Summary
In this review, we give an overview of the four large classes of small non-coding RNAs: miRNAs, piRNAs, snoRNA and the new class of tRNA-derived fragments describing their biogenesis and role in cancer development and progression, as well as in drug resistance.
Abstract
The ENCODE project has reported that at least 80% of the human genome is biologically active, yet only a small part of human DNA encodes for protein. The massive amount of RNA transcribed but not translated into protein can be classified as housekeeping RNA (such as rRNA, tRNA) and regulatory RNA (such as miRNA, piRNA, lncRNA). Small non-coding RNAs, in particular, have been the focus of many studies in the last 20 years and their fundamental role in many human diseases is currently well established. Inter alia, their role in cancer development and progression, as well as in drug resistance, is being increasingly investigated. In this review, focusing our attention on recent research results, we provide an overview of the four large classes of small non-coding RNAs, namely, miRNAs, piRNAs, snoRNA and the new class of tRNA-derived fragments, highlighting their fundamental role in cancer and their potential as diagnostic and prognostic biomarkers.
Introduction
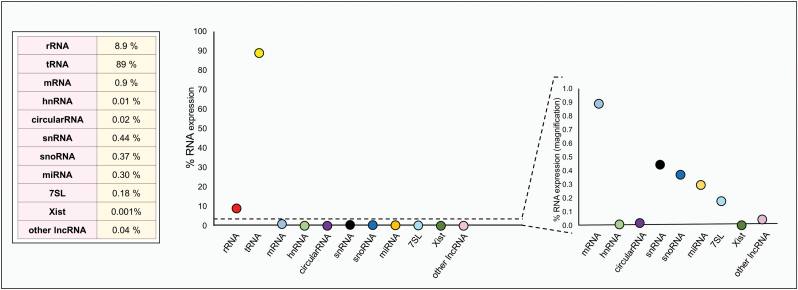
It is now clear that 98% of human DNA is non-protein coding. For many years, scientists considered this portion of the human genome as ‘junk’ DNA, while it actually is common thought that ‘junk is not discarded but stored away for some potential use later’ (1). The discovery milestones regarding DNA are many, from the first nucleic acid isolation and identification in 1869 by Johannes Friedrich Miescher, passing through Edward Tatum and George Beadle’s ‘one gene, one enzyme’ hypothesis in 1941, as well as James Watson and Francis Crick’s description of the DNA structure in 1953, ending in 2001 with the complete sequencing of the human genome, just to name a few. Nonetheless, only in 2012 the ENCODE project, in which researchers investigated deeper into the until-then-called ‘junk DNA’, uncovered the evidence proving that at least 80% of the human genome is biologically active (2). As cis/trans-regulatory elements, introns, pseudogenes, repeat sequences and telomeres are all part of this junk DNA, a consistent part of it is thus transcribed in non-coding RNA comprising functional RNA molecules. This class of RNA transcripts is composed by highly abundant ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs), as well as a small but fundamental fraction of other RNA types, such as small nucleolar RNA (snoRNAs), microRNAs (miRNAs), siRNAs, snRNAs, Piwi-interacting RNA (piRNAs) and long ncRNAs. Palazzo et al. (3) estimated that 99% of the total RNA content in mammalian cells is comprised of non-coding RNA (Figure 1). Specifically, we do not yet know the exact quantity of truly functional ncRNAs, as the list of validated ncRNA transcripts is growing year by year. Currently, we can classify ncRNAs by length (small 18–200 nt; long > 200 nt) or by function [housekeeping ncRNAs, such as rRNAs and transfer RNAs (tRNAs), as well as regulatory transcripts, such as miRNAs, piRNA, small non-coding transfer RNA-derived RNA fragments (tRFs), long non-coding RNAs (lncRNAs)] (4). The role of housekeeping ncRNAs (tRNAs and rRNA) is widely known and characterized. In the last two decades, a lot of emphasis has been given to the study of regulatory RNAs. The substantial progress made in the elucidation of the biogenesis and functions of these ncRNAs has produced ample evidence of the fundamental role played by these molecules in virtually all biological pathways. Deregulation of miRNAs, piRNA and tRFs, e.g. is implicated in several metabolic diseases as well as in cancer (5,6). In this review, we provide an overview of the most representative classes of sncRNA and their involvement in cancer, focusing on their role as regulatory RNAs as well as their biomarker potential.
Figure 1.
