We conducted a genome-wide gene-smoking interaction analysis in non-small cell lung cancer using genotype from 35737 individuals including both discovery and validation datasets. We identified three novel SNPs with significant interactions with tobacco smoking.
Abstract
Non-small cell lung cancer is the most common type of lung cancer. Both environmental and genetic risk factors contribute to lung carcinogenesis. We conducted a genome-wide interaction analysis between single nucleotide polymorphisms (SNPs) and smoking status (never- versus ever-smokers) in a European-descent population. We adopted a two-step analysis strategy in the discovery stage: we first conducted a case-only interaction analysis to assess the relationship between SNPs and smoking behavior using 13336 non-small cell lung cancer cases. Candidate SNPs with P-value <0.001 were further analyzed using a standard case–control interaction analysis including 13970 controls. The significant SNPs with P-value <3.5 × 10−5 (correcting for multiple tests) from the case–control analysis in the discovery stage were further validated using an independent replication dataset comprising 5377 controls and 3054 non-small cell lung cancer cases. We further stratified the analysis by histological subtypes. Two novel SNPs, rs6441286 and rs17723637, were identified for overall lung cancer risk. The interaction odds ratio and meta-analysis P-value for these two SNPs were 1.24 with 6.96 × 10−7 and 1.37 with 3.49 × 10−7, respectively. In addition, interaction of smoking with rs4751674 was identified in squamous cell lung carcinoma with an odds ratio of 0.58 and P-value of 8.12 × 10−7. This study is by far the largest genome-wide SNP-smoking interaction analysis reported for lung cancer. The three identified novel SNPs provide potential candidate biomarkers for lung cancer risk screening and intervention. The results from our study reinforce that gene-smoking interactions play important roles in the etiology of lung cancer and account for part of the missing heritability of this disease.
Introduction
Lung cancer is one of the most common cancers worldwide and the leading cause of cancer-related death in both men and women in the United States (1). Non-small cell lung cancer (NSCLC) contributes to ~80–85% of lung cancer cases (2). NSCLC has three major subtypes: adenocarcinoma, squamous cell carcinoma and large cell carcinoma. About 40% of NSCLC are adenocarcinoma, whereas squamous cell carcinoma (SQC) represents ~25–30% of NSCLC and is strongly related to a history of having ever smoked (3–5).
Genome-wide association studies have been successful in identifying common variants associated with lung cancer in the past decade. The identified susceptibility genes include the CHRNA5, CHRNA3 and CHRNB4 genes at 15q25, TERT at 5p15, the HLA region at 6p21, TP63 at 3q28 and several additional variants (6–13). Most of the identified common variants have a relatively small genetic effect [odds ratio (OR) <1.5] and together account for a fraction of the heritability of lung cancer. Gene–environment interactions are believed to explain part of the missing heritability (14). Tobacco smoking is the major risk factor associated with lung cancer risk and ~80–90% of European-descent lung cancer cases have a history of exposure to cigarette smoke (15). Interactions between genes and smoking behavior play important roles in the development of lung cancer (16–18). An interaction effect manifests itself when the disease risk associated with a genotype varies by smoking behavior. In 2014, Zhang et al. (16) detected two single nucleotide polymorphisms (SNPs), rs1316298 and rs4589502 (OR: 0.71, P-value of 6.73 × 10−6 and OR: 1.55, P-value of 3.84 × 10−6, respectively), in a genome-wide gene-smoking interaction scanning using genotype data from 3865 cases and 4566 controls from a Han Chinese population. Studies of gene-smoking interactions are important in deciphering the lung cancer etiology because they will reveal those genes involved in lung tumorigenesis that interacting with tobacco smoking that would not be discovered by main effect association analysis without jointly modeling with smoking status. The identified genetic variants with heterogeneous effects between subgroups defined by smoking behavior will contribute to lung cancer risk prediction and disease prevention.
However, genome-wide interaction scanning remains a challenge. Most genome-wide association studies were designed for main effect association analysis and have limited power for interaction analysis. Analyses of power show that a sample size at least a four-fold larger is required for interaction analysis if a standard case–control design is used and the power limitations are more extreme when the effect size is modest or the risk allele has a lower frequency (19). In the absence of gene–environment correlation, a case-only approach has been shown to be much more powerful than a standard case–control design (20,21). If the gene–environment independence assumption is not met, then false positives can be introduced when a case-only design is followed. A two-step test strategy was proposed by researchers for gene–environment interaction analysis: step 1, comprises a case-only test to test the association between SNPs and environmental risk factor; step 2, candidate SNPs from step 1 were further submitted to standard case–control logistic interaction analysis (21). There are two advantages using this two-step study design: first, the step 1 test allows us to filter the SNPs tested in step 2 thus reducing the power loss from multiple comparisons in the step 2 test; and second, standard case–control interaction analysis in step 2 is more stringent and is robust to the gene–environment requirement of the case-only design, thus reducing the false discovery rate that may otherwise plague the case-only design.
The current reports on genome-wide gene-smoking interaction analysis in lung cancer are still quite limited (16). To explore gene-smoking interactions in NSCLC lung cancer development in a European-decent population, we conducted a genome-wide interaction analysis based on ~500000 SNPs genotype data from ~27000 individuals of European descent. We tested the interactions between each SNP and the smoking status (never-smokers versus ever-smokers). The interaction analyses were further categorized by lung cancer histology subtypes including adenocarcinoma and SQC. The candidate SNPs were further validated using independent genotype data from another sample of ~8400 individuals. As far as we know, this is the largest genome-wide SNP-smoking interaction analysis in lung cancer study up to date.
Materials and methods
Study populations
The discovery genotype data in this study came from OncoArray consortium, which was designed to identify genetic variants associated with common cancers including breast, colon, lung, prostate and ovarian cancers (22). We restricted the analysis to individuals with European ancestry and valid information on smoking status and lung cancer histology (23). The smoking status was denoted as never- versus ever-smokers, and ever-smokers included current and former smokers based on self-reported information about smoking status when the samples were recruited. The large sample size (n > 25000) in the discovery phase derives from samples that were collected from 28 individual institutes. To minimize the potential for false-positive findings, we randomly grouped the data into three balanced datasets S1–S3 (Supplementary Table S1, available at Carcinogenesis Online). The three subsets serve as internal replication datasets for the associations and help to reduce the potential for spurious association findings. The sample size from the 28 sites varies from 146 to 3195. We ‘randomly’ distributed the sites to three groups following two criteria: (i) there are sites with sample size >1000 and sites with sample size <1000 in each group and (ii) the sample size of each group are balanced (within range of average ±500). There are 9480, 9059 and 8767 individuals in S1–S3, which sum to 13970 controls and 13336 patients with NSCLC lung cancer (Table 1). The NSCLC lung cancer cases include 7015 adenocarcinoma patients and 4529 SQC patients. All the samples were genotyped using the Illumina OncoArray-500K BeadChip (22). The independent replication data include 5377 controls and 3054 NSCLC cases genotyped on a separate Affymetrix array (24). The smoking statuses in the replication data were recorded following the same classification as in the discovery data. The percentage of never-smokers in the control samples are 32.14 and 29.85% in discovery and replication data; and 10.49 and 11.43% in the disease samples in the discovery and replication data, respectively (Table 1).
