Abstract
Neuronal survival and morphological maturation depends on the action of the transcription factor calcium responsive element binding protein (CREB), which regulates expression of several target genes in an activity-dependent manner. However, it remains largely unknown whether CREB-mediated transcription could play a role at early stages of neuronal differentiation, prior to the establishment of functional synaptic contacts. Here, we show that CREB is phosphorylated at very early stages of neuronal differentiation in vivo and in vitro, even in the absence of depolarizing agents. Using genetic tools, we also show that inhibition of CREB-signaling affects neuronal growth and survival in vitro without affecting cell proliferation and neurogenesis. Expression of A-CREB or M-CREB, 2 dominant-negative inhibitors of CREB, decreases cell survival and the complexity of neuronal arborization. Similar changes are observed in neurons treated with protein kinase A (PKA) and Ca2+/calmodulin-dependent protein kinase II (CaMKII) inhibitors, which also show decreased levels of pCREBSer133. Notably, expression of CREB-FY, a Tyr134Phe CREB mutant with a lower Km for phosphorylation, partly rescues the effects of PKA and CaMKII inhibition. Our data indicate that CREB-mediated signaling play important roles at early stages of cortical neuron differentiation, prior to the establishment of fully functional synaptic contacts.
Keywords: cortex, CREB signaling, immature neurons, dendritic differentiation, neuronal survival
Introduction
Neuronal dendrite morphology is key for the integration of synaptic inputs and defines many functional properties of individual neurons in the adult central nervous system (Hausser et al. 2000). The neuronal dendritic tree grows by the addition and elongation of branches: Primary dendrites emerge from the cell body and branch to form secondary and tertiary dendrites. Different cell types in the CNS display unique dendritic branching patterns, suggesting the existence of particular mechanisms controlling both branch addition and maintenance, as well as branch retraction and elimination (Scott and Luo 2001). Since neuronal morphology is deeply correlated with function, understanding the mechanisms that coordinate the overall growth and detailed patterning of dendritic arborization is fundamental to understand the basis of neurological diseases associated with defects in neuronal morphology, such as Down, Rett, and fragile X syndromes (Kaufmann and Moser 2000).
Activity-dependent mechanisms play a fundamental role in shaping the morphology of neurons, controlling both the addition and elimination of processes (Cline 2001; Wong and Ghosh 2002). In many instances, activity-dependent mechanisms act through activation of transcription mediated by CREB (calcium responsive element binding protein). Phosphorylation of CREB at serine133 (Ser133) is required for the recruitment of its co-activators CREB-binding protein and p300, which in turn mediate target gene induction via their association with RNA polymerase II complexes and via intrinsic histone acetyltransferase activities that mobilize promoter-bound nucleosomes (Mayr and Montminy 2001).
In cultured cortical neurons, membrane depolarization induced by KCl opens voltage-gated calcium channels, generating influx of calcium and activation of calcium/calmodulin-dependent protein kinase IV (CaMKIV). This kinase phosphorylates CREB at Ser133 (pCREBSer133) leading to an increase in neuronal dendritic complexity (Redmond et al. 2002). CaMKIV expression in the rat cerebral cortex reaches maximal levels during the second and third postnatal week, which coincides with the period of maximal dendritic growth and synaptic activity. Accordingly, dendritic growth of tectal neurons increases after visual light stimulation, and this effect is suppressed by both the NMDA receptor antagonist AP5 and the AMPA receptor antagonist CNQX, indicating that a glutamate-receptor-mediated signaling mechanism mediates dendritic growth (Sin et al. 2002).
Still, little is known about the possible roles of CREB-mediated gene transcription at early stages of neuronal differentiation, prior to the formation of synaptic contacts. Here, we show that phosphorylation of CREB at Ser133 occurs in immature cortical neurons at stages when no spontaneous activity is present and in the absence of induced depolarization. Genetic manipulations of CREB-mediated signaling though either expression of A-CREB, a dominant-negative inhibitor of CREB (Ahn et al. 1998), or M-CREB, a dominant-negative mutant CREB (Berger et al. 2011), lead to reduced cell survival and severe aberrations in neuronal morphology. These morphological changes could be mimicked by pharmacological blockade of specific kinases, which in turn was partly rescued by overexpression of CREB-FY, a Tyr134Phe CREB mutant with a lower Km for phosphorylation. Together, our data suggest that CREB-signaling is active during neuronal development, prior to the onset of synaptic activity, and that CREB-signaling play important roles in neuronal survival and growth.
Material and Methods
Animals
All procedures were performed according to international directives and were approved by the local committee for care and use of laboratory animals. Experiments used C57/Bl6 (mus musculus) pregnant females whose embryos were removed at distinct embryonic time points. We considered embryonic Day 0 (E0) the day of vaginal plug detection. Animals were housed under standard conditions with access to water and food ad libitum on a normal 12 h light/dark cycle.
Primary Cell Culture
Isolation and dissociation of dorsal telencephalic cells was performed as described previously (Costa et al. 2008). Briefly, brains were removed under sterile conditions and dorsolateral telencephalons of E14 mice were micro-dissected in Hank's Balanced Salt Solution at 4 °C. After a brief centrifugation to precipitate the tissue, 1 mL of trypsin-EDTA (0.05%) (GIBCO) was added to digest the tissue for 15 min at 37 °C. Trypsin activity was stopped by adding 2 mL of Dulbecco's Modified Eagle Medium (DMEM, GIBCO) supplemented with 10% fetal calf serum (FCS) (Sigma) and penicillin/streptomycin (100 U/mL penicillin, 100 mg/mL streptomycin; GIBCO). Next, cells were mechanically dissociated with a fire-polished and FCS-coated Pasteur pipette and washed twice in DMEM (10% FCS) and centrifuged for 5 min (at 340 × g). Next, cells were counted using a Neubauer chamber, plated at a density of 5 × 105 cells per well in a 24-well plate on poly-D-lysine-coated glass coverslips, and incubated at 37 °C and 5% CO2 for 7 days. Retroviruses carrying the plasmids of interest (see below) were added to the cultures 2 h after plating. This procedure allows transduction of actively dividing cells, thus being a suitable method to trace the lineage of individual progenitor cells and decrease the variability of morphology (due to difference in generation time among cultured neurons). For some experiments, adeno-associated virus (AAVs) were used. After 1 day in vitro (DIV), DMEM supplemented with 2% B27 was added in order to decrease serum concentration to 5%. Before fixation, cells were washed with PBS and then fixed in PFA 4% for 15 min. After 3 washes with PBS at room temperature, cells were prepared for immunocytochemistry.
Retrovirus
Retroviruses were produced as previously described (Jagasia et al. 2009) using expressing plasmids containing the CAG promoter, an internal ribosomal entry site and a green fluorescent protein (GFP). Control plasmid contains only the GFP gene sequence (Control-GFP) and test plasmids contain either A-CREB (A-CREB-GFP), which contains an acidic amphipathic extension onto the N-terminus of the CREB leucine zipper domain, or CREB-FY (CREBFY-GFP), a Tyr134Phe CREB mutant with a lower Km for phosphorylation. The influence of these later plasmids over CREB-mediated signaling in neural cells was previously described (Jagasia et al. 2009; Herold et al. 2011).
