Abstract
A unique population of cells, called “lot cells,” circumscribes the path of the lateral olfactory tract (LOT) in the rodent brain and acts to restrict its position at the lateral margin of the telencephalon. Lot cells were believed to originate in the dorsal pallium (DP). We show that Lhx2 null mice that lack a DP show a significant increase in the number of mGluR1/lot cells in the piriform cortex, indicating a non-DP origin of these cells. Since lot cells present common developmental features with Cajal–Retzius (CR) cells, we analyzed Wnt3a- and Dbx1-reporter mouse lines and found that mGluR1/lot cells are not generated in the cortical hem, ventral pallium, or septum, the best characterized sources of CR cells. Finally, we identified a novel origin for the lot cells by combining in utero electroporation assays and histochemical characterization. We show that mGluR1/lot cells are specifically generated in the lateral thalamic eminence and that they express mitral cell markers, although a minority of them express ΔNp73 instead. We conclude that most mGluR1/lot cells are prospective mitral cells migrating to the accessory olfactory bulb (OB), whereas mGluR1+, ΔNp73+ cells are CR cells that migrate through the LOT to the piriform cortex and the OB.
Keywords: accessory olfactory bulb, Cajal–Retzius cells, lateral thalamic eminence, lot cells, Tbr2
Introduction
Olfaction is one of the oldest sensory modalities in the phylogenetic history of mammals. Besides the detection of odorants, the olfactory system plays a significant role in the regulation of behavioral responses such as sexual behavior, the recognition of predators, and the care of offspring (Halpern and Martinez-Marcos 2003; Ashwell 2012). The olfactory system is the only sensory circuit lacking a relay station in the thalamus (Kay and Sherman 2007), and the information integrated by the principal neurons in the olfactory bulb (OB) is conveyed directly to the higher olfactory centers in the brain by axons that course along the lateral olfactory tract (LOT).
During development, mitral cells, the principal projection neurons of the OB, elongate their axons within a prospective LOT territory, which is populated by an early generated population of neurons, the lot cells. The lot cells are considered guidepost cells for mitral cell axons, since chemically induced ablation of those cells prevents LOT formation in organotypic cultures (Sato et al. 1998; Hirata et al. 2012).
Lot cells supposedly derive from progenitor cells in the dorsal pallium (DP) and migrate tangentially within the pallial marginal zone (MZ) to enter into the LOT [Sato et al. 1998; Tomioka et al. 2000; Kawasaki et al. 2006; for reviews, see Marin et al. (2010); Bielle and Garel (2013); Squarzoni et al. (2015)]. Lot cells are visualized using the monoclonal antibody (mAb) lot1 that recognizes the glutamate metabotropic receptor mGluR1 (Sato et al. 1998; Hirata et al. 2012). According to these authors, mGluR1 expression does not occur within the proposed migration territory of lot cells within the pallial MZ, but only once the lot cells arrive at the LOT territory. However, the absence of specific cell markers precludes the identification of lot cells during their migration within the pallial MZ. Thus, it is unclear whether mGluR1/lot cells have a pallial origin as previously suggested or, on the contrary, they are akin to the AP-2α/Lhx5 presumptive mitral cells destined for the posterior accessory OB (pAOB) that arise in the thalamic eminence (TE; Huilgol et al. 2013).
The pAOB is a structure that resides in the dorsocaudal region of the OB and forms a parallel yet independent pathway (Mohedano-Moriano et al. 2007) directly related to the neural control of aggressive and defensive behaviors, just as the anterior tier of the AOB (aAOB) serves sexual behaviors (Kumar et al. 1999). Unlike mitral cells of the main OB and the aAOB that are generated in the OB neuroepithelium, Lhx5/AP-2α presumptive pAOB mitral cells have their origin in the TE, a transient anatomical domain of the developing brain located at the rostral end of the diencephalon and topologically related to the telencephalic roof including the choroid plexus neuroepithelium and the cortical hem (Roy et al. 2014). In addition to being a source of Lhx5/AP-2α presumptive pAOB mitral cells, the TE has been proposed as a neurogenic source of Trp73+ Cajal–Retzius (CR) cells that will go to the entorhinal and piriform cortex (Meyer et al. 2002, 2004; Tissir et al. 2009; Meyer 2010).
Here, we use a combination of genetic fate mapping in mice, in utero electroporation experiments and histochemical characterization to clarify the origins and neurochemical features of mGluR1/lot cells. Our data indicate that mGluR1/lot cells originate in the lateral tier of the TE and migrate rostrally along the LOT. These cells are a heterogeneous population that includes a large group of prospective mitral cells that migrate to the pAOB and a small group of CR cells going to the piriform cortex and the OB.
Materials and Methods
Animals
ΔNp73 Cre-IRES-eGFP and B6.129P2-Grm1tm1Dgen/J mice and wild-type C57BL/6J and ICR mice were maintained in the animal facilities of Universidad Miguel Hernández, San Juan de Alicante, Spain. Fixed Lhx2−/− mouse embryos and wild-type littermates were from the TATA Institute of Fundamental Biology. Fixed Wnt3aIRES-Cre; Ai9td-Tomato embryos were provided by Sonia Garel, École Normale Supérieure Paris, France, and Dbx1Cre; ROSA26YFP embryos were provided by Alessandra Pierani, Institut Jacques Monod, CNRS, University of Paris Diderot, Paris, France. The care and handling of the animals prior or during the experimental procedures followed European Union (2010/63/UE) and NIH guidelines and were approved by the Animal Care and Use Committees of the participating institutions. The day a vaginal plug was detected was considered as embryonic day 0.5 (E0.5).
Immunohistochemistry and In Situ Hybridization
Embryos were fixed in 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer saline (PBS), pH 7.4, at room temperature for 2 h for immunohistochemistry or at 4 °C overnight for in situ hybridizations, and rinsed in PBS for 2 h. For thick sections (100 µm), embryonic brains were embedded in 4% agarose diluted in PBS and cut in a Vibratome (Leica VT1000S). For thinner sections, brains were cryoprotected in 30% sucrose in 0.1 M PB under agitation for 4 h and frozen in liquid N2-chilled isopentane. Immunohistochemical staining was performed as described (Espinosa et al. 2009) in 100 µm Vibratome sections. Primary antibodies used were: chicken anti-GFP (Aves Laboratories, 1 : 2000); rabbit polyclonal anti-GFP (Abcam, 1 : 1000); rabbit polyclonal anti-DsRed (Clontech, 1 : 500); mouse mAb anti-reelin G10 (provided by A. Goffinet, 1 : 2000); mouse mAb 3B5 anti-AP-2α (Santa Cruz, 1 : 20); mAb 4F2 that recognizes the LIM-homeobox proteins Lhx1 and Lhx5 (Developmental Studies Hybridoma Bank, 1 : 200); rabbit polyclonal anti-calretinin (Swant, 1 : 2000); rabbit polyclonal anti-Tbr1 (Millipore, 1 : 500); chicken anti-Tbr2 (Millipore, 1 : 500); rabbit polyclonal anti-COUP-TFI (provided by M. Studer, 1 : 500); rabbit polyclonal anti-β-galactosidase (Cappel, MP Biomedicals, 1 : 1000); rat mAb lot1 (provided by T. Hirata, 1 : 200); rabbit polyclonal anti-Vglut2 (Synaptic Systems, 1 : 500); and mouse mAb Tuj1 (Covance, 1 : 500). Different combinations of AlexaFluor secondary antibodies (Invitrogen, 1 : 500) were used.
In situ hybridization was performed as described (Martinez-Lopez et al. 2015). The following probes were used: BMP4 (Jones et al. 1991), Dbx1 (Lu et al. 1994), Emx2 (Simeone et al. 1992), Fgf15 (McWhirter et al. 1997), Gdf10 (Zhao, Lawler, et al. 1999), Grm1 (Zhu et al. 1999), Gsh2 (Hsieh-Li et al. 1995), Lhx5 (Zhao, Sheng, et al. 1999), Mash1 (Guillemot and Joyner 1993), Neurog2 (Gradwohl et al. 1996), Neuropilin 2 (NP2; Tamamaki et al. 2003), Pax6 (Walther and Gruss 1991), Sfrp2 (Kim et al. 2001), Tbet (Faedo et al. 2002), Trp73 (Tissir et al. 2009), Wnt3a (Roelink and Nusse 1991), and Wnt8b (Richardson et al. 1999).
Slice Cultures and CMFDA Injections
Timed pregnant dams were killed by cervical dislocation. Uterine horns were removed and isolated in cold Krebs solution. E12.5 brain embryos were embedded in 4% low melting point agarose (Sea Plaque Agarose, Cambrex) in PBS. Three hundred-micrometer vibratome coronal sections were transferred to Millicell CM culture plate inserts (Millipore) previously placed in 6-well culture plastic dishes (Nunc, Thermo Scientific) containing 1 mL of DMEM–F12 medium supplemented with N2 supplement (5 μL/mL), l-glutamine (0.1 mM), glucose (2.4 mg/mL), penicillin–streptomycin (500 U/mL), and 10% fetal bovine serum (all these reagents provided by Invitrogen). Slices were maintained at 37 °C in 5% CO2 in a standard sterile incubator for 1 h. Next, resin beads (Bio-Rad), previously soaked in CellTracker™ Green CMFDA [5-chloromethylfluorescein diacetate] (Molecular Probes–Invitrogen), were placed on selected microanatomical areas of the slices. Then, DMEM-F12 medium was replaced by Neurobasal medium supplemented with B27 (1×), glucose (2.4 mg/mL), penicillin–streptomycin (500 U/mL), and l-glutamine (0.1 mM), and the slices were kept in the incubator at 37 °C and 5% CO2 for 2 days (Stoppini et al. 1991). Slices were then fixed in 4% PFA in PBS for 2 h and mounted on glass slides.
Primary Cultures
TE and DP explants from E11.5 ICR wild-type embryos were carefully dissected in chilled L15 medium (Gibco) supplemented with glucose (6 g/L, Sigma), HEPES (5 mM; Gibco), glutamine (2 mM, Gibco), and penicillin–streptomycin (1×, Gibco). The TE and DP explants were incubated in 500 μL of differentiation medium DMEM/F12 (Gibco), glucose (6 g/L Sigma), HEPES (5 mM, Gibco), glutamine (2 mM, Gibco), penicillin–streptomicin (1×, Gibco), N2 (1 : 100, Gibco), B27 (1 : 50, Gibco), FBS (5%, Gibco), and mechanically dissociated by repeated pipetting to isolate individual cells. A total of 250 000 cells/well were incubated on laminin- (5 μg/mL, Sigma) and poly-lysine- (100 μg/mL, Sigma) coated coverslips and maintained in a sterile incubator at 37 °C and 5% CO2. Medium was daily replaced by 500 μL of fresh differentiation medium at 37 °C. After 4 days in vitro (DIV), cells were fixed in 4% PFA in PBS at 4 °C for 20 min.
In Utero Electroporation
The procedures were as previously described (Borrell et al. 2005; García-Frígola et al. 2007) with some modifications. Plasmid pCAG-GFP (Matsuda and Cepko 2004; Addgene, Teddington, Middlesex, UK) was purified with a Midiprep Endofree Kit (Macherey-Nagel, Düren, Germany). The DNA solution (2 µg/µL in PBS, with 0.05% Fast-green added) was injected in the third ventricle or in the lateral ventricle using pulled glass pipettes. Embryos were electroporated using tweezers-type electrodes. Five square electric pulses were passed at 1 s intervals (50 ms; 35 V for E11.5 embryos).
