The genetic causes of distal hereditary motor neuropathy (dHMN) are still to be fully elucidated. Using exome sequencing and functional studies, Tsai, Soong et al. identify a mutation in WARS, which alters aminoacylation and angiostatic functions of tryptophanyl-tRNA synthetase, as a novel cause of autosomal dominant distal hereditary motor neuropathy.
Keywords: distal hereditary motor neuropathy, dHMN, exome sequencing, WARS, tryptophanyl-tRNA synthetase
Abstract
Distal hereditary motor neuropathy is a heterogeneous group of inherited neuropathies characterized by distal limb muscle weakness and atrophy. Although at least 15 genes have been implicated in distal hereditary motor neuropathy, the genetic causes remain elusive in many families. To identify an additional causal gene for distal hereditary motor neuropathy, we performed exome sequencing for two affected individuals and two unaffected members in a Taiwanese family with an autosomal dominant distal hereditary motor neuropathy in which mutations in common distal hereditary motor neuropathy-implicated genes had been excluded. The exome sequencing revealed a heterozygous mutation, c.770A > G (p.His257Arg), in the cytoplasmic tryptophanyl-tRNA synthetase (TrpRS) gene (WARS) that co-segregates with the neuropathy in the family. Further analyses of WARS in an additional 79 Taiwanese pedigrees with inherited neuropathies and 163 index cases from Australian, European, and Korean distal hereditary motor neuropathy families identified the same mutation in another Taiwanese distal hereditary motor neuropathy pedigree with different ancestries and one additional Belgian distal hereditary motor neuropathy family of Caucasian origin. Cell transfection studies demonstrated a dominant-negative effect of the p.His257Arg mutation on aminoacylation activity of TrpRS, which subsequently compromised protein synthesis and reduced cell viability. His257Arg TrpRS also inhibited neurite outgrowth and led to neurite degeneration in the neuronal cell lines and rat motor neurons. Further in vitro analyses showed that the WARS mutation could potentiate the angiostatic activities of TrpRS by enhancing its interaction with vascular endothelial-cadherin. Taken together, these findings establish WARS as a gene whose mutations may cause distal hereditary motor neuropathy and alter canonical and non-canonical functions of TrpRS.
Introduction
Distal hereditary motor neuropathy (dHMN), also known as distal spinal muscular atrophy (dSMA), comprises a clinically and genetically heterogeneous group of inherited neuropathies characterized by slowly progressive distal limb muscle weakness and atrophy with no or minimal sensory involvement (Harding, 2005; Rossor et al., 2012). Phenotypically, dHMNs are similar to axonal Charcot–Marie–Tooth disease (CMT) and both entities may share common causative genes (Murphy et al., 2012; Rossor et al., 2013). Traditionally, dHMNs are divided into seven subtypes based on age at onset, mode of inheritance, and presence of additional features (i.e. vocal cord paralysis, predominate upper limbs involvement, and pyramidal signs) (Harding, 2005; Rossor et al., 2012). However, there are substantial genetic heterogeneities for a clinically homogenous dHMN subtype actually being attributed to mutations in different genes. To date, mutations in at least 15 genes have been implicated in dHMN (http://neuromuscular.wustl.edu), but the genetic diagnosis for more than 70% of patients with dHMN remains elusive (Dierick et al., 2008; Murphy et al., 2012; Rossor et al., 2012, 2013).
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed, essential enzymes responsible for the first step of translation process, in which ARSs charge amino acids to cognate tRNA molecules (Delarue, 1995). Mutations in six ARS genes have been implicated in hereditary polyneuropathies. These genes include glycyl- (GARS; MIM *600287; Antonellis et al., 2003), tyrosyl- (YARS; MIM *603623; Jordanova et al., 2006), alanyl- (AARS; MIM *601065; Latour et al., 2010), histidyl- (HARS; MIM *142810; Vester et al., 2013; Brozkova et al., 2015), methionyl- (MARS; MIM *156560; Gonzalez et al., 2013), and lysyl- (KARS; MIM *601421; McLaughlin et al., 2010) tRNA synthetase genes. All of them are associated with axonal or intermediate CMT, and three of them (GARS, AARS and HARS) are also linked to dHMN. Especially for GARS, YARS, AARS and HARS, the evidence in favour of pathogenicity is strong with multiple mutations and families reported, supporting a critical role of ARSs in maintaining peripheral nerve function. Except for KARS mutations leading to a recessively inherited CMT, neuropathies related to the other five ARS genes are autosomal-dominantly inherited and each of the implicated ARSs functions as dimers or tetramers (Ibba and Soll, 2000). These common features imply a dominant-negative mechanism underlying these ARS-related neuropathies (Wallen and Antonellis, 2013).