Adeno-Associated Virus
Serotype 2 AAV was produced as previously described (Berger et al. 2011) using the plasmid carrying the gene for M-CREB, in which Ser133 is replaced by alanine to prevent phosphorylation, fused to the enhanced GFP in the N-terminal (Addgene, cat # 68551).
Pharmacological Treatments
Twenty-four hours after plating, cell cultures were treated with the protein kinase A (PKA) inhibitor H-89 (10 µM, Sigma), CaM kinase II inhibitor KN-62 (10 µM, Sigma), mitogen-activated protein kinase (MAPK) inhibitor U0126 (10 µM, Sigma) or phosphoinositide 3-kinase (PI3K) inhibitor Wortmannin (100 nM, Sigma) (Redmond et al. 2002; Chevaleyre et al. 2007; Hu et al. 2008; Marcucci et al. 2010). All drugs were diluted in Dimethyl sulfoxide, which was added at the same concentration to control cultures.
Time-Lapse Video Microscopy
Time-lapse video microscopy of primary cultures was performed with a cell observer (Zeiss) at constant conditions of 37 °C and 5% CO2. Phase contrast images were acquired every 2 min up to 5 days using a 20× phase contrast objective (Zeiss) and an AxioCamHRm camera with a self-written VBA module remote controlling Zeiss AxioVision 4.7 software (Rieger et al. 2009). Cell cycle parameters, mode of cell division, and cell survival were analyzed using Timm's Tracking Tool software (Araújo et al. 2014). Mode of cell division was classified based on the behavior of daughter cells in: symmetric progenitor (SP, both daughter cells continue to proliferate), asymmetric (A, one daughter cell continues to proliferate and the other becomes postmitotic), or symmetric terminal (ST, both daughter cells become postmitotic). Cell cycle length was measured as the time spanned by proliferating cells between their generation and division. Cell survival was quantified for each cell lineage by dividing the number of cells alive at 12, 24, 36, 48, 60, 72, and 84 h for the total number of cells generated before these time points were reached within individual clones. A total of 556 trees (or clones) were analyzed: Control: n = 310; A-CREB: n = 106; CREBFY: n = 140.
Immunocytochemistry
Cell cultures or coronal slices (20 μm thick) of the telencephalon were incubated in primary antibodies overnight at 4 °C in 0.5% triton X-100 and 10% of normal goat serum in PBS 0.1 M. Primary antibodies used were anti-Microtubule Associated Protein 2 (MAP2, mouse IgG1 1:1000, Sigma), anti-betaIII tubulin (Tuj1, mouse IgG2b 1:1000, Sigma), anti-phospho-CREB Ser133 (pCREBSer133, rabbit 1:1000, Cell Signaling), and anti-GFP (chicken, AvesLab 1:500). Fluorescent secondary antibodies were used according to the manufacturer's recommendation (Life Technologies). Nuclei were visualized by incubating cells for 5 min with 0.1 μg/mL DAPI (4′6′-diamino-2-phenylindone, Sigma) in PBS 0.1 M. Cells and sections were mounted in Aqua Polymount (Polyscience) and analysed using epifluorescence microscopy (AxioImager, Zeiss) or confocal microscopy (LSM 710, Zeiss).
Morphological Analysis
Neuronal morphology was analyzed using ImageJ (v. 1.46e) and NeuronJ (v. 1.4.2). Only GFP- and MAP2-expressing neurons were analyzed. We quantified: 1) dendrite length; 2) the sum of length of all dendrites per neuron (total dendrite length); 3) the length of the longest process originating from the soma (primary dendrite); 4) the number of dendrites per neuron: 5) the number of primary dendrites per neuron; 6) the number of branches (all dendrites, except the primary dendrites); 7) the number of branching points per neuron and per dendrite.
Western Blotting
The dorsal telencephalon of E13, E16, and E18 mice was dissected as previously described. Tissues were homogenized in 500 μL of lysis buffer containing 150 mM sodium chloride 1.0% NP-40 or Triton X-100 0.5% sodium deoxycholate 0.1% SDS (sodium dodecyl sulfate) 50 mM Tris, pH 8.0. To confirm specificity of anti-pCREBSer133 antibody, half of samples were dephosphorylated using one unit of Calf Intestinal Protease (CIP, Abcam) per µg of protein in the CIP buffer, 60 min at 37 °C. Next, a 10% SDS-PAGE gel was loaded with 20 µg of protein with or without dephosphorylation treatment and ran for 1.5 h at 100 V. Cell cultures were solubilized with 30 μL of lysis buffer containing Tris-HCl (150 mM), NaCl (150 mM), 0.1% SDS, sodium orthovanadate (2 mM), sodium phosphate (5 mM), and protein inhibitor (4 mM). Twenty micrograms of protein from each sample were resolved on a polyacrylamide gel under denaturing conditions (SDS-PAGE) at 10%. Following electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Immun-Blot PVDF membrane, Bio Rad). Antibody staining was performed using antibodies against β-actin (goat 1:1000, Abcam), CREB (rabbit, 1:1000, Cell Signaling Technology), β-Tubulin Isotype III (mouse, 1:2000, Sigma), and pCREBSer133 (rabbit 1:1000, Cell Signaling). Optical densities were measured using ImageJ (v. 1.46e), Gel Analyzer plugin. Normalization was done with β-actin (whole brain analysis) or β-Tubulin Isotype III (cell cultures).
Statistical Analysis
For morphological analysis, data were derived from at least 7 independent batches of cell culture. The total number of cells and clones analyzed are provided throughout the results section. Statistical analysis of pCREBSer133 expression derived from 3 independent experiments of cell culture and 4 coverslips from each condition (3, 5, and 7 div) were analyzed. Data in the graphics are presented as Mean ± Standard Error of the Mean (SEM). For statistical analysis we used GraphPad Prism software version 5. Results were initially tested for normality using the Kolmogorov–Smirnov test. Parametric data were compared using One-Way ANOVA followed by Bonferroni's or Dunnett's post hoc test as indicated in figure legends. Non-parametric data were compared using Mann–Whitney test or Kruskal–Wallis test followed by Dunn's post hoc test. The confidence interval is set to 95%.
