Abstract
Background
Circular RNAs(circRNAs) have been reported as a diverse class of endogenous RNA that regulate gene expression in eukaryotes. Recent evidence suggested that many circular RNAs can act as oncogenes or tumor suppressors through sponging microRNAs. However, the function of circular RNAs in gastric cancer remains largely unknown.
Materials and methods
The circRNA levels in gastric carcinoma tissues and plasmas were detected by real-time quantitative reverse transcription-polymerase chain reaction. The correlation between the expression of circRNA and clinic pathological features was analyzed. Rate of inhibiting of proliferation was measured using a CCK-8 cell proliferation assay. Clone formation ability was assessed with a clone formation inhibition test. We used the bioinformatics software to predict circRNA-miRNA and miRNA-mRNA interactions. Relative gene expression was assessed using quantitative real-time polymerase chain reaction and relative protein expression levels were determined with western blotting. CircRNA and miRNA interaction was confirmed by dual-luciferase reporter assays.
Results
We characterized that one circRNA named circ-SFMBT2 showed an increased expression level in gastric cancer tissues compared to adjacent non-cancerous tissues and was associated with higher tumor stages of gastric cancer. Silencing of circ-SFMBT2 inhibited the proliferation of gastric cancer cells significantly. Importantly, we demonstrated that circ-SFMBT2 could act as a sponge of miR-182-5p to regulate the expression of CREB1 mRNA, named as cAMP response element binding protein 1, and further promote the proliferation of gastric cancer cells.
Conclusion
Our study reveals that circ-SFMBT2 participates in progression of gastric cancer by competitively sharing miR-182-5p with CREB1, providing a novel target to improve the treatment of gastric cancer. mutation-analysis-of-beta-thalassemia-in-east-western-indian-populatio-peer-reviewed-article-TACG for an example.
Keywords: tissues, plasmas, target, progression, AGO2, sponge
Introduction
Gastric cancer (GC) remains one of the most common malignant tumors with a poor prognosis all over the world.1 According to cancer statistics in 2015, the number of GC cases in China was approximately 679,000 and the number of deaths was 498,000, second only to lung cancer.2 At present, the diagnostic rate of early GC in clinical practice is <10% and approximately 50%–70% of GC patients relapse.3 As a result, precise exploration of the molecular mechanisms that lead to the development and progression of GC is crucial to identify novel targets to improve the treatment of this malignancy.
There is increasing evidence that circular RNAs (circRNAs), abundant and stable RNAs in mammalian cells, are closely associated with atherosclerotic vascular disease, neurological diseases and various types of cancers.4 Unlike the well known linear RNAs that begin with 5′ caps and are terminated with 3′ tails, cir-cRNAs form a closed continuous loop structure without 5′-3′ polarities or polyadenylated tails, showing potential ability to be resistant to exonuclease-mediated degradation.5 Numerous studies have shown that circRNAs are an important regulator of various biological processes involved in cancer, including cell proliferation, invasion and metastasis.6–8
To date, great efforts have been made to investigate the possible role of circRNAs in serving as the huge diagnostic and therapeutic potential in GC. For example, Lai et al examined differentially expressed circRNAs and mRNAs in GC tissues and paired noncancerous tissues using circRNA and mRNA microarrays and reported three newly upregulated circRNAs in GC, which were hsa_circ_0047905, hsa_ circ_0138960 and hsa-circRNA7690-15. In vitro experiment showed that knockdown of these three circRNAs suppressed GC cell proliferation and invasion significantly.9 Apart from tissues, the expression of circRNAs in plasma samples from patients with GC has also been well detected. Huang et al reported that hsa_circ_0000745 was downregulated in both GC tissues and plasma samples from GC patients. Clinical information analysis revealed that hsa_circ_0000745 expression in GC tissues was correlated with tumor differentiation, while the expression level in plasma was correlated with tumor-node-metastasis (TNM) stage.10 However, the function of circRNAs in GC remains in their infancy at present and is required to be further excavated.
The present study characterized one abundant circRNA named hsa_circ_0017639 (GSE78092 in the GEO database; hsa_circ_0017639 is derived from gene SFMBT2 and thus we named it as circ-SFMBT2 and investigated the potential modulation of it in GC progression. Importantly, we demonstrated that circ-SFMBT2 might act as a sponge for miR-182-5 p to modulate the mRNA expression of cAMP responsive element binding protein 1 (CREB1). Our findings indicate that circ-SFMBT2 takes part in GC progression through regulating CREB1 mRNA by competing for shared miR-182-5 p, which may provide a novel target to improve the treatment of GC.