Here, we report a recurrent p.His257Arg mutation in WARS (MIM *191050), initially identified by whole exome sequencing and segregating in two unrelated autosomal dominant dHMN Taiwanese pedigrees and later found in one additional dHMN family of European ancestry in Belgium. WARS encodes the human cytoplasmic tryptophanyl-tRNA synthetase (TrpRS), which is an essential enzyme responsible for charging tRNAtrp molecules with tryptophan (Shen et al., 2006; Yang et al., 2006) and also possesses a non-canonical effect on angiostasis (Wakasugi et al., 2002; Kise et al., 2004). We have characterized the phenotypic features and functional influences of the WARS mutation and postulate that WARS causes dHMN when mutated.
Materials and methods
Subjects
Eleven individuals of two unrelated Taiwanese families of Han Chinese origin afflicted with a molecularly unassigned autosomal-dominantly inherited dHMN were enrolled into the study, including seven patients and four unaffected individuals (Families A and B in Fig. 1A). An additional 78 unrelated Taiwanese index patients including 73 with molecularly unassigned axonal CMT and five with dHMN were also recruited. Genomic DNA was isolated from peripheral blood leucocytes following a standard protocol. Pathogenic variants in 20 genes known to be associated with dHMN or other hereditary motor neuropathies, including AARS, ATP7A, BICD2, BSCL2, DNAJB2, DYNC1H1, FBLN5, GARS, HARS, HINT1, HSPB1, HSPB3, HSPB8, IGHMBP2, MYH14, PLEKHG5, SBF1, SETX, SLC5A7 and TRPV4, had been excluded in the index patients by a targeted multi-gene next-generation sequencing (Hsiao et al., 2016). The lower limbs of Patient A-III-3 and one healthy control subject were studied with MRI by using a Signa EXCITE 1.5 T system (GE Medical Systems). T1-weighted images (repetition time 517–613 ms, echo time 7.98–8.34 ms, 10-mm slice thickness) were used to look for the presence of muscle atrophy with fatty substitution.
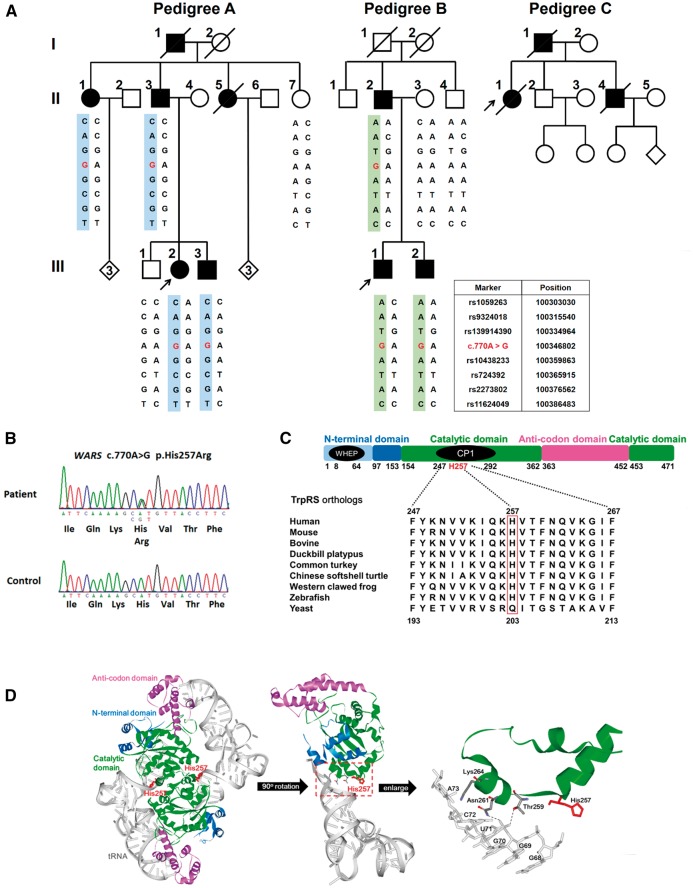
Figure 1.
