Summary
The methylation of subtelomeric repeat D4Z4 and pericentromeric repeat NBL2 in peripheral blood leukocytes was examined with pyrosequencing in prostate cancer. The methylation of D4Z4 was associated with BCR.
Abstract
Global DNA methylation may affect chromosome structure and genomic stability and is involved in carcinogenesis. In this study, we aimed to investigate whether methylation of pericentromeric repeat NBL2 and subtelomeric repeat D4Z4 in peripheral blood was associated with the aggressiveness of prostate cancer (PCa). We measured the methylation status of different CpG sites of NBL2 and D4Z4 in 795 PCa patients and compared their methylation levels among patients with different Gleason Score at diagnosis. We then analyzed the association of the NBL2 and D4Z4 methylation with the risk of biochemical recurrence (BCR) in patients receiving radical prostatectomy or radiotherapy using a multivariate Cox proportional hazards model. In addition, we used the Kaplan–Meier survival function and log-rank tests to assess BCR-free survival associated with D4Z4 methylation. There was no significant difference in methylation level of NBL2 and D4Z4 between clinically defined aggressive and non-aggressive PCa at diagnosis. However, the methylation of D4Z4 was associated with BCR, while the methylation of NBL2 was not. In tertile analysis, patients in the highest tertile of D4Z4 methylation had an increased risk of BCR (HR = 2.17, 95% CI 1.36–3.48) compared to patients in the lower tertiles after adjustment of age, body mass index, smoking status, pack year, D’Amico risk groups and treatments. Among the four CpG sites in this region, the association was mostly attributable to the methylation of the second CpG site of D4Z4. These data suggest that higher methylation in D4Z4 was associated with worse prognosis of localized PCa patients.
Introduction
Prostate cancer (PCa) is the most common cancer and the third leading cause of death from cancer among men in the United States (1). Although early detection of PCa due to the wide use of prostate-specific antigen (PSA) screening has greatly reduced the mortality of PCa, the overdiagnosis and overtreatment of PCa have become a major clinical problem (2). The majority of PSA-detected PCa are indolent and pose little of no threat to the survival of patients. However, about 90% of men with localized PCa receive aggressive treatment, which is often associated with significant morbidity. This clinical problem is largely attributed to the fact that clinical variables, such as Gleason Score (GS), PSA level and tumor stage, cannot accurately distinguish aggressive from indolent diseases at diagnosis while patients with similar clinical features can have dramatically different clinical outcomes. Therefore, biomarkers are urgently needed for more accurate prediction of individual tumor behavior and patient prognosis.
DNA methylation plays an important role in tumorigenesis and cancer progression (3). DNA hypermethylation is frequently observed in the promoter regions of tumor suppressor genes (4,5), leading to gene silencing, while genome-wide global hypomethylation events are common in human cancer, causing genomic instability (6,7). Global hypomethylation in peripheral blood leukocyte DNA has been associated with increased risk of various cancers (8–15). However, no study has evaluated the role of global leukocyte DNA methylation in the prognosis of localized PCa patients.
Most of the published studies measured the methylation of long-interspersed nucleotide elements (LINE-1) or Alu repeats as surrogate markers of the genome-wide methylation (8–15). These elements are highly abundant, randomly distributed throughout the genome, and are heavily methylated. Their methylations were estimated to account for one third of all DNA methylations (16). There are other types of DNA repeats that have more focused distributions, e.g. in centromeric, pericentromeric and subtelomeric regions. Previous studies have shown that the methylation level of tandem DNA repeats in these specific chromosome regions are different from that of LINE-1 and Alu sequences (17). There were also evidence suggesting that different chromosome regions, e.g. pericentromeric and subtemeric regions, plays differential roles in maintaining chromosome structure and function (18–21). We hypothesize that the methylation of different chromosome regions may have differential effect on disease development. In this study, we measured the methylation level of pericentromeric repeat (NBL2) and subtelomeric repeat (D4Z4) in peripheral blood leukocytes of a large group of localized PCa patients and analyzed their associations with GS as diagnosis and biochemical recurrence (BCR) in patients receiving radical prostatectomy or radiotherapy.
