinucleotide, is a common epigenetic mechanism associated with aberrant gene expression in cancer. Specifically, CpG hypermethylation in promoter regions is believed to play a crucial role in repressing gene expression, perhaps by blocking transcription factor binding (5). Grimm et al. and studies from our group identified aberrant methylation in a well-established Apc(min/+) intestinal tumorigenesis model using methylated DNA immunoprecipitation followed by next-generation sequencing (6,7). Katsurano et al. (8) observed that aberrant DNA methylation accumulates in epithelial cells during DSS-induced inflammation. More recently, Abu-Remaileh et al. (9) used whole-genome bisulfite sequencing to demonstrate that inflammatory signals establish a novel epigenetic landscape that silences gene sets important for cellular transformation in an AOM/DSS-induced CAC model. The early onset, reversible and dynamic nature of epigenetic mechanisms indicate their value as promising cancer chemoprevention targets. It would be interesting to investigate whether inflammation-induced methylome alterations are associated with colon cancer prevention.
Pyrosequencing, methylation-specific PCR, Sanger sequencing of bisulfite-converted DNA and methylated DNA immunoprecipitation PCR have been commonly used to examine the DNA methylation level at specific loci (10,11). However, our effort to understand the epigenetic landscape in diseases has been hindered by a lack of high throughput, quantitative and accurate methods. With the development of next-generation sequencing technologies, there are now multiple methods to characterize a genome-wide DNA methylation profile (12). Agilent SureSelect Methyl-seq is a comprehensive target enrichment system for focusing on regions where methylation is important for gene expression regulation. This system selectively captures cancer-specific, differentially methylated regions (DMRs), GENCODE promoters, CpG islands, CpG shores and shelves, DNase I hypersensitive sites and RefGenes and prepares bisulfite-conversion of samples to detect individual CpG methylation levels using the Illumina platform. This novel target enrichment system has the potential to reveal methylated regions that are undetected by traditional detection systems such as reduced representation bisulfite sequencing and methylated DNA immunoprecipitation sequencing.
Curcumin is the major active component in the golden spice Curcuma longa (also known as turmeric) and has been used as a common food spice for more than a thousand years. Curcumin is well known for its anti-inflammatory and antioxidative properties (13), and many studies have reported that curcumin can play a role in colon cancer prevention (14). One of our recent studies demonstrated that dietary administration of 2% (wt/wt) curcumin effectively suppressed colon tumor growth in AOM- and DSS-induced CAC in C57BL/6 mice. In addition, we obtained a global transcriptional profile from AOM- and DSS-induced tumors with or without curcumin treatment to understand the molecular targets modulated by curcumin in CAC prevention. Because epigenetic modifications are highly associated with dietary factors and occur at lower concentrations, the ability of curcumin to modulate epigenetic changes for colon cancer chemoprevention has received considerable attention. Studies from our laboratory have shown that curcumin decreased the CpG methylation of Nrf2, Neurog1 and DLEC1 and activated their expression in murine prostate cancer TRAMP-C1 cells, human prostate cancer LnCap cells and human colon cancer HT29 cells, respectively (15–17). A genome-wide methylation microarray has been performed to reveal significant DNA methylation in curcumin-treated colon cancer cell lines (18). However, alterations to the methylome upon curcumin treatment in a clinically relevant rodent colon cancer model have not been fully elucidated. Therefore, this study aimed to identify DNA methylation variants in response to AOM- and DSS-induced CAC and their prevention by curcumin administration.
Materials and methods
Chemicals, diets and animals
AOM, hematoxylin and eosin were purchased from Sigma–Aldrich (St. Louis, MO). DSS (molecular weight: 36 000–50 000) was obtained from MP Biomedicals (Solon, OH). Curcumin C3 complex, a kind gift from the Sabinsa Corporation (East Windsor, NJ), was blended into an AIN-93M rodent diet at a final concentration of 2% (wt/wt) by Research Diets (New Brunswick, NJ). The diet was stored at 4°C throughout the animal experimentation period. Four-week-old male C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME) and were maintained in a controlled environment (20–22°C, 12 h light and 12 h dark cycles, and 45–55% relative humidity) at the Rutgers Animal Care Facility. The diet and water were provided ad libitum. All of the animal experiments were performed under the relevant animal protocol (01-016), which was approved by the Institutional Animal Care and Use Committee (IACUC) of Rutgers University.
AOM- and DSS-induced colon tumor model
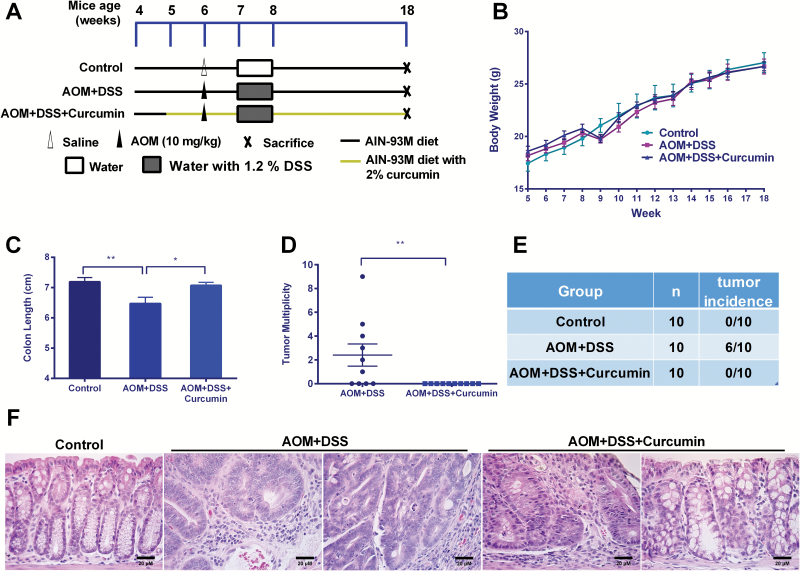
The AOM+DSS model was generated as described previously (19), and the procedure is presented in Figure 1A. Mice were randomly assigned to three experimental groups (n = 10) and were fed a specific (curcumin) diet from 5 weeks of age until the end of the experiment. All of the animals except those in the control group received an AOM injection and DSS-containing water (1.2%) for 1 week. Food and fluid consumption and body weight were recorded weekly. The mice were killed 12 weeks after the AOM injection (at the age of 18 weeks).
Figure 1.
Curcumin suppressed AOM- and DSS-induced colon cancer. (A) Experimental design of the animal study (n = 10). (B) The weekly recording of body weight during the experiment. (C) Curcumin attenuated colon shortening by AOM and DSS. The length differences were significant (One-way ANOVA; *P < 0.05, **P < 0.01). (D and E) The suppression of tumor multiplicity and tumor incidence by curcumin administration (Mann–Whitney; **P < 0.01). (F) Histopathological examination of colon tissues at ×400 magnification. Scale bars represent 20 μM.
Tissue preparation and isolation of colon epithelial cells
At necropsy, the colons of the mice were removed, and the length was measured. All of the colons were flushed with ice-cold saline to remove feces and were opened longitudinally on filter paper. The colons were carefully examined, and the number of tumors was counted. All of the isolated colons were cut into two halves along the main axis. The left portion of the tissue was fixed in 10% buffered formalin for 24 h and gradually dehydrated, embedded in paraffin and stored at 4°C for further histological analyses. The proximal segment and tumors of the right portion of the colon were removed, and then colon epithelial cells were isolated from the remaining tissues using a previously described scraping method (20). Briefly, tissues were scraped using a glass slide on ice, and the mucosal scrapings were resuspended in ice-cold phosphate-buffered saline. The remaining tissue after scraping was snap frozen and stored for further analyses. The epithelial preparations were centrifuged at low speed (150×g) and washed twice with ice-cold phosphate-buffered saline to reduce blood cell contamination. Isolated epithelial cells were pelleted, snap frozen with dry ice and stored at −80°C for further analysis.