Table 1.
The number of never- and ever-smokers in controls and lung cancer subtypes.
| Data | Controls | Never1 | Ever | Ade2 | Never | Ever | Sqc3 | Never | Ever | NSCLC4 | Never | Ever | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery | Subset 1 | 4463 | 1472 | 2991 | 2732 | 351 | 2381 | 1690 | 28 | 1662 | 5017 | 416 | 4601 |
| Subset 2 | 4490 | 1377 | 3113 | 2446 | 426 | 2020 | 1422 | 87 | 1335 | 4569 | 583 | 3986 | |
| Subset 3 | 5017 | 1641 | 3376 | 1837 | 320 | 1517 | 1417 | 44 | 1373 | 3750 | 400 | 3350 | |
| Combined | 13970 | 4490 | 9480 | 7015 | 1097 | 5918 | 4529 | 159 | 4370 | 13336 | 1399 | 11937 | |
| Replication | 5377 | 1605 | 3772 | 1759 | 275 | 1484 | 952 | 38 | 914 | 3054 | 349 | 2705 |
1, Never and ever denote smoking status and ever-smokers include current smokers and ex-smokers; 2, adenocarcinoma; 3, squamous cell carcinoma; 4, non-small cell lung cancer, including ade, sqc, large cell lung cancer. IBD analysis was performed to remove the duplicated between discovery and replication data.
Ethics statement
All subjects provided informed consent, and the institutional review boards of each participating institutes approved this collaborative study.
Genotype data quality control
In the discovery stage, we started with genotypes from 43959 samples on 517820 SNPs. We inferred ancestry information using the FastPop program and individuals with probability of European ancestry >0.8 were inferred as having European-descent population (25). IBD analysis and sex checking were conducted as quality control checks to identify close relatives or possible sample processing issues. Individuals and SNPs with genotype call rate <0.95 were excluded from the analysis. IBD analysis was further performed among samples between discovery and replication datasets, and duplicate samples included in discovery study were removed. A total of 27306 individuals including 13970 controls and 13336 patients with NSCLC lung cancer were included in the discovery study. FlashPCA was used for Principal Component Analysis (PCA) and we adjusted for the first three principal components in the interaction analysis (26). A total of 502933 SNPs were analyzed in the interaction analysis (23).
In the replication study, a total of 12651 individuals were genotyped using Affymetrix Array platform on 404740 SNPs (24). IBD analysis was conducted to remove duplicate samples or close relatives within the dataset. Individuals with genotype call rate <0.95 were excluded from the analysis. The Structure program was run to infer ancestry origin and 0.8 was used as the cutoff for European-descent population inference (27). A total of 8431 samples were included in replication study including 5377 controls and 3054 patients with NSCLC lung cancer. EIGENSTRAT was run for PCA analysis and we adjusted for the first three principal components in the analysis (28).
Statistical analysis
We conducted a genome-wide interaction analysis comprising a discovery stage in which candidate SNPs were identified, and then these SNPs were validated in a subsequent replication study using an independent set of cases and controls (Supplementary Figure S1, available at Carcinogenesis Online). A two-step analysis strategy was adopted in discovery stage: step 1, a genome-wide case-only logistic regression analysis was performed to assess the association between each SNP and smoking status using formula (1) (E denotes smoking status) using all the discovery data; SNPs with case-only P-value <0.001 were further submitted to step 2 analysis with a standard case–control logistic model as denoted in formula (2) (D denotes disease status).
| (1) |
| (2) |
The Bonferroni corrected cut-off P-value in the step 2 case–control analysis was set to 0.05 divided by the number of SNPs entering the step 2 analysis. For example, if 500 SNPs had case-only P-value <0.001 then the cutoff P-value in case–control analysis was 0.05/500=1 × 10−4. The significant candidate SNPs following the step 2 test were chosen for further study based on two additional criteria: (i) the SNPs have case–control interaction P-value <0.1 from each of three subsets in discovery data; and (ii) case–control interaction P-values less than the Bonferroni corrected P-value from the combined discovery data. The candidate SNPs were further submitted for verification in replication study. In the interaction analysis, SNPs were coded in an additive model (0, 1 or 2). There were three categories of reported smoking status, never-smoker, current smoker and ex-smoker, in the phenotype data. And ex-smoker was defined as time since last smoking more than 2 years. We grouped the samples into never-smokers (0) and ever-smokers (1, including both current smokers and ex-smokers). The first three principal components were adjusted in the interaction analysis.
The interaction analysis was further stratified by histology subtypes including adenocarcinoma and SQC. For those SNPs validated in replication study (case–control interaction P-value <0.05), we also performed a meta-analysis to combine the information from both discovery and replication data.
Genotype imputation
To increase the density of SNP markers at regions surrounding the significant SNPs verified in replication study, we used IMPUTE2 to impute the flanking SNPs in ~250 kb of the three validated SNPs rs6441286, rs17723637, and rs4751674 in the discovery data. Because of the limited overlap in SNP panels between discovery and replication data, we also conducted imputation to increase the SNP density and overlap in the replication data. The 1000 Genomes Project Phase 3 release was used as the reference dataset (29). The output dosage file from IMPUTE2 was used as input in logistic regression analysis and the first three PCs were adjusted in the imputed genotype analysis.