Quantification and Statistical Analysis
Images were captured with a digital camera coupled with a Leica MZ APO stereomicroscope or a Leica MD5000 fluorescence microscope. Confocal microscope analyses were carried out in a Leica TCS SP2 AOBS or an Olympus FluoView FV1200 Laser Scanning Confocal Microscope. Figures were prepared using Adobe Photoshop CS5 and Adobe Illustrator CS5, and 2D mosaic reconstructions were produced when needed using the Photomerge tool of Photoshop CS5 software package. A minimum of 3 animals and 3 slices of each animal were used for all the analyses and quantifications. InStat (GraphPad, San Diego, CA, USA) software was used for statistical analysis.
Results
Comparative Expression of Lot Cell Markers
To assess the origin of lot cells, we first compared the expression patterns of several markers with known expression in the TE and/or the LOT areas: (1) the LIM-homeobox transcription factor Lhx5, expressed in future mitral cells (Huilgol et al. 2013) and also in CR cells (Abellan et al. 2010; Miquelajáuregui et al. 2010), (2) the transcription factor Trp73, expressed by CR cells (Meyer et al. 2002, 2004; Yoshida et al. 2006; Tissir et al. 2009; Meyer 2010), and (3) the metabotropic glutamate receptor mGluR1, expressed in lot cells when already located in the LOT territory (Hirata et al. 2012).
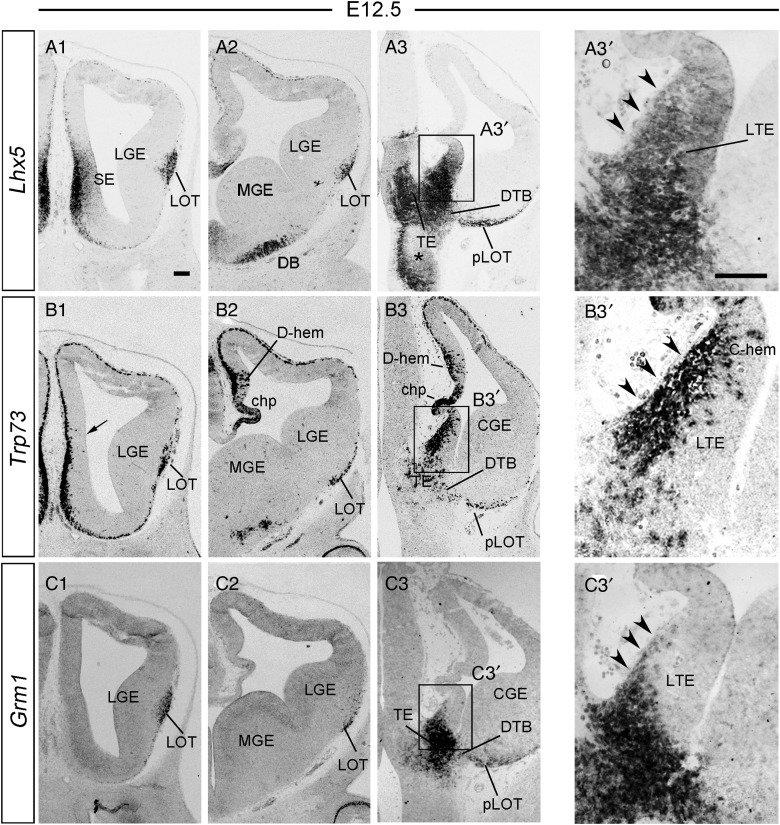
In situ hybridization in coronal sections of E12.5 wild-type embryos revealed that Lhx5 was expressed in the TE, in the diencephalic–telencephalic boundary (DTB), and in the LOT territory (Fig. 1A1–A3). Trp73 was expressed in the latera TE (LTE), a territory otherwise called strionuclear neuroepithelium (Altman and Bayer 1995; Meyer 2010) that is continuous with the caudal end of the cortical hem (hereafter, caudal hem), and showed a continuous distribution pattern from the LTE to the TE mantle, the DTB, the posterior part of the LOT (pLOT), and along the LOT territory (Fig. 1B1–B3). Trp73 was expressed in postmitotic cells and also in progenitor cells of the ventricular zone of both the LTE and the caudal hem.
Figure 1.
Comparing expression patterns of 3 genes expressed in the TE and the LOT territory: Lhx5, Trp73, and Grm1 (coding for mGluR1). Coronal sections of E12.5 wild-type embryos at 3 rostrocaudal levels. (A1–A3′) Lhx5 is expressed in the septum, the nuclei of the diagonal band in the basal forebrain, the whole TE, and the supraoptic–paraventricular domain of the hypothalamus (asterisk in A3). A3′ is a close-up image of the boxed area in A3 that displays Lhx5 expression in the LTE, including its ventricular zone. Arrowheads point to the subpial side of the LTE. (B1–B3′) Trp73 is expressed in the ventricular zone of the pallial septum (B1, arrow), the dorsal cortical hem (B2), the choroid plexus (B2 and B3), the caudal cortical hem, and the lateral TE (B3 and B3′). In B3, the ventricular zones of the dorsal hem, the caudal hem, and the LTE display Trp73-positive progenitors. Note subpial accumulations of transcripts in the dorsal and caudal hem and in the lateral TE (arrowheads in B3′). The distribution of discrete transcript clumps suggests the invasion of the TE mantle by p73 cells derived from the lateral TE (in B3). (C1–C3′) Compared with Lhx5 and Trp73, Grm1 expression is spatially much more limited, but is outstanding in the TE. Note in C3′ the absence of Grm1 expression in the ventricular zone of the LTE, and the modest accumulation of transcripts in the subpial side of the LTE (arrowheads in C3′). Note, for all these 3 genes, the strong hybridization signal in the LOT at all rostrocaudal levels, and the presence of the respective transcripts within the space between the lateral hypothalamus and the caudal ganglionic eminence (the DTB), suggesting a putative migrating path between the TE and the pLOT. CGE, caudal ganglionic eminence; C-hem, caudal cortical hem; chp, choroid plexus neuroepithelium; DB, diagonal band; D-hem, dorsal cortical hem; DTB, diencephalic – telencephalic border; LGE, lateral ganglionic eminence; LOT, lateral olfactory tract territory; LTE, lateral thalamic eminence; MGE, medial ganglionic eminence; pLOT, posterior part of the LOT territory; SE, septum; TE, thalamic eminence. Scale bars: A1–C3, 100 μm; A3′–C3′, 50 μm.
The expression of Grm1, coding for mGluR1, showed a pattern similar to Lhx5 and Trp73 in the TE and LOT (Fig. 1C1–C3) with the notable exception that Grm1 was not expressed in the TE ventricular zone. At E11.5, Grm1 was already expressed in the TE, DTB, and LOT territory (see Supplementary Fig. 1). Unlike Grm1, at E12.5, Lhx5 and Trp73 were also expressed in the MZ of the cortex, likely in CR cells as previously reported (Yamazaki et al. 2004; Tissir et al. 2009; Abellan et al. 2010; Miquelajáuregui et al. 2010). Most notably, Grm1 and Lhx5 in situ hybridization signals overlapped in the LOT territory at different rostrocaudal levels (see Supplementary Fig. 2). mAb lot1 recognizes mGluR1 (Sato et al. 1998; Hirata et al. 2012) and we observed that Grm1 (mGluR1) in situ hybridization (Fig. 1D1–D3) provided equivalent results to mAb lot1 immunostaining (Fig. 2A,B). Even further, reporter mGluR1 mice B6.129P2-Grm1tm1Dgen/J, where LacZ was inserted in frame with the mGluR1 coding sequence (Sachs et al. 2007), confirmed that mAb lot1 specifically recognizes the mGluR1 protein (see Supplementary Fig. 3).
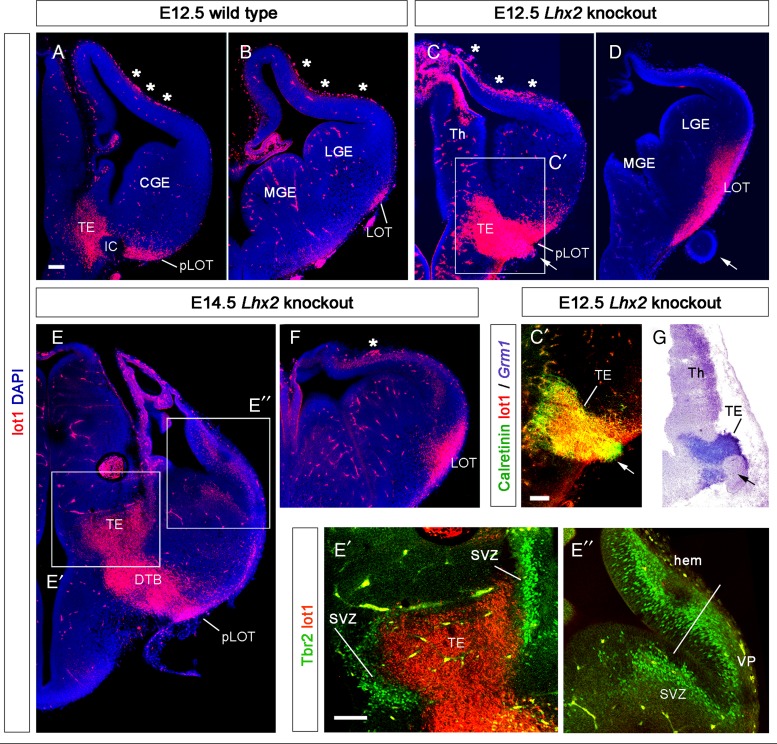
Figure 2.
Absence of DP in Lhx2−/− mice does not prevent an increased invasion of the piriform cortex by mGluR1/lot cells. (A–D) mGluR1 immunohistochemistry with mAb lot1 in coronal sections of E12.5 wild-type (A and B) and Lhx2 knockout littermates (C and D) at 2 different rostrocaudal levels. The immunohistochemical signal was more extended and more intense in the TE and LOT of the Lhx2−/− mice than in the wild-type embryo. Arrows in C, C′, D, and G indicate the abnormal optic-like vesicle located at the DTB, typical of these mutants. Asterisks signal the presence of artefactual staining over the meninges. (C′) From the boxed area in C. Calretinin immunostaining in the putative migration path from the TE to the pLOT in Lhx2−/− mice. Note the coexpression of mGluR1 and calretinin except in the optic-like vesicle, which is calretinin+ but mGluR1−. (G) In situ hybridization shows Grm1 transcripts surrounding the optic-like vesicle in a section similar to that in C′. (E and F) mGluR1 immunostaining in E14.5 Lhx2−/− mouse embryos shows the notable enlargement of the putative migration pathway of mGluR1 cells along the DTB to the posterior part of the LOT (pLOT, in E) and in the LOT at mid-rostrocaudal levels (in F). (E′) From the boxed area E′ in E. Tbr2-immunoreactive subventricular zones surround the mGluR1+ mantle of the TE and serve to corroborate its TE identity. (E″) From the boxed area E″ in E. Tbr2-immunoreactive secondary progenitor cells in the expanded VP and the cortical hem, both known features of Lhx2−/− mice. A white line signals the limit between these 2 structures. CGE, caudal ganglionic eminence; DTB, diencephalic – telencephalic border; IC, internal capsule; LGE, lateral ganglionic eminence; LOT, lateral olfactory tract territory (in B, D), or lateral olfactory tract (in F); MGE, medial ganglionic eminence; pLOT, posterior part of the LOT territory (in A, C), or posterior part of the LOT (in E) ; SVZ, subventricular zone; Th, thalamus; VP, ventral pallium. All calibration bars, 100 µm. Bar in A applies to A–F; in E′, the bar applies to E′–E″.
