This study reports that single-cell molecular analysis overcomes problems associated with the heterogeneity of bulk tumors for monitoring tumor subclonal evolution and molecular pathways relevant to GBM relapse.
Abstract
Glioblastoma multiforme (GBM) remains an incurable brain tumor. The highly malignant behavior of GBM may, in part, be attributed to its intraclonal genetic and phenotypic diversity (subclonal evolution). Identifying the molecular pathways driving GBM relapse may provide novel, actionable targets for personalized diagnosis, characterization of prognosis and improvement of precision therapy. We screened single-cell transcriptomes, namely RNA-seq data of primary and relapsed GBM tumors from a patient, to define the molecular profile of relapse. Characterization of hundreds of individual tumor cells identified three mutated genes within single cells, involved in the RAS/GEF GTP-dependent signaling pathway. The identified molecular pathway was further verified by meta-analysis of RNA-seq data from more than 3000 patients. This study showed that single-cell molecular analysis overcomes the inherent heterogeneity of bulk tumors with respect to defining tumor subclonal evolution relevant to GBM relapse.
Introduction
Glioblastoma multiforme (GBM) is a devastating malignant tumor of the brain with a dismal prognosis and high cellular heterogeneity. Historically, gliomas (including astrocytoma, oligodendroglioma and glioblastoma) were classified by histologic criteria (1). The modern classification of gliomas is based on the World Health Organization (WHO) Classification of Central Nervous System Tumors, initially published in 1979, which has since been revised four times (2). The current WHO classification of gliomas, released in 2016, is based not only on histopathologic appearance, but also on well-established molecular parameters. This reflects the clinical significance of understanding the molecular signature of GBM. After surgical excision of GBM, adjuvant radiation and chemotherapy are usually given. Despite the survival benefits associated with this type of treatment, the majority of patients relapse and disease continues to progress, ultimately resulting in the demise of the patient. Currently, when relapse occurs in a patient with GBM, both local and systemic therapies are treatment options. Localized therapies include debulking reoperations and irradiation. Carmustine polymer wafers can be placed during the reoperation for locally recurrent disease. Due to a lack of prospective data, the efficacy of reirradiation in recurrent GBM is uncertain. Stereotactic radiosurgery and brachytherapy have also been tried but there is no firm data to suggest they provide a significant benefit at this time. Systemic therapy for recurrent GBM may include bevacizumab, nitrosoureas and temozolomide rechallenge. No single agent has been shown to be clearly superior to another. Recent trials comparing combination therapy to single-agent therapy showed no difference in overall survival (3,4). Alternating electric fields are now also available for treatment of recurrent GBM after their recent approval by the FDA. Therefore, detailed genetic information enabling improved characterization and classification of GBM would open the door to individualized and targeted therapies.
Current protocols for the molecular characterization of GBM are inherently limited by the confounding variables associated with the characterization of ‘bulk’ clinical tumor specimens. These patient tumor biopsies not only contain diverse subpopulations of tumor cells, but also non-malignant cells such as normal brain tissues, inflammatory cell populations, vasculature, etc (5). Such shortcomings are now addressed by single-cell molecular analysis. A recently reported list of mutated genes identified in primary GBM with single-cell transcriptome analysis suggests that conventional analysis of bulk GBM biopsy specimens do not necessarily distinguish tumor subtype populations present within these gross samples (6). Single-cell transcriptome data offer a more nuanced perspective on the heterogeneity within GBM. In this study, we apply single-cell characterization technology to gain insight into tumor heterogeneity. With single-cell molecular analysis of individual patient’s tumor subpopulations, identification and subsequent targeting of the most clinically relevant subsets may be achieved. Herein, with the identification of three mutations related to RAS/GEF GTP-dependent signaling, prediction and therapeutic targeting of subtypes most likely to contribute to tumor relapse appears feasible and realistic.