Materials and methods
Patients and clinical samples
A total of 36 GC and corresponding adjacent non-tumorous tissue samples were obtained from GC patients. All tissue samples were from the Department of General Surgery, Nanjing Medical University Nanjing Hospital, Nanjing, China, from January 2014 to November 2017. All of the patients were naive-radiotherapy or -chemotherapy before enrollment, and their tissue specimens were immediately kept at −80°C in a refrigerator until analysis after removal from stomachs. The paired adjacent non-tumor tissues were localized at 5 cm away from the edge of the GC site and further confirmed by pathological analysis.
Peripheral blood (3 mL) of 26 GC patients was obtained before the operation and then the plasma was isolated. Normal plasma samples were collected from 18 healthy people at Nanjing Hospital, China in February 2017. Ethylenediami-netetraacetic acid was used to deal with blood samples as the anticoagulant. Written informed consent was obtained from each patient before recruitment, and the ethics committee of Nanjing First Hospital, Nanjing Medical University approved the study protocol.
Cell line, cell culture and transfection
Human GC cell lines MKN-45, BGC-823, MGC-803, SGC-7901 and AGS were bought from Shanghai Institutes for Biological Sciences, China. The human gastric epithelial cell line GES-1 was obtained from the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences (Beijing, China). MKN-45 and SGC-7901 cells were transfected with 100 nM si-circ-SFMBT2 or si-negative control (si-NC) using the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA). The si-circ-SFMBT2 sequences were as follows: si-1:GTCGGTGACTAAGCAATCAAA; si-2:GCGTCGGTGACTAAGCAATCA; si-3:CGGTGACTAAGCAATCAAAGA.
RNA isolation, reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA from paired tissues was extracted by using RNAsimple Total RNA Kit (TIANGEN, Beijing, China) and total RNA in plasma was extracted by TIANamp Virus RNA Kit (TIANGEN). RNA was reverse transcribed into cDNA using the Goldenstar™ RT6 cDNA Synthesis Kit (TSINGKE, Beijing, China). Circ-SFMBT2 expression level was detected using the following primer pair: 5′-GCGTCGGTGACTAAGCAATC-3′ (forward or F) and 5′- CCAATCCCACATAGCGAAGG-3′ (reverse or R). The primer pair of SFMBT2 is 5′-TCTGCGCTACTGCGGTTAC-3′ (F) and 5′-ACCAGTCAAGTCACGTATGAGAA-3′ (R). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control, with a primer pair 5′-GCACCGTCAAGGCTGAGAAC-3′ (F) and 5′- GGATCTCGCTCCTGGAAGATG-3′ (R). To accurately verify the expression of circ-SFMBT2, calculated Ct values were normalized against those of GAPDH that was amplified from the same sample (ΔCt = Cttested – CtGAPDH), and the −ΔCt method was used to estimate the difference value. Each sample was run in triplicates, and all reactions were repeated three times independently to ensure the reproducibility of all the data.
CCK-8 assay
The proliferation of MKN-45 and SGC-7901 cells was tested by CCK-8 kit (Dojindo, Kumamoto, Japan). Approximate 2×103 cells in 100 μL were incubated in triplicate in 96-well plates. At 0, 24, 48, 72 and 96 hours, the CCK-8 reagent (10 μL) was added to each well and incubated at 37°C for 2 hours. The optical density at 450 nm was measured using an automatic microplate reader.
Clone formation experiment
MKN-45 and SGC-7901 cells were transfected with 100 nM si-circ-SFMBT2 or si-NC. Each group of cells in the logarithmic growth phase was selected and digested with 0.25% trypsin and spun into single cells. The cells were suspended in RPMI-1640 containing 10% FBS and incubated in six-well plates at 37°C in 5% CO2 and saturated humidity for 2 weeks. The culture was terminated when a macroscopic clone appeared in the dish. We fixed the cells with methanol and stained them with 0.1% crystal violet. Then the colonies were well imaged and accurately counted in order to calculate the rate of clone formation.
Nucleus-cytoplasm fractionation
The GC cell layer was digested into single cells. We resuspended and washed them with 500 μL of PBS and centrifuged them for 5 minutes at 500×g at 4°C. Both nuclear and cytoplasmic RNAs from cultured GC cells were isolated by PARIS KIT 50 RXNS (AM1921; Life Technologies, Carlsbad, CA, USA) according to the manufacture’s instruction. U6 RNA and GAPDH-processed mRNA were detected in isolated RNAs as control for nuclear RNA and cytoplasm RNA, respectively. The abundance of circ-SFMBT2 and SFMBT2 mRNA was carried out using qRT-PCR. Each experiment was repeated for three times.
RNA fluorescence in situ hybridization (RNA-FISH)
The RNA FISH probe of circ-SFMBT2 was synthesized by Ribo Bio Technology Co. Ltd. (Guangzhou, China). Cells were first fixed with 4% paraformaldehyde for 10 minutes and then permeabilized in PBS with 0.5% Triton X-100 for 5 minutes. Next, the cells were hybridized with labeled FISH probe of circ-SFMBT2 at 37°C overnight. Afterwards, the cells were washed with 4× sodium citrate buffer containing 0.1% Tween-20 for 5 minutes and then washed with 1× SSC for 5 minutes. Finally, cells were stained with 4,6-diamidino-2-phenylindole for 10 minutes. All images were acquired by using confocal microscope.