Results
Discovery study
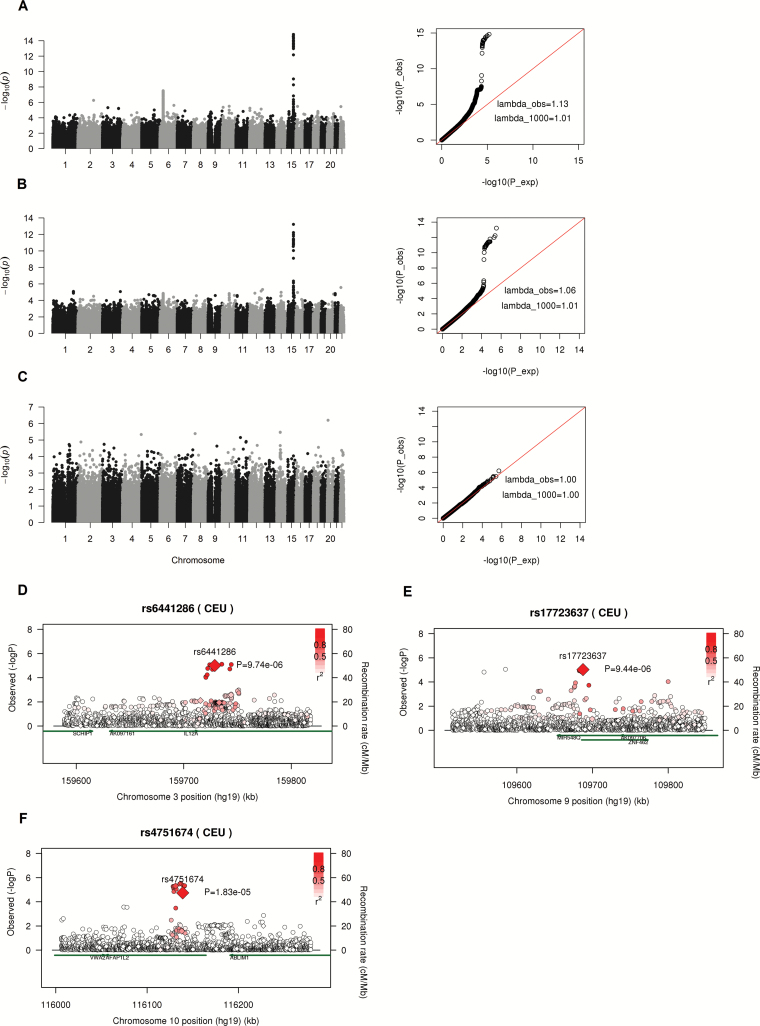
In discovery study, we first performed the genome-wide interaction analysis to test the association between SNPs and smoking behavior using only the lung cancer patients; the samples with P-value <0.001 were submitted to interaction analysis to test the association between SNP-smoking interaction and lung cancer risk using S1–S3 subset as well as the combined data in the discovery stage. Figure 1A–C displays the Manhattan plot of –log10(p) from the case-only studies including 13336 NSCLC cases, 7015 adenocarcinoma cases and 4529 SQC cases, respectively. The Q–Q plots displayed the observed P-values versus expected P-values, and the observed genomic inflation factor (λ, lambda) were 1.13, 1.06 and 1.00 for NSCLC, adenocarcinoma and SQC, respectively. Since the lambda value scales with sample size, we also computed the inflation factor for an equivalent study of 1000 cases (30). The scaled lambda values were 1.01, 1.01 and 1.00 for interaction analysis in NSCLC, adenocarcinoma and SQC, respectively (Figure 1A–C). No obvious inflation of type I error rate was detected in the study. In the association analysis between smoking behavior and SNPs using only cases, 1379, 867 and 468 SNPs, including the SNPs at the well-known chr15q24.3—chr15q25.1 region (the CHRNA5, CHRNA3, CHRNB4, IREB2, PSMA4 gene cluster) with P-value <0.001 were detected in NSCLC, adenocarcinoma and SQC case-only interaction analysis, respectively (Supplementary Table S2, available at Carcinogenesis Online). And these SNPs entered the step 2 test in discovery stage to test the associations between gene-smoking interactions and lung cancer disease using all the cases and controls data.
Figure 1.
(A–C) Manhattan plot (left) and Q–Q plot (right) of P-values from case-only genome-wide interaction analysis (step 1) in NSCLC, adenocarcinoma and SQC patients using discovery data. lambda_obs and lambda_1000 indicate genomic inflation factor from observed data and from an equivalent study of 1000 cases, respectively. λ_1000=1+(λ_obs-1)×(1/n_cases)/(1/1000). (D–F) Regional association plot around three significant SNPs validated in replication study using imputed SNPs from Oncoarray data. Diamonds and circles denote genotyped and imputed SNPs. Since the genotype data for imputation analysis went through additional QC procedures so the final sample size was a little smaller than that in genotype analysis. There were 12624 controls and 12979 NSCLC cases in imputed data analysis. So the signals at the genotyped SNPs (indicated by diamond) were a little bit different from that at genome-wide interaction analysis in discovery stage as shown in Table 2.
In step 2 test, the Bonferroni corrected P-values were computed by dividing 0.05 by the number of SNPs entered the analysis. And we got 3.63 × 10−5, 5.77 × 10−5 and 1.07 × 10−4 for NSCLC, adenocarcinoma and SQC subgroup studies, respectively (Supplementary Table S2, available at Carcinogenesis Online). For consistency, we used 3.5 × 10−5 as the cutoff in step 2 case–control interaction analysis across all the three studies by histology. The significant SNPs from step 2 test in discovery stage were chosen based on two criteria: (i) the association between disease status and gene-smoking interaction has a P-value <0.1 from each of the S1–S3 subset; and (ii) has a P-value < 3.5 × 10−5 from the combined data analysis. For example, 438, 766 and 925 SNPs had a case–control interaction P-value <0.1 from subset 1–3 in NSCLC cohort and 105 of them were common to all the three subsets. Among the 105 SNPs, 52 had a case–control interaction P-value <3.5 × 10−5 using the combined data (Supplementary Table S2, available at Carcinogenesis Online). In adenocarcinoma and SQC lung cancer cohort, 41 and 10 SNPs were selected as significant markers for further replication analysis (Supplementary Tables S2 and S3, available at Carcinogenesis Online); 33 and 26 SNPs at chr15q24.3—chr15q25.1 region had significant interaction P-values in discovery stage from NSCLC and adenocarcinoma interaction analysis, respectively. The CHRNA5 region had been extensively studied in several independent studies (7,12,18). However, these P-values were much less significant compared with that from main-effect-only association analysis, which means the association effect between disease status and SNPs were much more significant when only genetic main effect was considered in the model compared with the association effect between disease status and gene-smoking interactions (Supplementary Table S3, available at Carcinogenesis Online). These SNPs on chromosome 15q were not novel SNPs and the interaction effect between smoking and SNPs was not as striking as that found in main effect analysis.
Replication study and meta-analysis
The replication data came from a separate study so the genotype panel was different from that of discovery data. Some of the selected candidate SNPs from discovery study were not available in validation data but we still validated the signals at three novel SNPs using genotypes from replication data. In the cohort including all NSCLC cases, SNP rs6441286 on chromosome 3q25.33 had a P-value of 6.30 × 10−6 in gene and smoking behavior association analysis (step 1) and P-value of 1.16 × 10−5 in gene-smoking and disease status association analysis (step 2) using combined discovery data. The step 2 interaction P-values were 3.39 × 10−3, 1.67 × 10−2 and 4.79 × 10−3 for S1–S3 subset. The replication data produced an interaction P-value of 2.02 × 10−2. The interaction OR varied from 1.21 to 1.31 across the different subsets and the overall OR was 1.24. This SNP has a P-value of 6.96 × 10−7 in the meta-analysis to combine both the discovery and replication data. rs6441286 was located at the intron of IL12A-AS1 gene, which is an antisense RNA regulating IL12 gene, a key regulator in immune response.