Materials and methods
Study population
This study included 795 non-Hispanic white men with histologically confirmed adenocarcinoma of prostate from the University of Texas MD Anderson Cancer Center. Blood specimens were collected from the patients at diagnosis before any treatments. DNA and plasma were isolated and banked. Clinical and follow-up data were abstracted from patient medical records by clinical coding specialists; these data included date of diagnosis, performance status, clinical stage, histological grade and pathological stage, treatment (active surveillance, prostatectomy, radiotherapy and hormone therapy) and progression (BCR and metastasis). The MD Anderson Tumor Registry conducts annual vital status follow-ups for all cancer patients. Biochemical recurrence will be defined as a serum PSA level of at least 0.2 ng/ml with a second confirmatory PSA level of at least 0.2 ng/ml for patients who undergo a radical prostatectomy or with an increase in PSA level above 0.2 ng/ml and two consecutive increase over a minimum of 3 months for patients receiving external-beam radiotherapy. This study was approved by the MD Anderson Cancer Center Institutional Review Board, and written consent forms were obtained from each patient.
Methylation analysis with pyrosequencing
One microgram of genomic DNA was treated with sodium bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA) according to the manufacturer’s protocol. The samples were eluted in 40 μl of M-Elution Buffer, and transferred to DNA Methylation Analysis Core at The University of Texas MD Anderson Cancer Center for subsequent PCR and pyrosequencing analysis. PCR primers for the repetitive sequences NBL2 and D4Z4 were mostly as previously described with one exception for D4Z4 sequencing primer (17), which was designed using the Pyromark Assay Design SW 2.0 software (Qiagen, Hilden, Germany) to improve on the general quality of the pyrosequencing reaction. The primers used in this study are listed in Supplementary Table 1, available at Carcinogenesis Online. PCR reactions were performed with ZymoTaq DNA Polymerase kit (Zymo Research, Irvine, CA) using 2 μl of bisulfite-treated DNA in a total volume of 15 µl, and the entire volume was used for each pyrosequencing reaction. Controls for high methylation (SssI-treated DNA), low methylation (WGA-amplified DNA) and no-DNA template were included in each run. In preparation for the pyrosequencing reaction, PCR product purification was done with streptavidin-sepharose high-performance beads (GE Healthcare Life Sciences, Piscataway, NJ), and co-denaturation of the biotinylated PCR products and sequencing primer (3.6 pmol/reaction) was conducted following the PSQ96 sample preparation guide. Pyrosequencing was performed on a PSQ HS 96 system (Biotage AB, Uppsala, Sweden) with the PyroMark Gold Q96 CDT Reagents (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The DNA methylation of each repetitive sequence was determined as the average of the methylation of all CpG sites in the evaluated sample, and each sample was run in duplicate. Methylation level values were based on pyrosequencing analysis of polymorphic nucleotide C and T at any specific site generated by bisulfate treatment as described before (22). Bisulfite treatment of DNA converts all unmethylated cytosine to uracil. The pyrosequencing software calculates ratio of the signal from cytosine (that signifies a methylated cytosine) to the sum of cytosine plus thymidine for each specific CpG site, which was termed β value as a parameter of methylation percentage at that site. Usually a single pyrosequencing assay analyzes between 3 and 5 CpG sites at a selected sequence, and the methylation for each assay is presented as the average methylation measured for all sites within the sequenced region. We added this information in Methods.
Statistical analysis
To control the batch effect, we put similar numbers of GS 6, seven and eight cases in each run and batch was treated as a nuisance covariant in our analysis. We first compared the methylation of each individual CpG site as well as the mean methylation of each region among patients with different clinical characteristics at baseline using analysis of variance (ANOVA). We then used multivariate logistic regression to estimate the odds ratio (ORs) and 95% confidence intervals (CIs) for analyzing the association of NBL2 and D4Z4 methylation with the aggressiveness of PCa at diagnosis. We then analyzed the association of CpG sites with the risk of BCR in patients receiving radical prostatectomy or radiotherapy using a multivariate Cox proportional hazards model adjusting for age, body mass index, smoking status, pack year, D’Amico risk groups and treatments. In addition, we used the Kaplan–Meier survival function and log-rank tests to assess BCR-free survival associated with these markers.