Histopathological analysis
The histopathological analysis was performed as described previously (19). Tissue blocks were serially sectioned (4 µm) and mounted on glass slides. The sections were stained with hematoxylin and eosin and were carefully evaluated by a histopathologist. Images of hematoxylin and eosin-stained sections were captured at ×400 total magnification.
Isolation of the nucleic acid and next-generation sequencing
Total RNA and DNA were extracted from colon epithelial cells isolated from each animal using an AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA). The RNA and DNA from the AOM/DSS group colonic tumors were also prepared for further validation. The quality and concentration of extracted RNA and DNA were determined using an Agilent 2100 Bioanalyzer and a NanoDrop spectrophotometer, respectively. Twelve RNA samples (four samples per group) and six DNA samples (two samples per group) were subjected to RNA-seq and SureSelect Methyl-seq. In the AOM+DSS group, only mice with the most tumors were chosen. Specifically, mouse # C33, C34, C36 and C40 with four, nine, three and five tumors, respectively, were used for RNA-seq, and two of them (C34 and C40) were used for Methyl-seq. Library preparation and sequencing were performed by RUCDR Infinite Biologics. The RNA-seq procedures were the same as those described previously (21). The DNA samples were further processed using an Agilent Mouse SureSelect Methyl-seq Target Enrichment System (Agilent Technologies, Santa Clara, CA) and sequenced on an Illumina NextSeq 500 instrument with 76-bp single-end reads, generating 34–47 million reads per sample. For RNA-seq, 75-bp paired-end reads were produced at a rate of 15–36 million reads per sample.
Bioinformatics analyses
SureSelect Methyl-seq
The reads were aligned to the in silico bisulfite-converted mouse genome (mm9) with the Bismark (version 0.15.0) alignment algorithm (22). After alignment, DMRfinder (version 0.1) was used to extract methylation counts and cluster CpG sites into DMRs (23). Each DMR contains at least five CpG sites. Methylation differences >0.10 and with P < 0.05 were considered significant. Genomic annotation was performed with ChIPseeker (version 1.10.3) in R (version 3.4.0) (24).
RNA-seq
Cutadapt (25) was used to remove the Illumina Universal Adapter sequence. The reads were aligned by TopHat (version 2.1.1) to the mouse genome (mm9) (26), and PCR duplicates were removed. Reads overlapping with genomic features were counted by featureCounts (version 1.5.1) (27) and were analyzed for differential expression with DESeq2 (version 1.14.1) in R (version 3.4.0) (28).
Quantitative polymerase chain reaction
The mRNA expression of selected genes was determined using quantitative polymerase chain reaction (qPCR) analysis. First-strand cDNA was synthesized from 500 ng of total RNA using TaqMan® Reverse Transcription reagents (ThermoFisher, Waltham, MA) and was then analyzed by qPCR using a QuantStudio 5 Real-Time PCR System (ThermoFisher) with SYBR Green PCR Master Mix (ThermoFisher). Primers were designed by and purchased from Integrated DNA Technologies (IDT, Coralville, IA), and their sequences are available in Supplementary Table 1, available at Carcinogenesis Online. The mRNA expression was calculated as the fold change with normalization to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the 2−ΔΔCT method.
Bisulfite conversion and pyrosequencing
Pyrosequencing analysis was performed to validate the methylation status of two regions of Tnf observed in SureSelect Methyl-seq. The sequences of primers for amplifying and sequencing these two regions were designed using the PyroMark Assay Design Software 2.0 (Qiagen). Primers were obtained from Integrated DNA Technologies. First, 500 ng of genomic DNA was subjected to bisulfite conversion, as reported previously (15), using EZ DNA Methylation-Gold Kits (Zymo Research Corp., Orange, CA). The bisulfite-treated DNA was amplified by PCR using Platinum PCR Taq DNA polymerase (Invitrogen, Carlsbad, CA) with the forward and reverse primers listed in Supplementary Table 2, available at Carcinogenesis Online. Specifically, the reverse primers were biotinylated at the 3′ end. The PCR product was separated by agarose gel electrophoresis and was visualized by ethidium bromide staining using a Gel Documentation 2000 system (Bio-Rad, Hercules, CA) to ensure pure PCR products. Later, the biotinylated PCR product was captured using streptavidin-coated beads (GE Healthcare, Piscataway, NJ) and PyroMark Q24 Advanced CpG Reagents (Qiagen) and was washed in a vacuum prep workstation (Qiagen). After annealing with the sequencing primer at 80°C for 5 min, the single-stranded PCR product was pyrosequenced on a PyroMark Q24 advanced instrument (Qiagen).
Plasmids, transfection and luciferase reporter assay
The PCR primers used for amplifying region (amplicon) B of Tnf were purchased from IDT. Primer sequences are shown in Supplementary Table 2, available at Carcinogenesis Online. The vector expressing renilla luciferase was purchased from InVivogen (San Diego, CA; Catalog number: pcpgf-promlc). This vector has the human EF-1α promoter upstream of the luciferase gene and was engineered to contain no CpG dinucleotides in the backbone. The vector expressing firefly luciferase was purchased from Promega (pGL4.13, catalog number: E6681). The restriction enzymes HindIII and NcoI, T4 ligase and CpG methyltransferase M.SssI were purchased from Thermofisher. The dual-luciferase reporter assay kit was purchased from Promega (Catalog number: E1910). Mammalian cells were purchased from ATCC. The transfection reagent Lipo LTX was purchased from Life Technologies. Region B of the mouse Tnf gene was amplified by PCR and then was inserted into the pCpGfree-lucia vector at restriction sites HindIII and NcoI. Cytosines at all of the CpG sites were methylated by CpG methyltransferase M.SssI. Three plasmid samples were prepared for the transfection assay: control vector (no insertion), un-methylated or methylated vector with region B of Tnf inserted. Cells were seeded into 12-well plates at a density of 1.0E5 cells; 24 h later, the cells were co-transfected with 500 ng of the above plasmids and 50 ng of the pGL4.13 plasmid. Twenty-four hours after transfection, the cells were lysed, and the luciferase activity assay was performed on a Lucetta Luminometer (Lonza) following the manufacturer’s instructions. The luciferase levels of the pCpGfree plasmids were normalized to that of the pGL4.13 plasmid. The data were presented as the fold change relative to the control group (no insertion).
Statistical analysis
The data are presented as means ± SD. Comparisons of multiple groups were analyzed using one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test, and simple comparisons between two groups were analyzed using Student’s t-test. Tumor incidence was examined by Fisher’s exact test. Methylation differences were analyzed by Mann–Whitney U-test. A P-value ˂0.05 was considered statistically significant.