Another validated SNP in NSCLC cohort was SNP rs17723637 located in ZNF462 gene. It had a case-only interaction P-value of 4.92 × 10−4 in the gene and smoking behavior association analysis, a gene-smoking and disease status association P-value of 1.06 × 10−5 in combined discovery data and P-value of 9.76 × 10−3 in validation analysis. The interaction ORs were 1.40 [95% confidence interval (CI): 1.09, 1.79], 1.32 (95% CI: 1.06, 1.64), 1.45 (95% CI: 1.12, 1.86) and 1.43 (95% CI: 1.09, 1.88) for S1–S3 and replication data, respectively. The overall interaction OR was 1.37 with P-value of 3.49 × 10−7 in the meta-analysis.
In the genome-wide gene-smoking interaction analysis stratified by different tumor subtype, SNP rs4751674 had a case-only interaction P-value of 3.69 × 10−5 and the case–control interaction P-value of 1.07 × 10−5 in discovery stage and 2.62 × 10−2 in replication stage in SQC lung cancer group (Table 2). The interaction ORs were 0.59, 0.54, 0.68 and 0.58 for S1–S3 and replication data, respectively. The overall OR was 0.58 from the meta-analysis with a P-value of 8.12 × 10−7. We also identified another neighbor rs2244178 with a gene and smoking behavior association P-value of 2.23 × 10−4 and gene-smoking interaction and squamous cell lung cancer disease status association P-value of 3.14 × 10−5 from combined discovery data analysis. The OR of lung cancer risk associated with rs2244178 was 0.58 (95% CI: 0.45, 0.75). Unfortunately, rs2244178 was not available at the replication data and we could not verify it. Both rs2244178 and rs4751674 were located at gene AFAP1L2 (XB130), which is an adaptor that regulated signal transduction in lung.
Table 2.
Replicated signals from genome-wide joint analysis of main effect and interaction effect.
| MAF | Gene | Smoking status | Interaction | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytoband | Gene | P_casec | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | ||
| rs6441286 (Ca) | 3q25.33 | IL12A-AS1 | nsclcb | |||||||
| S1 | 0.404 | 0.76 (0.64, 0.90) | 1.34E-03 | 5.23 (4.37, 6.26) | 9.01E-73 | 1.31 (1.09, 1.57) | 3.39E-03 | |||
| S2 | 0.406 | 0.88 (0.77, 1.02) | 8.42E-02 | 2.58 (2.19,3.05) | 3.09E-29 | 1.21 (1.04, 1.41) | 1.67E-02 | |||
| S3 | 0.412 | 0.82 (0.70, 0.96) | 1.22E-02 | 3.47 (2.90, 4.16) | 6.02E-42 | 1.28 (1.08, 1.53) | 4.79E-03 | |||
| S1–S3 combined | 0.407 | 6.30 × 10−6 | 0.83 (0.76, 0.91) | 3.92E-05 | 3.51 (3.18, 3.88) | 2.68E-134 | 1.24 (1.13, 1.37) | 1.16E-05 | ||
| Replicate | 0.399 | 0.81 (0.68, 0.96) | 1.58E-02 | 2.77 (2.29, 3.34) | 3.25E-26 | 1.25 (1.03, 1.50) | 2.02E-02 | |||
| Meta | 0.83 | 1.92E-06 | 1.24 | 6.96E-07 | ||||||
| rs17723637 (G) | 9q31.2 | ZNF462 | nsclc | |||||||
| S1 | 0.149 | 0.75 (0.60, 0.94) | 1.38E-02 | 5.77 (5.02, 6.63) | 3.92E-134 | 1.40 (1.09, 1.79) | 7.95E-03 | |||
| S2 | 0.146 | 0.78 (0.64, 0.95) | 1.20E-02 | 2.79 (2.46, 3.16) | 5.81E-59 | 1.32 (1.06, 1.64) | 1.33E-02 | |||
| S3 | 0.158 | 0.71 (0.56, 0.90) | 4.17E-03 | 3.83 (3.33, 4.39) | 1.69E-81 | 1.45 (1.12, 1.86) | 4.14E-03 | |||
| S1–S3 combined | 0.151 | 4.92 × 10−4 | 0.75 (0.66, 0.85) | 6.31E-06 | 3.81 (3.53, 4.11) | 4.17E-262 | 1.36 (1.19, 1.56) | 1.06E-05 | ||
| Replicate | 0.143 | 0.73 (0.56, 0.94) | 1.40E-02 | 3.01 (2.61, 3.47) | 2.11E-51 | 1.43 (1.09, 1.88) | 9.76E-03 | |||
| Meta | 0.74 | 2.79E-07 | 1.37 | 3.49E-07 | ||||||
| rs4751674 (A) | 10q25.3 | AFAP1L2 | sqc | |||||||
| S1 | 0.271 | 1.68 (0.96, 2.93) | 6.72E-02 | 44.19 (25.12, 77.76) | 1.88E-39 | 0.59 (0.33, 1.03) | 6.34E-02 | |||
| S2 | 0.275 | 1.70 (1.24, 2.33) | 1.06E-03 | 9.90 (7.05, 13.90) | 5.13E-40 | 0.54 (0.39, 0.75) | 2.70E-04 | |||
| S3 | 0.265 | 1.57 (1.01, 2.44) | 4.60E-02 | 21.11 (13.61, 32.73) | 2.96E-42 | 0.68 (0.43, 1.06) | 9.10E-02 | |||
| S1–S3 combined | 0.270 | 3.69 × 10−5 | 1.68 (1.33, 2.12) | 1.21E-05 | 18.55 (14.58, 23.59) | 2.43E-125 | 0.58 (0.46, 0.74) | 1.07E-05 | ||
| Replicate | 0.269 | 1.80 (1.12, 2.87) | 1.44E-02 | 14.76 (8.97, 24.28) | 2.91E-26 | 0.58 (0.36, 0.94) | 2.62E-02 | |||
| Meta | 1.70 | 5.52E-07 | 0.58 | 8.12E-07 | ||||||
S1–S3 indicate independent subsets in discovery study. Meta-analysis was performed based on the results from combined discovery data analysis and replicate data analysis. First three principal components were adjusted in both discovery and replication study. In step 1 test at discovery stage, SNPs with case-only P-value <0.001 using all the patients in discovery data were selected. Bonferroni P-values were calculated by diving 0.05 by the number of selected SNPs from step 1 tests. For consistency, we used 3.5 × 10-5 as the cutoff P-value for all the subtype studies in step 2 test. In step 2 test at discovery stage, SNPs with case–control interaction P-value <0.1 from each subset S1–S3 and less than 3.5 × 10−5 from combined discovery data were submitted to validation analysis in replication stage. a, the allele in the brackets indicates the minor allele; b, abbreviation indicates disease histology subgroup, NSCLC for non-small cell lung carcinoma and SQC for squamous cell carcinoma. c, P_case indicates case-only P-value from association test between SNPs and smoking behavior using lung cancer patients (step 1 test).