The Vast Majority of the mGluR1/Lot Cells Do not Originate in the DP
It has been previously proposed that cells located at the LOT territory originate in the DP neuroepithelium (Tomioka et al. 2000; Kawasaki et al. 2006). We decided to re-examine the pallial origin of mGluR1/lot cells by analyzing mice lacking the transcription factor Lhx2 (Lhx2−/− mice). In this animal, whereas the DP is absent, the cortical hem and the ventral pallium (VP) are considerably enlarged (Bulchand et al. 2001; Monuki et al. 2001; Vyas et al. 2003; Mangale et al. 2008; Subramanian and Tole 2009; Subramanian et al. 2009; Roy et al. 2013, 2014), and the TE is aberrantly expanded (Roy et al. 2014). In Lhx2−/− mice, the OB mitral cells are specified but they reside in an ectopic OB-like structure (OBLS) in which mitral cells are not able to pioneer their axonal projections toward the LOT (Saha et al. 2007). Additionally to these previously reported observations, we found that AOB marker NP2, the aAOB marker Tbet, and the pAOB marker Lhx5 were all expressed in the OBLS region (see Supplementary Fig. 4). In contrast, the Lhx2−/− brain displayed a considerably enlarged lot1 (mGluR1) immunoreactive territory. Compared with E12.5 wild-type mice (Fig. 2A,B), Lhx2 mutants showed an intensely lot1-immunoreactive TE field that expanded along the DTB, the pLOT, and the LOT territories at mid-rostrocaudal levels (Fig. 2C,D). The expansion of the mGluR1 expression domains remained conspicuous in E14.5 Lhx2−/− mice (Fig. 2E,F). We based our identification of the enlarged TE in Lhx2−/− mutant mice first on the intense calretinin immunostaining of the TE mantle [Fig. 2C′; see Abbott and Jacobowitz (1999)] and, second, on the presence of a Tbr2-immunostained TE subventricular zone encircling the lot1+ TE mantle (Fig. 2E′). Tbr2 also decorated the subventricular zone of the VP and highlighted the enlarged cortical hem of these animals (Fig. 2E″).
As recently described by Roy et al. (2013), Lhx2−/− mice display an abnormal optic-like vesicle at the DTB (Fig. 2C,C′,D,G, arrows). This vesicle was calretinin-positive (Fig. 2C′), and combined lot1/calretinin immunostaining (Fig. 2C′) or Grm1 in situ hybridizations (Fig. 2G) showed that the putative migration path of mGluR1 TE-derived cells surrounded the optic-like vesicle, which in the mutants is anatomically very close to the pLOT territory.
In conclusion, because the Lhx2−/− mice lack a DP, it is very unlikely that the mGluR1/lot cells in the LOT territory originate in the neocortical primordium.
mGluR1/Lot Cells Do not Originate from Wnt3a Progenitors in the Hem, or from Dbx1 Progenitors in the Ventral Pallium or the Septum
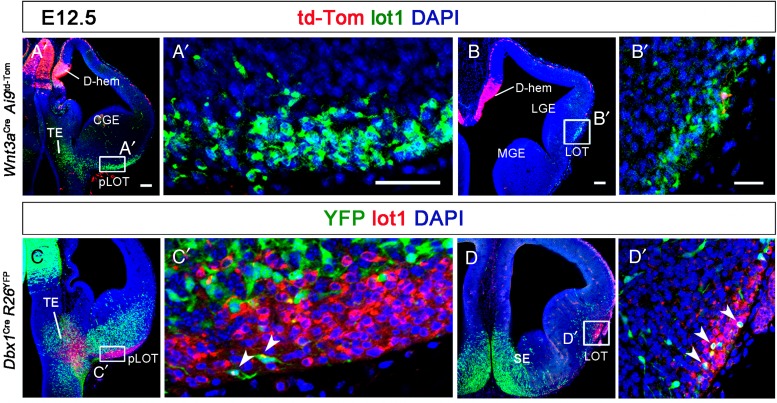
We then wondered whether mGluR1/lot cells originate from the hem. To test this hypothesis, we used a Wnt3aCre mouse line (Yoshida et al. 2006) crossed with Ai9Td-Tomato mice to drive the expression of the Td-Tomato reporter in Wnt3a-lineage cells. We did not find Wnt3a-lineage, td-Tomato cells in the TE mantle or in the LOT territory (Fig. 3A,A′). At E12.5, we observed strong expression of td-Tomato in the cortical hem and in the MZ of the DP that corresponds to cortical hem-derived CR cells (Fig. 3A,B). At rostral levels, there were scattered Wnt3a/td-Tomato cells in the LOT territory, but they were not mGluR1-positive (Fig. 3B,B′). This result demonstrates that Wnt3a hem progenitors are not the precursors of mGluR1/lot cells.
Figure 3.
mGluR1/lot cells do not derive from Wnt3a progenitors in the cortical hem or Dbx1 progenitors in the VP. (A and B) Coronal sections of an E12.5 Wnt3aCre;Ai9td-Tom mouse embryo. td-tom+ cells are Wnt3a-lineage neurons that concentrate in the dorsal cortical hem. Td-Tom+ cells were conspicuous in the pallial preplate, but virtually no td-Tom+ cells were found in the LOT territory. (A′ and B′) Close-ups of the boxed areas in A and B show the absence of td-Tom+ cells in the pLOT and occasional td-tom+ (but mGluR1−) cells at more rostral levels of the LOT, thus discarding that Wnt3a progenitors in the cortical hem could generate mGluR1/lot cells. (C and D) Coronal sections of an E12.5 Dbx1Cre;R26RYFP mouse embryo. Dbx1-lineage, YPF+ cells were abundant in the TE, in the basal telencephalon mantle, and in the septum, but very scarce in the LOT territory. (C′ and D′) High magnification images of the boxed areas in C and D show that occasional Dbx1-lineage cells may coexpress mGluR1 (white arrowheads) both in the pLOT and at rostral levels of the LOT (see text for details). CGE, caudal ganglionic eminence; D-hem, dorsal cortical hem; LGE, lateral ganglionic eminence; LOT, lateral olfactory tract territory; MGE, medial ganglionic eminence; pLOT, posterior part of the LOT territory; SE, septum; TE, thalamic eminence. Scale bars: A–D, 100 μm; A′–D′, 50 μm.
Another possible source of lot cells could be the VP or the septum. To test this possibility, we used a Dbx1Cre mouse line (Bielle et al. 2005) crossed with ROSA26RYFP mice to detect Dbx1-lineage cells. Dbx1 is a homeodomain transcription factor expressed in progenitor cells in the VP and pallial septum that generate glutamatergic neurons destined to the cortex and amygdala (Bielle et al. 2005; Hirata et al. 2009; Griveau et al. 2010; Teissier et al. 2010; Zimmer et al. 2010). In addition, the ventricular zone of the medial TE (MTE) displayed strong Dbx1 expression (see Supplementary Fig. 5C). Dbx1-lineage cells distributed widely in the hypothalamus and the basal telencephalon, but very few of them invaded the LOT territory (Fig. 3C). In the posterior LOT, we found that only 3.3 ± 0.85% (mean ± SEM) of mGluR1/lot cells displayed Dbx1-YFP immunoreactivity (Fig. 3C,C′, white arrowheads). At rostral levels, the percentage of mGluR1/lot cells displaying Dbx1-YFP immunoreactivity was also very low (4.16 ± 0.76%) (Fig. 3D,D′, white arrowheads). These results indicate that Dbx1 progenitors from the VP or the pallial septum are not the source of mGluR1/lot cells.
The TE Has the Potential to Produce Lot Cells
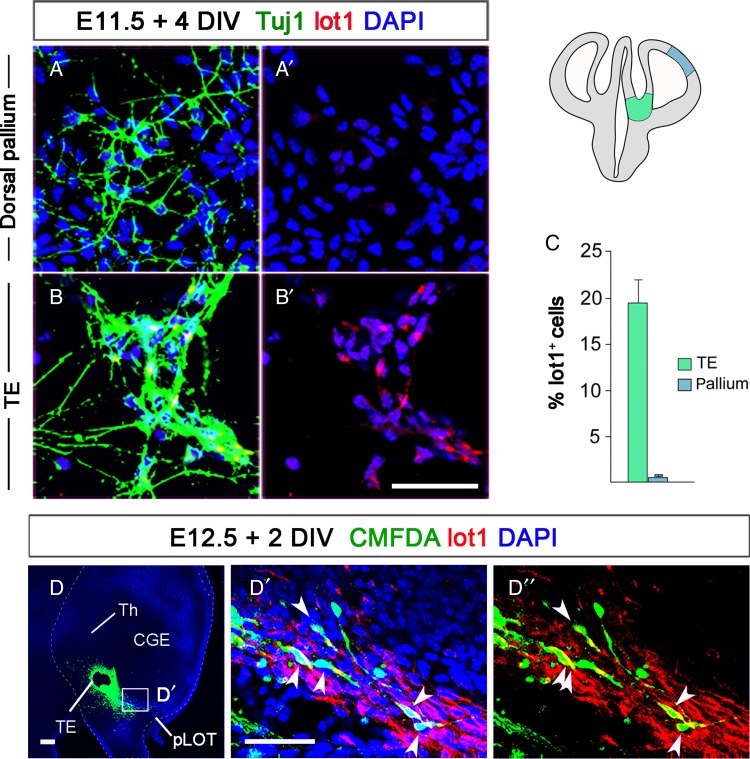
Given that the mGluR1/lot cells do not originate from the DP, hem, VP, or septum, we considered the possibility that they are generated in the TE. To explore the potential of the TE to produce lot cells, we dissociated cells from TE or DP from E11.5 embryos (n = 17) and put them in culture (n = 3). After 4 DIV, cells were immunostained for Tuj1 and mGluR1 using mAb lot1 (Fig. 4A,B) and a minimum of 500 Tuj1+ newly differentiated neurons were counted. In the case of TE-derived primary cultures, 19.5 ± 2.4% of the Tuj1+ cells also expressed mGluR1, whereas in the case of pallial cultures only 0.5% ± 0.2% of the Tuj1+ cells were also mGluR1+ (Fig. 4A–C). This experiment demonstrates that the E11.5 TE has the potential to generate mGluR1 cells and confirms that the DP has a very limited capacity of generating lot cells at this embryonic stage (Tomioka et al. 2000), consistent with the results obtained with the Lhx2−/− mice.
Figure 4.
The E11.5 TE has the potential to generate mGluR1 cells. (A and B) Primary cultures of E11.5 cells from DP (A and A′) and TE (B and B′) after 4 DIV. In both cases, we counted the percentages of cells coexpressing mGluR1+ and Tuj1+ cells over the total of Tuj1+ young neurons. (C) The percentages of mGluR1-expressing neurons were much higher in TE than in DP cultures. (D) Organotypic culture of a 300-μm thick coronal section at the level of the TE from one wild-type embryo. A tiny crystal of CellTracker CMFDA (green) was placed on the TE. (D′–D″) High magnification of the boxed area in D shows the migratory pathway of lot cells along the DTB after 2 DIV. The cells that had incorporated CMFDA while in the TE are mGluR1+. CGE, caudal ganglionic eminence; pLOT, posterior lateral olfactory tract territory; TE, thalamic eminence; Th, thalamus. Scale bars: A and B′, 100; D, 100 μm ; D′–D″, 25 μm.
Organotypic slice cultures (Stoppini et al. 1991) provided a further line of evidence in favor of a TE origin for mGluR1 cells in the LOT territory. We placed a minute crystal of CellTracker CMFDA over the cross-sectioned TE in coronal slices from E12.5 wild-type embryos and cultured them for 24 h. CMFDA-labeled cells were then detected in the DTB and in the LOT (Fig. 4D), and mAb lot1 immunostaining revealed that the CMFDA-labeled cells were mGluR1+ (Fig. 4D–D″).