Luciferase reporter assay
The circ-SFMBT2-wild type (circ-SFMBT2-WT), circ-SFMBT2-mutant (circ-SFMBT2-Mut), CREB1-WT and CREB1-Mut binding sites were inserted into the KpnI and SacI sites of the pGL3 promoter vector (Realgene, Nanjing, Shanghai, China). MKN-45 and SGC-7901 cells were seeded on a 96-well plate and cultured in a medium containing 10% FBS, incubated in a 37°C, 5% CO2 incubator for a period of time. They were co-transfected with luciferase reporters and miR-182-5 p mimics. After incubation for 48 hours, the firefly and Renilla luciferase activities were both measured with a dual-luciferase reporter assay (Promega, Madison, WI, USA) according to manufacturer’s instruction.
RNA immunoprecipitation (RIP)
All the steps were performed following the instructions of Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Approximately 1×107 cells were pelleted and resuspended with RIP lysis buffer plus protease and RNase inhibitors. The 100 mL cell lysates were incubated with human anti-AGO2 antibody (Abcam, Cambridge, MA, USA) or negative control (NC) mouse immunoglobulin G (IgG; Millipore) at 4°C overnight. The immunoprecipitated RNAs were extracted using the RNeasy MinElute Cleanup Kit (Qiagen, China) and reverse transcribed using the Golden-star™ RT6 cDNA Synthesis Kit (TSINGKE). The abundance of circ-SFMBT2 was detected by qRT-PCR assay.
Western blotting
Western blotting was performed following standard protocols with anti-CREB1 (1:1000; Abcam). Anti-GAPDH antibody (1:1000; Abcam) was used as a protein loading control. The electrochemiluminescence (ECL) was used to detect the amount of protein fluorescence. Before use, the ECL chromogenic solutions A and B were mixed in a ratio of 1:1 and uniformly added to the surface of the membrane. The fluorescein gel imaging system was developed, photographed and recorded.
Statistical analysis
An independent t-test was used to analyze the comparison of consecutive data. A receiver operating characteristic (ROC) curve was used to evaluate its diagnostic value. All statistical analyses were performed using SPSS for Windows version 17.0 and GraphPad Prism Software 5.0. For all results, P<0.05 was considered statistically significant.
Results
The biological structure of circ-SFMBT2
A total of three GC tissues and their matched non-GC tissues were collected and screened for dysregulated cir-cRNA using human circRNA microarray from the database (GSE78092).11 A total of 16 circRNAs were upregulated, and 84 circRNAs were downregulated in GC. We selected ten circRNAs and then used qRT-PCR to verify the expression levels of these dysregulated circRNAs in eight pairs of GC and noncancerous tissues. Results showed that the expression of circ-SFMBT2 was significantly upregulated in GC tissues compared to non-GC tissues, which was selected as a candidate target for GC for further study.
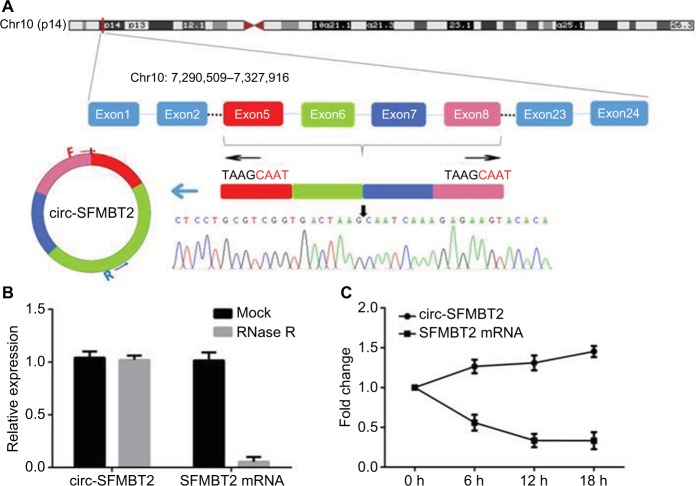
As shown in Figure 1A, circ-SFMBT2 was derived from exon 5-8 of the gene SFMBT2. Sanger sequencing was used to confirm the back-splice junction of circ-SFMBT2 and the specificity and correctness of the primers. Resistance to exonuclease as well as actinomycin D further confirmed that circ-SFMBT2 was a stable circRNA (Figure 1B and C).
Figure 1.