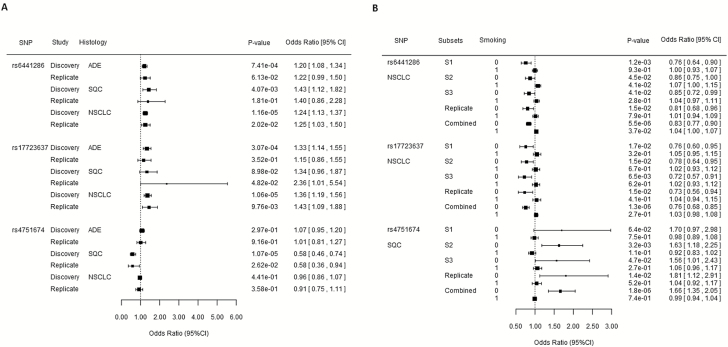
We further checked the interaction effect of the three SNPs at different lung cancer subtypes. Both SNP rs6441286 and rs17723637 had a marginal interaction effect in adenocarcinoma and squamous cell lung cancer. These interaction effect only achieved genome-wide significance in NSCLC analysis when both adenocarcinoma and SQC patients were included in the analysis to get a larger sample size (Figure 2A). The interaction effect between SNP rs4751674 and smoking behavior was only detected in squamous cell lung cancer subtype. This effect was not existing in adenocarcinoma subtype although there were 3293 more samples in adenocarcinoma than in squamous cell group.
Figure 2.
(A) Forest plot of interaction effect of three identified SNPs stratified by histology. (B) Forest plot of risk effect of SNPs in never-smokers and ever-smokers. 0 and 1 standing for never-smokers and ever-smokers. S1–S3 denote subsets 1–3 in discovery stage. ADE, cases are from patients with adenocarcinoma lung cancer; SQC, cases are from patients with squamous cell carcinoma; NSCLC, cases are from patients with non-small cell lung cancer.
Imputation analysis
To further verify the replicated interactions between SNPs and smoking behavior, we imputed ~250kb flanking regions around each of the three significant SNPs using the genotypes from the discovery data to increase the density of markers in the regions harboring the three target SNPs. We plotted the signals from 1000 up- and down-stream SNP markers of the target SNPs. Because the genotype data for imputation analysis went through additional QC procedures, the final sample size was a little smaller than that used in genotype analysis. There were 12624 controls and 12979 NSCLC cases in imputed data analysis. In the imputation analysis, we found another eight imputed SNPs with interaction P-values <3.5 × 10−5 around rs6441286 and the most significant SNP was rs66785795 with interaction P-value of 7.63 × 10−6 (Supplementary Table S4, available at Carcinogenesis Online and Figure 1D). All the most significant SNPs were within IL12A-AS1 gene. For SNP rs17723637, we found another 6 SNPs with interaction P-values <3.5 × 10−5 and all the most significant SNPs were within gene ZNF462 which encodes a zinc finger protein (Supplementary Table S4, available at Carcinogenesis Online and Figure 1E). For SNP rs4751674 from interaction analysis in squamous cell lung cancer, we detected 14 more SNPs with interaction P-values <3.5 × 10−5 harboring both rs2244178 and rs4751674. All of these SNPs were located within the gene AFAP1L2 and the most significant P-value came from imputed SNP rs2483911 with a P-value of 2.87 × 10−6 (Supplementary Table S4, available at Carcinogenesis Online and Figure 1F). The results from imputed genotype analysis strongly supported the identified SNPs in our gene-smoking interactions analysis.
Because the SNP panels between discovery and replication data were different and only limited number of SNPs were available in both panels, we also imputed the replication genotype data using 1000 Genome as the reference. We identified another three SNPs with case–control interaction P-value <0.05 in the imputed replication data. They are rs10477550 on chromosome 5, rs4557740 on chromosome 8 and rs11544453 on chromosome 22 (Supplementary Table S3, available at Carcinogenesis Online). rs10477550 was located within COMMD10 gene, and it has interaction P-values of 9.05 × 10−2, 7.69 × 10−4 and 4.30 × 10−2 with adenocarcinoma disease in the three subsets S1–S3 in discovery data and 4.33 × 10−3 in the imputed replication data. rs11544453 was within WNT7B gene and it has interaction P-values of 5.61 × 10−4, 7.07 × 10−2 and 2.17 × 10−3 with squamous cell lung cancer in three subsets in discovery data and 4.05 × 10−2 in imputed replication data (Supplementary Table S3, available at Carcinogenesis Online). The results from imputed replication data suggest the potential interaction effect with smoking behavior at these two genes but direct genotype data would be more reliable for further validation.
Risk effect of lung cancer at significant SNPs stratified by smoking status
For the replicated SNPs with significant interactions with smoking status, we further investigated the risk effect of the SNPs in smoking and never smoking groups separately. There were 5899 never-smokers in the NSCLC cohort and 1399 individuals were NSCLC patients in the never-smokers (Table 1). The minor allele at SNP rs6441286 had a protective effect on NSCLC in never-smokers and the overall OR was 0.83 (95% CI: 0.77, 0.90) when we combined both the discovery and replication dataset (Figure 2B). However, this protective effect did not exist in samples from only smokers (OR: 1.04; 95% CI: 1.00, 1.07) in the study combining both discovery and replication data. Similarly, SNP rs17723637 had a protective effect on NSCLC in non-smokers with the overall OR was 0.76 (95% CI: 0.68, 0.85) in never smoking group. No significant effect was identified in the smoking group.
SNP rs4751674 had a negative interaction with smoking behavior in SQC cohort. Among the 4649 never-smokers, only 159 of them were SQC lung cancer patients. SNP rs4751674 had a squamous cell lung cancer risk effect with the overall OR of 1.66 (95% CI: 1.35, 2.05) in non-smokers when we combined both discovery and replication data (Figure 2B). This risk effect for lung cancer did not exist in the smoking group (OR: 0.99; 95% CI: 0.94, 1.04). SNP rs4751674 had a risk allele A, and there was no significant difference between the allele frequencies in never-smokers versus ever-smokers in controls (OR: 1, P=0.98) (Supplementary Table S5, available at Carcinogenesis Online). In patients with squamous cell lung cancer, 63.52% of never-smokers have at least one risk allele, compared with 45.73% in ever-smoker patients (OR: 2.07, P-value=1.44 × 10−5). The neighbor SNP rs2244178 had a risk effect with OR of 1.60 (95% CI: 1.25, 2.04) in never-smoker group and no significant effect in ever-smoker group.