Materials and methods
Cohort and tumor diagnosis
The patient is an adult male who complained of progressive right-sided weakness as well as a decrease in mentation. Serial computed tomographic (CT) imaging showed persistent edema in the left parietofrontal region, with a left parietal intracerebral hemorrhage. Over 4 weeks, he had decreased mentation and speech. CT scan of the brain without contrast, 2 weeks after presentation, showed extensive edema that appeared as a hypodense area. The hypodensity had increased in size in the left region as confirmed with magnetic resonance imaging. A stereotactic craniotomy was performed and the left-side ventricle occipital horn tumor was debulked. In the tumor biopsy, EGFRviii expression was not detected; MGMT gene promoter methylation was detected; IDH1/IDH2 mutation was not detected. After surgical excision of the tumor, adjuvant radiation and chemotherapy were given, including temozolomide. Upon neurological assessment for possible relapse six months post-resection, magnetic resonance imaging was used to confirm progression of disease. A debulking re-operation was performed followed by re-irradiation and systemic therapy (bevacizumab, nitrosoureas and temozolomide re-challenge). Biopsies from both debulking operations were obtained with IRB approval for molecular analysis.
Organotypic slice culture sample preparation
We first isolated brain tumor stem cells from patient tumor biopsies with IRB approval as described previously (7) and then enriched for tumor stem cells by engineered organotypic slice culture (8). Briefly, brain slices ranging from 200 to 300μm in thickness were generated with a Leica VT1000 S Vibrating blade microtome (Leica Biosystems Inc., Buffalo Grove, IL) and washed three times in HBSS to remove any tissue debris and any potentially toxic substances (e.g. excitatory amino acids). The slices were then placed on culture plate inserts (0.4μm Millicell-CM, Millipore) in sterile-filtered slice culture medium. Slice culture medium was prepared by mixing Minimal Essential Medium (Invitrogen, Carlsbad, CA) containing 25% heat-inactivated horse serum (Invitrogen), 25mM HEPES, 25% HBSS, 6.4mg/ml glucose, 0.5mM glutamine, 10ng/ml insulin-like growth factor (IGF) and 1× penicillin–streptomycin–glutamine (Invitrogen). One millitre slice culture medium was added to each organotypic slice culture before incubation at 37°C and 5% CO2. Single cells were then isolated by using Human Indirect CD133 MicroBead kit (Mitenyi Biotec GmbH Germany, Auburn, CA) and RNA was extracted to perform single-cell RNA-seq on the cultured cells (enriched for tumor stem cells). Genomic DNA and mRNA were isolated from slides with 80% or more tumor cells using TRIzol® reagent (Life technologies).
Library construction and sequencing
For each sample, 1 μg gDNA and 5 μg RNA were prepared and submitted for sequencing. The DNA and RNA quality was evaluated and libraries were constructed with a library construction kit (Illumina). Libraries were sequenced on the Illumina HiSeq 2000 platform (Illumina). The raw reads generated were filtered according to sequencing quality and with regard to adaptor contamination and duplicated reads. Thus, only high-quality reads remained and were used in the genome assembly. Both RNA-seq and Exome-seq data were analyzed with Partek Flow version 4 (Partek Inc.). Bases with a Phred score less than 20 were trimmed from both ends of the raw sequencing reads, and trimmed reads shorter than 25 nt were excluded from downstream analyses. Both pre- and post-alignment QA/QC were carried out with default settings as part of the Flow workflow.
Gene expression analysis
For RNA-seq samples, trimmed reads were mapped onto human genome hg38 using Tophat 2.0.8 as implemented in Flow with default settings, and using Gencode 20 annotation as guidance. Gencode 20 annotation (www.gencodegenes.org) was used to quantify aligned reads to genes/transcripts using Partek E/M method (9). Read counts per gene in all samples were normalized using Upper Quartile normalization and analyzed for differential expression using Partek’s Gene Specific Analysis method (genes with less than 10 reads in any sample were excluded). To generate a list of significantly differentially expressed genes among different tissues, a cutoff of FDR adjusts P < 0.05 (Poisson regression) and fold change >|2| was applied.