The biological structure of circ-SFMBT2. (A) Schematic diagram shows that circ-SFMBT2 is derived from SFMBT2 exon 5–8. The amplification products were used for Sanger sequencing to determine the correctness of circ-SFMBT2 primers. The divergent primers were designed to confirm the back-splice junction of circ-SFMBT2. (B) qRT-PCR for the abundance of circ-SFMBT2 and SFMBT2 mRNA in GC cells treated with RNase R. (C) qRT-PCR for the abundance of circ-SFMBT2 and SFMBT2 mRNA in GC cells treated with actinomycin.
Abbreviations: GC, gastric cancer; qRT-PCR, quantitative real-time PCR.
The expression of circ-SFMBT2 in GC and the association with clinicopathological information in GC patients
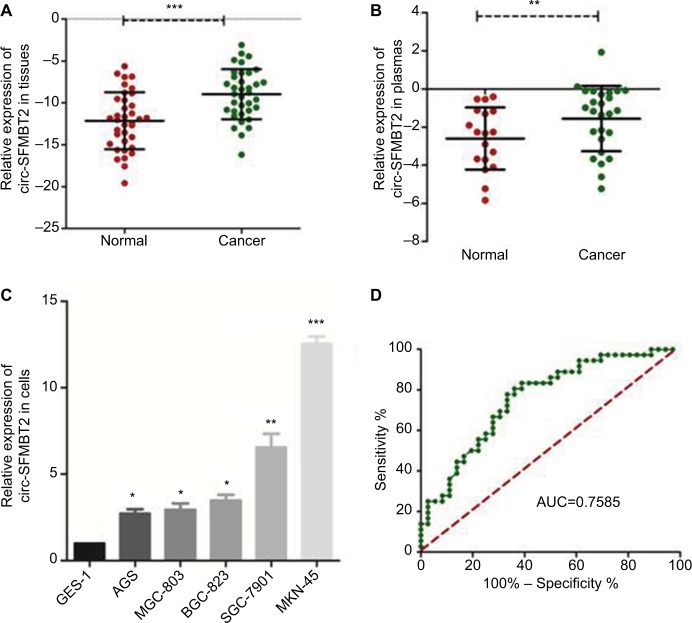
Using qRT-PCR, the circ-SFMBT2 expression level was tested within 36 paired cancerous and adjacent noncancerous tissues from GC patients, and the results showed that the expression of circ-SFMBT2 in GC tissues was significantly higher than that in adjacent noncancerous tissues (Figure 2A). In addition, the expression of circ-SFMBT2 in 26 GC plasma samples was measured. In addition, the expression of circ-SFMBT2 in 26 GC plasma samples and 18 healthy individuals’ plasmas were measured. The results indicated that circ-SFMBT2 expression was upregulated in GC plasma compared with healthy cases (Figure 2B). Then, by detecting the level of circ-SFMBT expression in five GC cell lines, we found that the expression levels in MKN-45 and SGC-7901 were the top two (Figure 2C). Furthermore, the area under the ROC curve of circ-SFMBT2 in distinguishing cancer tissues and normal ones was 0.7585 and the cutoff value was −11.46 with sensitivity of 80.56% and specificity of 63.89% (Figure 2D). Clinicopathological features showed that the circ-SFMBT2 level was not associated with age, gender, differentiation, lymphatic metastasis or common clinical biomarkers in patients with GC, whereas it was positively associated with TNM stage (Table 1).
Figure 2.
Relative circ-SFMBT2 expression and its clinical significance in GC.
Notes: (A) The expression of circ-SFMBT2 level in GC tissues was significantly higher than that of adjacent noncancerous tissues from GC patients. (B) The expression of circ-SFMBT2 level in GC plasmas was significantly higher than that of normal controls. (C) The expression of circ-SFMBT2 in AGS, BGC-823, SGC-7901 and MKN-45 cells was higher than that in GES-1 significantly. (D) The ROC curve has been used to evaluate circ-SFMBT2 potential diagnostic value; AUC was 0.7585. *P<0.05, **P<0.01, ***P<0.0001.
Abbreviations: GC, gastric cancer; ROC, receiver operating characteristic; AUC, area under the ROC curve.
Table 1.
The relationship of circ-SFMBT2 expression levels (−ΔCt) in gastric cancer tissues with clinicopathological factors of patients with gastric cancer
| Variables | No. of patients (%) | Mean ± SD | P |
|---|---|---|---|
|
| |||
| Age (years) | 0.491 | ||
| ≥65 | 15 | 2.57±4.07 | |
| <65 | 21 | 3.60±4.58 | |
| Gender | 0.064 | ||
| Female | 5 | −0.15±4.94 | |
| Male | 31 | 3.71±4.06 | |
| Diameter (cm) | 0.550 | ||
| ≥4 | 21 | 2.80±4.66 | |
| <4 | 15 | 3.69±3.92 | |
| Differentiation | 0.811 | ||
| Low/middle | 19 | 3.01±4.90 | |
| Well | 17 | 3.36±3.74 | |
| Lymphatic metastasis | 0.744 | ||
| Yes | 27 | 3.31±4.21 | |
| No | 9 | 2.75±4.92 | |
| TNM stage | 0.002* | ||
| I–II | 14 | 0.48±4.06 | |
| III–IV | 22 | 4.88±3.63 | |
| AFP | 0.284 | ||
| High | 16 | 4.16±3.98 | |
| Normal | 20 | 3.06±3.82 | |
| CEA | 0.624 | ||
| High | 13 | 3.71±3.05 | |
| Normal | 23 | 4.35±3.85 | |
Note:
P<0.05.