Joint analysis of SNP and smoking behavior in lung cancer
To better understand the interaction effect between the SNP and smoking status, we conducted a joint analysis with never-smoker without risk allele as the reference group (Table 3). For ever-smokers without risk allele group, the NSCLC risk at SNP rs6441286 was 3.51 (95% CI: 3.18, 3.88); for never-smokers with risk allele group, the risk was 0.79 (95% CI: 0.70, 0.90) and for ever-smokers with risk allele group, the disease risk was 3.66 (95% CI: 3.30, 4.06). The interaction effect between the risk genotype and smoking behavior was 1.33 (95% CI: 1.16, 1.52). A similar pattern was found at SNP rs17723637. For people carrying at least one risk allele, the OR of lung cancer was 0.77 (95% CI: 0.67, 0.88) in never-smokers and 3.94 (95% CI: 3.61, 4.30) in ever-smokers. The interaction between smoking and the SNP was 1.38 (95% CI: 1.18, 1.60). The joint analysis at these two SNPs displayed that for a person who was a carrier of the risk allele the lung cancer risk varied dramatically depending on the smoking behavior of the person. Cigarette smoking had a synergetic effect on the risk genotype and abstinence from smoking among those risk allele carrier population would significantly decrease their risk for NSCLC lung cancer.
Table 3.
Joint analysis of SNP and smoking behavior in lung cancer using combined discovery data.
| Never-smokers versus ever-smokers | ||||
|---|---|---|---|---|
| Risk allele | Smoking | OR (95% CI) | P-value | |
| rs6441286 | 0 | 0 | Reference | |
| NSCLC | 0 | 1 | 3.51 (3.18, 3.88) | <2.2 × 10−16 |
| 1 | 0 | 0.79 (0.70, 0.90) | 3.08 × 10−4 | |
| 1 | 1 | 3.66 (3.30, 4.06) | <2.2 × 10−16 | |
| Interaction | 1.33 (1.16, 1.52) | 4.83 × 10−5 | ||
| rs17723637 | 0 | 0 | Reference | |
| NSCLC | 0 | 1 | 3.83 (3.54, 4.13) | <2.2 × 10−16 |
| 1 | 0 | 0.77 (0.67, 0.88) | 2.15 × 10−4 | |
| 1 | 1 | 3.94 (3.61, 4.30) | <2.2 × 10−16 | |
| Interaction | 1.38 (1.18, 1.60) | 3.74 × 10−5 | ||
| rs4751674 | 0 | 0 | Reference | |
| SQC | 0 | 1 | 19.67 (15.08, 25.65) | <2.2 × 10−16 |
| 1 | 0 | 1.92 (1.38, 2.67) | 1.01 × 10−4 | |
| 1 | 1 | 19.04 (14.59, 24.85) | <2.2 × 10−16 | |
| Interaction | 0.48 (0.34, 0.67) | 2.11 × 10−5 | ||
Individuals with no risk allele genotype (0) and never-smoker (0) were used as reference group. The genotype with at least one risk allele was coded as 1. The first three PCs were adjusted in the analysis.
For SNP rs4751674, we found that smoking had a very big risk effect for squamous cell lung carcinoma which alone contributed an OR of 19.67 (95% CI: 15.08, 25.65). SNP rs4751674 was located at a potential tumor gene AFAP1L2 and the risk allele contributed an OR of 1.92 (95% CI: 1.38, 2.67). The OR was decreased to 19.04 (95% CI: 14.59, 24.85) when both risk factors occurred which meant that smoking behavior had an antagonistic effect on the risk allele and there was a negative interaction between the SNP and smoking behavior (OR: 0.48; 95% CI: 0.34, 0.67).
SNP rs4751674 is located at gene AFAP1L2 (alias: XB130) on chromosome 10 which encodes an adaptor protein that participates in many cellular functions, including cell proliferation and survival process in various cancers (31). AFAP1L2 is a potential oncogene and the knockdown of AFAP1L2 by RNAi was associated with induced cell death in human lung cancer cells (32). The results from our statistical analysis of SNP rs4751674 suggested that this gene was involved in SQC in non-smokers.
Discussion
Lung cancer has a complicated disease mechanism and both genetic and environmental factors affect the disease development. Tobacco smoking is the most important environmental risk factor associated with lung cancer. In this study, we conducted a genome-wide gene-smoking behavior interaction analysis on NSCLC lung cancer using genotype data from ~36000 samples including both discovery and validation datasets. As far as we know, this is by far the largest genome-wide SNP-smoking interaction analysis in lung cancer. The three subsets at the discovery stage and independent validation data at the replication stage allow us to identify SNPs with consistent effect in interaction analysis across different datasets and to provide solid evidence for interactions between reported SNPs and smoking behavior. The genome-wide association studies are designed for main effect association analysis and have limited power for interaction effect detection. We adopted a two-step test for the interaction analysis in discovery stage. The case-only interaction analysis at step 1 allowed us to filter the SNPs for further case–control interaction analysis at step 2. On the basis of the number of SNPs in standard interaction analysis at step 2, we computed the genome-wide interaction significant level in a conservative way, i.e. 0.05 divided by the number of SNPs tested in step 2 analysis and we chose 3.5 × 10−5 as the significance cutoff for all the analyses in discovery study. And the candidate SNPs were submitted to replication study using independent dataset.
Although the SNPs at the chr15q24.3—chr15q25.1 region passed the significance criteria at gene-smoking association analysis in case-only study and had a significant interaction effect with smoking behavior in disease status association analysis. The interaction effect was much less significant compared with the main effect association analysis when no interaction effect was considered. For example, rs7163730 and rs11638372, both at chr15q25 region, had a P-value of 5.66 × 10−32 and 5.24 × 10−25 in the main effect model, respectively. The interaction P-values were only 8.78 × 10−6 and 1.95 × 10−5 when SNP-smoking interaction was considered in the model (Supplementary Table S3, available at Carcinogenesis Online).