Mutation profiling
For Exome-seq samples, trimmed reads were mapped onto human genome hg38 using BWA-MEM 0.7.9a (10) as implemented in Flow with default settings. The aligned reads of each sample were then used to call variants among the samples using Samtools (11) with default settings as implemented in Flow. Identified variants were visually inspected using the Integrative Genomics Viewer. We utilized BioBase, which compares the variation sites against publicly available HGMD® (professional, 2015), COSMIC v71, GWAS (February 17 2015) EVS for known exon variants (cESP6500) ClinVar (2015-02), the pharmaco Genomic Mutation Database 2015.1(beta), allele frequency from 1000 Genomes (dbSNP141) and dbNSFP for non-synonymous functional prediction (v2.9), to identify putative oncogenes and variants that have been singled out as cancer risk genes. Before constructing the analysis set, we removed dubious short reading frames and obviously unrelated genes resulting from the filtering parameters.
Ethics statement
Fresh tissues from primary and relapsed tumors were obtained during surgery with IRB approval and patient consent as described previously (7). Surgical specimen isolation and handling was done in compliance with appropriate standards of clinical care.
Results
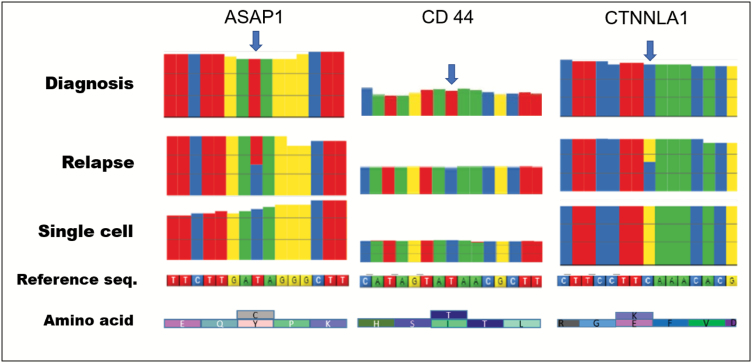
Because relapse tumors develop from a treatment-resistant primary tumor, mutations that are responsible for the relapse are enriched and dominate the relapse tumor, but may exist as a minority in the primary tumor. Using a microfluidic platform for single-cell transcriptome analysis (12,13), we obtained high-quality cDNA and performed deep single-cell RNA-seq (20 million reads/cell) to identify potential relapse-related mutations within single cells. Among the single cells we profiled, the majority carried multiple mutations which could be detected in the relapse biopsy but not in the original diagnostic biopsy (Figure 1). Among the detected mutations, there were three independent genes, all involved in RAS/GEF GTP-dependent signaling regulation, a pathway known to be involved in GBM malignancy and glioma cell migration (14).
Figure 1.
Single-cell transcriptome analysis reveals interactive mutations in relapse. RNA-seq detected protein-coding mutations in diagnosis bulk lysate, relapse bulk lysate and tumor stem cell line single-cells, respectively. The three identified missense mutations (indicated by blue arrows), CTNNAL1 (1579 C>G), ASAP1 (2182T>C) and CD 44 (1436T>C), are interactive mutations which are responsible for relapse in this patient. These mutations were all detected in the relapse tissue and single tumor stem cells cultured form the relapse tissue, but were not detectable in the primary tumor. The bulk lysate represents a mixture of cells, so both wild type and mutations are detected in relapse tissue for ASAP1 and CTNNLA1. Because these mutations occur within the same cell, they can interact with each other and lead to relapse. Corresponding amino acid changes are indicted under the reference sequence.
Specifically, through analyzing single-nucleotide polymorphisms (SNP), we identified 3 homozygous point mutations responsible for missense substitutions in three individual proteins that were detected in the single-cell and relapse biopsies, but not in the primary tumor. These mutations were at position E574Q in the vinculin domain of catenin α-like 1 protein (CTNNAL1); at position I594V in a structural region of ArfGAP with SH3 Domain of Ankyrin Repeat and PH Domain 1(ASAP1), also called GTPase-activating protein; and at position I230T in the structural region of CD44.