Abbreviations: AFP, α-fetoprotein; CEA, carcino-embryonic antigen; TNM, tumor-node-metastasis.
Silencing circ-SFMBT2 markedly inhibits GC cell proliferation
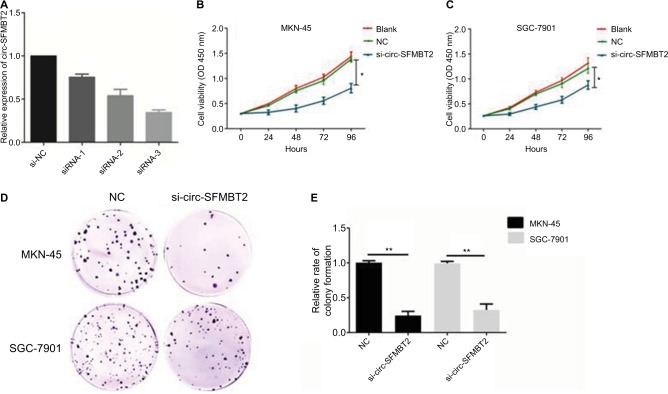
MKN-45 and SGC-7901 cells were transfected with si-circ-SFMBT2 and si-NC, respectively. At 48 hours after treatment, circ-SFMBT2 expression was effectively knocked down in GC cells (Figure 3A). Results showed that inhibiting circ-SFMBT2 expression significantly repressed proliferation in the CCK-8 assay (Figure 3B and C) and clone formation assay (Figure 3D). The invasion and migration ability of circ-SFMBT2 in GC cells was explored; however, no obvious difference was found.
Figure 3.
Circ-SFMBT2 promoted the proliferation of gastric cancer cells. (A) Knockdown of circ-SFMBT2 was confirmed via qRT-PCR, demonstrating the effective knockdown in GC cells. (B) Knockdown of circ-SFMBT2 inhibited cell proliferation significantly in MKN-45 cells. (C) Knockdown of circ-SFMBT2 inhibited cell proliferation significantly in SGC-7901 cells. (D) Clone formation assay showed that after knockdown of circ-SFMBT2, the population dependence and proliferation ability of gastric cancer cells were significantly decreased. (E) After knockdown of circ-SFMBT2, the relative rate of colony formation of gastric cancer cells was significantly decreased. *P<0.05, **P<0.01.
Abbreviations: NC, negative control; qRT-PCR, quantitative real-time PCR.
Circ-SFMBT2 competitively shares miR-182-5p with CREB1
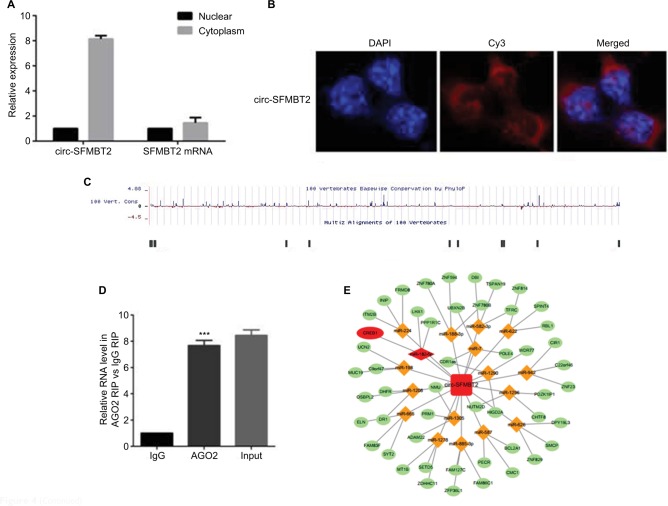
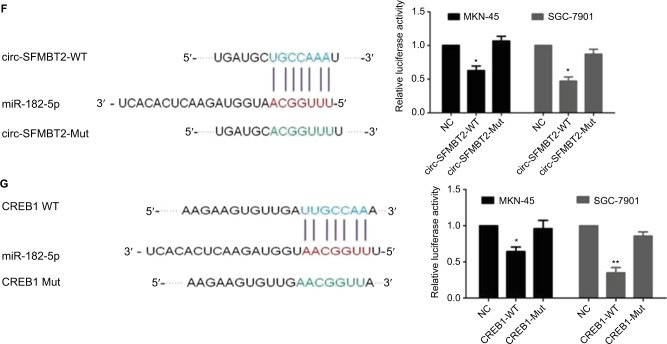
Increasing evidence indicates that circRNAs are able to regulate transcription and pathways by manipulating microRNAs in cancer cells. The qRT-PCR analysis of circ-SFMBT2 after nuclear and cytoplasmic separation experiment as well as FISH experiment demonstrated that circ-SFMBT2 was mainly localized in the cytoplasm (Figure 4A and B), providing the most basic premise for circ-SFMBT2 followup research. We downloaded the available data sets from doRiNA (http://dorina.mdc-berlin.de) and acquired the Ago2-binding sites of the circ-SFMBT2 genomic region. AGO2 immunoprecipitation analysis revealed circ-SFMBT2 had a high degree of AGO2 occupancy (Figure 4C). To validate this result, we conducted RIP for GC cells, and significant enrichment of AGO2 was observed compared with the IgG control (Figure 4D). Next, circRNA interactome (https://circinteractome.nia.nih.gov/) was used to predict circ-SFMBT2 binding sites with microRNAs (Figure 4E), and miR-182-5 p had a relatively higher context score percentile. To further validate the interaction between