Results
The characteristics of the 795 PCa patients are shown in Table 1. Over 90% of patients were diagnosed at age 55 or older. The median age of patients at diagnosis was 64 (range, 42–85) years. About 36% of patients were overweight and 32% were obese. There were 354 (44.5%) never smokers, 364 (45.8%) former smokers and 69 (8.7%) current smokers. Based on the biopsy before the treatment, 274 had GS = 6 PCa, 240 had GS = 7 PCa and 281 had GS ≥ 8 PCa. Using the D’Amico rick stratification criteria, 260 patients were classified as low risk, 217 patients had intermediate risk and 313 were in the high-risk group. Almost 80% of the individuals had PSA levels lower than 10 mg/ml. Among those subjects, 375 (47.2%) patients underwent definitive radical prostatectomy as the initial primary treatment and 133 (16.7%) received definitive radiotherapy. The overall methylation levels of NBL2 and D4Z4 across patients with different characteristics were similar with no significant differences among all strata, except that the methylation level of D4Z4 appeared to be higher when patients is older than 65 years compared with these younger than 65 years (P = 0.022) (Table 1). We also compared the methylation level of each individual CpG site of NBL2 and D4Z4 with GS (Table 2) and different D’Amico rick groups (data not shown). The methylation in the five CpG sites in NBL2 and four CpG sites were not significantly different among patients with different aggressiveness at diagnosis.
Table 1.
NBL4 and D4Z4 methylation stratified by PCa patient characteristics
| Characteristics | N (%) | NBL4, β (SD) | P value | D4Z4, β (SD) | P value |
|---|---|---|---|---|---|
| Age at diagnosis, years | |||||
| <55 | 66 (8.3) | 79.0 (2.6) | 68.5 (4.8) | ||
| 55–65 | 346 (43.5) | 78.6 (3.1) | 68.4 (4.1) | ||
| >65 | 383 (48.2) | 78.7 (3.0) | 0.493 | 69.2 (4.8) | 0.022 |
| Body mass index at diagnosis, kg/m2 | |||||
| <25 | 115 (14.5) | 78.9 (2.8) | 68.7 (4.3) | ||
| 25–29.99 (overweight) | 287 (36.1) | 78.8 (2.9) | 68.9 (4.4) | ||
| ≥30 (obese) | 252 (31.7) | 78.3 (3.0) | 68.5 (4.7) | ||
| Unknown | 141 (17.7) | 79.0 (3.4) | 0.787 | 69.0 (4.5) | 0.882 |
| Smoking status at diagnosis | |||||
| Non-smoker | 354 (44.5) | 78.7 (3.0) | 69.0 (4.4) | ||
| Former smoker | 364 (45.8) | 78.7 (3.0) | 68.7 (4.6) | ||
| Current smoker | 69 (8.7) | 78.7 (2.9) | 68.3 (4.7) | ||
| Unknown | 8 (1.0) | 77.3 (2.2) | 0.725 | 68.8 (5.1) | 0.473 |
| D’Amico risk group | |||||
| Low | 260 (32.9) | 78.6 (3.0) | 69.0 (4.6) | ||
| Intermediate | 217 (27.5) | 78.9 (3.0) | 68.5 (4.3) | ||
| High | 313 (39.6) | 78.7 (3.0) | 0.828 | 68.8 (4.6) | 0.525 |
| Total Gleason score | |||||
| ≤6 | 274 (34.5) | 78.5 (3.0) | 69.0 (4.5) | ||
| 7 | 240 (30.2) | 78.8 (3.0) | 68.5 (4.2) | ||
| ≥8 | 281 (35.3) | 78.7 (3.0) | 0.423 | 68.8 (4.7) | 0.403 |
| Clinical tumor stage | |||||
| T1 | 572 (71.9) | 78.7 (3.0) | 68.7 (4.4) | ||
| T2 | 54 (6.8) | 78.7 (3.1) | 68.6 (4.9) | ||
| T3–T4 | 169 (21.3) | 78.7 (2.9) | 0.810 | 69.2 (4.8) | 0.311 |
| PSA at diagnosis | |||||
| <10 ng/ml | 633 (79.6) | 78.7 (3.0) | 68.8 (4.5) | ||
| 10–20 ng/ml | 80 (10.1) | 78.5 (3.3) | 68.7 (4.3) | ||
| >20 ng/ml | 82 (10.3) | 78.5 (3.0) | 0.371 | 68.4 (5.0) | 0.244 |
| Initial primary treatment | |||||
| Radical prostatectomy | 375 (47.2) | 78.7 (3.0) | 68.8 (4.3) | ||
| Radiotherapy | 133 (16.7) | 78.9 (2.8) | 68.6 (4.3) | ||
| Surveillance or unknown | 268 (33.7) | 78.5 (3.1) | 68.7 (4.9) | ||
| Other treatment | 19 (2.4) | 80.0 (2.9) | 0.803 | 70.0 (4.5) | 0.763 |
Table 2.