52, 41 and 10 SNPs were identified from discovery study in NSCLC, adenocarcinoma lung cancer and SQC subgroups, respectively. Because of the limited overlap in SNP panel between discovery and replication genotype data, only 35, 26 and 1 of them were available in the replication genotype data, which limited our ability in replication study (Supplementary Table S2, available at Carcinogenesis Online). Three novel SNPs, rs6441286, rs17723637 and rs4751674, were validated in the replication analysis with significant interactions with smoking behavior in lung cancer development. The gene-smoking interaction and lung cancer disease association P-values from meta-analysis are 6.96 × 10−7, 3.49 × 10−7 and 8.12 × 10−7 for these three SNPs. The overall ORs combing both discovery and replication data are 1.24, 1.37 for SNP rs6441286 and rs17723637 in NSCLC, respectively; and 0.58 for SNP rs4751674 in SQC lung cancer. The large sample size in this study allow us to identify two SNP-smoking interactions with moderate effect (OR: 1.24 and 1.37). The minor alleles C at SNP rs64412866 and G at rs17723637 both have protective effect for NSCLC in never-smokers but these protective effects are not existing in smokers. SNP rs64412866 is located at gene IL12A-AS1, which encodes an antisense RNA of IL12A gene. Antisense RNA is widely transcribed in human genome and is an important regulatory mechanism human gene expression (33,34). Studies have shown that cigarette smoking affects non-coding RNA, such as microRNA and antisense RNA, expression in humans (35,36). One study found that some stress-induced non-coding RNAs were up-regulated by exposure to tobacco carcinogen nicotine-derived nitrosamine betone (NNK) in lung cancer and breast cancer cell lines (36). Xi et al. (35) found that the exposure of human respiratory epithelial cells and lung cancer cells to cigarette smoke increased the expression of microRNA miR-31 in both these two types of cells and overexpression of miR-31 was associated with increased lung cancer risk. IL12A encodes the subunit of IL12, which has been shown to be a potent cytokine with antitumor activity in human (37). Our results suggest smoking behavior interact with IL12A-AS1 gene and increase the risk of NSCLC lung cancer in smokers. The other SNP rs17723637 is located at gene ZNF462, which is a member of zinc finger protein transcription factor family in human. The functions of zinc finger proteins in human tumorigenesis vary in different cancers and the report about ZNF462 is still quite limited. Studies showed that ZNF462 could be involved in chronic obstructive pulmonary disease development (38). Our results suggested it had a protective effect for lung cancer in nonsmokers.
SNP rs6441286 and rs17723637 are common variants with minor allele frequency of 0.4 and 0.15, respectively. The lung cancer risk among individuals carrying the risk allele of each of these two SNPs varies drastically by smoking status (OR 0.79 versus 3.66 at SNP 6441286 and 0.77 versus 3.94 at SNP 17723637 between never- and ever-smokers, Table 3). The positive interactions between smoking behavior and these two SNPs illustrated the adverse effect of smoking behavior in NSCLC development again. The results at these two SNPs provided us another evidence that smoking is harmful to our health and quitting smoking will greatly reduce the risk for lung cancer in human.
In the interaction analysis stratified by disease subtype, we identified some significant interaction in adenocarcinoma cohort in discovery study but not validated successfully in replication study. In SQC cohort, we found two SNPs, rs2244178 and rs4751674, with P-values <3 × 10−5 in case–control interaction analysis in discovery study but only rs4751674 were available and successfully validated in replication study. The minor allele A at SNP rs4751674 has a strong interaction effect with smoking status and the OR is 0.58 in SQC lung cancer risk evaluation. The negative interaction effect between gene and smoking behavior, i.e. tobacco smoking decreases the genetic risk for lung cancer disease at a genetic locus, is rare but still existing in lung cancer development. For example, Zhang et al. (16) identified the negative interaction between rs1316298 and smoking behavior. This SNP has an OR of 1.12 (95% CI: 1.01–1.25) in non-smoking group, whereas an OR of 0.79 (95% CI: 0.71–0.87) is found among smokers (16). SNP rs1316298 is located within a potential tumor suppressor gene and close to genes with tumor-related functions as well. The SNP rs1316298 was not available in our genotype data so we could not validate their findings. In our analysis, rs4751674 is located at gene AFAP1L2 (alias: XB130), which is a member of actin filament-associated protein (AFAP) family. AFAP genes are adaptor proteins and have been shown to be related with tumorigenesis in prostate, lung and breast cancers (31,32,39). Study showed that XB130 regulated survival, cell cycle, migration and invasion of cancer by interacting with binging proteins (40). Our results support its oncogene function in never-smokers. Tobacco smoking has a complicated effect in the genome including impact on the signaling pathways, gene expression and induced methylation at many genes (41–44). A study on cadmium, one of the important toxic chemicals in cigarette, showed that cadmium suppressed AFAP1L2 gene expression (45). Tobacco smoking may reduce the AFAP1L2 gene expression, thus reduced its tumorigenesis risk effect in smokers.
SQC of the lung constitutes ~25% to lung cancers and is closely related with smoking history (46). In our squamous carcinoma cohort, only 3.51% and 3.99% of the patients are never-smokers in the discovery data and replication data, respectively, compared with 15.63% in adenocarcinoma cohort (Table 1). The large sample size in the study enabled us to identify the significant interactions in never-smoking SQC patients. However, the sample size in SQC cohort is still limited and there are only 159 and 38 never-smoker patients in the discovery and replication cohort, respectively. We are hoping that more never-smoker patients will be available in the future so these results can be further validated.
The three SNPs, rs6441286, rs17723637 and rs4751674, identified in our study stratify lung cancer risk by smoking behavior. The interaction ORs are 1.24, 1.37 and 0.58 for these three SNPs, respectively. rs6441286 and rs17723637 have increased risk effect for lung cancer in ever-smokers, whereas rs4751674 has a protective effect in ever-smokers compared with never-smokers. These three SNPs can be potential biomarkers used to improve the precision to which we can categorize an individual’s risk of lung cancer disease by smoking behavior. rs6441286 and rs17723637 have interaction effect with smoking behavior in NSCLC development, and rs4751674 only interacts with tobacco smoking in SQC lung cancer. These lung cancer subtype-specific biomarkers will further help us categorize the disease risk by tumor histology which is helpful for individualized prognosis and prediction of treatment plan.
All the three identified novel SNPs have little evidence for association with lung cancer risk in main-effect-only association analysis (P-values vary from 0.29 to 0.94 in main effect analysis), which displays that the gene–environment interaction analysis is an essential approach in exploring the missing heritability of lung cancer disease. There are significant gene-smoking interactions at well-known chr15q24.3—chr15q25.1 region in lung adenocarcinoma. But the interaction P-values are much less significant than that from the main-effect-only association analysis, which suggests the dominant roles of the main effect in lung cancer development (Supplementary Table S3, available at Carcinogenesis Online). The interaction effect at SNP rs4751674 only exists in SQC also suggests the difference of genomic features between SQC and adenocarcinoma in lung cancer from perspective of gene-smoking interaction analysis. This reported study was restricted to Caucasian population and the results may not be generalized to other ethnicities because of the different genetic backgrounds. The limited overlap between discovery genotype and replication genotype may have reduced the power in our validation study. We believe as more genotype data become available in the future, we can discover more important gene-smoking interaction in lung cancer disease.