Gene expression analysis of RAS/GEF GTP pathway
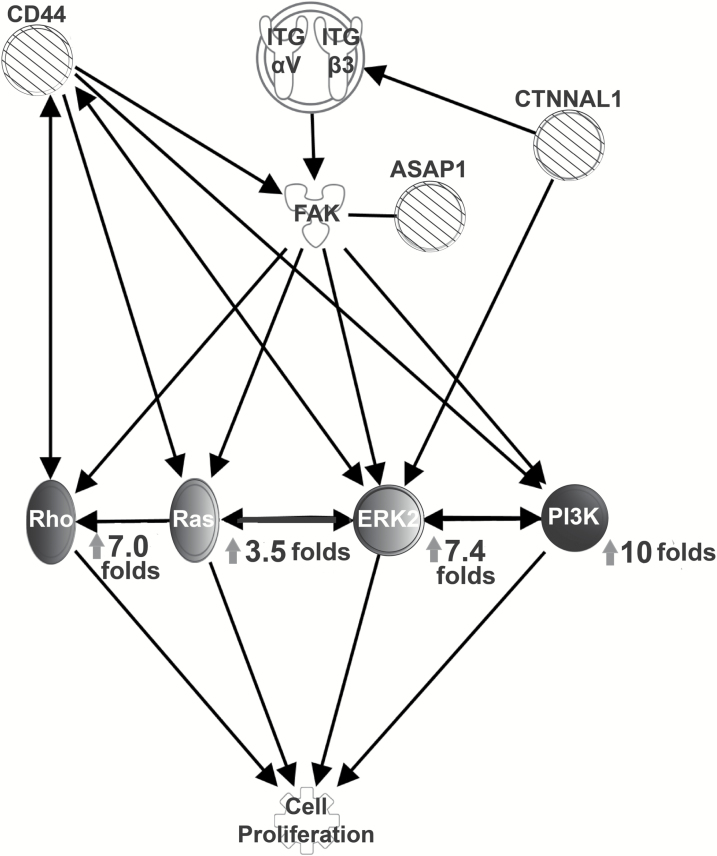
Besides the mutation analysis, we also analyzed the gene expression levels of the key downstream players (Rho, Ras, ERK2 and PI3K) in this RAS/GEF GTP pathway (Figure 2). Analysis of gene expression comparing the initial diagnostic biopsy with the recurrence sample also revealed a 64-fold increase in human estrogen receptor 2 (HER2) expression in the relapse sample, and there was also a 64-fold increase in S100A8, a calcium-binding protein that plays a prominent role in facilitating leukocyte arachidonic acid metabolism and traffic. It regulates inflammatory and immune processes by binding to TLR4 and AGER to activate the MAP-kinase and NF-κB pathways, initiating the amplification of pro-inflammatory cascades. The abnormal expression patterns of these members of the RAS/GEF GTP pathway further confirm its involvement in the relapse.
Figure 2.
Pathway analysis infers molecular pathway of relapse. Pathway analysis with QIAGEN Ingenuity Pathway Analysis software indicates that the three identified mutations, CTNNAL1 (1579 C>G), ASAP1 (2182T>C) and CD 44 (1436T>C), are members of the Rho/Gef signal pathway for cell proliferation, which leads to tumor relapse. Expression levels of related proliferation genes are presented with fold changes (relapse/primary). Striped circles: mutated genes; Grayscale circles: downstream members of the Rho/Gef signaling pathway with various degrees of upregulation (darker colour indicates higher expression).