circ-SFMBT2 and miR-182-5 p, luciferase reporter assay was conducted and the result revealed that the miR-182-5 p-mimics induced a decrease in relative luciferase expression in circ-SFMBT2-WT group compared to the NC (Figure 4F). However, there was no difference in the circ-SFMBT2-Mut group between the miR-182-5 p mimics and the control. The target genes of miR-182-5 p were predicted by MIRDB (http://mirdb.org/) and CREB1 had a relatively higher target score. As shown in Figure 4G, miR-182-5 p mimics induced a decrease in relative luciferase expression in CREB1-WT compared with the control. These results suggest that circ-SFMBT2 could competitively share miR-182-5 p with CREB1 via acting as a sponge.
Figure 4.
Circ-SFMBT2 may competitively share miR-182-5p with CREB1. (A) Circ-SFMBT2 was predominantly localized in the cytoplasm. (B) FISH experiment demonstrated that circ-SFMBT2 was mainly localized in the cytoplasm. (C) AGO2 followed by high-throughput sequencing data from doRiNA revealed a high degree of AGO2 occupancy in the region of circ-SFMBT2. (D) RNA RIP experiment in MKN-45 cells showed significantly different enrichment of circ-SFMBT2 between AGO2 and IgG. (E) Circ-SFMBT2 was predicted to have binding sites with various miRNAs. (F) MiR-182-5p mimics induced a reduction in relative luciferase expression in circ-SFMBT2-Wild compared with the negative control in GC cells. (G) MiR-182-5p mimics induced a reduction in relative luciferase expression in CREB1-Wild compared with the negative control in GC cells. *P<0.05, **P<0.01,***P<0.001.
Abbreviations: FISH, fluorescence in situ hybridization; GC, gastric cancer; Mut, mutant; NC, negative control; RIP, immunoprecipitation; WT, wild type.
Circ-SFMBT2 promotes the proliferation of GC through sponging miR-182-5p to enhance CREB1 expression
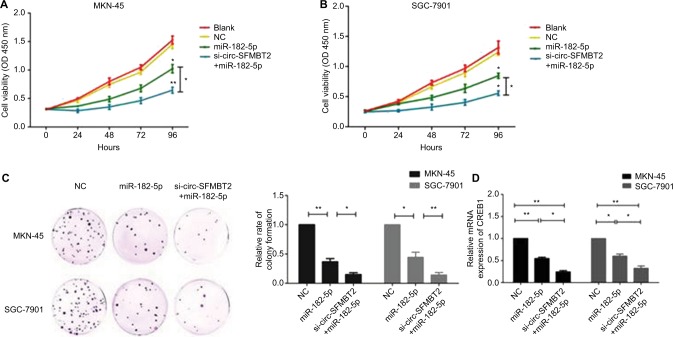
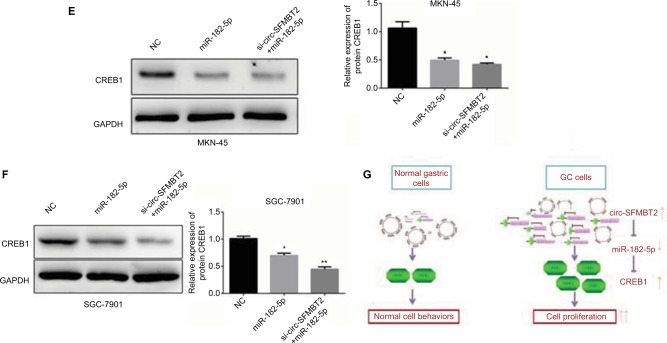
To explore whether circ-SFMBT2 participates in GC cell proliferation through sponging miR-182-5 p, we tested the proliferation ability of GC cells after adding miR-182-5 p mimics. Results showed that the proliferation ability of cells treated with si-circ-SFMBT2+ miR-182-5 p was significantly suppressed compared with those treated with miR-182-5 p in GC cells (Figure 5A–C). The mRNA and protein expression of CREB1 in the group treated with si-circ-SFMBT2+ miR-182-5 p was significantly decreased compared with the group treated with miR-182-5 p in GC cell lines (Figure 5D–F). These findings illustrate that circ-SFMBT2 takes part in the progress of GC through the cross talk with CREB1 by competing for miR-182-5 p (Figure 5G).