Methylation of repetitive sequences and the aggressiveness of PCa at diagnosis
| CpG sites | GS 6 | GS 7 | GS 8 | P for trend | |||
|---|---|---|---|---|---|---|---|
| Β | SD | β | SD | β | SD | ||
| NBL2 | |||||||
| Site 1 | 69.48 | 8.95 | 69.93 | 9.13 | 69.18 | 8.95 | 0.779 |
| Site 2 | 87.78 | 4.69 | 87.83 | 4.89 | 88.44 | 4.72 | 0.098 |
| Site 3 | 91.92 | 2.91 | 92.31 | 2.76 | 91.92 | 2.87 | 0.688 |
| Site 4 | 77.04 | 6.12 | 77.46 | 4.98 | 77.47 | 5.3 | 0.735 |
| Site 5 | 66.31 | 3.26 | 66.53 | 3.78 | 66.7 | 3.37 | 0.205 |
| Mean | 78.54 | 3 | 78.81 | 3.02 | 78.74 | 2.97 | 0.423 |
| D4Z4 | |||||||
| Site 1 | 64.02 | 4.64 | 63.3 | 4.02 | 63.69 | 4.71 | 0.230 |
| Site 2 | 65.89 | 5.76 | 65.54 | 5.23 | 66.09 | 5.77 | 0.822 |
| Site 3 | 84.58 | 4.52 | 84.42 | 4.47 | 84.23 | 5.07 | 0.294 |
| Site 4 | 61.58 | 5.4 | 60.58 | 5.39 | 61.18 | 5.65 | 0.415 |
| Mean | 69.02 | 4.53 | 68.46 | 4.22 | 68.8 | 4.75 | 0.403 |
β, methylation of CpG site (%); SD, standard deviation.
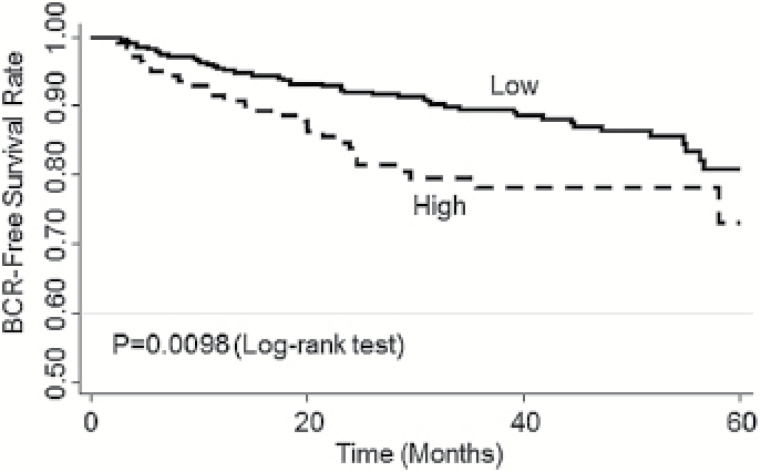
We then analyzed the association of the methylation of these CpG sites with BCR in all the localized PCa patients and in localized patients receiving active treatment (prostatectomy and radiotherapy). Since the rate of BCR was relatively low among patients with localized PCa, we combined the first and second tertiles for the analysis of BCR to increase statistical power. As shown in Table 3, compared to patients in the first and second tertile groups, patients in the third tertile (i.e. with the highest methylation level) had an increased risk of BCR (HR = 1.74, 95% CI, 1.14–2.65; P = 0.0096) after adjustment of age, body mass index, smoking status, pack-year, D’Amico risk groups and treatments. In detailed analysis of each CpG site in this region, the association was mostly attributable to Site 2 (HR = 1.80, 95% CI, 1.19–2.72). In patients receiving active treatments, D4Z4 methylation was associated with BCR (Table 4), Patients in the third tertile exhibited an increased risk of BCR (HR = 2.17, 95% CI, 1.36–3.48; P = 0.001) compared to those in the first and second tertile groups. The association was mostly due to Site 1 and Site 2 in this region (HR = 1.67 and 2.211, P = 0.044 and 0.0009, respectively). In Kaplan–Meier survival analysis, patients with higher D4Z4 methylation level had a significantly shorter BCR-free survival time than those with lower methylation level (log rank P = 0.0098) (Figure 1). The overall methylation of pericentromeric NBL2 and the methylation of each site in the NBL2 region were not associated with BCR (data not shown).