Clinical evidence of GBM relapse pathway
We further analyzed the role of the RAS/GEF GTP pathway by mining clinical data. Meta-analysis of NIH Genomic Data Commons confirmed that the genes with mutations we identified in the single-cell analysis are all significantly over expressed in GBM (Table 1). Liang et al. (15) showed the genes ASAP1 and CD44 were overexpressed by 1.777 and 4.142 folds, respectively, in patients with glioblastoma. Bredel et al. (16) indicated that CTNNAL1 expression increased by 1.432 folds in glioblastoma patients compared to normal controls, with a P-value of 0.000908; the glioblastoma CD44 gene was found to be overexpressed by a factor of 7.243 (p= 1.17 × 10−15) in their study as well.
Table 1.
Clinical evidence of RAS/GEF GTP pathway associated with glioblastoma (GB)
| Gene | Cancer type | Fold change (GB/Normal) |
P-value | Gene rank | Data set (number of patients) | Ref |
|---|---|---|---|---|---|---|
| CTNNAL1 | Glioblastoma versus normal | 2.177 | 1.51E−11 | 79 | TCGA Brain (557) | TCGA |
| Glioblastoma versus normal | 2.984 | 1.42E−16 | 266 | Sun Brain (180) | (17) | |
| Glioblastoma versus normal | 1.432 | 9.08E−04 | 1648 | Bredel Brain 2 (54) | (16) | |
| ASAP1 | Glioblastoma versus normal | 1.181 | 0.003 | 1321 | Freije Brain (85) | (30) |
| Glioblastoma versus normal | 1.777 | 0.036 | 293 | Liang Brain (38) | (15) | |
| Secondary glioblastoma versus normal | 1.157 | 0.002 | 1186 | DNA Beroukhim Brain (187) | (31) | |
| Glioblastoma versus normal | 1.034 | 1.79E−10 | 1605 | DNA TCGA Brain 2 (1,531) | TCGA | |
| Glioblastoma versus normal | 1.478 | 3.45E−06 | 2068 | Sun Brain (180) | (17) | |
| Glioblastoma versus normal | 4.916 | 5.15E−26 | 10 | TCGA Brain (557) | TCGA | |
| CD44 | Glioblastoma versus normal | 7.243 | 1.17E−15 | 21 | Bredel Brain 2 (54) | (16) |
| Glioblastoma versus normal | 4.142 | 3.36E−09 | 35 | Liang Brain (38) | (15) | |
| Glioblastoma versus normal | 6.456 | 3.30E−23 | 77 | Sun Brain (180) | (17) | |
| Glioblastoma versus normal | 3.592 | 3.46E−08 | 81 | Shai Brain (42) | (32) | |
| Glioblastoma versus normal | 2.591 | 7.09E−18 | 86 | Murat Brain (84) | (33) | |
| Glioblastoma versus normal | 3.14 | 1.47E−04 | 907 | Lee Brain (101) | (34) |
Although the fold changes of these genes (CTNNAL1, ASAP1 and CD44) do not represent dramatic changes to expression levels in GBM relative to normal tissues, their levels of expression are consistently and significantly (e.g. with very low P-values) different between GBM and normal tissues, as demonstrated by analysis of multiple GBM clinical databases with a combined total of 2951 patients. In the Sun Brain data set (italicized in Table 1) with 180 patients, all three genes are up-regulated significantly relative to normal tissues (17). Compared to normal tissues, CTNNAL1, ASAP1 and CD44 are overexpressed by 2.984, 1.478 and 6.456 folds with P-values of 1.42 × 10−16, 3.45 × 10−16 and 3.30 × 10−23, respectively. In another database, TCGA Brain (italicized in Table 1), which has data from 557 patients, CTNNAL1 and ASAP1 both are upregulated with P-values of 1.51E−11 and 5.15E−26, respectively. Considering that these data are from bulk lysates with cancer and normal cells mixed together, such low P-values suggest that these three genes are interacting with each other to form a molecular pathway in GBM.