Genetic analysis of families with distal hereditary motor neuropathy (dHMN). (A) Pedigrees of the two Taiwanese and one Belgian dHMN families harbouring the WARS c.770A > G (p.His257Arg) mutation. Haplotype analysis of seven SNP markers flanking the WARS variant revealed two distinct disease-associated haplotypes in the two Taiwanese families. Open symbol: unaffected; filled symbol: affected; symbol with a diagonal line: deceased; arrow: proband. (B) Sanger sequencing traces confirming the c.770A > G (p.His257Arg) variant. (C) Alignment of multiple TrpRS orthologues showing conservation of the His257 residue throughout vertebrate species, but not yeast. The domain diagram of human TrpRS shows that the His257 residue is localized in the connective polypeptide 1 insertion (CP1). WHEP: a helix-turn-helix motif, responsible for protein–protein interaction. (D) The structural model of human TrpRS homodimers (PDB ID: 2AKE) showing that the His257 residue is situated at the dimer interface (left) as well as in the catalytic domain (green) which is close to the recognition sites (Thr259 and Asn261) for the tRNA acceptor arm (grey) (middle and right).
In addition, we screened for mutations in the candidate gene in the index patients of 82 European, 57 Australian and 24 Korean families with molecularly unassigned dHMN, in whom pathogenic variants in genes implicated in inherited neuropathy had been excluded by exome sequencing. The Ethical Review Boards of the participating institutions approved this study. All patients or their legal representatives signed informed consent prior to enrolment.
Genetic analyses
In Family A, exome sequencing was performed in two affected and two unaffected individuals using Agilent SureSelect™ Human All Exon v2 kit for exome enrichment and the Illumina HiSeq2000 platform with a paired-end 2 x100 bp protocol for sequencing. As the dHMN in Pedigree A is inherited in an autosomal dominant manner, only heterozygous variants shared by the two patients but absent in any of the two unaffected individuals were taken for further analyses. Variants present in the Single Nucleotide Polymorphism Database (dbSNP144; https://www.ncbi.nlm.nih.gov/snp), the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2012), Exome Aggregation Consortium (ExAC version 0.3) (Lek et al., 2016), the 24 in-house exome controls, and those that do not alter coding sequences were excluded. Sanger sequencing was then performed to determine whether any of the remaining variants completely co-segregated with dHMN in Pedigree A (Supplementary Table 1). MutationTaster (Schwarz et al., 2014) and Combined Annotation Dependent Depletion (CADD) (Kircher et al., 2014) were used to predict the pathogenicity of the identified variants. Sanger sequencing of the coding exons and the exon-intron boundaries of WARS were performed in additional 79 unrelated Taiwanese index patients with axonal CMT or dHMN, including the one from Family B. Haplotype analysis was conducted in Families A and B by genotyping seven single nucleotide polymorphism (SNP) markers flanking WARS c.770A > G and covering a region of 83.5 kb (Fig. 1A).
Exome sequencing in the additional cohorts of total 163 dHMN index patients was performed using Illumina Nextera Rapid Capture Expanded exome kit, Agilent SureSelect Human All Exon kit v2, 4, 5 or Roche NimbleGen SeqCap EZ v3.0 for exome enrichment and Illumina HiSeq2000 or HiSeq2500 platform for sequencing. The exome data of 58 European and 10 Australian patients with dHMN were imported and reannotated into the GENESIS (Gem.app) platform, a web-based tool for next generation sequencing data analysis (http://thegenesisprojectfoundation.org/) (Gonzalez et al., 2015). The WARS coding variants revealed by exome sequencing were further confirmed by Sanger sequencing.