Estimated expression percentage of different RNA species in typical mammalian cells (Adapted from Palazzo et al.).
microRNAs
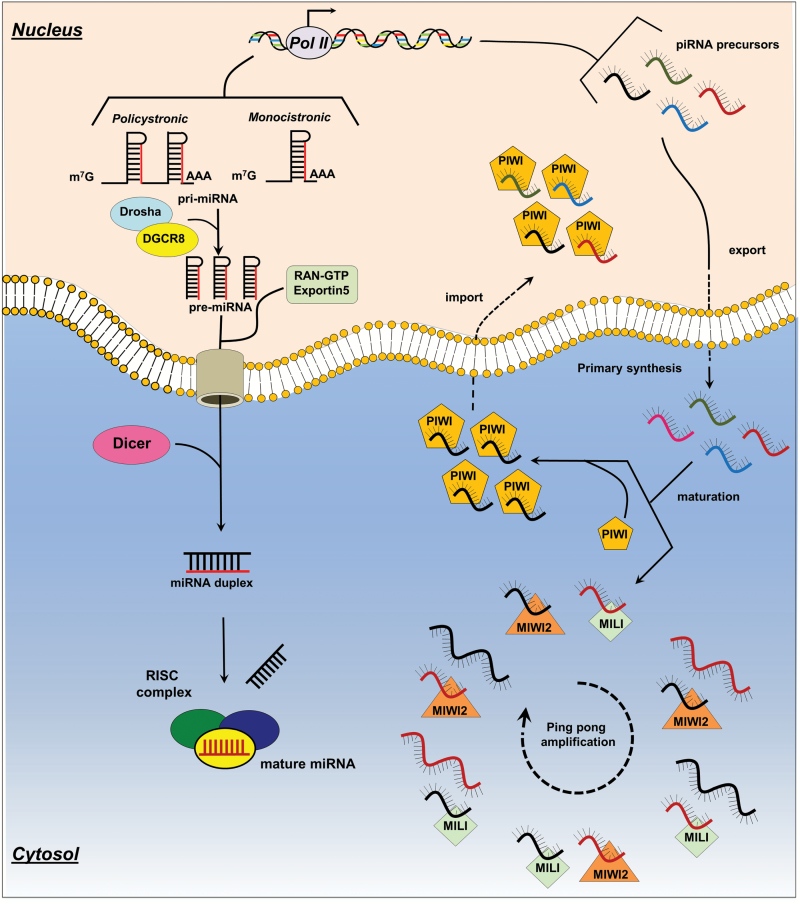
microRNA are 22nt-long non-coding RNA molecules, highly conserved and present in all eukaryotic cells. As of June 2014, in the latest release of miRBase (v.21), the official reference knowledge base on miRNAs, the miRNA class comprises 2588 mature miRNAs in human, approximately 1915 in mouse, 434 miRNAs in Caenorhabditis elegans, and 466 in Drosophila melanogaster. Canonically, miRNAs are encoded by introns of coding or non-coding transcripts, while some miRNAs are encoded by exonic regions (7). They are transcribed as large mono- or polycistronic primary miRNA precursors (pri-miRNAs) by RNA Pol. II (8) (Figure 2) For this reason, canonical pri-miRNAs contain an m7G cap at the 5′ end, and a poly(A) tail at the 3′ untranslated region (9). Additionally, some pri-miRNAs are transcribed by RNA Pol. III as C19MC, a polycistronic pri-RNA containing 46 miRNA genes (10). miRNA maturation is a multistep process which starts in the nucleus and ends in the cytoplasm: miRNAs are excised by RNase III Drosha in complex with DGCR8, a double-strand RNA-binding protein acting as ruler to measure the cleavage point. At this point, the small hairpin-shaped precursor RNA of 65 nucleotides (pre-miRNA) (11) is subsequently recognized and exported in the cytosol by Exportin5 (Exp5) (12), where it is cleaved by a RNase III/ double-strand RNA-binding protein complex, represented, in mammalian cells, by the RNase III DICER and by two double-strand RNA-binding protein, transactivation-response element RNA-binding protein (TRBP) and protein activator of the interferon-induced protein kinase (PACT) (13). Dicer is then immediately loaded onto an Argonaute protein complex termed RNA-induced silencing complex (RISC). Through the RISC complex, the mature miRNA, after specific target recognition, induces post-transcriptional gene silencing canonically by binding to target sites found within the 3′ untranslated region of the targeted mRNA (14). miRNA/mRNA interaction hinders protein synthesis and may initiate mRNA degradation. For miRNA target recognition, a fundamental component is represented by the ‘seed sequence’, a conserved Watson–Crick pairing miRNA region centered on nucleotides 2–7 (15). The natural structure of miRNAs allows them to target up to several hundreds of transcripts revealing them to be very powerful regulators whose aberrant expression can perturb a great number of cell signaling pathways thus having a profound impact on cancer onset and progression. Since the publication of the first evidences of the role of miRNAs in cancer (16,17), about 15 years ago, thousands of papers have assessed that miRNA play a fundamental role in the development and progression of cancer. Currently, we can affirm that the dysregulation in miRNAs signatures is implicated in virtually all stages of this disease: development, progression, metastasis and drug resistance (5). It has been established that miRNA signatures can discriminate between normal and cancer tissue, as well as different subtypes of a particular cancer (18). They have also been proven to play a fundamental role in drug resistance and as biomarkers for early diagnosis. In the last few years, miRNA studies have deeply investigated their role in drug resistance: the elucidation of chemo-resistant mechanisms in which they are involved will allow to improve patient treatment either by employing more specific drugs or by increasing the synergic effect of the combination of multiple drugs. The miRNA expression pattern in chemo-resistant cancer cells often differs when compared with that in their parental chemo-sensitive cells (19). miR-130, miR- mir-494 can regulate cell survival and TNF-related apoptosis-inducing ligand (TRAIL) resistance in non-small cell lung cancer (NSCLC) cell lines (20,21). Fang et al. (22) found that miR-17 was consistently over expressed in a group of chemo-resistant colon cancer patients compared with a sensitive one, also revealing how its ectopic over expression in sensitive colon cell lines turns them into resistant cells. Furthermore, the miR-221/222 cluster has been proven to be implicated in drug resistance in different tumors, being upregulated in lung (TRAIL resistance) (23) and breast (Tamoxifen and Fulvestrant) (24,25). Also, miR-34 has shown to regulate drug resistance in gastric, prostate and breast cancers as well as in CLL (19). Finally, there is emerging evidence that miRNAs undergo sequence modification through the RNA editing machinery (26,27). All of this evidence