Discussion
The clinical challenges in treating GBM are mainly due to a changing genetic landscape arising from intratumoral spatial and temporal heterogeneity and are further compounded by genetic drift as the tumor microenvironment coevolves with the tumor. Recent clinical trials in GBM include molecular profiling and targeted treatments such as topoisomerase I inhibitor, vascular endothelial growth factor (VEGF) inhibitor, tyrosine kinase inhibitor and immune checkpoint inhibitor. Aberrant expression-based molecular classification of EGFR, NF1 and PDGFRA/IDH1 define the classical, mesenchymal and proneural subtypes of GBM, respectively (18). Characterizing H3F3A mutations, affecting two critical amino acids (K27 and G34) of histone H3.3, further differentiate GBMs into six epigenetic subgroups with distinct global methylation patterns and predictable anatomical predispositions (19). These subtypes are clinically relevant for predicting prognosis and assisting in treatment planning for GBM patients. However, subtype-specific targeting strategies have not yet improved clinical outcomes within these subtypes. Tumor heterogeneity is a major hurdle that must be overcome in order to profile the molecular signature of GBM. Relapse tumors are generated from single cells carrying particular mutations, known as tumor stem cells, which are rare in the initial (diagnosis) tumor. These tumor stem cells may be detectable, and indeed prevalent, in the relapse biopsy, despite being too rare to be detected in the initial diagnostic biopsy. Here, we found that a relapse tumor of a GBM patient carried three missense mutations, specifically in CTNNAL1 (1579 C>G), ASAP1 (2182T>C) and CD 44 (1436T>C) (indicated by blue arrows in Figure 1). All three genes are downstream members of the Rho/Gef signal pathway, and may interact in a way that is responsible for the relapse.
This putative relapse pathway is known to be involved in GBM malignancy and glioma cell migration. In particular, guanine-nucleotide exchange factors (GEFs) encourage active Ras-GTP formation through their stimulation of Ras-mediated GDP/GTP exchange activity (10). Activated Ras-GTP exerts control over diverse signaling networks important for the regulation of cell proliferation, survival, differentiation, vesicular trafficking and gene expression (Figure 2). A key component in this relapse pathway, the membrane receptor Cluster of Differentiation 44 (CD44), is a well-recognized stem cell biomarker expressed in many tumor cells (20). In patients with laryngeal and pharyngolaryngeal cancer, CD44 was shown in a meta-analysis to be associated with advanced T categories (larynx: RR = 1.33, 95% CI 1.01–1.76; larynx and pharynx RR = 1.21, 95% CI 1.08–1.35), worse N categories (larynx: RR = 2.53, 95% CI 1.99–3.21; larynx and pharynx RR = 1.95, 95% CI 1.35–2.82), higher tumor grades (larynx and pharynx RR = 1.71, 95% CI 1.04–2.79), and worse 5-year OS rates (larynx: RR = 0.62, 95% CI 0.47–0.83; larynx and pharynx RR = 0.66, 95% CI 0.47–0.94) (21). Working with CD 44 and ASAP1, CTNNAL1 regulates the molecular pathway of relapse by serving as a scaffold for the Rho-specific guanine nucleotide exchange factor lymphoid blast crisis in lymphoid blast crisis-induced serum response factor activation (22). Clinically, high expression levels of CTNNAL1 and ILK are associated with poor overall survival in patients with non-small cell lung cancer (23), most likely due to their interaction with the NF-κB component, NF-κB kinase (IKK)-beta (24), which inhibits Ras-mediated signals to the cyclin D1 promoter (25). As CTNNAL1 contributes to the invasive behavior of metastatic cells, it may serve as a prognostic marker and future therapeutic target for cancer patients (26).