Results
Curcumin suppresses colon carcinogenesis in an AOM- and DSS-induced mouse model
The animal study was carried out according to scheme shown in Figure 1A. During the feeding period, we did not observe noticeable body weight loss or sickness in mice fed with a 2% (wt/wt) curcumin-supplemented diet (Figure 1B). Twelve weeks after AOM injection, the mice were killed to determine curcumin’s inhibitory effects on colon cancer. As shown in Figure 1C, curcumin significantly attenuated the colon shortening caused by AOM and DSS. Of the 10 animals, six were identified to have tumor growth in the colon with a tumor multiplicity of 2.4 ± 0.9 (Figure 1D and E). Of note, no animals in the AOM+DSS+Curcumin group had developed tumors by the end point. The decrease in tumor incidence and tumor multiplicity by curcumin was statistically significant. Hematoxylin and eosin staining showed that AOM- and DSS-induced animals developed crypt adenomas with inflammation, crypt dilatation and leukocyte infiltration into the lumen, leading to an increase in the nucleus to cytoplasm ratio, nuclear crowding, mitosis and nuclear hyperchromasia. Some animals in the AOM+DSS group exhibited adenocarcinoma with inflammation including cribriform glands, nuclear hyperchromasia and mitosis, as well as a nuclear polarity loss with respect to the basement membrane. Dietary administration of curcumin attenuated the inflammation severity, and only one animal was observed to have hyperplasia with inflammation. No adenoma or adenocarcinoma was observed in the AOM+DSS+Curcumin group. Taken together, our data strongly suggested a chemopreventive effect by curcumin in AOM+DSS-induced colon cancer.
Observations from RNA-seq data
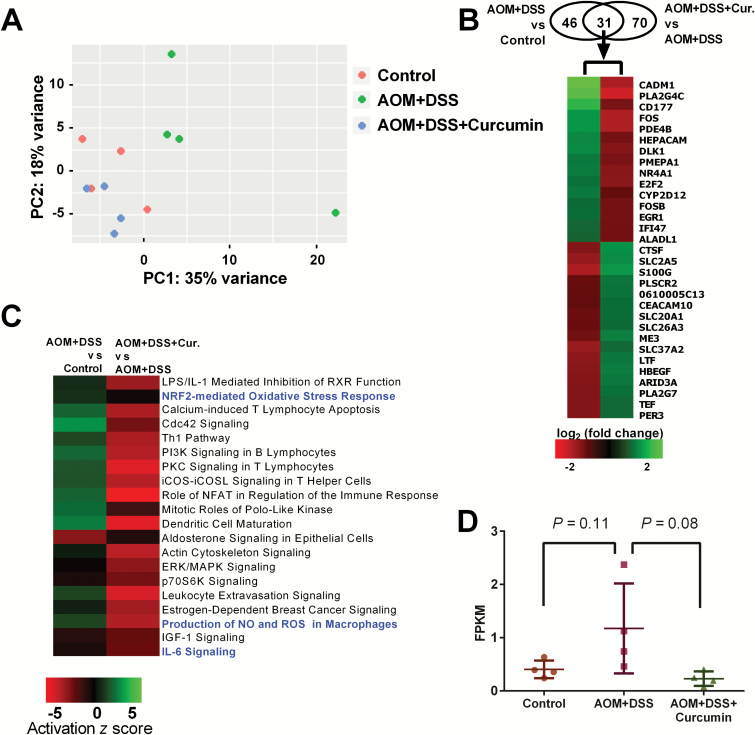
Epithelial cell samples from 12 mice in 3 groups were used to obtain RNA-seq data. Using these data, we first performed principal component analysis (PCA) to determine the differences between samples and observed that AOM- and DSS-induced mice were plotted distinctly from the other two groups of mice (Figure 2A). It was noted that the PCA result showed one mouse in the AOM+DSS group was clustered far away from the other three mice in the same group on the PCA plot. We further confirmed that it showed a different gene expression pattern for a wide range of genes compared with the other three animals in the same group. Thus, this animal was considered an outlier and was excluded from further RNA-seq data analyses (Supplementary Figure 1, available at Carcinogenesis Online). Since only three mice were included in the AOM+DSS group, we anticipated that fewer genes with statistically significant expression change in pairwise comparisons to AOM+DSS group would be discovered. We then used pairwise comparison to gain insights from the RNA-seq data. Only differentially expressed genes with a log2-fold change ˃1 and a false discovery ˂0.05 were considered. As shown in Figure 2B, we identified 77 differentially expressed genes upon comparing the AOM+DSS and control groups, among which 35 were upregulated, and 42 were down-regulated. Curcumin administration changed the expression of 101 genes compared with the AOM+DSS group, with 50 genes increased and 51 genes decreased. In our recently published study, we identified 1291 differentially expressed genes in tumors induced by AOM and DSS at 28 weeks compared with age-matched colonic tissue in the control group. We also identified 189 genes in tumors from the AOM+DSS+Curcumin group that were differentially expressed compared with tumors from AOM- and DSS-induced animals (21). Fewer differentially expressed genes were identified in this study. This could be due to RNAs being extracted from colonic epithelial cells in this study, whereas tumors were used in the previous study. In addition, more genes might be involved in carcinogenesis over a longer period, as the animals were killed at 28 weeks in the previous study and at 18 weeks in the current study. Although fewer differentially expressed genes were identified in this study, there were 31 common genes in these two data sets. The expression of all 31 genes showed opposite regulation changes, suggesting the involvement of these molecular targets in curcumin-mediated prevention of AOM- and DSS-induced CAC. We further performed ingenuity pathway analysis with RNA-seq data to identify the pathways regulated by AOM/DSS and/or curcumin administration. To achieve this goal, we used a broader list of differentially regulated genes defined only by the false discovery rate (q < 0.5) and analyzed 1740 genes from AOM+DSS versus control and 1708 genes from AOM+DSS+Curcumin versus AOM+DSS with ingenuity pathway analysis. We then obtained two lists of regulated pathways from the two comparisons. Based on the −log (P-value) of pathways generated by ingenuity pathway analysis, the top 20 pathways in common, all of which had a −log (P-value) > 2 (P < 0.01; Supplementary Tables 3 and 4, available at Carcinogenesis Online), were selected, and their ‘activation z scores’ are shown in a heatmap (Figure 2C). Among these pathways, 15 were up-regulated in the AOM+DSS group compared with the control group but were down-regulated in the AOM+DSS+Curcumin group compared with the AOM+DSS group. Interestingly, three of them were anti-inflammatory and/or anti-oxidative pathways (highlighted in blue). These findings suggest that curcumin administration can reverse the gene expression patterns modified by AOM+DSS, especially for genes in anti-inflammatory and anti-oxidative pathways. The expression of Tnf, an inflammation-related gene, was 4.3 times higher in the AOM+DSS group than in the control group but was decreased to 0.6-fold in the AOM+DSS+Curcumin group compared with the AOM+DSS group. However, these differences were not significant in our analyses, with P-values of 0.11 and 0.08 (One-way ANOVA) for AOM+DSS versus control and AOM+DSS+Curcumin versus AOM+DSS, respectively (Figure 2D).
Figure 2.
Overview of differentially expressed genes and regulated pathways from RNA-seq data. (A) PCA of 12 RNA-seq samples. One sample in AOM+DSS group was an outlier and was excluded from further analyses. (B) Venn diagram showing differentially expressed genes from both comparisons and heatmap showing differentially expressed genes in common in these two comparisons. (C) Heatmap showing top 20 regulated pathways in common in both comparisons. Anti-inflammatory and anti-oxidative stress pathways are highlighted in blue. (D) RNA level of Tnf in the three groups from RNA-seq data. P-values, One-way ANOVA.