reveals how miRNAs possess a high potential as diagnostic and prognostic biomarkers in cancer. Taking into account that miRNAs have also been found in plasma and serum along with a substantial deregulation of their expression in cancer patient samples even in distinct early stages of the disease, a new perspective in cancer prevention and early detection has opened (28,29). It is possible, indeed, to employ miRNA signatures as clinical biomarkers for diagnostic, predictive and prognostic purposes. Finally, by virtue of the profound knowledge acquired regarding miRNA modus operandi gained in the last 15 years, it has been possible to exploit their molecular characteristics to design artificial miRNAs able to target multiple genes in multiple sites (30), thus having the possibility to employ such artificial molecules to downregulate multiple proteins in the same pathway of interest, reducing off-target effects.
Figure 2.
Schematic representation of miRNA (left) and piRNA (right) biogenesis. Primary miRNAs (pri-miRNAs) product by RNA POL II can be monocistronic if they carry just one mir or polycistronic if they contain multiple miRNAs. While in the nucleus, pri-miRNAs are cleaved by the Drosha/DGR8 complex into 65nt precursor miRNAs (pre-miRNAs) and then exported by the Exportin5/RAN-GTP complex into the cytoplasm where they are in turn cleaved by the Dicer/TRBP/PACT complex which thus produces a miRNA duplex ready to be incorporated into the RISC complex; only one strand of the miRNA is preferentially selected to be coupled with the RISC complex. On the right, piRNA precursors are produced through the primary processing pathway, transcribed by POL II and exported into the cytosol where they are cut into mature piRNAs. Then, mature piRNAs in complex with PIWI proteins migrate to the nucleus where they silence TEs. In addition, MIWI2 and MILI coupled with mature piRNAs, cleave transcripts bearing sites complementary to the piRNA sequence, thus amplifying mature piRNA species through the ping-pong pathway.
piRNAs
PIWI-interacting RNAs (piRNAs) are single-stranded ncRNAs of 26–31 nucleotides that interact with P-element-induced wimpy testis (PIWI) proteins, a germ line-specific Argonaute family. piRNAs display a very diverse set of nucleotide sequences when compared with any other known cellular RNA family, comprising also the largest known class of ncRNAs. Discovered in 2006 in mouse testes independently by four groups (31–34), piRNAs have been shown to be implicated in the silencing of retrotransposons, both at the post-transcriptional and epigenetic levels, as well as of other genetic elements in germ lines, particularly those during spermatogenesis (35). They are 5′ monophosphated and 2′-O-methyl modified in the 3′ terminal, characteristics which have been proposed to increase piRNA stability. Although in the last few years, the molecular mechanisms of piRNA biogenesis and function have been thoroughly studied, there are still some gaps that need to be addressed, inter alia the production of mature piRNA and the elucidation of their cellular functions (36). We can classify piRNAs on the basis of their origin: (1) transposon-derived piRNA, (2) lncRNA-derived piRNAs, (3) mRNA-derived piRNAs, with only the function of the first group being well understood (37). Although it is not fully clear how precursors are processed into mature piRNAs, two mechanisms have been characterized: (1) primary synthesis and (2) ping pong amplification. The vast majority of piRNAs are clustered in relatively short genomic loci, on chromosomes 17, 5, 4 and 2 (38). They are transcribed by RNA polymerase II as long transcripts which are then exported to the cytoplasm and processed into smaller sequences (mature piRNAs) by unknown protein complexes in a still unclear DICER-independent manner (36). Most of the studies have investigated piRNAs in Drosophila, but several mammalian orthologs were also identified. It is currently known that in the piRNA primary maturation complex some mitochondrial proteins are essential, namely Zuc (mitoPLD in mouse), Mino (GPAT1/2 in mouse), GasZ (GASZ in mouse) and Armi (MOV10L1 in mouse). Although mitochondrial proteins have been identified to be essential for piRNA primary maturation, it is still unclear whether the mitochondrial activity is actually required for piRNAs biogenesis (39). Mature piRNAs in cytoplasm form a complex with PIWI proteins and migrate back into the nucleus, reaching their target transcripts and mobilizing the silencing machinery to block the transcription of transposable elements, maintaining genome integrity (40). The “ping-pong” amplification mechanism occurs