Our data suggest a novel drug cocktail of multiple inhibitors for the identified target genes. This is of particular interest for the highly lethal brain cancer glioblastoma, which shows intratumoral heterogeneity of signaling networks. Indeed, single-cell phosphoproteomics revealed that alterations in mTOR kinase pathway result in targeted cancer therapy resistance in patient-derived in vivo GBM models (27). Such spatiotemporal alterations can be resolved as early as 2.5 days after treatment with combination therapies, resulting in complete and sustained tumor suppression in vivo (27). Tracking single cell-derived clonal evolution has proven to be essential for treatment of human glioblastoma (28). With the advancement of technology, single-cell transcriptome analysis has become a powerful tool to investigate molecular pathways. Tumor biopsies are heterogeneous tissues (normal tissues mixed with tumor cells). Bulk lysates are the physical averages of such heterogeneous biopsies. Therefore, traditional assays of bulk lysates only detect the most obvious correlations among genes. The subtle correlations among carcinogenesis genes are masked by normal cells in bulk lysates due to the effect of averaging. Single-cell transcriptome analysis reveals the gene expression correlation within a cell, in which proteins are directly interacting with each other, to provide a clear relationship among carcinogenesis genes.
It should be noted that single-cell study does not mean only studying one cell. One must be careful to confirm that the cells investigated are indeed tumor cells, and hundreds of individual cells must be studied to ensure the correlations are not experimental artifacts. In our case, we screened more than 200 individual cells. Finally, the inferred pathways must be confirmed with clinical data. We confirmed the pathway we identified with a meta-analysis of thousands of patients. It is also not surprising that some mutated genes found in our study were not discovered in the single-cell analyses of other patients (6). Individual patients are expected to carry different mutations, which underscores the heterogeneity of GBM. In fact, our finding is consistent with a recent study of combining 14 226 single-cell RNA sequencing (scRNA-seq) profiles from 16 patient samples with bulk RNA-seq profiles from 165 patient samples (29). They found that isocitrate dehydrogenase (IDH)-mutant astrocytoma and oligodendroglioma share molecular signatures showing the enhanced proliferation of malignant cells, larger pools of undifferentiated glioma cells and an increase in macrophage over microglia expression programs evolved with the tumor microenvironment over time. The strength of single-cell transcriptome analysis lies in the spatial-temporal identification of such molecular signatures at the single-cell level, which in turn helps to identify the dynamic changes in various types of tumor-associated cells, thereby delineating the lineage-specific genetic changes in a tumor and the influence of the tumor microenvironment. However, single-cell transcriptome analysis is not without limitations. Single-cell RNA-seq of a limited number of representative tumors must be combined with bulk data from large cohorts to decipher differences between tumor subclasses for clinical applications. Our single-cell study provides evidence for a novel relapse pathway, suggesting that finding these mutations could contribute predictive power to potential strategies to improve efficacy by enabling clinicians to deliver the right drugs to the right patient at the right moment.
Conflict of Interest Statement: None declared.
Acknowledgements
We thank Dr. Zhi-Wei Ma for constructive suggestions and editing the manuscript. This work was supported by grants R01CA197903 and R01CA164509 from the National Institutes of Health, USA (JFZ), and CHE1213161 from the National Science Foundation, USA (JFZ). Grants of cstc2016shms-ztzx10003 (XZ) and NSFC81270569(XZ) from National Natural Science Foundation of China, China.