The TE Expresses Genes Associated with Glutamatergic Neurogenesis
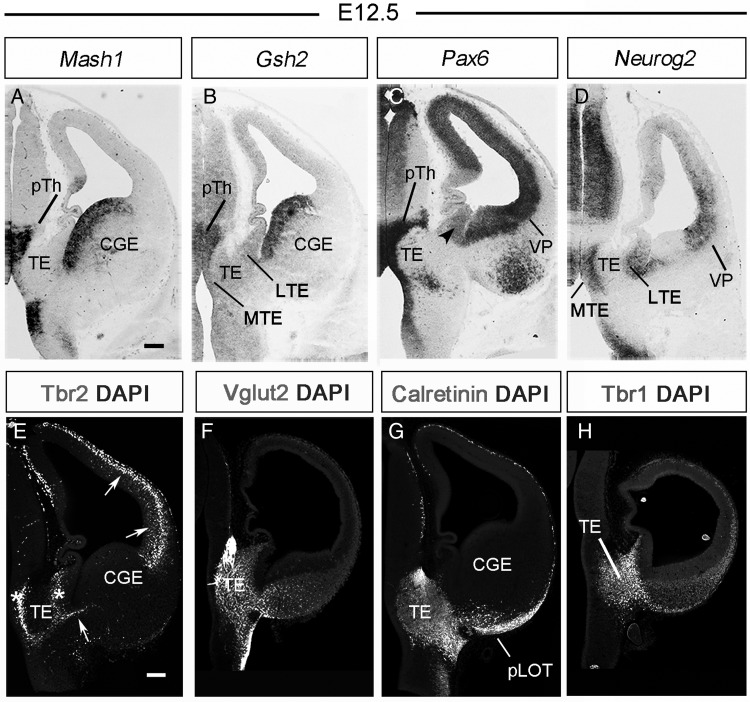
The prethalamus is the neurogenic site of GABAergic neurons that will populate the thalamus in the adult brain (Inamura et al. 2010) and thus expresses genes encoding transcription factors involved in GABAergic neurogenesis such as Mash1 and Gsh2 (Fig. 5A,B). However, we observed that, unlike the prethalamus, the TE is negative for genes related to GABAergic neurogenesis (Fig. 5A,B).
Figure 5.
The TE is a source of glutamatergic cells. (A and B) In situ hybridizations for Mash1 and Gsh2 in coronal sections of an E12.5 wild-type embryo. Both transcription factors, implicated in GABAergic neurogenesis, are expressed in the ventricular zone of the caudal ganglionic eminence and prethalamus but not in the TE. (C and D) In situ hybridization for Pax6 and Neurog2 in coronal sections of an E12.5 wild-type embryo. Both transcription factors, implicated in glutamatergic neurogenesis, are expressed in the ventricular zone of the TE. Note that Pax6 is also expressed in the prethalamus, and that it is expressed with low intensity in the LTE and more intensely in the MTE. (E–H) Immunolabeling for different markers of glutamatergic neurons in coronal sections of E12.5 wild-type embryos. Tbr2 (in E) is expressed in the subventricular zone of both the TE (asterisks) and the pallium (arrows). Vglut2 (F), calretinin (G), and Tbr1 (H), 3 glutamatergic neuron markers, are expressed in the TE mantle, indicating that the TE produces glutamatergic neurons. Note that VGlut2, calretinin, and Tbr1 are expressed in the migration path between the TE and the pLOT along the DTB. CGE, caudal ganglionic eminence; LTE, lateral thalamic eminence; MTE, medial thalamic eminence; pLOT, posterior lateral olfactory tract territory; pTh, prethalamus; TE, thalamic eminence; VP, ventral pallium. Scale bar, 100 μm.
The TE was, in contrast, positive for genes that participate in glutamatergic neurogenesis such as Pax6, Neurog2, and Tbr2 (Schuurmans and Guillemot 2002; Mattar et al. 2004; Englund et al. 2005; Hevner et al. 2006). The paired domain transcription factor Pax6 was expressed in the ventricular zones of prethalamus, TE (with a medialhigh–laterallow gradient), and pallium (Fig. 5C). The proneural gene Neurog2 was expressed in the TE ventricular zone (Fig. 5D), and the T-box transcription factor Tbr2 was expressed in the TE subventricular zone (Fig. 5E). We also observed that cell populations in the TE mantle expressed Vglut2, calretinin, and Tbr1 (Fig. 5F–H), 3 markers of glutamatergic neurons (Bulfone et al. 1995; Abbott and Jacobowitz 1999; Ina et al. 2007). All these traits of molecular identity are also compatible with the idea that the TE is a potential source of lot cells.
mGluR1 Cells Are Generated in the Lateral TE
While analyzing the expression of glutamatergic markers in the TE, we realized that the TE encompasses 2 well-differentiated neuroepithelial compartments, that is, the medial TE (MTE) and the LTE, which surround a centrally located mantle zone. To properly characterize the TE, we decided to carefully define the expression patterns of genes that have been previously identified in this brain region. We found that certain genes were expressed in the entire TE, while some others highlighted specific TE subdomains, sometimes also encompassing hem subregions (see Supplementary Figs 5 and 6). In particular, Wnt3a expression delimited the complete cortical hem and its expression domain ceased at the point where the caudal hem continues with the LTE (see Supplementary Fig. 5B,D1–D3). Similarly, BMP4 expression highlighted the cortical hem (see Supplementary Fig. 6A). In contrast, Wnt8b (see Supplementary Fig. 5E1–E3) and Emx2 (see Supplementary Fig. 6B) expressions revealed the existence of a genoarchitectonic continuum between the caudal hem and the LTE, although the Emx2 expression domain extended far more dorsally than Wnt8b. Other genes, such as Sfrp2, a secreted modulator of Wnt signaling (see Supplementary Fig. 5C,F1–F3), and the transcription factor Dbx1 (see Supplementary Fig. 6C), delimited the neuroepithelium of the MTE. Interestingly, Sfrp2 and Dbx1 also labeled the VP at the pallium–subpallium boundary. Gdf10 (BMP3b) (see Supplementary Fig. 5G1–G3; Shimogori et al. 2010) and Fgf15 (see Supplementary Fig. 6D) were detected in both the LTE and the MTE ventricular zones but not so in the cortical hem.
Therefore, the differential anatomical distributions of these markers indicate that the MTE and the LTE have different molecular identities and support the hypothesis that they might be programmed to generate distinct sets of neuronal populations.
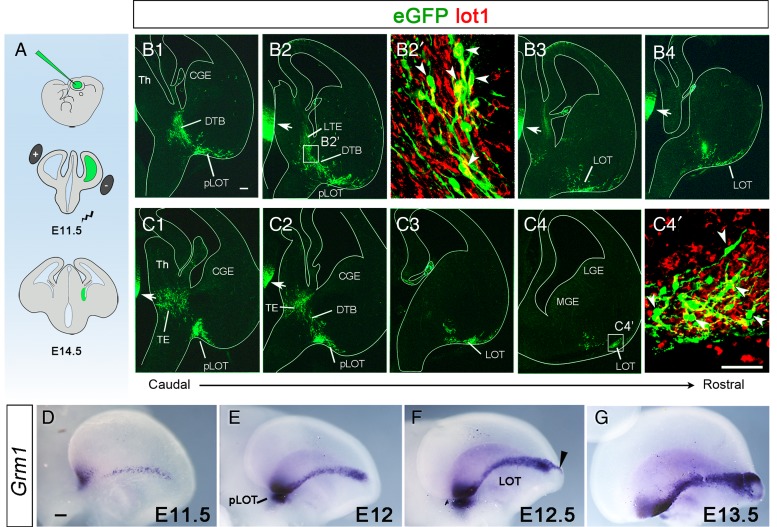
In utero electroporation allows gene targeting to specific locations of the developing brain (Borrell et al. 2005; García-Frígola et al. 2007). Therefore, we used this approach to express eGFP in progenitors residing in the developing TE of E11.5 embryos. By manipulating the position and polarity of the electrodes, different subdomains of the TE were targeted (Fig. 6 and Supplementary Fig. 7). When the MTE region was targeted by electroporation, no eGFP+ cells were detected in the LOT territory or the ventral telencephalon at E14.5 (see Supplementary Fig. 7). However, when progenitors in the LTE were targeted, groups of eGFP+ cells in the TE mantle were systematically found at E14.5 together with a larger population of labeled cells scattered along the DTB migratory path, the pLOT (Fig. 6B2,C2), and the LOT (Fig. 6B4,C3,C4). eGFP+ cells derived from the TE localized around the LOT and those located in the DTB and in the LOT were mGluR1+ (Fig. 6B2′,C4′). In summary, these experiments showed that mGluR1/lot cells generated in the LTE at E11.5 migrate to the TE mantle and then ventrally to the posterior LOT to subsequently turn rostrally along the LOT. In agreement with this interpretation, whole-mount in situ hybridizations for Grm1 (Fig. 6D–G) revealed the progressive invasion of the pAOB by mGluR1+ cells between E11.5 and E13.5.
Figure 6.
mGluR1 cells are specifically generated in the LTE. (A) Schemata of the in utero electroporation experiments. Plasmid pCAG-GFP was injected in the lateral ventricle (green) of E11.5 embryos, and tweezers-type electrodes were oriented to electroporate progenitor cells in the ventricular zone of LTE, facing the lateral ventricle. Three days after electroporation, at E14.5, the embryos were removed, fixed, and processed for immunohistochemistry. (B1–B4 and C1–C4) Coronal sections of E14.5 embryos in which the LTE was targeted at E11.5. The sections show that the cells electroporated in the LTE express eGFP (green) and have migrated to the TE mantle, the DTB, and the LOT in the piriform cortex at successive caudorostral levels. Note that the contralateral thalamus was also labeled (arrows) due to the direction of the current applied. (B2′ and C4′) Close-ups of the boxed areas in B2 and C4, showing eGFP+ cells coexpressing mGluR1 (white arrowheads) while they are migrating along the DTB (B2′) and also when they locate in the LOT (C4′). (D–G) Whole-mount in situ hybridizations for Grm1 at different embryonic ages, from E11.5 to E13.5. Note the progressive invasion of the rostral levels of the LOT and the pAOB (arrowhead in F) by mGluR1/lot cells as revealed by the distribution of Grm1 transcripts. CGE, caudal ganglionic eminence; DTB, diencephalic – telencephalic border; LGE, lateral ganglionic eminence; LOT, lateral olfactory tract; LTE, lateral thalamic eminence; MGE, medial ganglionic eminence; pLOT, posterior part of the LOT (in B1,B2,C1,C2), or posterior part of the LOT territory (in E); Th, thalamus. Scale bars: B1–B4, C1–C4, 100 μm; B2′ and C4′, 50 μm; D–G, 200 μm.
mGluR1/Lot Cells Exhibit Neurochemical Features Typical of pAOB Mitral Cells
Our observations suggested that a majority of mGluR1/lot cells may be prospective pAOB mitral cells, similar to the cells described by Huilgol et al. (2013) and therefore, we explored whether mGluR1/lot cells have neurochemical properties in common with mitral cells. To this end, we performed double immunostainings for mGluR1 and reelin or Tbr2, which are general markers of mitral cells, and for mGluR1 and COUP-TFI, Lhx5, or AP-2α, which are selective markers of pAOB mitral cells [see Supplementary Fig. 8; see also Huilgol et al. (2013)].