fully in the cytoplasm. Specifically, piRNAs, in association with AGO3 or AUB proteins in Drosophila (MILI and MIWI2 in mouse), recognize transposon transcripts containing clusters of identical (or near identical) sequences and, after annealing such sequences through AGO3/AUB slicer activity, it allows the cleavage of these active transposable elements, producing new antisense piRNAs which can, in turn, displaying a loop mechanism, target other piRNA-cluster transposons (41). This general ping-pong framework is similar in various species, as established in some studies (42,43) (Figure 2). Though the biogenesis and function of piRNAs are still not perfectly clear, through deep sequencing technology many studies have managed to compare different expression profiles of these small non-coding RNAs (sncRNAs), in different tissue. Particularly, researchers have sought to analyze a possibly differential expression of these sncRNAs in tumor tissues compared with normal tissues, along with investigating their implication in metastatic disease as well as their presence in peripheral blood. For example, in NSCLC, piR-L-163 was found downregulated in such cancer cell line compared with an epithelial cell line, also proving to be able to regulate migration, invasion and also cell proliferation by accelerating G2-M accumulation (44). In breast, piR-34736, piR-36249, piR-35407, piR-36318, piR-34377, piR-36743, piR-36026 and piR-31106 were found significantly differentially expressed between tumor and matched non-malignant tissue (45). In another study, piR-4987, piR-20365, piR-20485 and piR-20582 were found upregulated in tumor tissue compared with normal (46). Also, piR-651 was found overexpessed in gastric cancer paired tissue, colon, lung and breast cancer tissues as well as hepatic carcinoma, mesothelioma, cervical, breast and lung cancer cell lines (47). Finally, piR-32051, piR-39894, piR-43607 belong to the same cluster on chr 17 and they were found up-regulated in kidney cancer tissue compared with normal (48). The role of miR-piR-823 in tumorigenesis is a moot point as it was found upregulated in multiple myeloma patients, while downregulated in gastric patient samples (49,50). Nevertheless, piR-651 and piR-823 were both investigated as potential plasma biomarkers in gastric cancer having been proven to discriminate between healthy control and tumor patients, being also associated to tumor stage and distant metastasis (51).
tRNA-derived fragments: mature tRNA-related and tsRNAs?
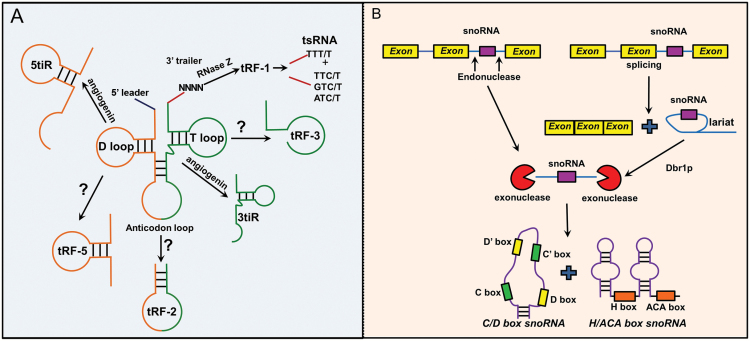
Recently, researchers, while sequencing size-selected short RNAs (<40nt) in the attempt to discover novel miRNAs, have realized that a number of sequencing reads actually map to RNA fragments derived from the cleavage of tRNA transcripts. Emphasis on this new class of small RNAs has grown in the last few years as their potential implication in the suppression of gene expression as well as many other cell functions, such as regulation of apoptosis and trans-generational epigenetic inheritance (52), is emerging. In the beginning, researchers had misclassified these ncRNAs as piRNAs or miRNAs without realizing they were in front of a novel ncRNA species. In reality, the classification of this rather new class of ncRNA is based on their length and the location they map to on the primary or mature tRNA transcript. The first group of tRNA-derived fragments (tRFs) discovered comprises the stressed- and starvation-induced tRNA fragments (tiR), or tRNA halves, 31–40 nt in length, cleavage product of the anticodon loop of mature tRNAs. The cleavage of these ncRNA fragments is performed by angiogenin, a ribonuclese (RNAse A or T), as tRNAs possess a 5′ hydroxyl rather than a 5′ phosphate as miRNAs, which are instead cleaved by Dicer, and as all RNAs cleaved by RNase type III (53). There are two subclasses of tRNA halves, namely, 5tiR and 3tiR, according to which segment of the anticodon they include, whether 5′ or 3′ (54). Several tiRs are found highly expressed in a sex hormone-dependent manner, respectively, in estrogen receptor-positive breast and androgen receptor-positive prostate cancer cell lines. Additionally, they have been found to be significantly functionally involved in cell proliferation, thus confirming a novel tRNA-engaged pathway in the tumorigenesis of hormone-dependent cancers and promoting tiRNAs as potential candidates for biomarkers and therapeutic targets (55). Another class of tRFs, which has received a lot of attention due to its similarity to miRNAs, is represented by 14–30-nt long ncRNAs mapping to the ends of mature tRNA or primary tRNA transcripts, displaying a 5′ phosphate and 3′ hydroxyl group (56). They are classified on the basis of their mapping positions: tRF-5, tRFs-3 and tRFs-1, with the latter originating from the cleavage of 3′ ends of primary tRNA transcripts, while tRF-5 and tRFs-3 originating from 5′ and 3′ ends of mature tRNA, respectively. They can be also subclassified by their length as: tRFs-5a (14–16 nt), tRFs-5b (22–24 nt), tRFs-5c (28–30 nt), tRFs-3a (18 nt) and tRFs-3b (22nt). The enzymes responsible of cleaving tRF-5s and tRF-3s from the mature tRNA are currently unknown. The possible role of Dicer and/or Drosha has been excluded due to evidence proving the accumulation of these tRNA fragments also in DICER/ DROSHA knock down cells. Furthermore, the existence of the 5′ phosphate and 3′ hydroxyl groups at their extremities excludes the possible involvement of RNase type A endonucleases (52). tRF-1s, instead, are derived from the cleavage of the trailer sequence of tRNA precursors by RNase Z, before the addition of the ‘CCA’ sequence to the mature tRNA. As the location of the stop signal for RNA Pol III for the 3′ end of pre-tRNAs differs substantially among distinct tRNA species, the length distribution of tRF-1s is not discrete as with tRF-3s and tRF-5s (57). A final subclass of tRFs is represented by the less common tRF-2s, essentially generated only from the anticodon stem loop of tRNAs (58) (Figure 3). tRFs have different localization, with tRF-1s and tRF-3s apparently mostly localized in the cytoplasm, conversely to tRF-5s which are found in the nucleus. Inasmuch as tRNA maturation is a nuclear event, at least tRF-1s seem to be selectively transported into the cytoplasm through some yet unknown mechanism (59,60). Similarly, the existence of a mechanism that transports tRFs-5s back into nucleus has also been hypothesized. tRFs have been detected in several organisms, from viruses to human, going through bacteria, protozoa, plants, mice and chicken (52), in most of which tRFs have been associated to a biological role. Additionally, a specific tRF-1, tRF-1001, has been positively correlated with cell proliferation through promotion of the M-G2 transition in prostate cancer cell lines. In 2013, a tRF-3, tRF-3027 (named by the authors ‘CU1276’), was found associated with Ago proteins and expressed in memory B cells but not in transformed B cells, with lymphoma biopsies indicating a downregulation of this ncRNA during malignant transformation of B cells. The authors also found that tRF-3027 repressed the expression of RPA1, a DNA-binding protein implicated in DNA repair modulation and in cell proliferation (61). tRFs-5030 showed to repress a target mRNA (in a miRNA-like fashion) in the cytoplasm, promoting virus replication (62), while, in association with a PIWIL4 protein (in piRNA-like fashion) it is able to repress CD1a expression in monocytes (63). Finally, in 2015 Goodarzi et al. (58) have shown how a set of tRF-2s can act as tumor suppressors, binding and sequestering YBX1, an RNA-binding protein able to stabilize relevant oncogenic transcripts. In a very recent paper, researchers identified a differential signature for a category of pre-tRNA derived fragments termed tRNA-derived small RNAs (tsRNAs) in CLL and lung cancer, proving that such molecules are dysregulated in human cancers (64). Moreover, they also provided evidence that tsRNAs interact with Ago1, Ago2 and PiwiL2 (64). Two years before, the same group found that miR-3676 (now identified as a tsRNA instead) targets TCL1 and results co-deleted, along with p53, in CLL, such deletion contributing significantly to the malignant phenotype (65).
Figure 3.
Schematic representation of t-RF and snoRNA biogenesis: (A) All t-RF-1s (including tsRNAs) derive from the maturation of tRNA. After Pol III transcription, the tRNA 5′ sequence (leader) is removed by RNase P, whereas the 3′ end (trailer) is cleaved by the tRNase Z enzyme. The stress (and starvation)-induced tRNA fragments termed tiRNA (tiRs) (or tRNA halves) are the result of the cleavage carried out by angiogenin and they are called 5’- or 3’- according to which segment of the anticodone loop they include. The enzymes responsible of cleaving all other tRFs are still unknown. (B) Small nucleolar RNAs (snoRNAs) are predominantly located in introns. The snoRNA biogenesis pathway is essentially represented by endonuclease cleavage after mRNA splicing. Lariats generated after mRNA splicing, and carrying snoRNA, are linearized by the Debranching RNA Lariats 1 protein (DBR1). snoRNAs can alternatively be directly excised by endonucleases from pre-mRNA before splicing. In both cases, exonucleases finally release the mature snoRNA.