Observations from the Methyl-seq data
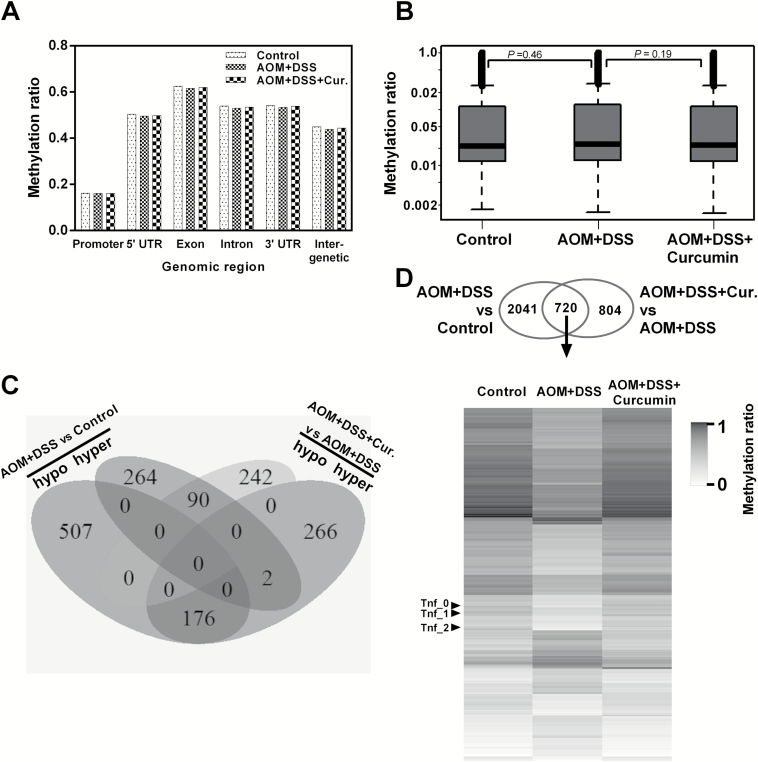
To understand the involvement of DNA methylation in colon tumorigenesis, we determined the single-base-resolution DNA methylation of colon epithelial cell samples from the three groups as in the above RNA-seq section. A comparison of the methylation landscape of the three groups showed a high degree of similarity for global methylation among the three datasets (Supplementary Figure 2, available at Carcinogenesis Online). Previous studies have indicated that AOM+DSS treatment induces localized DNA methylation aberrations rather than large-scale global DNA methylation changes (9). Thus, we segmented our datasets according to gene substructure and found that the methylation levels were unevenly distributed across the genome. The methylation ratios were much higher in the 5′-UTR, exon, intron, 3′-UTR and intergenic regions than in the promoter regions (Figure 3A). However, the overall methylation levels in these three datasets throughout these regions were not significantly different. When we compared the methylation levels of all the annotated DMRs in our datasets, we did not observe considerable differences between AOM+DSS and either of the other two groups (Figure 3B). We then focused on gene promoters (<3 kb) containing DMRs in the following two comparisons: AOM+DSS versus control and AOM+DSS+Curcumin versus AOM+DSS using a cutoff for the methylation ratio difference of greater than or equal to 0.1 and a P-value of less than or equal to 0.05. The comparison between the AOM+DSS and control groups showed the greatest difference: 683 DMRs exhibited a lower methylation level in epithelial cells from AOM- and DSS-induced animals compared with control animals, whereas 346 DMRs showed higher methylation. The dietary administration of curcumin in the AOM+DSS model caused a lower methylation level in 332 DMRs and a higher methylation in 444 DMRs (Figure 3C). Of the 268 DMRs identified in both comparisons, 176 were hypomethylated in the AOM+DSS group but were hypermethylated in the AOM+DSS+Curcumin group, and 90 were hypermethylated in the AOM+DSS group but were hypomethylated in the AOM+DSS+Curcumin group. Interestingly, Tnf was one of the genes among the 176 hypomethylated DMRs, suggesting that the Tnf methylation level decreased in AOM- and DSS-induced carcinogenesis was reversed by curcumin treatment. When looking at DMRs for all genomic locations except distal intergenic regions with a cutoff methylation level at 0.1 and a P-value of 0.05, we identified 2761 DMRs with lower methylation levels in the AOM+DSS group than in the control group and 1524 DMRs with higher methylation in the AOM+DSS+Curcumin group than in the AOM+DSS group. These two comparisons had 720 DMRs in common (Figure 3D, Venn diagram). We then compared the methylation level of these DMRs from all three groups and found that the methylation pattern of the AOM+DSS group was dramatically different from that of the other two groups (Figure 3D, heatmap). Again, Tnf was one of the genes containing these 720 DMRs. These findings indicate that curcumin administration prevented DNA methylation pattern changes in AOM- and DSS-induced mice.
Figure 3.
Overview of DNA methylation level changes. (A) Methylation level of gene substructures in control, AOM+DSS and AOM+DSS+Curcumin groups. (B) Box plots showing the distribution of DMRs in control, AOM+DSS and AOM+DSS+Curcumin groups. Only DMRs with >12 CpG sites were included in this analysis to ensure comparability and analytical robustness. Differences between groups were not significant. P-values, Mann–Whitney. (C) Venn diagram showing the number of DMRs within promoter regions in the comparisons of control versus AOM+DSS and AOM+DSS versus AOM+DSS+Curcumin. (D) Venn diagram showing the number of DMRs in all genomic locations except distal intergenic regions, in the comparisons of control versus AOM+DSS and AOM+DSS versus AOM+DSS+Curcumin. Methylation ratio of DMRs from both comparisons is shown in heatmap for all three groups.