Results
Phospho-CREB is Expressed in Immature Cortical Neurons
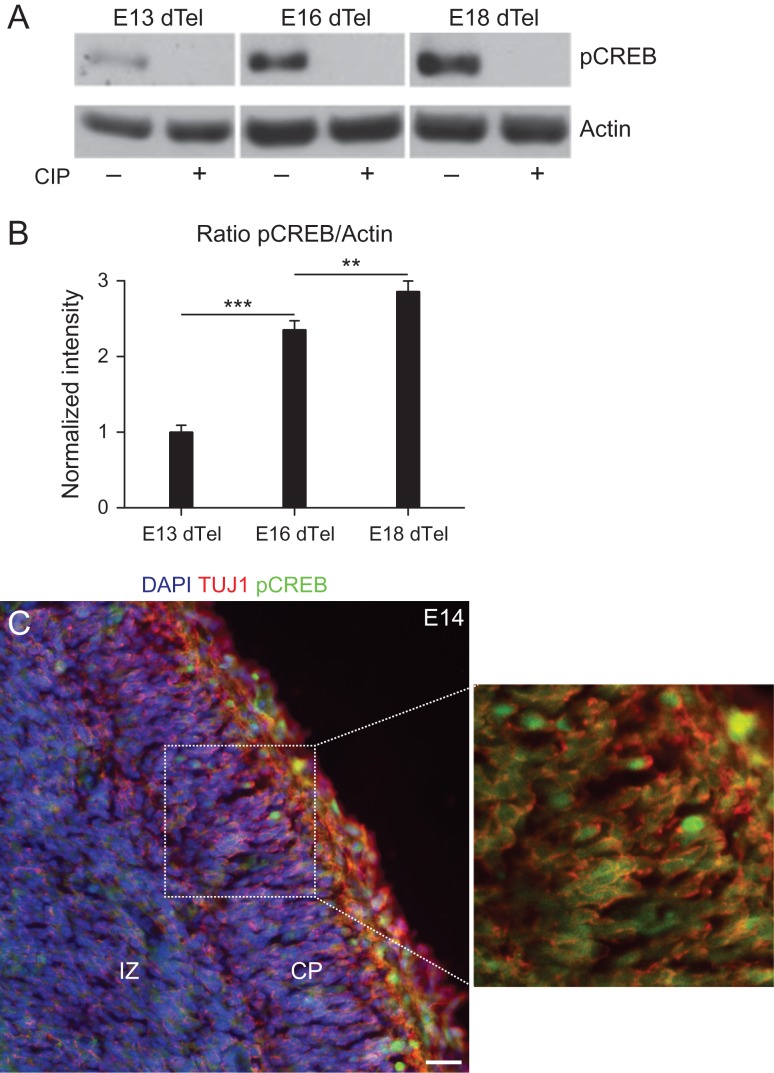
Signaling mediated by phospho-CREB is involved in the morphological maturation of excitatory cortical neurons upon KCl induced depolarization (Redmond et al. 2002). However, early stages of neuronal differentiation lacks spontaneous electrical activity (Zona et al. 1994) (Supplementary Fig. 1 and Supplementary Movie 1), suggesting that phosphorylation of CREB would not be present at early stages of cortical development. To test this possibility, we performed western blotting and immunohistochemistry analysis of the dorsal telencephalon using an antibody specific for CREB phosphorylated at Ser133 (Fig. 1). Surprisingly, we observed that pCREBSer133 expression could be detected in the dorsal telencephalon at embryonic Day 13 and increased at E16 and E18 (Fig. 1A,B). Antibody staining could be abolished by treatment with CIP indicating the specificity of the staining (Fig. 1A). Similarly, immunohistochemical analysis of the dorsal telencephalon revealed a widespread expression of pCREBSer133 in the intermediate zone and cortical plate at E14 (Fig. 1B) and E17 (Supplementary Fig. 2). Together, these findings suggest that phosphorylation of CREB occurs in early postmitotic neurons.
Figure 1.
Phosphorylation of CREB in the developing dorsal telencephalon. (A) Western blot analysis of pCREBSer133 expression in the dorsal telencephalon at early (E13), mid (E16), and late (E18) cortical neurogenesis. Observe the increase in pCREBSer133 levels at E16/E18 and the absence of antibody staining after treatment of samples with CIP. (B) Quantification of pCREBSer133 expression by western blotting optical densitometry (One-way ANOVA followed by Bonferroni's post hoc test; **P < 0.01 and ***P < 0.001). (C) Coronal section of E14 mouse brain showing the expression of pCREBSer133 (green) in TU1+ neurons (red). Nuclei are labeled with DAPI (blue). Dashed box is shown in higher magnification to highlight the expression of pCREBSer133 in neurons. CP, cortical plate; IZ, intermediate zone. Calibration bar: 20 μm.
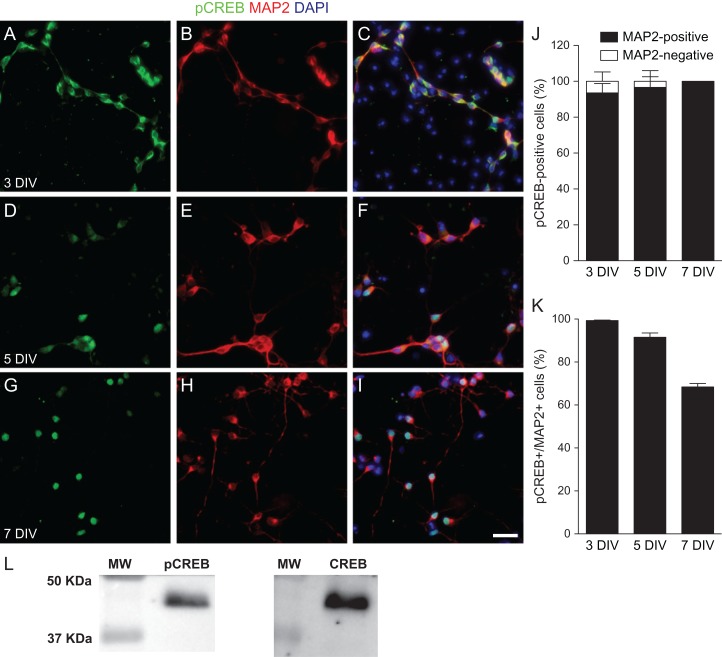
To further characterize the cells expressing pCREBSer133 in the absence of induced depolarization, we cultured embryonic Day 14 (E14) cortical cells and analyzed the expression of pCREBSer133 at Days 3, 5, and 7 in culture (Fig. 2). We observed pCREBSer133 expression mostly in MAP2+ neuron at the examined days in vitro (div). Notably, however, pCREBSer133 expression was diffuse in neurons at 3 div, including both cytoplasm and nucleus, and became mostly nuclear at 7 div (Fig. 2A–I). At all time points, the vast majority of cells expressing pCREBSer133 also expressed the neuronal protein MAP2 (3 div: 93.56 ± 5.20%; 5 div: 96.61 ± 5.96%; 7 div: 100%) (Fig. 2J). Moreover, the majority of neurons at 3, 5, and 7 div expressed pCREBSer133 (Fig. 2K). Total CREB protein and pCREBSer133 were also detected in cultured cortical neurons at 5 div by western blotting (Fig. 2L). At this stage, the ratio pCREBSer133/CREB was 81.4 ± 11.48% (mean ± SEM), indicating that most CREB protein is phosphorylated in early differentiating cerebral cortex neurons. Together, these results indicate that immature cortical neurons, but not progenitors or other neural cell types, phosphorylate CREB at Ser133 even in the absence of spontaneous or induced electrical activity.
Figure 2.
Phosphorylation of CREB in immature cortical neurons. (A–I) Epifluorescence images of E14 cortical cell cultures at 3, 5, and 7 DIV immunostained for pCREBSer133 (A, D, and G, green) and MAP2 (B, E, and H, red). Nuclei are satined with DAPI (C, F, and I, blue). Scale bar: 25 µm. Note that expression of pCREBSer133 is mostly cytoplasmic at 3 DIV (A–C), and becomes predominantly nuclear at 5 DIV (D–F) and 7 DIV (G–I). (J) Percentage of cells expressing pCREBSer133 with a neuronal (MAP2-positive) or non-neuronal (MAP2-negative) phenotype at different days in vitro. (K) Percentage of neurons expressing pCREBSer133 at different days in vitro. (L) Western blotting analysis of pCREBSer133 and total CREB expression in cortical cell cultures at 5 div. MW, molecular weight marker.