snoRNAs
The snoRNAs are a class of small (60–300 nts) non-coding RNAs implicated in the chemical modification of rRNA. They function as a guide for the post-transcriptional modification of rRNA but in recent years a new role in the regulation of other cellular pathways has emerged (66). There are two main classes of snoRNAs identified on the basis of sequence/structure/function: box C/D snoRNA and box H/ACA snoRNA. A third less represented class are the small Cajal body-specific RNAs (scaRNAs) that are associated to Cajal bodies, small membrane-less sub-compartments of the nucleus, and are involved principally in post-transcriptional small RNA modification (66). The box C/D snoRNA are 60–200nt characterized by two highly conserved boxes: the C box (with RUGAUGA canonical motif) and the D box (with CUGA canonical motif), their principal role being that of carrying out the 2′-O-ribose methylation of specific rRNA residues (67,68). The H/ACA box snoRNAs are 120–250 nt RNAs characterized by a structure with two harpins connected by a region with an H box (ANANNA where N can be any nucleotide). The ACA box is located three residues upstream of the 3′ untranslated region of the molecule. Their principal role is pseudouridylation of rRNAs (67,68). Finally, scaRNAs are longer then the other two and display both C/D and H/ACA boxes, in addition to a CAB box (UGAG) which represents the Cajal body localization signal. They are implicated both in pseudouridylation and 2′-O-ribose methylation (69). All snoRNAs work in complex with specific protein partners in order to form small nucleolar ribonucleoprotein (snoRNPs) complexes. Practically, snoRNAs recognize and bind complementary sequences on target rRNAs and signal to partner proteins the exact base to modify. They are hosted in introns of coding and non-coding transcripts (at times in introns of pseudogenes of the protein partners in snoRPN complexes) (70) (Figure 3). The role of snoRNAs in rRNA biogenesis has been well documented but in the last few years some studies have highlighted other possible roles of these sncRNA in cellular regulation as well as a role in cancer development and progression (71). Additionally, there is some evidence that snoRNAs can act in miRNA-like post-transcriptional gene silencing, the first evidence of which is represented by ACA42, a H/ACA scaRNA able to downregulate CDC2L6 in a mirna-like manner (72). Brameier et al. (73) identified numerous snoRNA-derived molecules with miRNA-like functions, including H/ACA box snoRNAs and C/D box snoRNAs. In 2009, Dong et al. proved the implication of snoRNA U50 in breast cancer. Indeed, this snoRNA happens to map in 6q14.3 which has also been reported to include the translocation breakpoint in large B cell lymphoma. U50 is also downregulated in prostate cancer (74). SNORA42 is frequently upregulated in NSCLC and its down regulation reduces colony formation, induces apoptosis in NSCLC as well as inhibit tumor formation in a mouse model. Furthermore, in clinical cancer samples, high expression of this snoRNA is correlated to a poor prognosis (75). In 2010, Liao et al. screened surgical specimens from 22 stage I NSCLC patients and found 6 snoRNAs deregulated: SNORD33, SNORD66, SNORD73B, SNORD76, SNORD78 and SNORA42. Three of them (SNORD33, SNORD66 and SNORD76) displayed higher plasma expressions in NSCLC patients compared with cancer-free individuals (76). In a more recent study, Mannor et al. (77) found 22 snoRNAs to be deregulated in tumor-initiating cells (TICs) of NSCLC, particularly with the expression of two snoRNAs (snoRA3 and snoRA42) being inversely associated with survival of NSCLC patients.
Conclusion
In the past 20 years, many studies have deeply investigated the role of ncRNAs in cell biology and, particularly, in cancer development and progression, as well as their involvement in drug resistance. Now we know that they participate in practically all cellular processes, playing a fundamental role at all cancer stages, many of them having been selected as good biomarkers for early cancer detection or drug response. In particular, the role of miRNAs in post-transcriptional regulation has been very well characterized and currently their involvement in cell–cell communication has also started to be thoroughly studied (78). The new frontier with miRNAs is now represented by the implementation of effective delivery methodologies for these small RNAs for cancer therapy. At present, there is a wide gap between the in vitro and in vivo applications. The main issues stem from biological barriers (i.e. cellular nuclease or physical barriers such as the blood–brain barrier) and the specificity of delivery (to a specific tissue). Currently, RNA modification and the employment of particular vectors and targeted conjugates are the elected strategies. At the present moment, there are two miRNA-based drugs in clinical trial: LNA-modified-anti-miR-122 (SPC3649; http://www.santaris.com) for the treatment of the Hepatitis C Virus (HCV) (ClinicalTrials.gov Identifier: NCT00979927), and Mirna Therapeutic’s MRX34 in treatment of renal cell carcinoma, acral melanoma and hepatocellular carcinoma (ClinicalTrials.gov Identifier: NCT01829971). The other classes of sncRNAs were less deeply investigated than miRNAs and their biogenesis and functions are still far from being fully elucidated. In the last few years, new technologies, such as next-generation sequencing, have allowed to collect a substantial amount of data at lower cost in a shorter period of time than previously, making it easier to perform comparisons between different samples (79). For this reason, many recent studies focalized on finding a diagnostic/prognostic signature of sncRNAs in different types of cancer, or in cancer versus normal tissue, or even among patients at different cancer stages, as well as in drug resistance, taking the emphasis away from biogenesis and function studies. Moreover, the concurrent publication of new studies by different laboratories on new sncRNAs (as it took place with tRFs and tsRNA) has hindered the establishment of a standardized nomenclature. In addition, the challenges posed by the specific genomic contexts characterizing certain sncRNA species has lead to high debate regarding the appropriate strategy to employ regarding the correct identification and quantification of said species amongst the immense amount of next-generation sequencing data produced (79,80). In conclusion, although much has been learned regarding sncRNAs, it is just the tip of the iceberg as we still need to shed light on many aspects of their biogenesis and function, as well as address the necessity of an appropriate methodology for the therapeutic delivery of these small RNAs.