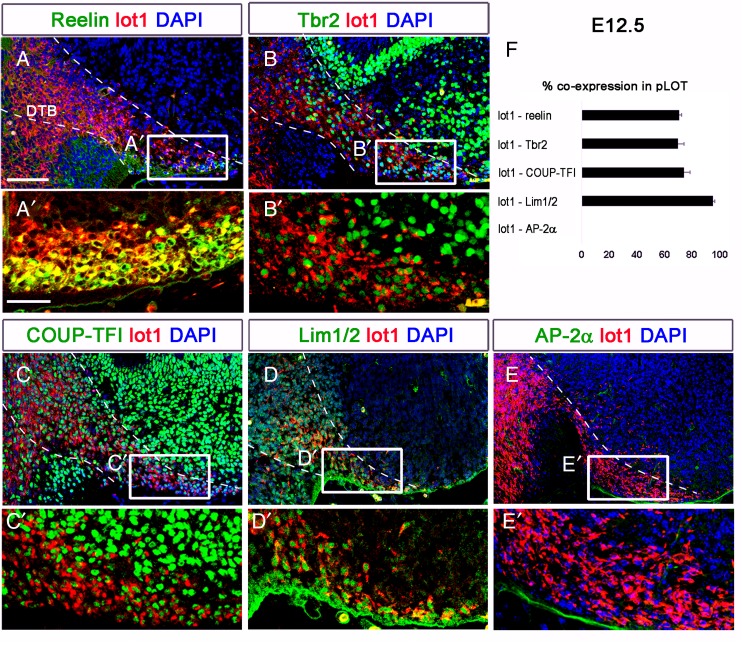
Coronal sections at different rostrocaudal levels of E12.5 embryos showed that reelin was strongly expressed in the LOT territory, but not along the migratory route between the TE and the pLOT (Fig. 7A). In the pLOT, 70.7 ± 1.5% of the mGluR1/lot cells coexpressed reelin (Fig. 7A′,F), whereas in the anterior part of the LOT (aLOT) the percentage was 89.74 ± 3.04% (data not shown). mGluR1/lot cells along the DTB coexpressed Tbr2 (Fig. 7B). In the pLOT territory, 69.8 ± 4.25% of the mGluR1/lot cells coexpressed Tbr2 (Fig. 7B′,F), similar to the aLOT (76.10 ± 4.04%; data not shown). Interestingly, Tbr2 expression discriminated the Tbr2+ mGluR1/lot cells from the presumed Tbr2− CR cells also present in the LOT (Figs 8A′ and 9A′,B′,C,C′).
Figure 7.
Immunohistochemical characterization of the mGluR1+ cells in the pLOT. (A–E) Immunohistochemistry of mGluR1 and additional markers of lot cells in coronal sections of E12.5 embryo. The dashed lines highlight the pathway between the TE and the pLOT through the DTB. Note that reelin started its coexpression in mGluR1+ cells when in the pLOT territory, that Tbr2 was coexpressed in mGluR1+ cells along the DTB and in the LOT, and mGluR1+ cells started to coexpress COUP-TFI and Lhx5 within the TE. AP-2α was not detected in the pLOT territory at E12.5. (A′–E′) High magnifications of the pLOT territory (boxed areas in A–E) showing in detail the coexpression of different markers with mGluR1. Note the coexpression of mGluR1 and COUP-TFI in C′ and of mGluR1 and Lhx5 in D′. (F) Percentages of mGluR1 cells that express additional markers in the pLOT territory. DTB, diencephalic – telencephalic border. Scale bars: A–E, 100 μm in A; A′–E′, 50 μm in A′.
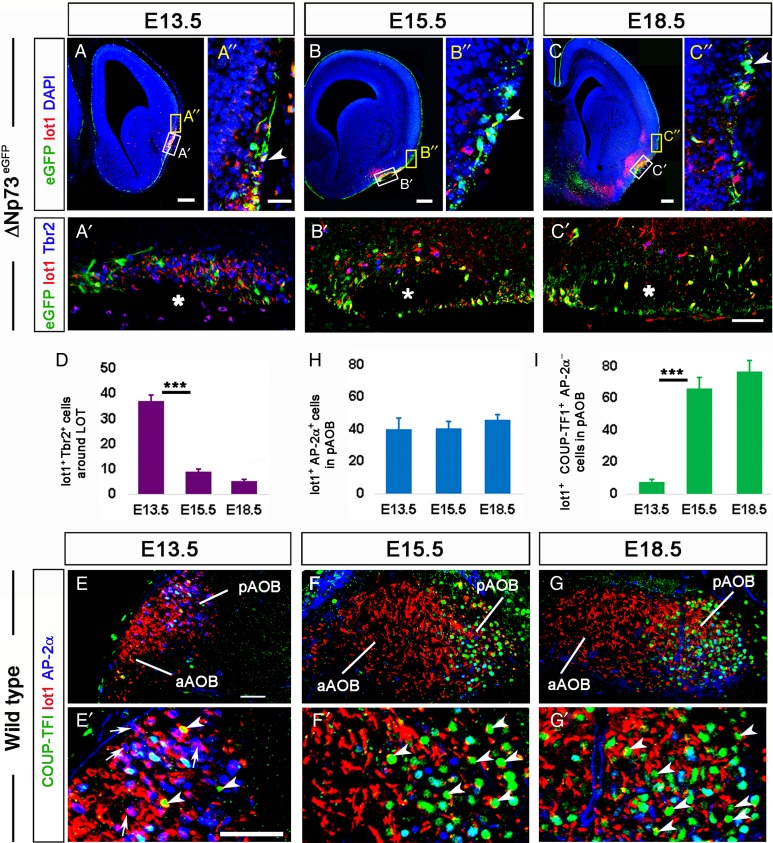
Figure 8.
mGluR1/lot cells follow a rostral migratory path along the LOT. (A–C) Immunostaining for eGFF, mGluR1, and Tbr2 in ΔNp73eGFP embryos in rostral coronal sections at E13.5, E15.5, and E18.5. (A′–C′) High magnifications of the white-boxed areas in A–C that contain the LOT. Asterisks indicate the space occupied by the (unstained) mitral cell axons. Most of the mGluR1+ cells at E13.5 were Tbr2+, whereas only a small proportion of them were ΔNp73+ but Tbr2−. Note the apparent descent in the number of mGluR1+/Tbr2+ cells with age (see graph in D). (A″–C″) High magnifications of the yellow-boxed areas in A–C showing details of the mGluR1+/ΔNp73+ cells in the pallial marginal zone. The arrowheads point to mGluR1/ΔNp73 cells that have reached the boundary between the piriform cortex and the neocortex. (D) Graph representing the average numbers per section of mGluR1+/Tbr2+ cells in the LOT territory at E13.5, and in the LOT at E15.5 and E18.5. The number of cells descended significantly between E13.5 and E15.5 (P < 0.001; Tukey–Kramer multiple comparisons test). (E–G) Sagittal sections of E13.5, E15.5, and E18.5 embryos showing mitral cells in the pAOB immunostained for COUP-TFI, mGluR1, and AP-2α. Note in E the presence of AP-2α+ cells in the pAOB already at E13.5, and the drastic increase in the number of COUP-TFI+ cells between E13.5 and E15.5, in E and F′. (E′–G′) High magnification views of the pAOB in sagittal sections to highlight the presence of coexpressions of mGluR1, COUP-TFI, and AP-2α. Arrows point to mGluR1+/AP-2α+ cells, and white arrowheads point to COUP-TFI+/AP-2α− cells. (H) Graph indicating the number of mGluR1+/AP-2α+ cells in the pAOB between E13.5 and E18.5. Note the number of these cells reaches a plateau already at E13.5. I. Graph indicating the statistically significant increase in the number of COUP-TFI+/AP-2α− mitral cells in the pAOB from E13.5 and E15.5 (P < 0.001; Tukey–Kramer multiple comparisons test). aAOB, anterior accessory olfactory bulb; pAOB, posterior accessory olfactory bulb. Scale bars, A–G and E, 100 μm; A′–C′ (in C′), A″ and B″ (in A″), and E′–G (in G), 50 μm.
Figure 9.
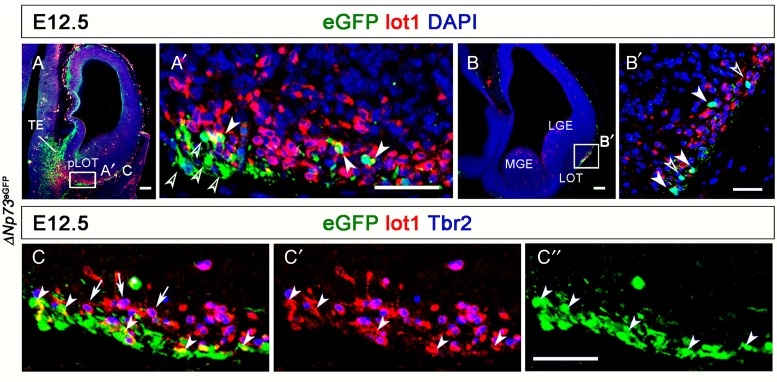
A small percentage of mGluR1/lot cells express p73. (A and B) Coronal sections of an E12.5 ΔNp73eGFP embryo. ΔNp73-eGFP+ expressing cells were observed in the TE, the DTB, and the LOT territory. (A′ and B′) Close-up images of the boxed areas in (A and B). A small number of mGluR1+ cells in the LOT territory were ΔNp73-eGFP+ (white arrowheads). Hollow arrowheads point to ΔNp73-expressing, mGluR1-negative cells. See text for details. (C and C′) Close-up image of an area similar to that boxed in A. mGluR1/lot cells (arrows) were Tbr2+. Note that both the mGluR1+/ΔNp73-eGFP+ cells and the mGluR1−/ΔNp73-eGFP+cells were Tbr2-negative. LGE, lateral ganglionic eminence; LOT, lateral olfactory tract territory; MGE, medial ganglionic eminence; pLOT, posterior part of the LOT territory; TE, thalamic eminence. Scale bars: A and B, 100 μm; A′, B′, and C–C″, 50 μm.
The Chicken Ovoalbumin Upstream Promoter Transcription Factor 1 (COUP-TFI) is expressed in the pAOB at E15.5 (see Supplementary Fig.8B and Fig. 8F,F′) and E18.5 (Fig. 8G,G′). At E12.5, 74.25 ± 4.37% of mGluR1/lot cells in the pLOT territory coexpressed COUP-TFI (Fig. 7C,C′,F) versus 59.56 ± 3.17% in the aLOT (data not shown). We also observed that the majority of the lot cells were positive for the mAb 4F2 that recognizes Lhx1/5 in both the pLOT (95.41 ± 0.91%; Fig. 7D,D′,F) and the aLOT (92.39 ± 1.47%; data not shown).
At E12.5, we found mGluR1/lot cells positive for AP-2α in the aLOT (see Supplementary Fig. 9B′,C′, white arrowheads), but not in the pLOT (Fig. 7E,E′ and Supplementary Fig. 9A,A′). Within the AP-2α population in the aLOT, we also found cells coexpressing AP-2α and COUP-TFI (see Supplementary Fig.9B′,C′, yellow arrowheads).
Altogether, these results indicate, first, that the Tbr2+ mGluR1/lot cells coexpress reelin and pAOB mitral cell markers such as COUP-TFI, Lhx5, or AP-2α and, second, that the AP-2α cells are a subpopulation of the mGluR1/lot cells.
Most mGluR1/Lot Cells Follow a Rostral Migratory Path to Finally Populate the pAOB
The number of Tbr2+/mGluR1+ cells in the LOT decreased from E13.5 to E18.5 (Fig. 8A′–C′,D). By E13.5, when the axons of the OB mitral cells have already formed the LOT, there was an average of 37.2 ± 2.25 Tbr2+ mGluR1/lot cells per section around this axonal tract at mid-rostrocaudal levels. This number significantly dropped to 8.9 ± 1.03 (q = 20.15, P < 0.001) by E15.5 (Fig. 8A′,D), a result compatible with the hypothesis that these cells migrate rostrally.
AP-2α and COUP-TFI immunolabeling distinguished 2 groups of mGluR1+ mitral cells in the AOB. At E13.5, when the OB evagination was already apparent, we detected abundant mGluR1+/AP-2α+ cells in the pAOB (Fig. 8E,E′, arrows, and Fig. 8H). At this early stage, there were also some (8.83 ± 1.80 cells per section) mGluR1+/COUP-TFI+ cells negative for AP-2α in the pAOB (Fig. 8E′, white arrowheads). At subsequent stages, while the amount of AP-2α+/mGluR1+ cells was constant in the pAOB (Fig. 8H), the number of AP-2α-negative/mGluR1+/COUP-TFI+ cells (marked in Fig. 8F′,G′ with white arrowheads) increased significantly from E13.5 to E15.5 (Fig. 8F′,G′,I). These results strongly suggest the existence of an early arriving wave of mGluR1/AP-2α expressing neurons that complete their migration to the pAOB by E13.5–E14.5 [in agreement with the data by Huilgol et al. (2013)], and a second wave of mGluR1/COUP-TFI-expressing cells that reach the pAOB at later stages, between E15.5 and E18.5.