CREB Signaling Affects Morphology of Immature Cortical Neurons
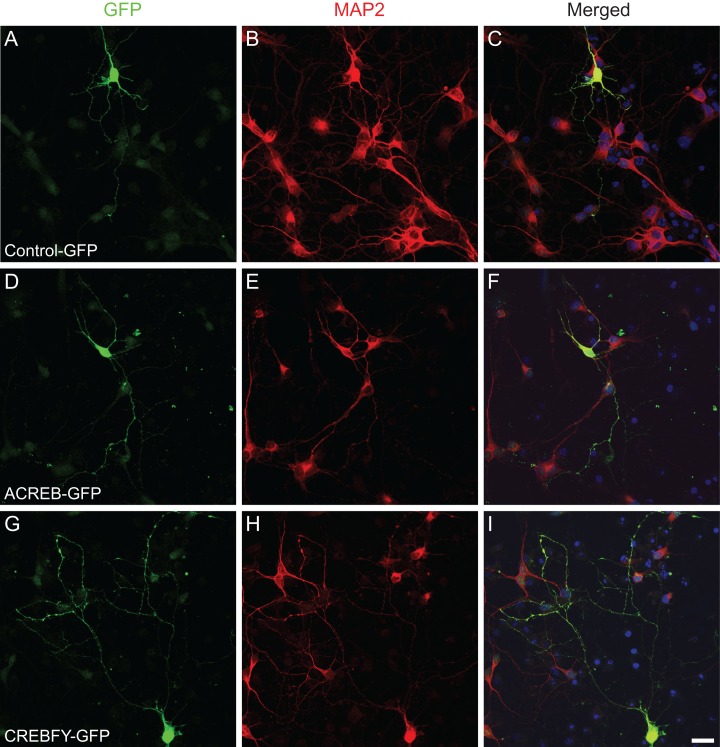
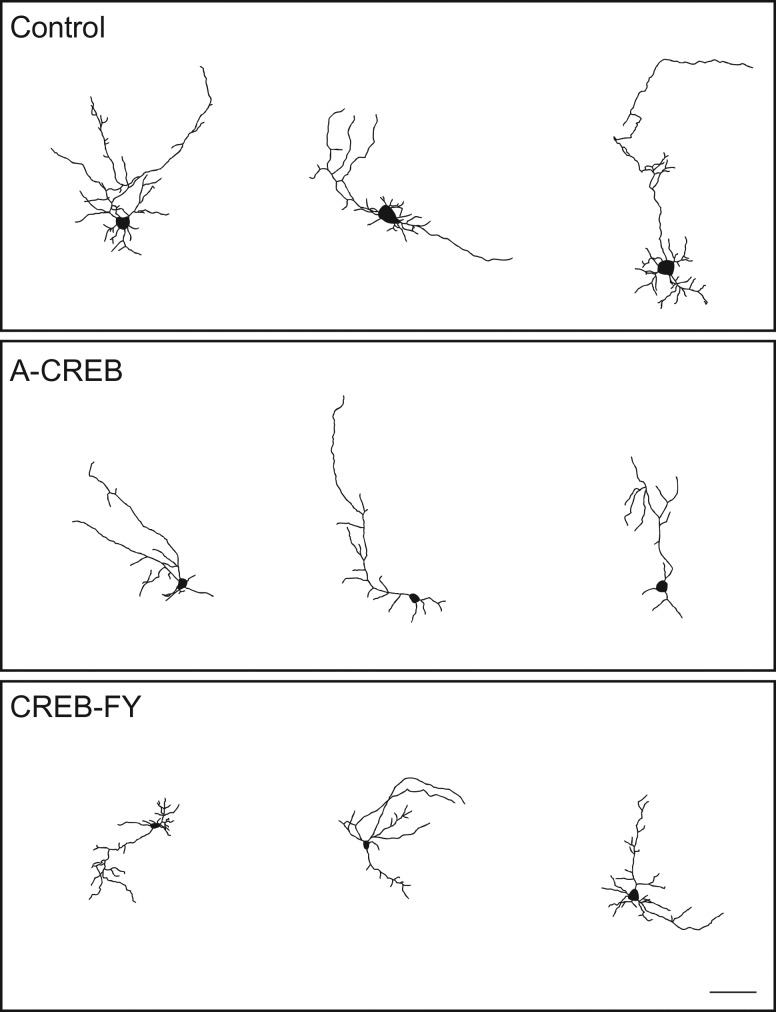
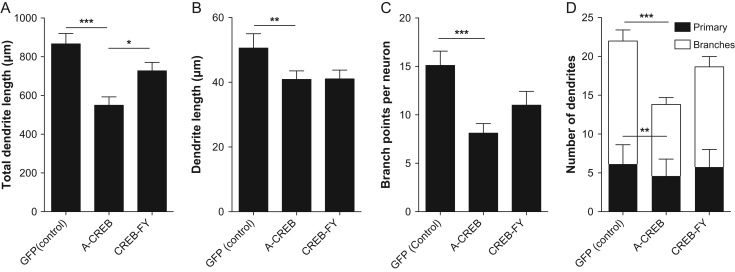
Phosphorylation of CREB in electrically immature neurons could be involved in the acquisition of initial morphological features, such as primary dendrites and dendrite branches. To directly test whether CREB-mediated signaling in electrically immature cortical neurons could affect these early steps of morphological maturation, set out to manipulate CREB-dependent gene expression using genetic manipulations. Towards this aim, we transduced cortical progenitors using retroviruses carrying plasmids encoding for the expression of A-CREB-GFP, CREBFY-GFP or only GFP (control). After 7 div, the morphology of individual neurons was assessed by the expression of GFP (Fig. 3). Using ImageJ software, single GFP-positive neurons were traced allowing the quantification of several hallmarks of neuronal morphology (Figs 4–6). We observed a significant reduction in the total dendrite length of neurons transfected with A-CREB as compared to controls and to neurons transfected with CREB-FY (Fig. 5A). This reduction was caused by both a decrease in the length of individual dendrites (Fig. 5B) and fewer dendritic branches (Fig. 5C,D). Similarly, AAV-mediated transfection of cultured cortical neurons with M-CREB also reduced the total neuronal dendrite length, due to a decrease in the number of branches (Supplementary Fig. 3). These observations indicate that CREB-mediated signaling in immature neurons is important to establish the initial neuronal dendritic complexity by controlling the addition and elongation of dendrites.
Figure 3.
Genetic manipulation of CREB-mediated signaling in cortical cell culture. (A–I) Confocal images of neurons at 7 DIV transfected with control-GFP (A–C), A-CREB-GFP (D–F) and CREBFY-GFP (G–I) plasmids. Transfection was performed 2 h after plating cortical cells using retroviral vector. Cultures were immunostained for GFP (A, D, and G) and MAP2 (B, E, and H). Nuclei are stained with DAPI (C, F, and I). Observe the less elaborated morphology of the neuron transfected with A-CREB-GFP (D–F). Scale bar: 20 µm.
Figure 4.