Conflict of Interest Statement: None declared.
Abbreviations
- miRNA
microRNA
- NSCLC
non-small cell lung cancer
- piRNA
Piwi-interacting RNA
- rRNA
ribosomal RNA
- sncRNA
small non-coding RNA
- snoRNA
small nucleolar RNA
- snoRNP
small nucleolar ribonucleoprotein
- sncRNA
small non-coding RNA
- tRNA
transfer RNA
- tRFs
transfer RNA-derived RNA fragments
- tsRNA
tRNA-derived small RNAs
References
- 1. Elgar G., et al. (2008) Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet., 24, 344–352. [DOI] [PubMed] [Google Scholar]
- 2. Pennisi E. (2012) Genomics. ENCODE project writes eulogy for junk DNA. Science, 337, 1159–1161. [DOI] [PubMed] [Google Scholar]
- 3. Palazzo A.F., et al. (2015) Non-coding RNA: what is functional and what is junk? Front. Genet., 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dozmorov M.G., et al. (2013) Systematic classification of non-coding RNAs by epigenomic similarity. BMC Bioinformatics, 14 (Suppl. 14), S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Acunzo M., et al. (2015) MicroRNA and cancer–a brief overview. Adv. Biol. Regul., 57, 1–9. [DOI] [PubMed] [Google Scholar]
- 6. Esteller M. (2011) Non-coding RNAs in human disease. Nat. Rev. Genet., 12, 861–874. [DOI] [PubMed] [Google Scholar]
- 7. Ha M., et al. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol., 15, 509–524. [DOI] [PubMed] [Google Scholar]
- 8. Lee Y., et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J., 23, 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chu C.Y., et al. (2007) Small RNAs: regulators and guardians of the genome. J. Cell. Physiol., 213, 412–419. [DOI] [PubMed] [Google Scholar]
- 10. Borchert G.M., et al. (2006) RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol., 13, 1097–1101. [DOI] [PubMed] [Google Scholar]
- 11. Lee Y., et al. (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature, 425, 415–419. [DOI] [PubMed] [Google Scholar]
- 12. Lund E., et al. (2004) Nuclear export of microRNA precursors. Science, 303, 95–98. [DOI] [PubMed] [Google Scholar]
- 13. Chendrimada T.P., et al. (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature, 436, 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 15. Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Costinean S., et al. (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA, 103, 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Calin G.A., et al. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA, 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iorio M.V., et al. (2012) microRNA involvement in human cancer. Carcinogenesis, 33, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma J., et al. (2010) MicroRNA and drug resistance. Cancer Gene Ther., 17, 523–531. [DOI] [PubMed] [Google Scholar]
- 20. Acunzo M., et al. (2012) miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene, 31, 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romano G., et al. (2012) MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc. Natl. Acad. Sci. USA, 109, 16570–16575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fang L., et al. (2014) MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget, 5, 2974–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garofalo M., et al. (2009) miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell, 16, 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Xin F., et al. (2009) Computational analysis of microRNA profiles and their target genes suggests significant involvement in breast cancer antiestrogen resistance. Bioinformatics, 25, 430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao J.J., et al. (2008) MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J. Biol. Chem., 283, 31079–31086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Nigita G., et al. (2015) A-to-I RNA editing: current knowledge sources and computational approaches with special emphasis on non-coding RNA molecules. Front. Bioeng. Biotechnol., 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishikura K. (2016) A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol., 17, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rabinowits G., et al. (2009) Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer, 10, 42–46. [DOI] [PubMed] [Google Scholar]
- 29. Taylor D.D., et al. (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol., 110, 13–21. [DOI] [PubMed] [Google Scholar]
- 30. Laganà A., et al. (2014) miR-Synth: a computational resource for the design of multi-site multi-target synthetic miRNAs. Nucleic Acids Res., 42, 5416–5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aravin A., et al. (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature, 442, 203–207. [DOI] [PubMed] [Google Scholar]
- 32. Girard A., et al. (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature, 442, 199–202. [DOI] [PubMed] [Google Scholar]
- 33. Grivna S.T., et al. (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev., 20, 1709–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Watanabe T., et al. (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev., 20, 1732–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Siomi M.C., et al. (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol., 12, 246–258. [DOI] [PubMed] [Google Scholar]
- 36. Weick E.M., et al. (2014) piRNAs: from biogenesis to function. Development, 141, 3458–3471. [DOI] [PubMed] [Google Scholar]
- 37. Assumpção C.B., et al. (2015) The role of piRNA and its potential clinical implications in cancer. Epigenomics, 7, 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim V.N. (2006) Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev., 20, 1993–1997. [DOI] [PubMed] [Google Scholar]
- 39. Iwasaki Y.W., et al. (2015) PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem., 84, 405–433. [DOI] [PubMed] [Google Scholar]
- 40. Han B.W., et al. (2014) piRNAs. Curr. Biol., 24, R730–R733. [DOI] [PubMed] [Google Scholar]