A Small Percentage of mGluR1/Lot Cells Expresses p73 and Do not Integrate as Mitral Cells in the AOB
A recent study by Dixit et al. (2014) suggests that mGluR1/lot cells are p73 immunoreactive and thus proposes that lot cells are a subset of CR cells associated with the LOT. However, our results show that most mGluR1/lot cells express Tbr2 (Fig. 9C–C″), which is not a marker of CR cells and therefore, we decided to assess the degree of coexpression of p73 in mGluR1/lot cells to clarify this issue. To this end, we used a ΔNp73Cre-IRES-eGFP reporter mouse line (Tissir et al. 2009) in which eGFP expression recapitulates Trp73. It has been shown that ΔNp73, the N-truncated isoform of Trp73 (Meyer et al. 2004; Tissir et al. 2009), is expressed in cortical hem, septum (Meyer et al. 2002, 2004; Hernandez-Acosta et al. 2010), and TE-derived CR cells (Tissir et al. 2009), but it is not so in VP-derived CR cells (Hanashima et al. 2007). So, in the TE, ΔNp73-eGFP was expressed in the ventricular zone and the subpial side of the LTE, the TE mantle, and the putative migration path along the DTB to the pLOT (Fig. 9A), reflecting the distribution of the in situ hybridization signals in the same territories (Fig. 1B3,B3′). ΔNp73-eGFP immunoreactivity and Trp73 transcripts were also observed in the caudal cortical hem (Figs 1B,B′). We found that in ΔNp73Cre-IRES-eGFP mice, only a minority of E12.5 mGluR1/lot cells coexpressed ΔNp73, indicating that most mGluR1/lot cells are not CR cells. In the pLOT territory, only 15.36 ± 2.76% of the mGluR1/lot cells were ΔNp73-eGFP immunoreactive, quite similar to the 17.97 ± 3.13% detected in the LOT territory at mid-rostrocaudal levels (Fig. 9A′,B′, white arrowheads). Moreover, ΔNp73-eGFP cells were in all instances Tbr2-negative unlike mGluR1/lot cells destined to the pAOB (Fig. 9C–C″). While mGluR1/lot cells tended to locate deeper within the LOT territory, most ΔNp73-eGFP+ CR cells accumulated near the subpial side of the LOT at E12.5 (Fig. 9A,B). At subsequent developmental stages (E15.5 and E18.5), mGluR1/ΔNp73 cells extended into the piriform cortex, but they never invaded the neocortex primordium (Fig. 8B–B″,C–C″). At E15.5, some of the mGluR1/ΔNp73 cells (see Supplementary Fig. 10A′,B′, white arrowheads) reached the OB and located around (but not within) the mitral cell layers (see Supplementary Fig. 10A–C).
We also noticed that nearly half of the ΔNp73-eGFP-positive cells did not coexpress mGluR1 (52.98 ± 1.98%; Fig. 9A′,B′, hollow arrowheads) and, therefore, by definition, they should not be considered lot cells. However, p73 cells of any of these 2 groups, mGluR1-positive or mGluR1-negative, likely are CR cells. These data reveal that in contrast to what has been previously proposed, only a small percentage of the mGluR1/lot cells arising in the LTE are CR cells.
Discussion
As demonstrated by Sato et al. (1998), an early generated population of neurons, the lot cells, acts as guidepost cells for mitral cell axons along the LOT. Here, we demonstrate that, in contrast to the current belief, the lot cells predominantly originate from the TE, a transient structure localized in the boundary between the telencephalon and the diencephalon, and specifically from its lateral region, the LTE. The present findings highlight the TE as a neurogenic hub of diverse neuronal sets destined to different telencephalic domains that include CR cells (Meyer et al. 2002; Takiguchi-Hayashi et al. 2004; Tissir et al. 2009; Roy et al. 2014), Lhx5/AP-2α prospective pAOB mitral cells (Huilgol et al. 2013), and COUP-TFI+ mGluR1/lot cells that also invade the pAOB (present results).
The TE as a Primary Source of mGluR1/Lot Cells
Tomioka et al. (2000) and Kawasaki et al. (2006) cultured pallial and subpallial progenitor cells at E10.5 and found that the pallial progenitors, unlike the subpallial ones, could differentiate into the mGluR1-positive lot cells described previously by Sato et al. (1998). We replicated these experiments at E11.5 including cells derived from the TE. Similar to Tomioka et al. (2000), we detected a small proportion of cells derived from the E11.5 pallium that expresses mGluR1 in culture. However, the percentage of mGluR1 cells derived from the TE was almost 20-fold higher than that of mGluR1 cells derived from the pallium. Thus, our results show that the E11.5 TE progenitor cells have the potential to produce mGluR1 cells.
Migration experiments performed in whole embryo cultures (Tomioka et al. 2000) and in organotypic cultures (Kawasaki et al. 2006; Ito et al. 2008) supported the view that mGluR1/lot cells are generated in the DP and then migrate tangentially within the pallial MZ to reach the LOT territory in the piriform cortex, prior to the arrival of mitral cell axons. Lot cells share this purported pallial migration path with several early differentiated neurons such as preplate cells from the rostromedial telencephalic wall (Pedraza et al. 2014), CR cells from the cortical hem and pallial septum (Yoshida et al. 2006; Zhao et al. 2006; Garcia-Moreno et al. 2007; Zimmer et al. 2010), and olfactory cortex neurons from DP (Nomura et al. 2006; Garcia-Moreno et al. 2008). All these neurons migrate ventrally within the pallial MZ to reach the LOT territory, that is, the external pallial–subpallial boundary. However, although Tomioka et al. (2000), Kawasaki et al. (2006), and Ito et al. (2008) clearly demonstrated a ventrally directed tangential migration within the pallium MZ at E10.5–E11.5, the hypothesis that these migrating cells were lot cells could not be ascertained in their experiments because these cells were nonimmunoreactive to mAb lot-1. Moreover, the present study confirms the absence of mGluR1 expression in the pallial MZ at the stages considered.
In contrast, we report here that mGluR1 expression occurs in a continuous domain that comprises the TE, the DTB, and the LOT territory as early as E11.5 (see Supplementary Fig. 1), which strongly suggests a TE origin for these cells.
Lhx2 null mice, which lack a DP and display an enlarged TE and VP (Vyas et al. 2003; Roy et al. 2014), provided further evidence that mGluR1/lot cells originate from the TE similar to the prospective pAOB mitral cells described by Huilgol et al. (2013). In Lhx2−/− mice, the spread of mGluR1/lot cells in the piriform cortex was considerably larger than normal and, therefore, it is unlikely that these cells come from the DP. In a similar vein, the OB is specified as an OBLS in Lhx2−/− mice, in which mitral cell axons failed to pioneer the LOT projection. This anomaly is compounded by defects in axon guidance associated with the described mislocalization of the lot guidepost cells (Saha et al. 2007). However, the present findings show that lot cells are exuberant in Lhx2−/− mice. Intriguingly, Saha et al. (2007) detected in these mutants a notable overexpression of Sema6A in a rostrolateral region of the telencephalon that topographically corresponds to the expanded mGluR1 immunoreactive domain described in the present report, and we found that lot cells express Sema6A in wild-type mice (not shown). It remains to be seen in a future study how lot cell exuberance, the overexpression of Sema6A or other anomalies affect axon guidance along the LOT in Lhx2−/− mice.
An argument that has also been used to suggest a DP origin for the lot cells was the phenotype exhibited by the Gli3 null mice Gli3Xt/Xt. At E13.5, these animals show subpial clusters of mGluR1/lot cells limited to the DP (Tomioka et al. 2000) together with a convincing reduction of tangential cell migrations in the pallial surface attributed to the absence of Gli3 function (Kawasaki et al. 2006). The authors proposed that the lack of Gli3 function arrested the migration of lot cells originated in the pallium, causing these cells to stall abnormally within the pallium itself instead of reaching the LOT territory. However, contrasting with this interpretation, it has been amply demonstrated that the choroid plexus, the cortical hem, and the medial pallium are missing in Gli3 null mice juxtapositioning the DP and the TE (Grove et al. 1998; Theil et al. 1999; Tole et al. 2000). Moreover, the poorly developed DP contains a mixture of dorsal pallial, ventral pallial, and TE cells (Fotaki et al. 2006; Friedrichs et al. 2008). Furthermore, lot cells are correctly positioned in the piriform cortex in a conditional Gli3 mouse (Emx1Cre;Gli3fl/fl) where the pallium lacks Gli3 expression (Amaniti et al. 2015). This finding supports the interpretation that lot cells originate from outside the Emx1+ DP. Even further, our own unpublished data indicate that the Grm1+ clusters in the pallium of Gli3 null mice correspond to TE ectopias resulting from a faulty specification of the TE (N.R.-R., T.T., A.F., manuscript in preparation). Thus, these new findings clearly discard the cortical hem and the medial pallium, but also the DP, as a possible origin of mGluR1/lot cells.
Additionally, we explored the hem and the VP as potential sources of lot cells using 2 reporter mouse lines for Wnt3a and Dbx1 progenitors in the hem, VP, and septum, and demonstrated that mGluR1 cells do not arise from these progenitors. Finally, in utero electroporation provided decisive evidence that mGluR1/lot cells derive from neural progenitors in the LTE.
The mGluR/Lot Cells Are a Heterogeneous Cell Population
Our results indicate that most mGluR1/lot cells are akin to the cells described by Huilgol et al. (2013), because they both express mitral cell markers and incorporate into the pAOB. Moreover, the in utero electroporation experiments together with our immunohistochemical results suggest that there are at least 2 waves of lot cell generation: An early wave of cells that reach the pAOB at E13.5, which seems to be the AP-2α/Lhx5 cell population previously described by Huilgol et al. (2013), and a late wave of mGluR1/COUP-TFI cells that reach the pAOB at later stages.
On the other hand, we also found a population of mGluR1/lot cells in the LOT territory that express p73 and, therefore, may be considered as CR cells. These LOT-associated CR cells migrate rostrally to reach the AOB, but they never integrate into this structure as mitral cells. Instead, most of these cells spread over the paleocortical primordium by E18.5 without invading the neocortical primordium. This result differs from that of Dixit et al. (2014), who proposed that almost all the mGluR1/lot cells are p73 positive and, therefore, that lot cells are a subset of CR cells. The evidence shown by these authors was based on p73 immunohistochemical experiments performed with a p73 antibody whose specificity was not adequately tested. In contrast, we have performed cell lineage experiments using the ΔNp73eGFP mice and shown that eGFP labeling in these mice closely matched the in situ hybridization images for Trp73. Previous studies defining the expression and functional roles of p73 in CR cell biology (Meyer et al. 2002, 2004) have noted that the expression of p73 is a characteristic of most subtypes of CR cells, and only CR cells derived from the VP lack p73 expression (Hanashima et al. 2007).
In summary, we conclude that the mGluR1/lot cells are heterogeneous and form at least 2 different cell populations. First, a large group of Tbr2+ cells that are prospective pAOB mitral cells and that are divided into at least 2 subpopulations that migrate into the pAOB in 2 partially overlapping waves: The early arriving AP-2α+ cells and the late arriving COUP-TFI+ cells (Fig. 10). Second, ΔNp73+/mGluR1+ cells positioned around the LOT territory and in the MZ of the piriform cortex that together with the ΔNp73+/mGluR1− cells likely are CR cells.