A-CREB reduces complexity of cortical neuronal arborization. Drawings of cortical neurons transfected with Control, A-CREB or CREB-FY plasmids after 7 DIV. Observe the low number of process and overall less elaborated dendritic morphology of neurons transfected with A-CREB as compared with control and CREB-FY. Calibration bar: 100 μm.
Figure 6.
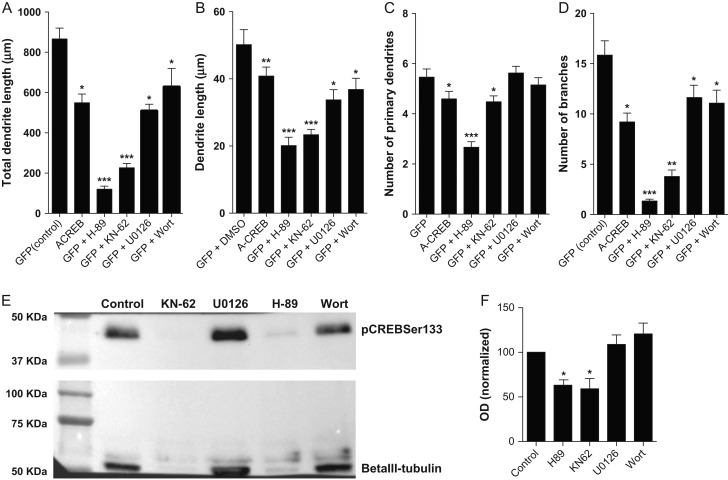
Inhibition of PKA, Ca2+/calmodulin-dependent protein kinase II (CaMKII), and MAPK mimics the effects of A-CREB expression on neuronal morphology. (A–D) Morphological features of E14 cortical neurons treated with H-89, KN-62, U0126, and Wortmannin (PKA, CaMKII, MAPK, and PI3K inhibitors, respectively), compared with neurons transfected with control or A-CREB plasmids without kinase inhibition. (A) Total dendrite length of neurons. (B) Average length of individual dendrites. (C) Number of primary dendrites per neuron. (D) Number of branches per neuron. Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, **P < 0.01, and ***P < 0.001). Analyzed neurons: Control n = 51; A-CREB n = 52; GFP + H-89 n = 50; GFP + KN-62 n = 51; GFP + U0126 n = 63; GFP + Wort n = 39. (E) Western blotting analysis of pCREBSer133 and betaIII-tubulin expression in cortical neurons at 5 div after treatment with different kinase inhibitors. (F) Normalized optical density of immunoblotting bands. Observe that only H-89 and KN-62 significantly decrease the expression of pCREBSer133 in cultured cortical neurons.
Figure 5.
A-CREB reduces neuronal branching complexity by reducing the total number of dendrites rather than the dendrites length. (A) Sum of all individual dendrite lengths of neurons transfected with control, A-CREB or CREB-FY plasmids. (B) Average length of individual dendrites. (C) Total number of branches per neuron. (D) Number of primary dendrites and branches. Observe that A-CREB expression affects the total dendrite length by both decreasing the number and extension of individual processes. Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, **P < 0.01, and ***P < 0.001). Analyzed neurons: Control n = 56; A-CREB n = 53; CREB-FY n = 54.
PKA and CaMKII Inhibitors Mimic the Effects of A-CREB Expression on Neuronal Dendrite Morphology
Although calcium-dependent phosphorylation of CREB in electrically active cortical neurons requires activation of CaMKIV (Redmond et al. 2002), other kinases are involved in CREB phosphorylation, such as PKA, MAPK, and PI3K (Mayr and Montminy 2001). To evaluate possible roles of these kinases in early morphological maturation of cortical neurons, we treated cell cultures with PKA, Ca2+/calmodulin-dependent protein kinase II (CaMKII), MAPK, and PI3K inhibitors H-89, KN-62, U0126, and Wortmannin, respectively. We observed that inhibition of all 4 kinases decreased the individual and total dendrite length (Fig. 6A,B), similar to A-CREB expression. Furthermore, PKA and CaMKII inhibitors mimicked the effect of A-CREB expression by decreasing the number of primary neuronal processes and branches (Fig. 6C,D). Thus, pharmacological inhibition of PKA and CaMKII affect both dendrite addition and elongation, whereas MAPK and PI3K affect only dendrite elongation, in a similar fashion to genetic blockade of pCREBSer133-mediated transcription.
In order to understand differences in the effects of kinase inhibition and A-CREB expression we set out to investigate whether treatment with kinase inhibitors interfered with phosphorylation of CREB at Ser133 in cultured cortical neurons. We found that PKA and CaMKII inhibitors significantly decreased the levels of pCREBSer133 in cells at 5 div, whereas MAPK and PI3K inhibitors had a trend of slightly increasing levels of pCREBSer133, however below statistical significance (Fig. 6E,F). Altogether these observations suggest that only the effects of PKA and CaMKII inhibition on the dendrite morphology of immature cortical neurons could be attributed to a reduction in CREB-mediated signaling.
CREB-FY Rescues the Effects of PKA and CaMKII Inhibition of Dendrite Length
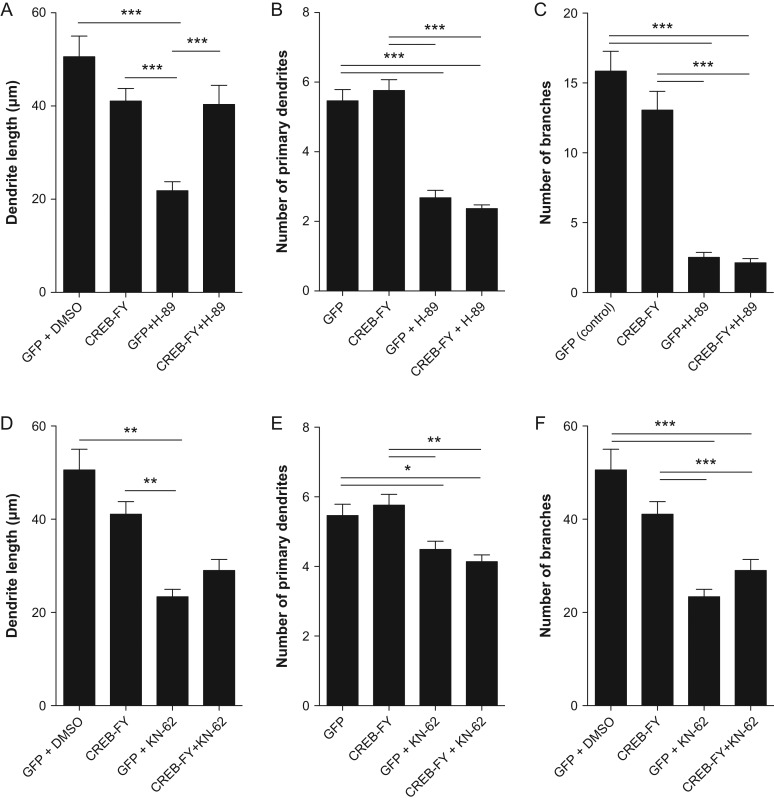
Next, we set out to test whether CREB-FY expression could rescue the effects of pharmacological kinase inhibition on neuronal morphology. Toward this aim, we combined genetic manipulation of CREB-mediate signaling using a CREB-FY-GFP plasmid and treatments with the blockers H-89 or KN-62 (Fig. 7). We observed that CREB-FY expression partly rescued the effect of H-89 and KN-62 on dendrite length (Fig. 7A,D). In contrast, CREB-FY expression was not sufficient to revert the effects of kinase inhibition on the number of primary dendrites and branches (Fig. 7B,C and E,F). Consistent with the absence of changes of pCREBSer133 levels in cultures treated with MAPK or PI3K inhibitors (Fig. 6E,F), we observed no reversion of the effects of these drugs on neuronal morphology after CREB-FY expression (Supplementary Fig. 4).