Supplementary material
Supplementary data are available at Carcinogenesis online.
Softwares used in the analysis
1. FlashPCA: https://github.com/gabraham/flashpca
2. STRUCTURE: https://webs.stanford.edu/group/pritchardlab/structure.html
3. EIGENSTRAT: https://github.com/DReichLab/EIG/tree/master/EIGENSTRAT
4. IMPUTE2: http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
Supplementary Material
Acknowledgment
This study was supported by grant U19CA148127.
Conflict of Interest Statement: None declared.
Abbreviations
- NSCLC
non-small cell lung cancer
- OR
odds ratio
- SQC
squamous cell carcinoma
References
- 1. American Cancer Society. (2017)Cancer Facts and Figures, 2017. American Cancer Society, Atlanta, GA. [Google Scholar]
- 2. Govindan R., et al. (2006) Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: Analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol., 24, 4539–4544. [DOI] [PubMed] [Google Scholar]
- 3. Wynder E.L., et al. (1995)The changing epidemiology of smoking and lung cancer histology. Environ. Health Perspect., Suppl 8, 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The Cancer Genome Atlas Research Network(2014)Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Cancer Genome Atlas Research Network(2012)Comprehensive genomic characterization of lung squamous cell lung cancers. Nature, 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buttitta F., et al. (2006)Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int. J. Cancer, 119, 2586–2591. [DOI] [PubMed] [Google Scholar]
- 7. Amos C.I., et al. (2008)Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Marchand L., et al. (2008)Smokers with the CHRNA lung cancer- associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res., 68, 9137–9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKay J.D., et al. ; EPIC Study (2008)Lung cancer susceptibility locus at 5p15.33. Nat. Genet., 40, 1404–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Landi M.T., et al. (2009)A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am. J. Hum. Genet., 85, 679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Y., et al. (2008)Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Truong T., et al. (2010)Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J. Natl. Cancer Inst., 102, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Miki D., et al. (2010)Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat. Genet., 42, 893–896. [DOI] [PubMed] [Google Scholar]
- 14. Maher B. (2008)Personal genomes: The case of the missing heritability. Nature, 456, 18–21. [DOI] [PubMed] [Google Scholar]
- 15. Ezzati M., et al. (2003)Estimates of global mortality attributable to smoking in 2000. Lancet, 362, 847–852. [DOI] [PubMed] [Google Scholar]
- 16. Zhang R., et al. (2014)A genome-wide gene-environment interaction analysis for tobacco smoke and lung cancer susceptibility. Carcinogenesis, 35, 1528–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thorgeirsson T.E., et al. (2010)Commentary: gene-environment interactions and smoking-related cancers. Int. J. Epidemiol., 39, 577–579. [DOI] [PubMed] [Google Scholar]
- 18. CanderWeele T.J., et al. (2012)Genetic variants on 15q25.1, smoking and lung cancer: an assessment of mediation and interaction. Am. J. Epidemiol., 175, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith P.G., et al. (1984)The design of case-control studies: the influence of confounding and interaction effects. Int. J. Epidemiol., 13, 356–365. [DOI] [PubMed] [Google Scholar]
- 20. Kraft P., et al. (2007)Exploiting gene-environment interaction to detect genetic associations. Hum. Hered., 63, 111–119. [DOI] [PubMed] [Google Scholar]
- 21. Murcray C.E., et al. (2009)Gene-environment interaction in genome-wide association studies. Am. J. Epidemiol., 169, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amos C.I., et al. (2017)The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol. Biomarkers Prev., 26, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McKay J.D., et al. (2017)Large scale genetic analysis identifies novel loci and histological variability in susceptibility to lung cancer. Nat genetics, 49, 1126–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kachuri L., et al. (2016)Fine mapping of chromosome 5p15.33 based on a targeted deep sequencing and high density genotyping identifies novel lung cancer susceptibility loci. Carcinogenesis, 37, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y., et al. (2016)FastPop: a rapid principal component derived method to infer intercontinental ancestry using genetic data. BMC Bioinformatics, 17, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abraham G., et al. (2014)Fast principal component analysis of large-scale genome-wide data. PLoS One, 9, e93766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pitchard J.K., et al. (2000)Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price A.L., et al. (2006)Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 29. Howie B.N., et al. (2009)A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Bakker P.I., et al. (2008)Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet., 17, R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shiozaki A., et al. (2011)Roles of XB130, a novel adaptor protein, in cancer. J. Clin. Bioinforma., 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lodyga M., et al. (2005)P-080 Prognostic expression of a novel adaptor protein XB130 innon-small cell lung cancer. Lung Cancer, 49, S1352005. [Google Scholar]
- 33. Pelechano V., et al. (2013)Gene regulation by antisense transcription. Nat. Rev. Genet., 14, 880–893. [DOI] [PubMed] [Google Scholar]
- 34. Balbin P.A., et al. (2015)The landscape f antisense gene expression in human cancers. Genome Res., 25, 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xi S., et al. (2010)Cigarette smoke induces C/EBP-β-mediated activation of miR-31 in normal human respiratory epithelia and lung cancer cells. PLoS One, 5, e13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Silva J.M., et al. (2010)Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics, 95, 355–362. [DOI] [PubMed] [Google Scholar]
- 37. Tugues S., et al. (2015)New insights into IL-12-mediated tumor suppression. Cell Death Differ., 22, 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boueiz A.E., et al. (2016)Clinical and genetic characteristics of emphysema distribution subtypes identified by unsupervised learning analysis. Am. J. Respir. Crit. Care Med., 193, A7480. [Google Scholar]
- 39. Zeng Z., et al. (2016)AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol., 37, 729–737. [DOI] [PubMed] [Google Scholar]
- 40. Shiozaki A., et al. (2011)Roles of XB130, a novel adaptor protein, in cancer. J. Clin. Bioinforma., 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zeilinger S., et al. (2013)Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One, 8, e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang T., et al. (2015)Meta-analysis of gene methylation and smoking behavior in non-small cell lung cancer patients. Sci. Rep., 5, 8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vink J.M., et al. (2017)Differential gene expression patterns between smokers and non-smokers: cause or consequence?Addict. Biol., 22, 550–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Birrell M.A., et al. (2008)Impact of tobacco-smoke on key signaling pathways in the innate immune response in lung macrophages. J. Cell. Physiol., 214, 27–37. [DOI] [PubMed] [Google Scholar]
- 45. Benton M.A., et al. (2011)Comparative genomic analyses identify common molecular pathways modulated upon exposure to low doses of arsenic and cadmium. BMC Genomics, 12, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kenfield S.A., et al. (2008)Comparison of aspects of smoking among the four histological types of lung cancer. Tob. Control, 17, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.