Table 3.
The association of D4Z4 methylation with BCR
| D4Z4 methylation | No BCR | BCR | Adjusted HRa (95% CI) | P value |
|---|---|---|---|---|
| N (%) | N (%) | |||
| Overall | ||||
| First and second tertile | 488 (88.25) | 65 (11.75) | Reference | N/A |
| Third tertile (highest) | 195 (84.42) | 36 (15.58) | 1.74 (1.14–2.65) | 0.0096 |
| Site 1 | ||||
| First and second tertile | 494 (87.43) | 71 (12.57) | Reference | N/A |
| Third tertile (highest) | 189 (86.30) | 30 (13.70) | 1.38 (0.89–2.15) | 0.144 |
| Site 2 | ||||
| First and second tertile | 478 (88.68) | 61 (11.32) | Reference | N/A |
| Third tertile (highest) | 205 (83.67) | 40 (16.33) | 1.80 (1.19–2.72) | 0.0055 |
| Site 3 | ||||
| First and second tertile | 471 (86.90) | 71 (13.10) | Reference | N/A |
| Third tertile (highest) | 212 (87.60) | 30 (12.40) | 1.30 (0.84–2.01) | 0.246 |
| Site 4 | ||||
| First and second tertile | 489 (87.32) | 71 (12.68) | Reference | N/A |
| Third tertile (highest) | 194 (86.61) | 30 (13.39) | 1.24 (0.80–1.92) | 0.326 |
aAdjusted for age, body mass index, smoking status, pack-year, D’Amico risk groups and primary treatment.
Table 4.
The association of D4Z4 methylation with BCR among localized PCa patients who received active treatments
| D4Z4 methylation | No BCR | BCR | Adjusted HRa (95% CI) | P value |
|---|---|---|---|---|
| N (%) | N (%) | |||
| Overall | ||||
| First and second tertile | 344 (86.00) | 50 (13.85) | Reference | |
| Third tertile (highest) | 116 (80.00) | 29 (20.00) | 2.17 (1.36–3.48) | 0.001 |
| Site 1 | ||||
| First and second tertile | 318 (85.03) | 56 (14.97) | Reference | N/A |
| Third tertile (highest) | 109 (82.58) | 23 (17.42) | 1.67 (1.01–2.75) | 0.044 |
| Site 2 | ||||
| First and second tertile | 305 (86.65) | 47 (13.35) | Reference | N/A |
| Third tertile (highest) | 122 (79.22) | 32 (20.78) | 2.21 (1.38–3.52) | 0.0009 |
| Site 3 | ||||
| First and second tertile | 294 (84.24) | 55 (15.76) | Reference | N/A |
| Third tertile (highest) | 133 (84.71) | 24 (15.29) | 1.44 (0.88–2.37) | 0.150 |
| Site 4 | ||||
| First and second tertile | 304 (84.92) | 54 (15.08) | Reference | N/A |
| Third tertile (highest) | 123 (83.11) | 25 (16.89) | 1.50 (.92–2.42) | 0.103 |
aAdjusted for age, body mass index, smoking status, pack-year, D’Amico risk groups and primary treatment.