Abbreviation
- GBM
glioblastoma multiforme
References
- 1. MacKenzie D.J. (1926)A classification of the tumours of the glioma group on a histogenetic basis with a correlated study of prognosis. Can. Med. Assoc. J., 16, 872–872. [Google Scholar]
- 2. Louis D.N., et al. (2016)The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol., 131, 803–820. [DOI] [PubMed] [Google Scholar]
- 3. Wick W., et al. (2017)Lomustine and bevacizumab in progressive glioblastoma. N. Engl. J. Med., 377, 1954–1963. [DOI] [PubMed] [Google Scholar]
- 4. Brada M., et al. (2010)Temozolomide versus procarbazine, lomustine, and vincristine in recurrent high-grade glioma. J. Clin. Oncol., 28, 4601–4608. [DOI] [PubMed] [Google Scholar]
- 5. Li S.C., et al. (2014)Cancer genomic research at the crossroads: realizing the changing genetic landscape as intratumoral spatial and temporal heterogeneity becomes a confounding factor. Cancer Cell Int., 14, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee J.K., et al. (2017)Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet., 49, 594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li S.C., et al. (2012)Cancer stem cells from a rare form of glioblastoma multiforme involving the neurogenic ventricular wall. Cancer Cell Int., 12, 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li S.C., et al. (2011)Increase developmental plasticity of human keratinocytes with gene suppression. Proc. Natl. Acad. Sci. USA, 108, 12793–12798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harrow J., et al. (2012)GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res., 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andrade W.A., et al. (2010)Early endosome localization and activity of RasGEF1b, a toll-like receptor-inducible Ras guanine-nucleotide exchange factor. Genes Immun., 11, 447–457. [DOI] [PubMed] [Google Scholar]
- 11. Li H. (2011)A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan J.B., et al. (2012)Highly parallel genome-wide expression analysis of single mammalian cells. PLoS One, 7, e30794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhong J.F., et al. (2008)A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip, 8, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee H.K., et al. (2015)RasGRP3 regulates the migration of glioma cells via interaction with Arp3. Oncotarget, 6, 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang Y., et al. (2005)Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc. Natl. Acad. Sci. USA, 102, 5814–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bredel M., et al. (2005)Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res., 65, 8679–8689. [DOI] [PubMed] [Google Scholar]
- 17. Sun L., et al. (2006)Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell, 9, 287–300. [DOI] [PubMed] [Google Scholar]
- 18. Verhaak R.G., et al. ; Cancer Genome Atlas Research Network (2010)Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sturm D., et al. (2012)Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell, 22, 425–437. [DOI] [PubMed] [Google Scholar]
- 20. Jaggupilli A., et al. (2012)Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin. Dev. Immunol., 2012, 708036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J., et al. (2014)Significance of CD44 expression in head and neck cancer: a systemic review and meta-analysis. BMC Cancer, 14, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park B., et al. (2002)Association of Lbc Rho guanine nucleotide exchange factor with alpha-catenin-related protein, alpha-catulin/CTNNAL1, supports serum response factor activation. J. Biol. Chem., 277, 45361–45370. [DOI] [PubMed] [Google Scholar]
- 23. Liang C.H., et al. (2013)α-Catulin drives metastasis by activating ILK and driving an αvβ3 integrin signaling axis. Cancer Res., 73, 428–438. [DOI] [PubMed] [Google Scholar]
- 24. Wiesner C., et al. (2008)Alpha-catulin, a Rho signalling component, can regulate NF-kappaB through binding to IKK-beta, and confers resistance to apoptosis. Oncogene, 27, 2159–2169. [DOI] [PubMed] [Google Scholar]
- 25. Merdek K.D., et al. (2004)Distinct activities of the alpha-catenin family, alpha-catulin and alpha-catenin, on beta-catenin-mediated signaling. Mol. Cell. Biol., 24, 2410–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cao C., et al. (2012)α-Catulin marks the invasion front of squamous cell carcinoma and is important for tumor cell metastasis. Mol. Cancer Res., 10, 892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wei W., et al. (2016)Single-cell phosphoproteomics resolves adaptive signaling dynamics and informs targeted combination therapy in glioblastoma. Cancer Cell, 29, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meyer M., et al. (2015)Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. USA, 112, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Venteicher A.S., et al. (2017)Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science, 355. doi:10.1126/science.aai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freije W.A., et al. (2004)Gene expression profiling of gliomas strongly predicts survival. Cancer Res., 64, 6503–6510. [DOI] [PubMed] [Google Scholar]
- 31. Beroukhim R., et al. (2007)Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. USA, 104, 20007–20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shai R., et al. (2003)Gene expression profiling identifies molecular subtypes of gliomas. Oncogene, 22, 4918–4923. [DOI] [PubMed] [Google Scholar]
- 33. Murat A., et al. (2008)Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol., 26, 3015–3024. [DOI] [PubMed] [Google Scholar]
- 34. Lee J.K., et al. (2007)A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc. Natl. Acad. Sci. USA, 104, 13086–13091. [DOI] [PMC free article] [PubMed] [Google Scholar]