DNA methylation of Tnf is decreased by AOM and DSS induction but increased by curcumin administration
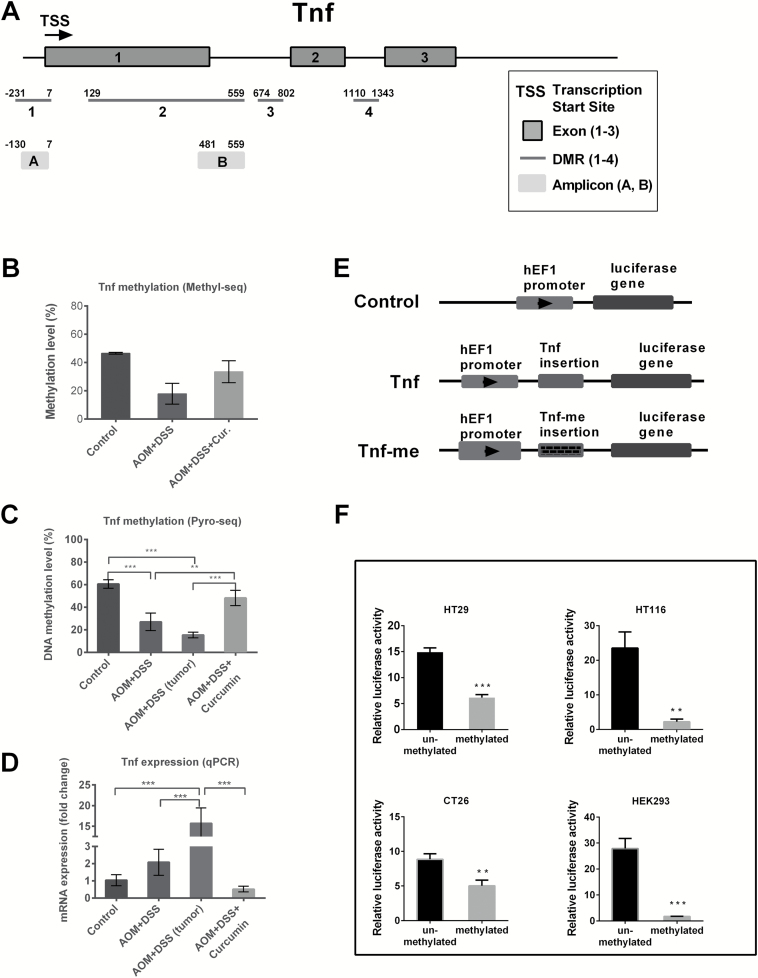
Considering the general observations from SureSelect Methyl-seq and RNA-seq, we later focused on the methylation status of Tnf, which is a key inflammatory mediator and is secreted by most tumors to facilitate cellular survival and enhance neoplastia (29). Using DMRfinder, four DMRs were identified in the Tnf gene, and their locations are shown in Figure 4A. DMR 1 (−231 to 7 to transcription start site, TSS) is located in the Tnf promoter and contains 11 CpG sites. DMR 2 (129–559), DMR 3 (674–802) and DMR 4 (1110–1343) span from exon to intron and contain 16, 5 and 8 CpG sites, respectively. The Tnf methylation level was calculated by averaging the methylation ratio of these four regions and is shown in Figure 4B. The Tnf methylation level was decreased in the AOM+DSS group but remained at the same level in the control and AOM+DSS+Curcumin groups. We postulated that AOM- and DSS-induced colon carcinogenesis decreased the Tnf DNA methylation. To validate our observations from Methyl-seq, we performed pyrosequencing analysis of the Tnf gene with two amplicons. Amplicon A (−130 to 7 to TSS) and amplicon B (481 to 559) both contain six CpG sites. Supplementary Figure 3, available at Carcinogenesis Online presents the single-base resolution CpG methylation status of amplicons A and B in each experimental group. The right panel (pyrosequencing results) shows a similar trend as the left panel (Methyl-seq results), confirming the quantitative properties of the Methyl-seq method and the accuracy of the bioinformatics analysis. Specifically, all of the six CpG sites in amplicon A were not methylated in all three groups. The methylation level of region B was similar in the control and AOM+DSS+Curcumin groups but was much lower in the AOM+DSS group (Supplementary Figure 3, available at Carcinogenesis Online, lower half). Using pyrosequencing, the average methylation level of amplicon B exhibited a similar trend as the methylation level calculated from the Methyl-seq results (Figure 4C). Tumor samples from the AOM+DSS group also underwent pyrosequencing to see whether Tnf methylation was altered in tumors. As shown in Figure 4C (the third bar), the methylation of Tnf in epithelial cells dropped from 60.5% in the control group to 27.1% in the AOM+DSS group, and the level was even lower in tumor samples in the AOM+DSS group (15.4%). Furthermore, curcumin treatment restored the methylation level to 48.2%. Tnf mRNA expression was examined using qPCR analysis and is shown in Figure 4D. The expression levels of Tnf in epithelial cells from the AOM+DSS group, in tumor samples from the AOM+DSS group, and in epithelial cells from the AOM+DSS+Curcumin group were 2.1-, 15.7- and 0.5-fold higher, respectively. Thus, the mRNA expression trend for Tnf in all of the groups was opposite to that of the DNA methylation trend. To further confirm the functional role of DNA methylation in regulating Tnf transcriptional activity, we constructed a luciferase reporter vector with Tnf inserted (amplicon B, 481–559 bp to TSS). The structure of this plasmid is shown in Figure 4E. In vitro CpG methylation of the plasmid by M.SssI CpG methyltransferase resulted in a significant decrease in the transcriptional activity of the luciferase gene in the human colon cancer cell lines HT29 and HCT116, the mouse colon cancer cell line CT26 and the human embryonic kidney cell line HEK293 (Figure 4F). Together with our results from AOM+DSS mice, these findings showed that DNA methylation plays a functional role in regulating Tnf and that dietary administration of curcumin effectively restored Tnf DNA methylation and suppressed its expression.
Figure 4.
Alterations of methylation level in Tnf. (A) Schematic diagram showing the structure of Tnf gene, the locations of DMRs and amplicons investigated in this study. (B and C) The methylation level of Tnf gene as observed from SureSelect Methyl-seq and Pyrosequencing. (D) The mRNA expression of Tnf measured by qPCR in the same groups as above. (E) The luciferase reporter vectors used in this study. Control vector has no insertion. Tnf and Tnf-me indicate unmethylated and methylated insertion of amplicon B of Tnf gene, respectively. (F) Luciferase activity of reporter vectors with unmethylated or methylated Tnf fragment. Data are from three independent experiments and are presented as fold change to control vector with normalization to firefly luciferase vector pGL4.13. Methylation of Tnf fragment (amplicon B) significantly decreased the luciferase activity. One-way ANOVA test for (B–D) and t-test for (F); *P < 0.05; **P < 0.01; ***P < 0.001.
DNA methylation and gene expression of a set of genes associated with inflammation
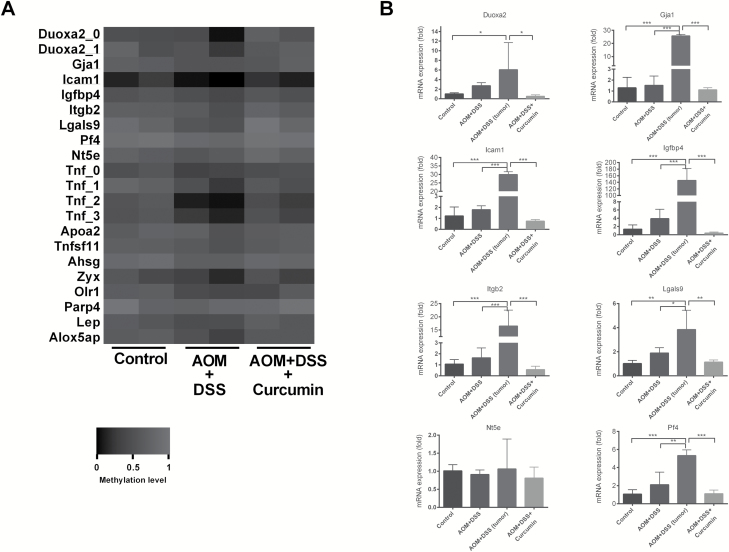
Given the important role of Tnf in inflammation, we turned our focus to the methylation profile of other genes important in the inflammatory response. First, we extracted a set of DMRs showing methylation ratio changes greater than 0.1 in the comparison of AOM+DSS versus control and AOM+DSS+Curcumin versus AOM+DSS in opposite directions. Next, we used gProfiler (30) to cluster the DMRs identified from Methyl-Seq to different functions. Specifically, 17 genes (Duoxa1 had 2 DMRs, and Tnf had 4 DMRs) were characterized based on their function in inflammation. The methylation levels are shown as a heatmap (Figure 5A). Overall, the color in the AOM+DSS columns was darker than in the control and AOM+DSS+Curcumin columns, indicating reduced methylation of these genes in the AOM+DSS group. We next examined the mRNA expression of randomly selected genes from this set using qPCR (Supplementary Table 1, available at Carcinogenesis Online). As shown in Figure 5B, the mRNA expression of Duoxa2, Gja1, Icam1, Igfbp4, Itgb2, Lgals9 and Pf4 was higher in epithelial cells from AOM- and DSS-induced mice. Gene expression was dramatically elevated in the tumor samples from AOM- and DSS-induced mice, whereas curcumin administration suppressed the abnormally high expression of these genes. The RNA-seq results of these genes are shown in Supplementary Figure 4, available at Carcinogenesis Online. Among the eight genes tested, only Nt5e showed no change among the control, AOM+DSS and AOM+DSS+Curcumin groups. The change in DNA methylation of Nt5e was also minimal. Collectively, our results suggest that a larger set of genes associated with the inflammatory response showed a similar modification trend as Tnf. AOM and DSS induction decreased their methylation level and increased their transcription, whereas curcumin administration blocked these changes.