Figure 7.
CREB-FY expression rescues some effects of kinase inhibition on dendrite morphology. (A–F) Mean individual dendrite length (A, D), number of primary dendrites (B, E), and branches (C, F) of neurons transfected with control-GFP or CREBFY-GFP plasmids and treated with H-89 or KN-62. Note that CREB-FY expression partly rescues the effects of PKA and CaMKII inhibition on the length of individual dendrites (A, D), but shows no significant effect in the number of dendrites. Kruskal–Wallis followed by Dunn's post hoc test (*P < 0.05, **P < 0.01, and ***P < 0.001). Analyzed neurons: Control n = 56; CREB-FY n = 54; GFP + H-89 n = 50; CREB-FY + H-89 n = 46; GFP + KN-62 n = 51; CREB-FY + KN-62 n = 51.
Collectively, our findings indicate that both PKA- and CaMKII-mediated phosphorylation of CREB at Ser133 is important for the morphological maturation of immature cortical neurons.
Mode of Cell Division and Cell Cycle Length of Progenitor Cells are Not Affected by CREB Signaling
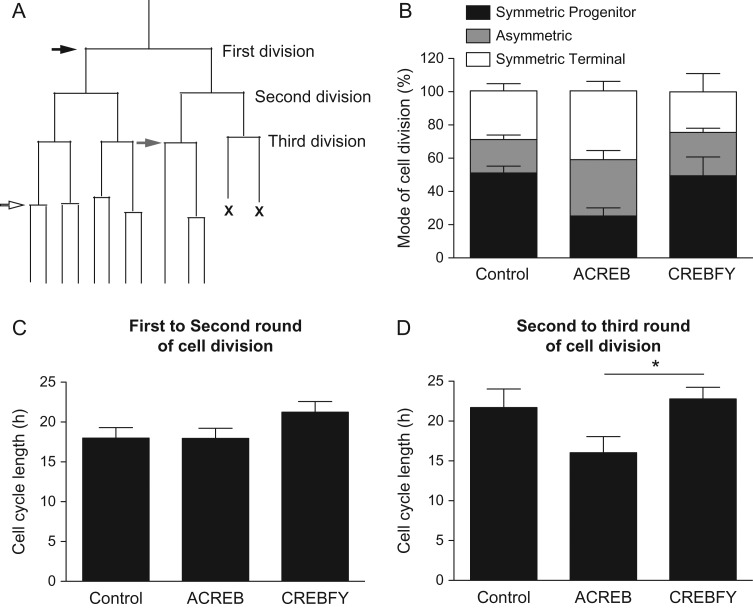
Expression of pCREBSer133 in MAP2-negative cells could indicate that at least some neural progenitor cells phosphorylate CREB. As a consequence, genetic manipulation of CREB-mediated transcription could affect progenitor behavior, delaying neuronal differentiation and, therefore, contribute to the malformation of dendrites. To rule out this possibility, we used time-lapse video microscopy to track the behavior of individual progenitor cells transfected with A-CREB-GFP, CREB-FY-GFP, and control-GFP plasmids (Fig. 8, Supplementary Fig. 5 and Supplementary Movies 3 and 4). First, we quantified the proportion of cells undergoing symmetric proliferative (both daughter cells are proliferating cells), ST (both daughter cells are non-proliferating, postmitotic cells) or asymmetric cell division (one daughter cell proliferates and the other becomes postmitotic). We observed that the expression of A-CREB or CREB-FY was not sufficient to significantly affect the mode of cell division of individual cortical progenitors (Fig. 8B). Next, we measured the cell cycle of individual progenitor cells generated in the first round (Fig. 8C) and second round of cell division (Fig. 8D). We observed that neither A- CREB or CREB-FY expression change the cell cycle length compared with controls. Thus, CREB-mediated signaling does not contribute to the control of cell cycle progression of cortical progenitors.
Figure 8.
CREB signaling does not affect mode of cell division and cell cycle of neural cortical progenitors. (A) Schematic lineage tree indicating the mode of cell division (black arrow: SP—both daughter cells undergo new cycle of cell division; gray arrow: asymmetric—one daughter cell undergo new cycle of cell division and the other becomes postmitotic; white arrow: ST—both daughter cells become postmitotic) and rounds of cells division (first, second, and third division). “X” indicates cell death. (B) Mode of cell division of individual progenitor cells after transfection with GFP (control), A-CREB-GFP, or CREBFY-GFP (see also Supplementary Fig. 5 and Supplementary Movies 3 and 4). (C–D) Cell cycle length of individual progenitor cells followed by time-lapse video microscopy. Data show the time in hours between first and second rounds of cell divisions (C) or between second and third rounds cell divisions (D). Note that only in the later A-CREB-GFP expression shortens the cell cycle length as compared with CREBFY-GFP (One-Way ANOVA followed by Tukey's post hoc test; *P < 0.05). Number of trees analyzed—Control: n = 310; A-CREB: n = 106; CREBFY: n = 140.
CREB-Mediated Transcription Regulates Early Neuronal Survival
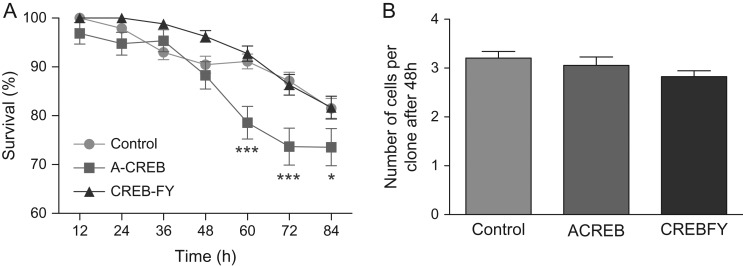
CREB-responsive genes have been shown to be involved in cell survival (Mantamadiotis et al. 2002; Ao et al. 2006). To test whether CREB signaling in immature neurons regulates neuronal survival, we quantified the rate of cell death of neurons transfected with A-CREB-GFP, CREBFY-GFP, or control-GFP using time-lapse video microscopy (Fig. 9). Cell survival, within individual cell lineages, was quantified by dividing the number of existing cells by the number of cells generated by a single progenitor each 12 h. We observed that the proportion of transfected cells surviving the first 48 h was similar among groups. However, after 60 h a significant decrease in cell survival of A-CREB transfected cells was observed (Fig. 9A). These results suggest that CREB signaling does not affect progenitor cells during the first 48 h of observation but is important for survival of immature neurons generated in the first rounds of cell division. Accordingly, the mean number of cells within clones after 48 h of observation was similar among control, A-CREB and CREB-FY transfected cells (Fig. 9B), suggesting that both proliferation and survival of progenitor cells is unaffected by CREB signaling disturbance.