Figure 5.
Circ-SFMBT2 promotes the proliferation of GC cells through sponging miR-182-5p to enhance CREB1 expression. (A) The proliferation ability of cells treated with si-circ-SFMBT2+miR-182-5p was significantly suppressed compared with those treated with miR-182-5p in MKN-45 cells. (B) The proliferation ability of cells treated with si-circ-SFMBT2+miR-182-5p was significantly suppressed compared with those treated with miR-182-5p in SGC-7901 cells. (C) Clone formation assay showed that the population dependence and proliferation ability of gastric cancer cells treated with si-circ-SFMBT2+miR-182-5p was significantly decreased compared with those treated with miR-182-5p in GC cells. (D) The mRNA expression level of CREB1 in cells treated with si-circ-SFMBT2+miR-182-5p was significantly decreased compared with those treated with miR-182-5p in GC cell lines. (E) The protein level of CREB1 in cells treated with si-circ-SFMBT2+miR-182-5p was significantly decreased compared with those treated with miR-182-5p in MKN-45 cells. (F)The protein level of CREB1 in cells treated with si-circ-SFMBT2+miR-182-5p was significantly decreased compared with those treated with miR-182-5p in SGC-7901 cells. (G) Circ-SFMBT2 takes part in the progress of GC through the crosstalk with CREB1 by competing for shared miR-182-5p.*P<0.05, **P<0.01.
Abbreviations: GC, gastric cancer; NC, negative control.
Discussion
Recent studies have shown that circRNAs have several notable characteristics. First, circRNAs have a covalently closed loop structure, less susceptible to degradation by RNA exonuclease.12 Second, circRNAs are mainly composed of exons and contain miRNA response elements.13 Third, circRNAs often exhibit tissue-specific expression.14 Fourth, circRNAs can sequester RNA binding proteins (RBPs), leading to the formation of large RNA-protein complexes.15 These properties indicate that circRNAs have the potential to become an ideal biomarker for diagnosing cancers. The current study provided the first report of the clinical significance of circ-SFMBT2 in GC. We found circ-SFMBT2 was overexpressed in both GC tissues and plasma samples and could induce proliferation. Importantly, patients with higher circ-SFMBT2 expression had more serious tumor stage than patients with low circ-SFMBT2 expression, suggesting that circ-SFMBT2 plays a vital role in the progression of GC.
Accumulating evidence suggests that circRNAs are able to serve as microRNAs sponge in orthopedic system disorders, cardiovascular disease and diabetes.16–18 Our results showed that circ-SFMBT2 was mainly localized in the cytoplasm and had a high degree of AGO2 occupancy. Luciferase reporter assay was conducted and confirmed that circ-SFMBT2 could competitively share miR-182-5 p with CREB1 mRNA acting as a sponge. Previous study showed that miR-182-5 p was down-expressed in breast cancer stem cells, normal mammary stem cells and embryonic carcinoma and could deregulate RGS17 mRNA by targeting its 3′-UTR to suppress the occurrence of lung cancer.19,20 Most importantly, Kong et al reported that miR-182-5 p could suppress cell proliferation by directly targeting the CREB1 mRNA 3′-UTR in GC, which was consistent with our results.21
CREB1, encoding a transcription factor in the leucine zipper family of DNA binding proteins, is involved in glucose homeostasis, growth factor-dependent cell survival and memory, and can bind as a homodimer to the cAMP-responsive element, an octameric palindrome.22–24 Activated CREB1 recognizes conserved cAMP response elements and regulates downstream gene expression including bcl-2, cyclinA1, cyclin B1 and signal transduction proteins, which are linked with cell proliferation, differentiation and survival signaling pathways.25,26 Therefore, CREB1 is thought to be an oncogene that promotes the growth and proliferation of tumor cells.27 In our study, the proliferation of cells treated with si-circ-SFMBT2+ miR-182-5 p was significantly inhibited compared with those treated with NC in GC cells. Both the mRNA and protein level of CREB1 in cells treated with si-circ-SFMBT2+ miR-182-5 p was significantly decreased compared with those treated with NC in GC cell lines. These findings reveal that high expression of circ-SFMBT2 leads to an abnormally high expression of CREB1 by binding miR-182-5 p, consequently accelerating cell proliferation in GC.