- 41. Czech B., et al. (2016) One loop to rule them all: The Ping-Pong Cycle and piRNA-guided silencing. Trends Biochem. Sci., 41, 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Fazio S., et al. (2011) The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature, 480, 259–263. [DOI] [PubMed] [Google Scholar]
- 43. Wang W., et al. (2014) The initial uridine of primary piRNAs does not create the tenth adenine that Is the hallmark of secondary piRNAs. Mol. Cell, 56, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mei Y., et al. (2015) A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat. Commun., 6, 7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hashim A., et al. (2014) RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget, 5, 9901–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang G., et al. (2013) Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin. Transl. Oncol., 15, 563–568. [DOI] [PubMed] [Google Scholar]
- 47. Cheng J., et al. (2011) piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin. Chim. Acta., 412, 1621–1625. [DOI] [PubMed] [Google Scholar]
- 48. Li Y., et al. (2015) Piwi-interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol. Med., 21, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cheng J., et al. (2012) piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett., 315, 12–17. [DOI] [PubMed] [Google Scholar]
- 50. Yan H., et al. (2015) piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia, 29, 196–206. [DOI] [PubMed] [Google Scholar]
- 51. Cui L., et al. (2011) Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin. Biochem., 44, 1050–1057. [DOI] [PubMed] [Google Scholar]
- 52. Kumar P., et al. (2016) Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci., 41, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fu H., et al. (2009) Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett., 583, 437–442. [DOI] [PubMed] [Google Scholar]
- 54. Li S., et al. (2012) Emerging role of angiogenin in stress response and cell survival under adverse conditions. J. Cell. Physiol., 227, 2822–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Honda S., et al. (2015) Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. USA, 112, E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Couvillion M.T., et al. (2010) A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev., 24, 2742–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumar P., et al. (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol., 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Goodarzi H., et al. (2015) Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell, 161, 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liao J.Y., et al. (2010) Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3’ trailers. PLoS One, 5, e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lee Y.S., et al. (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev., 23, 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Maute R.L., et al. (2013) tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA, 110, 1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Deng J., et al. (2015) Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther., 23, 1622–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang X., et al. (2016) IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA To upregulate CD1a molecules on monocytes/dendritic cells. J. Immunol., 196, 1591–1603. [DOI] [PubMed] [Google Scholar]
- 64. Pekarsky Y., et al. (2016) Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. USA, 113, 5071–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Balatti V., et al. (2015) TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA, 112, 2169–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Scott M.S., et al. (2011) From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie, 93, 1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reichow S.L., et al. (2007) The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res., 35, 1452–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Darzacq X., et al. (2002) Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. EMBO J., 21, 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dieci G., et al. (2009) Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics, 94, 83–88. [DOI] [PubMed] [Google Scholar]
- 71. Williams G.T., et al. (2012) Are snoRNAs and snoRNA host genes new players in cancer? Nat. Rev. Cancer, 12, 84–88. [DOI] [PubMed] [Google Scholar]
- 72. Ender C., et al. (2008) A human snoRNA with microRNA-like functions. Mol. Cell, 32, 519–528. [DOI] [PubMed] [Google Scholar]
- 73. Brameier M., et al. (2011) Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res., 39, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dong X.Y., et al. (2009) Implication of snoRNA U50 in human breast cancer. J. Genet. Genomics, 36, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mei Y.P., et al. (2012) Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene, 31, 2794–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liao J., et al. (2010) Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol. Cancer, 9, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mannoor K., et al. (2014) Small nucleolar RNA signatures of lung tumor-initiating cells. Mol. Cancer, 13, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang W.T., et al. (2014) Circulating miRNAs in cancer: from detection to therapy. J. Hematol. Oncol., 7, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Veneziano D., et al. (2015) Computational approaches for the analysis of ncRNA through deep sequencing techniques. Front. Bioeng. Biotechnol., 3, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Veneziano D., et al. (2016) Noncoding RNA: current deep sequencing data analysis approaches and challenges. Hum. Mutat., 37, 1283–1298. [DOI] [PubMed] [Google Scholar]