Figure 10.
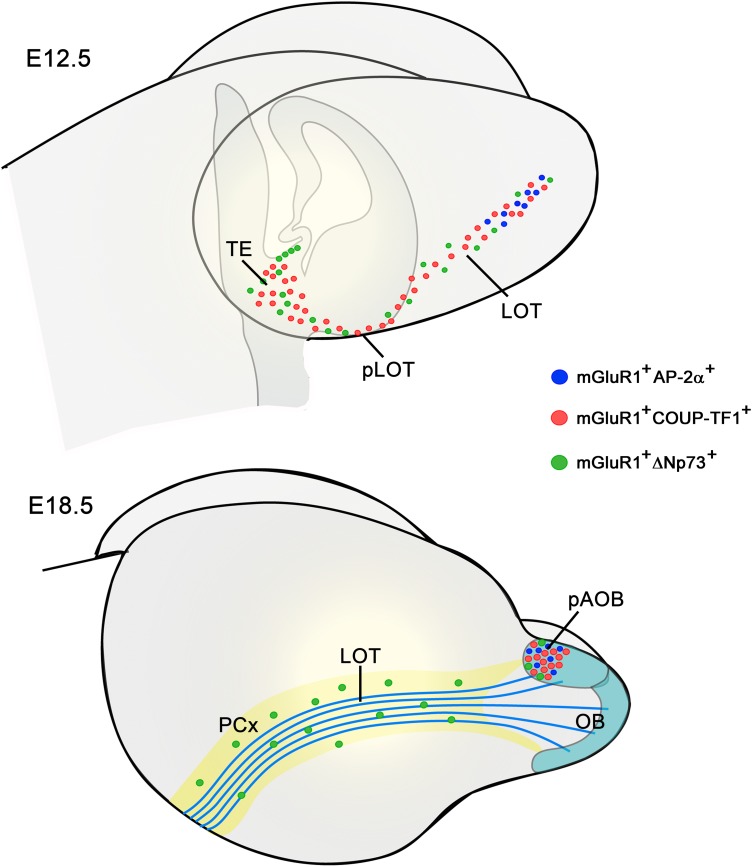
Temporal evolution of early generated cells in the LOT: mGluR1/lot cells and CR cells. Schemata summarizing the temporospatial changes in the distribution of early generated cells associated with the LOT. At E12.5, mGluR1/lot cells positive for AP-2α (blue dots) localize at rostral levels of the LOT, close to the prospective OB. The mGluR1/lot cells positive for COUP-TFI but negative for AP-2α (red dots) concentrate in the TE and extend rostrally along the LOT territory. Less abundant are the LTE-derived, mGluR1-positive and ΔNp73-positive CR cells (green dots). At E18.5, the mGluR1+/AP-2α+ cells and the mGluR1+/COUP-TFI+ cells have advanced rostrally to be conspicuous in the pAOB, whereas the mGluR1+/ΔNp73+ CR cells have extended dorsally within the paleocortex (PCx) and rostrally within the AOB, where they do not differentiate as mitral cells. Mitral cell axons were depicted in blue. LOT, lateral olfactory tract territory (in A), or lateral olfactory tract (in B); OB, olfactory bulb; pAOB, posterior accessory olfactory bulb; PCx, paleocortical primordium; pLOT, posterior LOT territory (in A), or posterior lateral olfactory tract (in B); TE, thalamic eminence.
Have the Lot Cells a Diencephalic or a Telencephalic Origin?
All our results unambiguously demonstrate that most lot cells originate in the TE. However, it is not clear whether the TE should be considered as a diencephalic or a telencephalic structure. Our chemoarchitectonic observations summarized in Figure 5 and Supplementary Figures 6 and 9 highlight several remarkable properties that the TE has in common with the telencephalic pallium. On the one hand, the TE displays a Tbr2-positive subventricular zone that is anatomically continuous with the pallial SVZ (Fig. 5E), and the TE mantle expresses Tbr1 as the cohorts of pallium-generated earliest neurons (Englund et al. 2005). Second, the LTE and the telencephalic caudal cortical hem share anatomically continuous gene expression domains such as Wnt8b (see Supplementary Fig. 5E1–E3) and Emx2 (see Supplementary Fig. 6B). Third, the MTE also displays gene expression features in common with the pallium such as Pax6, Neurog2, Dbx1, and Sfrp2. In fact, the caudal hem and the LTE are one long piece of Wnt2b- and Neurog2-expressing tissue that generates, and is divided by, the lateral ventricle choroid plexus [see Figs 1 and 2 in Grove et al. (1998)]. Such an anatomical continuum caudal hem–LTE was also suggested by Gorski et al. (2002) in their Supplementary Figure 1. Moreover, Yoshida et al. (2006) found that crossing a Wnt3a-floxed dt-a mouse with an Emx1-Cre mouse produced progeny lacking the hem—except in a very caudal region [see Yoshida et al. (2006) in their Fig. 3B] that likely corresponds to the LTE–caudal hem continuum. A very caudal tip of choroid plexus in the lateral ventricle is also present in the mutant, most likely generated from this region that could deserve to be called telencephalic. However, given that these arguments only suggest, but do not prove, a telencephalic identity for the TE, we conclude that the attribution of a telencephalic origin to the LTE-derived cells present in the LOT cannot be considered definitive and needs further research.
Supplementary Material
Supplementary material can be found at http://www.cercor.oxfordjournals.org/.
Funding
N.R.-R. held a FPI fellowship from the Spanish Government. N.R.-R. short stays in Dr Theil laboratory was also supported by the Spanish Government. This work was supported by grants from Queen Elisabeth Medical Foundation and JED-Belgique Foundation to F.T., the Tata Institute of Fundamental Research, Department of Atomic Energy, Government of India to S.T.; Medical Research Council (MR/K013750/1) to T.T.; Valencia Regional Government (Prometeo 2012-005), Spanish Government (BFU2010-16563), and the European Research Council (ERC2011-StG201001109) to E.H.; Spanish Government (BFU2007-60263 and BFU2010-17305) to A.F. The Instituto de Neurociencias is a Severo Ochoa Excellence Center (SEV-2013-0317).
Supplementary Material
Notes
We thank Alessandra Pierani and Sonia Garel for providing Dbx1Cre; ROSA26YFP and Wnt3aIRES-Cre; Ai9td-Tomato fixed mouse embryos, respectively. We also thank Tatsumi Hirata for mAb lot1; Michèle Studer for COUP-TFI antibody; André Goffinet for mAb G10 to reelin; and Se-Jin Lee, Suzie Chen, Salvador Martinez, and Oscar Marin for plasmids. mAb 4F2 to <Lhx1 and Lhx5 was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa (Department of Biological Sciences, Iowa City, IA, USA). Special thanks are due to Joan Galcerán and Jesús Martínez-López for their help with in situ hybridizations, and K. Hasenpusch-Theil for technical support. Conflict of Interest: None declared.
References
- Abbott LC, Jacobowitz DM. 1999. Developmental expression of calretinin-immunoreactivity in the thalamic eminence of the fetal mouse. Int J Dev Neurosci. 17:331–345. [DOI] [PubMed] [Google Scholar]
- Abellán A, Vernier B, Rétaux S, Medina L. 2010. Similarities and differences in the forebrain expression of Lhx1 and Lhx5 between chicken and mouse: insights for understanding telencephalic development and evolution. J Comp Neurol. 518:3512–3528. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. 1995. Atlas of the prenatal rat brain development. Ann Arbor: CRC Press. [Google Scholar]
- Amaniti EM, Kelman A, Mason JO, Theil T. 2015. Cerebral cortex expression of Gli3 is required for normal development of the lateral olfactory tract. PLoS ONE. 10(10):e0141525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell K. 2012. The olfactory system. In: Watson C, Paxinos G, Puelles L, editors. The mouse nervous system. Amsterdam: Academic Press; p. 653–660. [Google Scholar]
- Bielle F, Garel S. 2013. Neuronal migration of guidepost cells. In: Rubenstein J, Rakic P, editors. Comprehensive developmental neuroscience: cellular migration and formation of neuronal connections, Chapter 25. Amsterdam: Elsevier; p. 458–479. [Google Scholar]
- Bielle F, Griveau A, Narboux-Neme N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. 2005. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 8:1002–1012. [DOI] [PubMed] [Google Scholar]
- Borrell V, Yoshimura Y, Callaway EM. 2005. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 143:151–158. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. 2001. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 100:165–175. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JL. 1995. T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron. 15:63–78. [DOI] [PubMed] [Google Scholar]
- Dixit R, Wilkinson G, Cancino GI, Shaker T, Adnani L, Li S, Dennis D, Kurrasch D, Chan JA, Olson EC et al. 2014. Neurog1 and Neurog2 control two waves of neuronal differentiation in the piriform cortex. J Neurosci. 34:539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. 2005. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 25:247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa A, Gil-Sanz C, Yanagawa Y, Fairén A. 2009. Two separate subtypes of early non-subplate projection neurons in the developing cerebral cortex of rodents. Front Neuroanat. 3:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faedo A, Ficara F, Ghiani M, Aiuti A, Rubenstein JL, Bulfone A. 2002. Developmental expression of the T-box transcription factor T-bet/Tbx21 during mouse embryogenesis. Mech Dev. 116:157–160. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Yu T, Zaki PA, Mason JO, Price DJ. 2006. Abnormal positioning of diencephalic cell types in neocortical tissue in the dorsal telencephalon of mice lacking functional Gli3. J Neurosci. 26:9282–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs M, Larralde O, Skutella T, Theil T. 2008. Lamination of the cerebral cortex is disturbed in Gli3 mutant mice. Dev Biol. 318:203–214. [DOI] [PubMed] [Google Scholar]
- Garcia-Frigola C, Carreres MI, Vegar C, Herrera E. 2007. Gene delivery into mouse retinal ganglion cells by in utero electroporation. BMC Dev Biol. 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Moreno F, López-Mascaraque L, de Carlos JA. 2008. Early telencephalic migration topographically converging in the olfactory cortex. Cereb Cortex. 18:1239–1252. [DOI] [PubMed] [Google Scholar]
- García-Moreno F, Lopez-Mascaraque L, De Carlos JA. 2007. Origins and migratory routes of murine Cajal-Retzius cells. J Comp Neurol. 500:419–432. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. 2002. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 22:6309–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, Guillemot F. 1996. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 180:227–241. [DOI] [PubMed] [Google Scholar]
- Griveau A, Borello U, Causeret F, Tissir F, Boggetto N, Karaz S, Pierani A. 2010. A novel role for Dbx1-derived Cajal-Retzius cells in early regionalization of the cerebral cortical neuroepithelium. PLoS Biol. 8:e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. 1998. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 125:2315–2325. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Joyner AL. 1993. Dynamic expression of the murine Achaete-Scute homologue Mash-1 in the developing nervous system. Mech Dev. 42:171–185. [DOI] [PubMed] [Google Scholar]
- Halpern M, Martínez-Marcos A. 2003. Structure and function of the vomeronasal system: an update. Prog Neurobiol. 70:245–318. [DOI] [PubMed] [Google Scholar]
- Hanashima C, Fernandes M, Hebert JM, Fishell G. 2007. The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J Neurosci. 27:11103–11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Acosta NC, Cabrera-Socorro A, Morlans MP, Delgado FJ, Suárez-Solá ML, Sottocornola R, Lu X, González-Gómez M, Meyer G. 2011. Dynamic expression of the p53 family members p63 and p73 in the mouse and human telencephalon during development and in adulthood. Brain Res. 1372:29–40. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. 2006. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 55:223–233. [DOI] [PubMed] [Google Scholar]
- Hirata T, Kumada T, Kawasaki T, Furukawa T, Aiba A, Conquet F, Saga Y, Fukuda A. 2012. Guidepost neurons for the lateral olfactory tract: expression of metabotropic glutamate receptor 1 and innervation by glutamatergic olfactory bulb axons. Dev Neurobiol. 72:1559–1576. [DOI] [PubMed] [Google Scholar]
- Hirata T, Li P, Lanuza GM, Cocas LA, Huntsman MM, Corbin JG. 2009. Identification of distinct telencephalic progenitor pools for neuronal diversity in the amygdala. Nat Neurosci. 12:141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Witte DP, Szucsik JC, Weinstein M, Li H, Potter SS. 1995. Gsh-2, a murine homeobox gene expressed in the developing brain. Mech Dev. 50:177–186. [DOI] [PubMed] [Google Scholar]
- Huilgol D, Udin S, Shimogori T, Saha B, Roy A, Aizawa S, Hevner RF, Meyer G, Ohshima T, Pleasure SJ et al. 2013. Dual origins of the mammalian accessory olfactory bulb revealed by an evolutionarily conserved migratory stream. Nat Neurosci. 16:157–165. [DOI] [PubMed] [Google Scholar]