Bold values indicate P < 0.05.
Figure 1.
Kaplan–Meier curve comparing the probability of the BCR-free survival in patients receiving active treatments between higher D4Z4 methylation (third tertile of D4Z4 methylation) and D4Z4 methylation groups (first and second tertile of D4Z4 methylation). Log-rank P = 0.0098.
Discussion
In this study, we evaluated the association of methylation of subtelomeric repeat D4Z4 and pericentromeric repeat NBL2 in peripheral blood leukocytes with the clinical features of PCa patients at diagnosis and prognosis. We found that higher methylation of D4Z4 is associated with an increased risk of BCR in localized PCa patients receiving definitive therapies. The methylation of D4Z4 and NBL2 was not associated with clinical features at diagnosis. This is the first time to report an association between methylation of subtelomeric repeat D4Z4 and prognosis of PCa patients.
The association between cancer risk and global DNA methylation has been investigated in both tumor tissues and peripheral blood. LINE-1 was the most commonly studied global methylation marker and has been previously shown to be hypomethylated in cancer tissues (8,23). In a systematic review of global DNA methylation in PCa compared with non-tumor prostate tissue, DNA methylation was found associated with PCa development and progression (24). Zhu et al. (25) found that LINE-1 hypomethylation was associated with lung cancer prevalence, but not with prostate, colorectal or other cancers. A prior large nested case–control study used pyrosequencing to quantify DNA methylation levels at LINE-1 and Alu repetitive elements in pre-diagnostic blood samples from 694 PCa cases and 703 controls from the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial, and did not observe a significant association with PCa for overall LINE-1 or Alu methylation levels (14). A recent nested control study used the Illumina Infinium HM450K BeadChip to estimate genome-wide DNA methylation in leukocytes and did not find significant association between global DNA methylation and the risk of PCa or aggressive PCa (26). The current literature indicates that overall global methylation level in leukocytes may not be associated with the risk of PCa overall or aggressive PCa.
In contrast to previous studies that predominantly used LINE-1 or Alu repetitive sequences for global methylation analysis, we used relatively region-specific repetitive sequences, i.e. subtelomeric repeat D4Z4 and pericentromeric repeat NBL2. Previous studies have shown that the methylation level of tandem DNA repeats in these specific chromosome regions are different from those of LINE-1 and Alu sequences (17). A previous study even reported inter-locus and intra-locus heterogeneity in specific LINE-1 methylation sites among different cancers (27). There have been ample evidence supporting that these different repetitive sequences play distinct physiological and pathological roles through various mechanisms including methylation regulation and responses to cellular stress (28,29). It is therefore not surprising that we only found a significant association between subtelomeric repeat D4Z4 methylation and PCa prognosis. Subtelomeric regions are adjacent to telomere. Telomeres are protective structure at the chromosome ends that prevent the degradation of chromosome and unnecessary DNA repair activities and recombination (30). Subtelomeric regions are considered part of heterochromatic regions (31). The methylation changes of subtelomeric regions have been related to the development or human diseases (32). For example, subtelomeric DNA methylation at the end of Chr. 7q, 8q, 18p, 21q and XpYp was higher in glioma patients compared to control group (33). Similarly, hypermethylation of subtelomeric region of Chr.18p and 21q was found in hepatocarcinomas (HCCs) compared to their adjacent non-HCCs (34). The subtelomeric DNA methylation was also associated with telomere length in HCC (34). These observations were consistent with our finding that higher methylation of subtelomeric region D4Z4 was associated with worse prognosis of PCa patients, likely due to higher chromosome instability and weak DNA repair activity of patients with higher subtelomeric methylation.