Limitations
Several limitations should not be ignored when interpreting the results. First, all experimental samples are from one hospital; it is recommended that the samples from different hospitals and even different races could be studied. Second, although circ-SFMBT2 could combine with miR-182-5 p, we may guess whether or not circ-SFMBT2 could regulate the occurrence and development of GC through combining with more other miRNAs. Third, further study on whether or not circ-SFMBT2 regulates the occurrence and development of GC through other ways such as binding to RBPs is recom mended. It has been shown that 34% of the single circular exon contain the start codon in human fibroblasts, suggesting a widespread role of circRNAs as mRNA traps.28,29 As for circ-SFMBT2, an intriguing discovery is that we searched the circbank database (http://www.circbank.cn/) and found that circ-SFMBT2 has the potential of encoding proteins with a higher score of 0.9521. We look forward to further research to explore whether circ-SFMBT2 can indeed encode proteins, which will provide a deeper understanding of its capabilities.
Conclusion
In summary, our findings suggest that circ-SFMBT2 promotes the proliferation of GC through the cross talk with CREB1 mRNA by competing for miR-182-5 p. The clarification of the function of circ-SFMBT2 helps us to further understand the mechanisms of gastric carcinoma initiation and progression.
Consent for publication
All the authors agree to publish the manuscript.
Availability of data and material
All the data and material have been agreed by authors and Nanjing First Hospital.
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC) (No. 81672423, No. 81372656), Jiangsu Natural Science Foundation (BK20151292), Jiangsu Provincial Medical Innovation Team of the Project of Invigorating Health Care through Science, Technology and Education, 333 Project Foundation of Jiangsu Province (No. BRA2014355) for Xiufeng Cao and the Nanjing Science and Technology Fund (YKK16131) for Pengcheng Xi.
Footnotes
Disclosure
Handong Sun and Pengcheng Xi are the co-first authors. The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Isobe Y, Nashimoto A, Akazawa K, et al. Gastric cancer treatment in Japan: 2008 annual report of the JGCA nationwide registry. Gastric Cancer. 2011;14(4):301–316. doi: 10.1007/s10120-011-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capel B, Swain A, Nicolis S, et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73(5):1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 5.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li JF, Song YZ. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. 2017;39(7):1010428317709991. doi: 10.1177/1010428317709991. [DOI] [PubMed] [Google Scholar]
- 7.Liang HF, Zhang XZ, Liu BG, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–1576. [PMC free article] [PubMed] [Google Scholar]
- 8.Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai Z, Yang Y, Yan Y, et al. Analysis of co-expression networks for circular RNAs and mRNAs reveals that circular RNAs hsa_circ_0047905, hsa_circ_0138960 and has-circRNA7690-15 are candidate oncogenes in gastric cancer. Cell Cycle. 2017;16(23):2301–2311. doi: 10.1080/15384101.2017.1380135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang M, He YR, Liang LC, Huang Q, Zhu ZQ. Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol. 2017;23(34):6330–6338. doi: 10.3748/wjg.v23.i34.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang YS, Jie N, Zou KJ, Weng Y. Expression profile of circular RNAs in human gastric cancer tissues. Mol Med Rep. 2017;16(3):2469–2476. doi: 10.3892/mmr.2017.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. Embo J. 2013;32(7):923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Zhang X, Hu X, et al. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 ‘Sponge’ in Human Cartilage Degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 18.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimono Y, Zabala M, Cho RW, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, Fang R, Li C, et al. Hsa-mir-182 suppresses lung tumorigenesis through down regulation of RGS17 expression in vitro. Biochem Biophys Res Commun. 2010;396(2):501–507. doi: 10.1016/j.bbrc.2010.04.127. [DOI] [PubMed] [Google Scholar]
- 21.Kong WQ, Bai R, Liu T, et al. MicroRNA-182 targets cAMP-responsive element-binding protein 1 and suppresses cell growth in human gastric adenocarcinoma. FEBS J. 2012;279(7):1252–1260. doi: 10.1111/j.1742-4658.2012.08519.x. [DOI] [PubMed] [Google Scholar]
- 22.Shankar DB, Cheng JC, Sakamoto KM. Role of cyclic AMP response element binding protein in human leukemias. Cancer. 2005;104(9):1819–1824. doi: 10.1002/cncr.21401. [DOI] [PubMed] [Google Scholar]
- 23.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 24.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 25.Montminy M, Brindle P, Arias J, Ferreri K, Armstrong R. Regulation of somatostatin gene transcription by cyclic adenosine monophosphate. Metabolism. 1996;45(8 Suppl 1):4–7. doi: 10.1016/s0026-0495(96)90068-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Odom DT, Koo SH, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankar DB, Cheng JC, Kinjo K, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7(4):351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]