- Ina A, Sugiyama M, Konno J, Yoshida S, Ohmomo H, Nogami H, Shutoh F, Hisano S. 2007. Cajal-Retzius cells and subplate neurons differentially express vesicular glutamate transporters 1 and 2 during development of mouse cortex. Eur J Neurosci. 26:615–623. [DOI] [PubMed] [Google Scholar]
- Inamura N, Ono K, Takebayashi H, Zalc B, Ikenaka K. 2011. Olig2 lineage cells generate GABAergic neurons in the prethalamic nuclei, including the zona incerta, ventral lateral geniculate nucleus and reticular thalamic nucleus. Dev Neurosci. 33:118–129. [DOI] [PubMed] [Google Scholar]
- Ito K, Kawasaki T, Takashima S, Matsuda I, Aiba A, Hirata T. 2008. Semaphorin 3F confines ventral tangential migration of lateral olfactory tract neurons onto the telencephalon surface. J Neurosci. 28:4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Lyons KM, Hogan BL. 1991. Involvement of bone morphogenetic protein-4 (BMP-4) and Vgr-1 in morphogenesis and neurogenesis in the mouse. Development. 111:531–542. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Ito K, Hirata T. 2006. Netrin 1 regulates ventral tangential migration of guidepost neurons in the lateral olfactory tract. Development. 133:845–853. [DOI] [PubMed] [Google Scholar]
- Kay LM, Sherman SM. 2007. An argument for an olfactory thalamus. Trends Neurosci. 30:47–53. [DOI] [PubMed] [Google Scholar]
- Kim AS, Anderson SA, Rubenstein JL, Lowenstein DH, Pleasure SJ. 2001. Pax-6 regulates expression of SFRP-2 and Wnt-7b in the developing CNS. J Neurosci. 21:RC132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Dudley CA, Moss RL. 1999. Functional dichotomy within the vomeronasal system: distinct zones of neuronal activity in the accessory olfactory bulb correlate with sex-specific behaviors. J Neurosci. 19:RC32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Wise TL, Ruddle FH. 1994. Mouse homeobox gene Dbx: sequence, gene structure and expression pattern during mid-gestation. Mech Dev. 47:187–195. [DOI] [PubMed] [Google Scholar]
- Mangale VS, Hirokawa KE, Satyaki PR, Gokulchandran N, Chikbire S, Subramanian L, Shetty AS, Martynoga B, Paul J, Mai MV et al. 2008. Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science. 319:304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Valiente M, Ge X, Tsai LH. 2010. Guiding neuronal cell migrations. Cold Spring Harb Perspect Biol. 2(2):a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lopez JE, Moreno-Bravo JA, Madrigal MP, Martinez S, Puelles E. 2015. Mesencephalic basolateral domain specification is dependent on Sonic Hedgehog. Front Neuroanat. 9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. 2004. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 101:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattar P, Britz O, Johannes C, Nieto M, Ma L, Rebeyka A, Klenin N, Polleux F, Guillemot F, Schuurmans C. 2004. A screen for downstream effectors of Neurogenin2 in the embryonic neocortex. Dev Biol. 273:373–389. [DOI] [PubMed] [Google Scholar]
- McWhirter JR, Goulding M, Weiner JA, Chun J, Murre C. 1997. A novel fibroblast growth factor gene expressed in the developing nervous system is a downstream target of the chimeric homeodomain oncoprotein E2A-Pbx1. Development. 124:3221–3232. [DOI] [PubMed] [Google Scholar]
- Meyer G. 2010. Building a human cortex: the evolutionary differentiation of Cajal-Retzius cells and the cortical hem. J Anat. 217:334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Cabrera Socorro A, Perez Garcia CG, Martinez Millan L, Walker N, Caput D. 2004. Developmental roles of p73 in Cajal-Retzius cells and cortical patterning. J Neurosci. 24:9878–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Perez-Garcia CG, Abraham H, Caput D. 2002. Expression of p73 and Reelin in the developing human cortex. J Neurosci. 22:4973–4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquelajáuregui A, Varela-Echavarría A, Ceci ML, García-Moreno F, Ricaño I, Hoang K, Frade-Pérez D, Portera-Cailliau C, Tamariz E, De Carlos JA et al. 2010. LIM-homeobox gene Lhx5 is required for normal development of Cajal-Retzius cells. J Neurosci. 30:10551–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohedano-Moriano A, Pro-Sistiaga P, Ubeda-Bañón I, Crespo C, Insausti R, Martinez-Marcos A. 2007. Segregated pathways to the vomeronasal amygdala: differential projections from the anterior and posterior divisions of the accessory olfactory bulb. Eur J Neurosci. 25:2065–2080. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA. 2001. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 32:591–604. [DOI] [PubMed] [Google Scholar]
- Nomura T, Holmberg J, Frisen J, Osumi N. 2006. Pax6-dependent boundary defines alignment of migrating olfactory cortex neurons via the repulsive activity of ephrin A5. Development. 133:1335–1345. [DOI] [PubMed] [Google Scholar]
- Pedraza M, Hoerder-Suabedissen A, Albert-Maestro MA, Molnár Z, De Carlos JA. 2014. Extracortical origin of some murine subplate cell populations. Proc Natl Acad Sci USA. 111:8613–8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M, Redmond D, Watson CJ, Mason JO. 1999. Mouse Wnt8B is expressed in the developing forebrain and maps to chromosome 19. Mamm Genome. 10:923–925. [DOI] [PubMed] [Google Scholar]
- Roelink H, Nusse R. 1991. Expression of two members of the Wnt family during mouse development—restricted temporal and spatial patterns in the developing neural tube. Genes Dev. 5:381–388. [DOI] [PubMed] [Google Scholar]
- Roy A, de Melo J, Chaturvedi D, Thein T, Cabrera-Socorro A, Houart C, Meyer G, Blackshaw S, Tole S. 2013. LHX2 is necessary for the maintenance of optic identity and for the progression of optic morphogenesis. J Neurosci. 33:6877–6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Gonzalez-Gomez M, Pierani A, Meyer G, Tole S. 2014. Lhx2 regulates the development of the forebrain hem system. Cereb Cortex. 24:1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AJ, Schwendinger JK, Yang AW, Haider NB, Nystuen AM. 2007. The mouse mutants recoil wobbler and nmf373 represent a series of Grm1 mutations. Mamm Genome. 18: 749–756. [DOI] [PubMed] [Google Scholar]
- Saha B, Hari P, Huilgol D, Tole S. 2007. Dual role for LIM-homeodomain gene Lhx2 in the formation of the lateral olfactory tract. J Neurosci. 27:2290–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Hirata T, Ogawa M, Fujisawa H. 1998. Requirement for early-generated neurons recognized by monoclonal antibody lot1 in the formation of lateral olfactory tract. J Neurosci. 18:7800–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Guillemot F. 2002. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol. 12:26–34. [DOI] [PubMed] [Google Scholar]
- Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M et al. 2010. A genomic atlas of mouse hypothalamic development. Nat Neurosci. 13:767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Gulisano M, Acampora D, Stornaiuolo A, Rambaldi M, Boncinelli E. 1992. Two vertebrate homeobox genes related to the Drosophila empty spiracles gene are expressed in the embryonic cerebral cortex. EMBO J. 11:2541–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarzoni P, Thion MS, Garel S. 2015. Neuronal and microglial regulators of cortical wiring: usual and novel guideposts. Front Neurosci. 9:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. 1991. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 137:173–182. [DOI] [PubMed] [Google Scholar]
- Subramanian L, Remedios R, Shetty A, Tole S. 2009. Signals from the edges: the cortical hem and antihem in telencephalic development. Semin Cell Dev Biol. 20:712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Tole S. 2009. Mechanisms underlying the specification, positional regulation, and function of the cortical hem. Cereb Cortex. 19(Suppl 1):i90–i95. [DOI] [PubMed] [Google Scholar]
- Takiguchi-Hayashi K, Sekiguchi M, Ashigaki S, Takamatsu M, Hasegawa H, Suzuki-Migishima R, Yokoyama M, Nakanishi S, Tanabe Y. 2004. Generation of reelin-positive marginal zone cells from the caudomedial wall of telencephalic vesicles. J Neurosci. 24:2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Fujimori K, Nojyo Y, Kaneko T, Takauji R. 2003. Evidence that Sema3A and Sema3F regulate the migration of GABAergic neurons in the developing neocortex. J Comp Neurol. 455:238–248. [DOI] [PubMed] [Google Scholar]
- Teissier A, Griveau A, Vigier L, Piolot T, Borello U, Pierani A. 2010. A novel transient glutamatergic population migrating from the pallial-subpallial boundary contributes to neocortical development. J Neurosci. 30:10563–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Rüther U. 1999. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 126:3561–3571. [DOI] [PubMed] [Google Scholar]
- Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet AM. 2009. DeltaNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci USA. 106:16871–16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. 2000. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J). Dev Biol. 217:254–265. [DOI] [PubMed] [Google Scholar]
- Tomioka N, Osumi N, Sato Y, Inoue T, Nakamura S, Fujisawa H, Hirata T. 2000. Neocortical origin and tangential migration of guidepost neurons in the lateral olfactory tract. J Neurosci. 20:5802–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas A, Saha B, Lai E, Tole S. 2003. Paleocortex is specified in mice in which dorsal telencephalic patterning is severely disrupted. J Comp Neurol. 466:545–553. [DOI] [PubMed] [Google Scholar]
- Walther C, Gruss P. 1991. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 113:1435–1449. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Sekiguchi M, Takamatsu M, Tanabe Y, Nakanishi S. 2004. Distinct ontogenic and regional expressions of newly identified Cajal-Retzius cell-specific genes during neocorticogenesis. Proc Natl Acad Sci USA. 101:14509–14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Assimacopoulos S, Jones KR, Grove EA. 2006. Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development. 133:537–545. [DOI] [PubMed] [Google Scholar]
- Zhao C, Guan W, Pleasure SJ. 2006. A transgenic marker mouse line labels Cajal-Retzius cells from the cortical hem and thalamocortical axons. Brain Res. 1077:48–53. [DOI] [PubMed] [Google Scholar]
- Zhao R, Lawler AM, Lee SJ. 1999. Characterization of GDF-10 expression patterns and null mice. Dev Biol. 212:68–79. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. 1999. Control of hippocampal morphogenesis and neuronal differentiation by the LIM homeobox gene Lhx5. Science. 284:1155–1158. [DOI] [PubMed] [Google Scholar]
- Zhu H, Ryan K, Chen S. 1999. Cloning of novel splice variants of mouse mGluR1. Mol Brain Res. 73:93–103. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Lee J, Griveau A, Arber S, Pierani A, Garel S, Guillemot F. 2010. Role of Fgf8 signalling in the specification of rostral Cajal-Retzius cells. Development. 137:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.