Figure 5.
Methylation level and mRNA expression of a set of inflammatory genes. (A) Heatmap showing the methylation level of genes in the pathway of inflammatory response. (B) The mRNA expression of selected genes measured by qPCR. Note, only epithelial cells were used for Methyl-seq, whereas tumor samples were also included in qPCR experiments. One-way ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001.
Discussion
Colorectal cancer is known to be mostly sporadic, with no clear hereditary basis; it sometimes has hereditary or familial causes but much less often arises from solely inflammatory conditions in the bowel (31). More than 20% of inflammatory bowel disease (IBD) patients develop CAC, and 50% of these will die from CAC, making CAC the most serious complication of IBD (32). Although overt chronic inflammation is not the driving force for sporadic and hereditary cases, treatment with anti-inflammatory drugs is effective in preventing or delaying these colon cancer types (33), suggesting the involvement of inflammatory processes in colon cancer progression. Therefore, it is crucial to understand the molecular events as well as the epigenetic modifications that govern the transition from inflammation to malignancy. With the rapid development of techniques for the investigation of the cancer methylome, several studies have identified candidate risk loci or DMRs for IBD and CAC (9,34,35). However, it is not clear whether these epigenetic marks are associated with colon cancer prevention. Our recent study demonstrated for the first time that curcumin exhibits a superior preventive effect compared with aspirin against AOM- and DSS-induced CAC. Thus, we focused on the preventive potential of curcumin and investigated the changes of DNA methylation and transcriptional activity in AOM- and DSS-induced CAC with or without curcumin treatment. In this study, epithelial cells scraped from the colon were used to provide DNA and RNA samples for RNA-seq and SureSelect Methyl-seq, respectively. It should be noted that isolating epithelial cells by scraping could also collect cells of other type such as lymphocytes. We used epithelial cells instead of tumor samples for two reasons: First, no tumor growth was observed in the AOM+DSS+Curcumin group (Figure 1). Second, cancer epigenomic studies indicated that DNA methylation abnormalities in malignant tumors have already accumulated in precancerous tissues, and these early epigenetic marks further determine tumor development and patient outcomes (36,37). Of note, we included tumor samples from the AOM+DSS group when validating the Tnf DNA methylation ratio and the mRNA expression of multiple genes that are important for inflammatory responses (Figures 4 and 5). The alterations of those markers in tumor samples showed the same trend as in epithelial cells from the AOM+DSS group with more dramatic fold changes. We postulated that the DNA methylation patterns identified in epithelial cells from the control, AOM+DSS and AOM+DSS+Curcumin groups would reveal the early epigenetic events and preventive targets during colon carcinogenesis in the AOM- and DSS-induced mouse model. It should be noted that the statistical significance of the list of differentially expressed genes could be dampened due to the exclusion of the outlier in the AOM+DSS group.
We used Tnf as one of the examples to show the observed methylation changes and transcription changes in this study. Tnf initiates and propagates many biological activities, such as the production of other cytokines and chemokines, the activation of the transcription factors AP-1 and NF-κB and the promotion of tumor growth (38). Elevated Tnf levels have been detected in IBD animal models, as well as in patients with IBD (39). In this study, using SureSelect Methyl-seq and pyrosequencing, we showed that a list of inflammatory genes was hypomethylated in scraped colon epithelial cells and colonic tumors in AOM- and DSS-induced mice. It would be interesting to examine the sequential loss of DNA methylation of these genes during colon cancer progression. Further studies are also needed to investigate the interplay of other epigenetic regulators, such as histone methylation, histone readers and histone erasers, in the establishment of Tnf hypomethylation and dramatically elevated Tnf expression in colon cancer. It should be noted that other pathways could also play a role in curcumin-induced Tnf inhibition. For example, the transcription factor NF-kB could regulate the expression of Tnf, and curcumin could also regulate Tnf expression through inhibition of IkK, an upstream regulator of NF-kB (40,41).
A set of candidate genes was identified and showed that curcumin attenuated the loss of methylation compared with AOM and DSS alone (Figure 5). We tested the mRNA expression of eight selected genes from this list in epithelial cells as well as in tumor samples from AOM and DSS alone using qPCR analysis (Figure 5). The alterations of DNA methylation and mRNA expression of some genes, such as Itgb2 (integrin beta 2), agreed with previous reports. ITGB2, an important gene in immune responses and defects, was hypomethylated in human pancreatic adenocarcinoma and human gastric cancer tissue compared with paired normal tissues (42,43). Overexpression of ITGB2 was also detected in colon adenocarcinomas versus normal mucosa (44). For most of the genes on this list, aberrant DNA methylation has not been identified in colon cancer before. For example, hypermethylation-mediated downregulation of IGFBP4 (insulin-like growth factor binding protein 4) has been implicated in lung adenocarcinomas and giant cell tumors (45,46). In our current study, however, observed hypomethylation in Igfbp4 in the CAC model and the expression of Igfbp4 was increased in epithelial cells and tumor samples (Figure 5). Similarly, DUOXA2 (dual oxidase maturation factor 2) was also down-regulated and hypermethylated in lung cancer cell lines and lung cancer specimens (47), which differentiated from what we observed in the CAC model. Interestingly, MacFie et al. (48) found that DUOXA2 was upregulated in inflamed tissue in ulcerative colitis and adenomas associated with colitis, an observation that was consistent with our findings. Gja1 (gap junction protein, alpha 1) is a member of the connexin gene family. The encoded protein of this gene is a component of gap junction and is considered pro-metastatic (49). Biscanin et al. (50) found that GJA1 expression was higher in mucosa surrounding adenomas with high-grade dysplasia, which is in agreement with our findings that its expression was higher in normal tissue in AOM+DSS group and was even higher in tumors in AOM+DSS group (Figure 5B). The Icam1 (intercellular adhesion molecule 1) gene encodes a cell surface glycoprotein which is typically expressed in endothelial cells. Lee et al. (51) found that expression of Icam1 was upregulated by Tnf in an LPS-induced inflammation model in mouse. Im et al. (52) also found that expression of Icam1 and other genes were elevated in alcohol-induced inflammatory response. In our current study, we also observed elevated Icam1 expression in both normal and tumor tissues in AOM+DSS group (Figure 5B). Lgals9 (lectin, galactoside-binding, soluble, 9) is a member of galectin gene family. The encoded protein of this gene involved in modulating cell–cell and cell–matrix interactions. Lee et al. (53) found that Lgals9 was upregulated in the liver of tumor-bearing mice. Here, we observed elevated transcription levels in AOM+DSS group and decreased DNA methylation levels in accordance with its transcription levels. Lep (Leptin) encodes a gene that plays a major role in the regulation of immune and inflammatory response. Penrose et al. (54) found that leptin was up-regulated in high-fat diet induced tumors in a mouse model. Another study by Hansen et al. (55) showed that increased Lep gene expression was associated with decreased Lep promoter methylation in offspring of women with diabetes. In agreement with these findings on Lep, in this study, we found that DNA methylation of this gene was decreased in AOM+DSS group and curcumin administration restored its methylation level (Figure 5A). Notably, the expression of these inflammatory genes was slightly elevated in epithelial cells from the AOM+DSS group, whereas their expression was dramatically increased in tumor samples from the AOM+DSS group (Figure 5B). This might indicate that slight changes in DNA methylation and transcription in precancerous epithelial cells might predict the outcome of tumors. Further validation and investigation in a clinical setting are required to draw the conclusion that these genes are involved in colon carcinogenesis and colon cancer prevention.