Figure 9.
CREB signaling is important for cortical cell survival. (A) Survival of cells transfected with control-GFP (control), A-CREB-GFP, or CREBFY-GFP plasmids and followed by time-lapse video microscopy. Data show the mean frequency of surviving cells within individual clones at each 12 h. Observe the significant decrease in cell survival after 60 h (2-way ANOVA, *P < 0.05 and ***P < 0.001). (B) Total number of cells in individual clones after 48 h of observation in time-lapse video microscopy. Number of trees analyzed—Control: n = 310; A-CREB: n = 106; CREBFY: n = 140.
Discussion
In this work, we show that the transcription factor CREB is phosphorylated at Ser133 in immature neurons prior to the establishment of synaptic currents and that CREB-dependent transcription is important for early neuronal survival and dendritic tree formation. Genetic inhibition of CREB-mediated transcription using A-CREB impairs the growth of neuronal processes, especially by decreasing the length and number of processes. Similar reductions of morphological parameters occur after pharmacological inhibition of PKA and CaMKII, which are partly rescued by CREB-FY expression. Together, our results indicate that activity-independent signaling affect early neuronal differentiation in the developing cerebral cortex through activation of CREB-dependent transcription.
It is widely accepted that neuronal activity is necessary for appropriate dendrite branching and growth (Rajan and Cline 1998; Lohmann et al. 2002; Redmond et al. 2002). In fact, removal of sensorial input (i.e. light) hampers the dendritic arbor growth of optic tectal neurons in Xenopus tadpoles (Sin et al. 2002). Light-induced dendritic arbor growth requires glutamate-receptor mediated synaptic transmission, decreased RhoA activity and increased Rac and Cdc42 activity, indicating that Rho GTPases are important for activity-dependent neuronal plasticity (Sin et al. 2002). Depolarization of cortical neurons using KCl increases dendritic growth (Redmond et al. 2002). This effect is mediated by calcium influx through L-type voltage-sensitive calcium channels, activation of CaMKIV and phosphorylation of CREB. In hippocampal neurons, calcium influx though the canonical transient receptor potential channel 6 also activates CaMKIV and CREB phosphorylation, enhancing the dendritic growth (Tai et al. 2008). Collectively, these work indicate that electrical activity effects neuronal growth by increasing calcium influx, CaMKIV activation and CREB phosphorylation. According to this notion, pCREBSer133 regulates transcription of the gene for brain-derived neurotrophic factor, which is an important signal for neuronal growth (McAllister et al. 1997).
Interestingly, our data suggest that CREB-mediated transcription is regulated in cortical neurons prior to expected establishment of synaptic activity. Previously, isolated cortical neurons were shown to lack spontaneous synaptic currents during the first week in culture (Zona et al. 1994) also confirmed in our work by calcium imaging. Inward excitatory currents first appeared after 10 days in culture, with a peak in probability of spontaneous currents at 3 weeks in culture, and currents appears to be AMPA mediated (Zona et al. 1994). Thus, the phosphorylation of CREB at Ser133 described in our work is unlikely to depend on synaptic currents and is more likely mediated by signaling through metabotropic receptors.
We demonstrated that, at basal conditions, blockade of CREB-signaling by overexpression of A-CREB reduces the total dendrite length by 50%, due to a significant decrease in the length of dendrites, as well as the number of primary processes and branches. Similar decrease in total dendrite length and number of processes were obtained after expression of M-CREB. The different effects of A-CREB and M-CREB on the extension of individual dendrites can be explained by the fact that retrovirus and AAVs target different neuronal populations or by the distinct mechanisms of action of mutated CREB forms. A-CREB contains an acidic amphipathic extension onto the N-terminus of the CREB leucine zipper domain, which prevents the basic region of wild-type CREB from binding to DNA (Ahn et al. 1998). In the M-CREB, Ser133 is replaced by alanine to prevent phosphorylation (Berger et al. 2011). Previous work has shown the ability of A-CREB and M-CREB to block CREB-mediate signaling in neuronal cells both in vitro and in vivo (Jagasia et al. 2009; Berger et al. 2011; Herold et al. 2011), however, to the best of our knowledge, no direct comparison between these 2 dominant-negatives has been published.
Morphological changes observed in cortical neurons after A-CREB expression are mimicked, and somewhat impaired, by pharmacological inhibition of PKA and CaMKII. In contrast, inhibition of MAPK or PI3K also leads to a decrease in total dendrite length, due to a decrease in the extension of individual processes and in the number of branches, but with no significant effect in the number of primary dendrites. These observations suggest that different signaling pathways control the addition and elongation of processes in differentiating neurons. Therefore, the final neuronal morphology begins to be assembled at very early stages, prior the establishment of synaptic contacts, and it is likely the resultant of several signaling mechanisms, where some may be acting through CREB.
Indeed, expression of CREB-FY, a Tyr134Phe CREB mutant with a lower Km for PKA could rescue the effect of PKA-inhibition on dendrite length. Expression of CREB-FY could also partly rescue the effects of CaMKII on dendrite length, suggesting that at least part of the morphological malformations observed after kinase inhibition is dependent on CREB signaling. According to this interpretation, H-89 treatment decreases CRE-dependent transcription, whereas overexpression of the PKA catalytic subunit increases CRE-dependent transcription in cortical cell culture (Lau et al. 2004; Liang et al. 2008).
Different environmental signals could be involved in the activation of CREB phosphorylation and morphological changes in cortical neurons. For instance, dopamine or quinpirole (a dopamine agonist) treatment enhances dendrite growth in cortical neurons (Reinoso et al. 1996). This effect is mediated by the dopamine receptor subunit D2 and activation of protein kinase C and CaMKII, leading to CREB phosphorylation (Reinoso et al. 1996; Yan et al. 1999). Interestingly, dopamine also increases MAPK phosphorylation in this system, but this kinase does not seem to be necessary for CREB phosphorylation, indicating that MAPK and CREB signaling could result in the activation of different downstream targets (Yan et al. 1999). Together with our results showing that MAPK inhibition does not decrease CREB phosphorylation at Ser133, these observations suggest that MAPK-signaling pathways controls neuronal morphology by CREB-independent mechanisms.
Noteworthy, cocaine exposure during gestational stages leads to an increased dendritic growth of neurons in the anterior cingulate cortex, but not in the primary visual cortex, which lacks dopaminergic innervation (Jones et al. 2000). It is tempting to speculate that this effect of cocaine in neuronal morphology is mediated by CREB-signaling. Future experiments should clarify whether inhibition of CREB-mediated transcription could rescue these effects of cocaine exposure during pregnancy.
Notch signaling could also contribute to control early CREB-mediated neuronal growth. Indeed, Notch1 is expressed by immature cortical neurons (Sestan et al. 1999; Redmond et al. 2000) and restricts the dendrite length and branching (Redmond et al. 2000). Interestingly, Notch1 represses CREB-dependent gene expression in primary cortical neurons (Hallaq et al. 2015), indicating a potential link between Notch signaling and CREB-mediated changes in neuronal dendrite morphology. Moreover, Semaphorins, netrins, slits, wnt (wingless), and ephrins are also expressed in the developing cerebral cortex and have been implicated in neuronal dendrite morphogenesis (Valnegri et al. 2015). Future studies should clarify whether these and other extrinsic signals may be regulating dendritic growth through the phosphorylation of CREB at Ser133.