The major strength of this study is the relatively large patient population who were clinically managed at a single institution with comprehensive clinical and follow-up data. We measured the methylation of two distinct repetitive sequences. The pyrosequencing technique we applied is robust and reproducible. There are a couple of limitations. We could only evaluate the association with BCR, not mortality, due to the rare death events of localized PCa patients that limited our statistical power. Therefore, it is unknown whether global methylations of these repetitive sequences are associated with PCa-specific mortality. We only measured one time methylation at diagnosis, not longitudinal changes of methylation level. A recent pilot study suggested that longitudinal changes of global methylation were associated with PCa incidence and overall cancer mortality (15). It would be valuable to investigate whether longitudinal changes of global methylation are associated with recurrence and prostate-specific mortality. Finally, we measured the overall methylation in mixed blood cells. Blood cell heterogeneity has been found to affect the DNA methylation measurements at specific locus (35,36). Previous studies have indicated that global DNA methylation is less likely to be affected by blood cell population as measured by pyrosequencing (37,38), but no study has specifically compared NBL2 and D4Z4 methylation level among different blood cell types. We cannot rule out that the differences in blood cell types may confound our observed association with BCR. Future study is warranted to analyze specific blood cell types.
In summary, our study showed that the methylation of leukocyte subtelomeric repeats D4Z4 is a biomarker of BCR in localized PCa patients receiving definitive therapies. Future studies are warranted to validate our results in independent patient cohorts and evaluate the association of longitudinal changes of global DNA methylation in leukocytes with the prognosis of PCa.
Supplementary material
Supplementary material is available at Carcinogenesis online.
Funding
This study was financially supported by an individual researcher award (RP140556) from the Cancer Prevention and Research Institute of Texas (CPRIT) and a National Cancer Institute Specialized Program of Research Excellence (SPORE) grant (CA140388).
Supplementary Material
Acknowledgements
We thank the DNA Methylation Analysis Core of MD Anderson Cancer Center for performing the pyrosequening and Dr. Marcos Estecio for helpful discussion.
Conflict of Interest Statement: None declared.
Abbreviations
- BCR
biochemical recurrence
- GS
Gleason Score
- HCC
hepatocarcinoma
- LINE-1
long-interspersed nucleotide elements
- NBL2
methylation level of pericentromeric repeat
- PCa
prostate cancer
- PSA
prostate-specific antigen
References
- 1. Siegel R.L., et al. (2017) Cancer statistics, 2017. CA. Cancer J. Clin., 67, 7–30. [DOI] [PubMed] [Google Scholar]
- 2. Schröder F.H., et al. ; ERSPC Investigators. (2009) Screening and prostate-cancer mortality in a randomized European study. N. Engl. J. Med., 360, 1320–1328. [DOI] [PubMed] [Google Scholar]
- 3. Kulis M., et al. (2010) DNA methylation and cancer. Adv. Genet., 70, 27–56. [DOI] [PubMed] [Google Scholar]
- 4. Lee W.H., et al. (1994) Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc. Natl. Acad. Sci. USA, 91, 11733–11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jerónimo C., et al. (2004) A quantitative promoter methylation profile of prostate cancer. Clin. Cancer Res., 10, 8472–8478. [DOI] [PubMed] [Google Scholar]
- 6. Eden A., et al. (2003) Chromosomal instability and tumors promoted by DNA hypomethylation. Science, 300, 455. [DOI] [PubMed] [Google Scholar]
- 7. Breivik J., et al. (1999) Genomic instability, DNA methylation, and natural selection in colorectal carcinogenesis. Semin. Cancer Biol., 9, 245–254. [DOI] [PubMed] [Google Scholar]
- 8. Ardeljan D., et al. (2017) The human long interspersed element-1 retrotransposon: an emerging biomarker of neoplasia. Clin. Chem., 63, 816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lou Y.T., et al. (2014) LINE-1 methylation status correlates significantly to post-therapeutic recurrence in stage III colon cancer patients receiving FOLFOX-4 adjuvant chemotherapy. PLoS One, 10, e0123973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harada K., et al. (2015) LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann. Surg. Oncol., 22, 1280–1287. [DOI] [PubMed] [Google Scholar]