The investigation of the epigenetic effect of curcumin has been focused on the inhibition of DNA methylation in the promoter regions of genes. Studies from our laboratory have demonstrated that curcumin treatment effectively decreased promoter methylation of Nrf2, Neurog1 and DLEC1 in TRAMP-C1 cells, LNCaP cells and HT29 cells (15–17). Curcumin treatment was also shown to hypomethylate the promoter regions of the tumor suppressor genes Rassf1a (56), Rarβ (57) and p15INK4B (58), thereby increasing their expression in different in vitro cancer cell lines. In addition, these studies indicated that the expression of DNA methyltransferases was inhibited by curcumin treatment. In contrast, the effect of curcumin in increasing the DNA methylation level has rarely been investigated. Lewinska et al. (59) reported that decreased AgNOR expression in HeLa cells upon curcumin treatment might be mediated by global DNA hypermethylation. In contrast to other findings that curcumin worked as a demethylation agent and increased the expression of tumor suppressor genes; in this study, we found that curcumin could reverse the hypomethylation status of Tnf and other inflammatory genes and inhibit their expression. It has been reported that curcumin could modulate many epigenetic writers/erasers, such as histone deacetylases, DNA methyltransferases, histone methyltransferases and histone demethylases, etc. (60,61). The specific effect in increasing methylation level in a list of genes by curcumin in our current study may be mediated by curcumin’s effect in one or a combination of these epigenetic enzymes. In addition, curcumin may interact with other loci-specific binding factors which in turn block or facilitate the recruitment of epigenetic writers/erasers to these genes (62,63). Nevertheless, more investigations are needed to understand the exact mechanism how curcumin alters methylation of specific loci (64).
In summary, this study demonstrated the chemopreventive effect of curcumin against AOM- and DSS-induced CAC. Using SureSelect Methyl-seq and RNA-seq, we provided a quantitative global profile of the methylome and transcriptome in epithelial cells from colon cancer with or without curcumin treatment. Importantly, we identified that DNA methylation of a list of inflammatory genes was decreased in AOM- and DSS-induced cancer, and curcumin effectively restored the loss of DNA methylation, thereby suppressing their expression. These findings provide novel insights into the understanding of how epigenetic modifications affect the progression of CAC and the preventive effect of curcumin.
Accession codes
Transcript sequencing data have been deposited under GenBank Gene Expression Omnibus (GEO) accession GSE102342. Bisulfite methyl sequencing data have been deposited under GEO accession GSE102645.
Supplementary material
Supplementary Tables 1–4 and Figures 1–4 can be found at Carcinogenesis online.
Funding
National Center for Complementary and Alternative Medicines (NCCAM) (R01AT007065); Office of Dietary Supplements (ODS).
Conflict of Interest Statement: None declared.
Acknowledgements
We thank Dr Guangxun Li for his assistance in the histology evaluation. We are thankful to Drs Mou-Tuan Huang and Tin Oo Khor for their kind help in the design of the animal studies. The authors are grateful to Sabinsa Corporation for providing Curcumin C3 Complex®. We thank all of the members of Dr Kong’s laboratory for their helpful discussions and the preparation of this manuscript.
Abbreviations
- ANOVA
analysis of variance
- AOM
azoxymethane
- CAC
colitis-accelerated colon cancer
- DMRs
differentially methylated regions
- DSS
dextran sulfate sodium
- IBD
inflammatory bowel disease
- NO
nitric oxide
- PCA
principal component analysis
- qPCR
quantitative polymerase chain reaction
- ROS
reactive oxygen species
References
- 1. Söderlund S., et al. (2009)Decreasing time-trends of colorectal cancer in a large cohort of patients with inflammatory bowel disease. Gastroenterology, 136, 1561–1567; quiz 1818–1819. [DOI] [PubMed] [Google Scholar]
- 2. Delker D.A., et al. (1998)The role of alcohol dehydrogenase in the metabolism of the colon carcinogen methylazoxymethanol. Toxicol. Sci., 45, 66–71. [DOI] [PubMed] [Google Scholar]
- 3. Perše M., et al. (2012)Dextran sodium sulphate colitis mouse model: traps and tricks. J. Biomed. Biotechnol., 2012, doi:10.1155/2012/718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lao V.V., et al. (2011)Epigenetics and colorectal cancer. Nat. Rev. Gastroenterol. Hepatol., 8, 686–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robertson K.D. (2005)DNA methylation and human disease. Nat. Rev. Genet., 6, 597–610. [DOI] [PubMed] [Google Scholar]
- 6. Guo Y., et al. (2015)Association of aberrant DNA methylation in Apc(min/+) mice with the epithelial-mesenchymal transition and Wnt/β-catenin pathways: genome-wide analysis using MeDIP-seq. Cell Biosci., 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grimm C., et al. (2013)DNA-methylome analysis of mouse intestinal adenoma identifies a tumour-specific signature that is partly conserved in human colon cancer. PLoS Genet., 9, e1003250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katsurano M., et al. (2012)Early-stage formation of an epigenetic field defect in a mouse colitis model, and non-essential roles of T- and B-cells in DNA methylation induction. Oncogene, 31, 342–351. [DOI] [PubMed] [Google Scholar]
- 9. Abu-Remaileh M., et al. (2015)Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res., 75, 2120–2130. [DOI] [PubMed] [Google Scholar]
- 10. Kreutz M., et al. (2013)Pyrosequencing: powerful and quantitative sequencing technology. Curr. Protoc. Mol. Biol., 104, Unit 7.15. [DOI] [PubMed] [Google Scholar]
- 11. Parrish R.R., et al. (2012)Direct bisulfite sequencing for examination of DNA methylation with gene and nucleotide resolution from brain tissues. Curr. Protoc. Neurosci., 60, 7.24.1–7.24.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Laird P.W. (2010)Principles and challenges of genomewide DNA methylation analysis. Nat. Rev. Genet., 11, 191–203. [DOI] [PubMed] [Google Scholar]
- 13. Sahebkar A., et al. (2016)Curcumin downregulates human tumor necrosis factor-α levels: a systematic review and meta-analysis ofrandomized controlled trials. Pharmacol. Res., 107, 234–242. [DOI] [PubMed] [Google Scholar]
- 14. Patel V.B., et al. (2010)Colorectal cancer: chemopreventive role of curcumin and resveratrol. Nutr. Cancer, 62, 958–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo Y., et al. (2015)Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1. Biochem. Pharmacol., 94, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu T.Y., et al. (2011)Anti-inflammatory/Anti-oxidative stress activities and differential regulation of Nrf2-mediated genes by non-polar fractions of tea Chrysanthemum zawadskii and licorice Glycyrrhiza uralensis. AAPS J., 13, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shu L., et al. (2011)Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J., 13, 606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Link A., et al. (2013)Curcumin modulates DNA methylation in colorectal cancer cells. PLoS One, 8, e57709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo Y., et al. (2016)The epigenetic effects of aspirin: the modification of histone H3 lysine 27 acetylation in the prevention of colon carcinogenesis in azoxymethane- and dextran sulfate sodium-treated CF-1 mice. Carcinogenesis, 37, 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chahar S., et al. (2014)Chromatin profiling reveals regulatory network shifts and a protective role for hepatocyte nuclear factor 4α during colitis. Mol. Cell. Biol., 34, 3291–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo Y., et al. (2017)Mechanisms of colitis-accelerated colon carcinogenesis and its prevention with the combination of aspirin and curcumin: transcriptomic analysis using RNA-seq. Biochem. Pharmacol., 135: 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krueger F., et al. (2011)Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaspar J.M., et al. (2017)DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics, 18, 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu G., et al. (2015)ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics, 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- 25. Martin M. (2011)Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet, 17, 10–12. [Google Scholar]