Finally, our results also indicate that CREB-mediated transcription is important for survival of neurons at early times after cell generation. Our findings are in agreement with previous observations in the adult hippocampus and olfactory bulb, where survival of newly born neurons is reduced by A-CREB expression (Jagasia et al. 2009; Herold et al. 2011).
Altogether, our results indicate that CREB-mediated transcription is activated in cortical neurons at developmental stages most likely lacking synaptic activity, and that CREB-mediated transcription controls important steps of neuronal differentiation and survival.
Supplementary Material
Notes
We thank Dr Vinícius Ribas for helping with Western Blotting experiments and Dr Emelie Katarina Svahn Leão for carefully reading our manuscript. Conflict of Interest: None declared.
Supplementary Material
Supplementary material is available at Cerebral Cortex online.
Funding
Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação de Amparo a Pesquisa do Rio Grande do Norte (FAPERN).
References
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. 1998. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 18:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao H, Ko SW, Zhuo M. 2006. CREB activity maintains the survival of cingulate cortical pyramidal neurons in the adult mouse brain. Mol Pain. 2:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo GL, Araújo JA, Schroeder T, Tort AB, Costa MR. 2014. Sonic hedgehog signaling regulates mode of cell division of early cerebral cortex progenitors and increases astrogliogenesis. Front Cell Neurosci. 8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AK, Green T, Siegel SJ, Nestler EJ, Hammer RP Jr. 2011. cAMP response element binding protein phosphorylation in nucleus accumbens underlies sustained recovery of sensorimotor gating following repeated D(2)-like receptor agonist treatment in rats. Biol Psychiatry. 69:288–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevaleyre V, Heifets BD, Kaeser PS, Sudhof TC, Castillo PE. 2007. Endocannabinoid-mediated long-term plasticity requires cAMP/PKA signaling and RIM1alpha. Neuron. 54:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline HT. 2001. Dendritic arbor development and synaptogenesis. Curr Opin Neurobiol. 11:118–126. [DOI] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. 2008. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 135:11–22. [DOI] [PubMed] [Google Scholar]
- Hallaq R, Volpicelli F, Cuchillo-Ibanez I, Hooper C, Mizuno K, Uwanogho D, Causevic M, Asuni A, To A, Soriano S, et al. . 2015. The Notch intracellular domain represses CRE-dependent transcription. Cell Signal. 27:621–629. [DOI] [PubMed] [Google Scholar]
- Hausser M, Spruston N, Stuart GJ. 2000. Diversity and dynamics of dendritic signaling. Science. 290:739–744. [DOI] [PubMed] [Google Scholar]
- Herold S, Jagasia R, Merz K, Wassmer K, Lie DC. 2011. CREB signalling regulates early survival, neuronal gene expression and morphological development in adult subventricular zone neurogenesis. Mol Cell Neurosci. 46:79–88. [DOI] [PubMed] [Google Scholar]
- Hu Y, Lund IV, Gravielle MC, Farb DH, Brooks-Kayal AR, Russek SJ. 2008. Surface expression of GABAA receptors is transcriptionally controlled by the interplay of cAMP-response element-binding protein and its binding partner inducible cAMP early repressor. J Biol Chem. 283:9328–9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia R, Steib K, Englberger E, Herold S, Faus-Kessler T, Saxe M, Gage FH, Song H, Lie DC. 2009. GABA-cAMP response element-binding protein signaling regulates maturation and survival of newly generated neurons in the adult hippocampus. J Neurosci. 29:7966–7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LB, Stanwood GD, Reinoso BS, Washington RA, Wang HY, Friedman E, Levitt P. 2000. In utero cocaine-induced dysfunction of dopamine D1 receptor signaling and abnormal differentiation of cerebral cortical neurons. J Neurosci. 20:4606–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. 2000. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 10:981–991. [DOI] [PubMed] [Google Scholar]
- Lau GC, Saha S, Faris R, Russek SJ. 2004. Up-regulation of NMDAR1 subunit gene expression in cortical neurons via a PKA-dependent pathway. J Neurochem. 88:564–575. [DOI] [PubMed] [Google Scholar]
- Liang MH, Wendland JR, Chuang DM. 2008. Lithium inhibits Smad3/4 transactivation via increased CREB activity induced by enhanced PKA and AKT signaling. Mol Cell Neurosci. 37:440–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann C, Myhr KL, Wong RO. 2002. Transmitter-evoked local calcium release stabilizes developing dendrites. Nature. 418:177–181. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, et al. . 2002. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 31:47–54. [DOI] [PubMed] [Google Scholar]
- Marcucci H, Paoletti L, Jackowski S, Banchio C. 2010. Phosphatidylcholine biosynthesis during neuronal differentiation and its role in cell fate determination. J Biol Chem. 285:25382–25393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B, Montminy M. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2:599–609. [DOI] [PubMed] [Google Scholar]
- McAllister AK, Katz LC, Lo DC. 1997. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 18:767–778. [DOI] [PubMed] [Google Scholar]
- Rajan I, Cline HT. 1998. Glutamate receptor activity is required for normal development of tectal cell dendrites in vivo. J Neurosci. 18:7836–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. 2002. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 34:999–1010. [DOI] [PubMed] [Google Scholar]
- Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. 2000. Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci. 3:30–40. [DOI] [PubMed] [Google Scholar]
- Reinoso BS, Undie AS, Levitt P. 1996. Dopamine receptors mediate differential morphological effects on cerebral cortical neurons in vitro. J Neurosci Res. 43:439–453. [DOI] [PubMed] [Google Scholar]
- Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. 2009. Hematopoietic cytokines can instruct lineage choice. Science. 325:217–218. [DOI] [PubMed] [Google Scholar]
- Scott EK, Luo L. 2001. How do dendrites take their shape? Nat Neurosci. 4:359–365. [DOI] [PubMed] [Google Scholar]
- Sestan N, Artavanis-Tsakonas S, Rakic P. 1999. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 286:741–746. [DOI] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. 2002. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 419:475–480. [DOI] [PubMed] [Google Scholar]
- Tai Y, Feng S, Ge R, Du W, Zhang X, He Z, Wang Y. 2008. TRPC6 channels promote dendritic growth via the CaMKIV-CREB pathway. J Cell Sci. 121:2301–2307. [DOI] [PubMed] [Google Scholar]
- Valnegri P, Puram SV, Bonni A. 2015. Regulation of dendrite morphogenesis by extrinsic cues. Trends Neurosci. 38:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. 2002. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 3:803–812. [DOI] [PubMed] [Google Scholar]
- Yan Z, Feng J, Fienberg AA, Greengard P. 1999. D(2) dopamine receptors induce mitogen-activated protein kinase and cAMP response element-binding protein phosphorylation in neurons. Proc Natl Acad Sci USA. 96:11607–11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zona C, Palma E, Brancati A, Avoli M. 1994. Age-dependent appearance of synaptic currents in rat neocortical neurons in culture. Synapse. 18:1–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.