- 11. Li J., et al. (2014) The prognostic value of global DNA hypomethylation in cancer: a meta-analysis. PLoS One, 9, e106290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S.Y., et al. (2014) Alu and LINE-1 hypomethylation is associated with HER2 enriched subtype of breast cancer. PLoS One, 9, e100429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ikeda K., et al. (2013) Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann. Thorac. Surg., 96, 1790–1794. [DOI] [PubMed] [Google Scholar]
- 14. Barry K.H., et al. (2015) Prospective study of DNA methylation at LINE-1 and Alu in peripheral blood and the risk of prostate cancer. Prostate, 75, 1718–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joyce B.T., et al. (2016) Prospective changes in global DNA methylation and cancer incidence and mortality. Br. J. Cancer, 115, 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang A.S., et al. (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res., 32, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi S.H., et al. (2009) Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int. J. Cancer, 125, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deng W., et al. (2012) Pericentromeric regions are refractory to prompt repair after replication stress-induced breakage in HPV16 E6E7-expressing epithelial cells. PLoS One, 7, e48576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsuda H., et al. (2002) Correlation of DNA hypomethylation at pericentromeric heterochromatin regions of chromosomes 16 and 1 with histological features and chromosomal abnormalities of human breast carcinomas. Am. J. Pathol., 161, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richards R.I. (2001) Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet., 17, 339–345. [DOI] [PubMed] [Google Scholar]
- 21. Durkin S.G., et al. (2007) Chromosome fragile sites. Annu. Rev. Genet., 41, 169–192. [DOI] [PubMed] [Google Scholar]
- 22. Colella S., et al. (2003) Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques, 35, 146–150. [DOI] [PubMed] [Google Scholar]
- 23. Barchitta M., et al. (2014) LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PLoS One, 9, e109478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zelic R., et al. (2015) Global DNA hypomethylation in prostate cancer development and progression: a systematic review. Prostate Cancer Prostatic Dis., 18, 1–12. [DOI] [PubMed] [Google Scholar]
- 25. Zhu Z.Z., et al. (2011) Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: the Normative Aging Study. Cancer Causes Control, 22, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. FitzGerald L.M., et al. (2017) Genome-wide measures of peripheral blood DNA methylation and prostate cancer risk in a prospective nested case-control study. Prostate, 77, 471–478. [DOI] [PubMed] [Google Scholar]
- 27. Nüsgen N., et al. (2015) Inter-locus as well as intra-locus heterogeneity in LINE-1 promoter methylation in common human cancers suggests selective demethylation pressure at specific CpGs. Clin. Epigenet., 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li T.H., et al. (2001) Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene, 276, 135–141. [DOI] [PubMed] [Google Scholar]
- 29. Biémont C., et al. (2006) Genetics: junk DNA as an evolutionary force. Nature, 443, 521–524. [DOI] [PubMed] [Google Scholar]
- 30. Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 31. Riethman H., et al. (2005) Human subtelomere structure and variation. Chromosome Res., 13, 505–515. [DOI] [PubMed] [Google Scholar]
- 32. Wang T., et al. (2013) Subtelomeric hotspots of aberrant 5-hydroxymethylcytosine-mediated epigenetic modifications during reprogramming to pluripotency. Nat. Cell Biol., 15, 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choudhury S.R., et al. (2015) Selective increase in subtelomeric DNA methylation: an epigenetic biomarker for malignant glioma. Clin. Epigenet., 7, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oh B.K., et al. (2011) Frequent changes in subtelomeric DNA methylation patterns and its relevance to telomere regulation during human hepatocarcinogenesis. Int. J. Cancer, 128, 857–868. [DOI] [PubMed] [Google Scholar]
- 35. Reinius L.E., et al. (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One, 7, e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adalsteinsson B.T., et al. (2012) Heterogeneity in white blood cells has potential to confound DNA methylation measurements. PLoS One, 7, e46705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu H.C., et al. (2011) Global methylation profiles in DNA from different blood cell types. Epigenetics, 6, 76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bjornsson H.T., et al. (2008) Intra-individual change over time in DNA methylation with familial clustering. JAMA, 299, 2877–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.