- 26. Kim D., et al. (2013)TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol., 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liao Y., et al. (2014)featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- 28. Love M.I., et al. (2014)Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balkwill F. (2009)Tumour necrosis factor and cancer. Nat. Rev. Cancer, 9, 361–371. [DOI] [PubMed] [Google Scholar]
- 30. Reimand J., et al. (2007)g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res., 35(Web Server issue), W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lasry A., et al. (2016)Inflammatory networks underlying colorectal cancer. Nat. Immunol., 17, 230–240. [DOI] [PubMed] [Google Scholar]
- 32. Lakatos P.L., et al. (2008)Risk for colorectal cancer in ulcerative colitis: changes, causes and management strategies. World J. Gastroenterol., 14, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burn J., et al. ; CAPP2 Investigators (2011)Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet, 378, 2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin Z., et al. (2011)Identification of disease-associated DNA methylation in intestinal tissues from patients with inflammatory bowel disease. Clin. Genet., 80, 59–67. [DOI] [PubMed] [Google Scholar]
- 35. Häsler R., et al. (2012)A functional methylome map of ulcerative colitis. Genome Res., 22, 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamanoi K., et al. (2015)Epigenetic clustering of gastric carcinomas based on DNA methylation profiles at the precancerous stage: its correlation with tumor aggressiveness and patient outcome. Carcinogenesis, 36, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arai E., et al. (2010)DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics, 2, 467–481. [DOI] [PubMed] [Google Scholar]
- 38. Al Obeed O.A., et al. (2014)Increased expression of tumor necrosis factor-α is associated with advanced colorectal cancer stages. World J. Gastroenterol., 20, 18390–18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kollias G. (2004)Modeling the function of tumor necrosis factor in immune pathophysiology. Autoimmun. Rev., 3(Suppl 1), S24–S25. [PubMed] [Google Scholar]
- 40. Bachmeier B.E., et al. (2008)Curcumin downregulates the inflammatory cytokines CXCL1 and -2 in breast cancer cells via NFkappaB. Carcinogenesis, 29, 779–789. [DOI] [PubMed] [Google Scholar]
- 41. Killian P.H., et al. (2012)Curcumin inhibits prostate cancer metastasis in vivo by targeting the inflammatory cytokines CXCL1 and -2. Carcinogenesis, 33, 2507–2519. [DOI] [PubMed] [Google Scholar]
- 42. Vincent A., et al. (2011)Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin. Cancer Res., 17, 4341–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lim B., et al. (2014)Integrative genomics analysis reveals the multilevel dysregulation and oncogenic characteristics of TEAD4 in gastric cancer. Carcinogenesis, 35, 1020–1027. [DOI] [PubMed] [Google Scholar]
- 44. Cavalieri D., et al. (2007)Analysis of gene expression profiles reveals novel correlations with the clinical course of colorectal cancer. Oncol. Res., 16, 535–548. [DOI] [PubMed] [Google Scholar]
- 45. Sato H., et al. (2011)Insulin-like growth factor binding protein-4 gene silencing in lung adenocarcinomas. Pathol. Int., 61, 19–27. [DOI] [PubMed] [Google Scholar]
- 46. Fellenberg J., et al. (2013)Rescue of silenced UCHL1 and IGFBP4 expression suppresses clonogenicity of giant cell tumor-derived stromal cells. Cancer Lett., 336, 61–67. [DOI] [PubMed] [Google Scholar]
- 47. Luxen S., et al. (2008)Silencing of DUOX NADPH oxidases by promoter hypermethylation in lung cancer. Cancer Res., 68, 1037–1045. [DOI] [PubMed] [Google Scholar]
- 48. MacFie T.S., et al. (2014)DUOX2 and DUOXA2 form the predominant enzyme system capable of producing the reactive oxygen species H2O2 in active ulcerative colitis and are modulated by 5-aminosalicylic acid. Inflamm. Bowel Dis., 20, 514–524. [DOI] [PubMed] [Google Scholar]
- 49. Zuo X., et al. (2017)Metastasis regulation by PPARD expression in cancer cells. JCI Insight, 2, e91419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bišćanin A., et al. (2016)CX43 Expression in colonic adenomas and surrounding mucosa is a marker of malignant potential. Anticancer Res., 36, 5437–5442. [DOI] [PubMed] [Google Scholar]
- 51. Lee A.S., et al. (2016)Paricalcitol attenuates lipopolysaccharide-induced myocardial inflammation by regulating the NF-κB signaling pathway. Int. J. Mol. Med., 37, 1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Im H.J., et al. (2016)A preclinical model of chronic alcohol consumption reveals increased metastatic seeding of colon cancer cells in the liver. Cancer Res., 76, 1698–1704. [DOI] [PubMed] [Google Scholar]
- 53. Lee A., et al. (2011)Liver membrane proteome glycosylation changes in mice bearing an extra-hepatic tumor. Mol. Cell. Proteomics, 10, M900538MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Penrose H.M., et al. (2017)High-fat diet induced leptin and Wnt expression: RNA-sequencing and pathway analysis of mouse colonic tissue and tumors. Carcinogenesis, 38, 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hansen N.S., et al. (2017)Fetal hyperglycemia changes human preadipocyte function in adult life. J. Clin. Endocrinol. Metab., 102, 1141–1150. [DOI] [PubMed] [Google Scholar]
- 56. Du L., et al. (2012)Reactivation of RASSF1A in breast cancer cells by curcumin. Nutr. Cancer, 64, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jiang A., et al. (2015)Curcumin reactivates silenced tumor suppressor gene RARβ by reducing DNA methylation. Phytother. Res., 29, 1237–1245. [DOI] [PubMed] [Google Scholar]
- 58. Yu J., et al. (2013)Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS One, 8, e55934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lewinska A., et al. (2014)Curcumin-mediated decrease in the expression of nucleolar organizer regions in cervical cancer (HeLa) cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen., 771, 43–52. [DOI] [PubMed] [Google Scholar]
- 60. Li W., et al. (2016)Dietary phytochemicals and cancer chemoprevention: a perspective on oxidative stress, inflammation, and epigenetics. Chem. Res. Toxicol., 29, 2071–2095. [DOI] [PubMed] [Google Scholar]
- 61. Ramirez C.N., et al. (2017)In vitro-in vivo dose response of ursolic Acid, Sulforaphane, PEITC, and Curcumin in Cancer Prevention. AAPS J., 20, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Novak E.M., et al. (2003)Downregulation of TNF-alpha and VEGF expression by Sp1 decoy oligodeoxynucleotides in mouse melanoma tumor. Gene Ther., 10, 1992–1997. [DOI] [PubMed] [Google Scholar]
- 63. Falvo J.V., et al. (2010)Transcriptional control of the TNF gene. Curr. Dir. Autoimmun., 11, 27–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Falvo J.V., et al. (2013)Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation. Adv. Immunol., 118, 37–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.