Expression plasmids
A 1.4 kb PCR-amplified fragment containing the full-length coding region of WARS (NM_004184.3) was cloned into pcDNA3.1/myc-His (Invitrogen) or pEGFP-N1 (Clontech). The CDH5 cDNA clone (BC096363) was purchased from transOMIC. The expression vector for Flag-tagged vascular endothelial-cadherin (VE-cadherin) was generated by subcloning the coding sequence of CDH5 into pFlag-CMV5a (Sigma-Aldrich). For the production of recombinant His-tagged TrpRS in E. coli, the coding sequences of WARS encoding full-length and T2-TrpRS (residues 94-471 of full-length TrpRS) were separately cloned into pQE-30 vector (Qiagen). QuikChange Site-Directed Mutagenesis kit (Stratagene) was used to introduce the c.770A > G (p.His257Arg) mutation into the expression plasmids. All the primers and plasmids used in this study are listed in Supplementary Tables 2 and 3, respectively.
Cell culture, growth and transfection
Human embryonic kidney 293 (HEK293) cells and human SH-SY5Y neuroblastoma cells were grown in Dulbecco’s modified Eagle medium (HyClone) supplemented with 10% foetal bovine serum (FBS; Invitrogen). Mouse Neuro-2a (N2a) neuroblastoma cells were maintained in minimal essential medium (MEM; Invitrogen) supplemented with 10% FBS. Human umbilical vein endothelial cells (HUVECs) were maintained in medium M200 (Invitrogen) containing low serum growth supplement and 5% FBS. Rat ventral spinal cord neurons, extracted and isolated from Wistar rat ventral spinal cords at the 14th embryonic day, were purchased from ScienCell Research Laboratories, grown according to the supplier’s instructions, and used for primary motor neuron culture. Specificity of the motor neurons was attested by immunocytochemistry with antibodies against the motor neuron marker, non-phosphorylated neurofilament H (SMI-32), choline acetyl transferase (ChAT) and islet I. The motor neuron culture medium consisted of neuronal medium (ScienCell) supplemented with neuronal growth supplement, 10 ng/ml brain derived neurotrophic factor (BDNF), 50 ng/ml nerve growth factor (NGF), and 2% FBS. Cells were transfected using Lipofectamine 2000 (Invitrogen) in the following transfection experiments. For endogenous WARS knockdown, small interfering RNAs (siRNAs) specifically targeting the 3’ untranslated region (UTR) of WARS (Supplementary Table 2) were transfected into cells by using Lipofectamine® 2000 (Invitrogen). N2a cells were differentiated by incubation in MEM supplemented with 1 mM dibutyryl cAMP, 10 μM retinoic acid and 1% FBS. For differentiation, SH-SY5Y cells were treated with 10 μM retinoic acid.
Reporter assays for assessing protein synthesis
HEK293 cells were co-transfected with wild-type or mutant TrpRS expression constructs along with beta-galactosidase (β-Gal) reporter plasmid pCMV-lacZ (Clontech) or firefly luciferase reporter plasmid pCMV-Red Firefly Luc (Thermo Scientific). At 48 h after transfection, cell lysates containing equal amounts of protein (2.5 μg) were analysed for β-Gal or luciferase activities by using β-Galactosidase Reporter Gene Activity Detection Kit (Sigma-Aldrich) or Pierce Firefly Luciferase Glow Assay Kit (Thermo Scientific), respectively.
Production and purification of wild-type and His257Arg TrpRSs
Wild-type and His257Arg TrpRSs with a C-terminal in-frame His tag were expressed in E. coli M15 and purified by affinity chromatography with nickel resin according to the manufacturer’s instructions (Novagen). Protein concentration was determined by the Bradford method (Bio-Rad) using bovine serum albumin as a standard.
Aminoacylation activities of wild-type and His257Arg TrpRSs
The aminoacylation activities were estimated by quantifying the release of pyrophosphate and adenosine monophosphate (AMP) during aminoacylation. The reactions were performed in 50 µl of the reaction mixture containing TrpRS proteins (crude protein extract or purified protein) with 100 mM Tris-HCl (pH7.6), 10 mM MgCl2, 40 mM KCl, 1 mM dithiothreotol, 40 U/ml bovine liver tRNA (Sigma-Aldrich), 1 mM ATP, 0.2 mM tryptophan, and 5 U/ml of yeast inorganic pyrophosphatase (Sigma-Aldrich) at 37°C for 1 h. The pyrophosphate produced in the reaction was hydrolysed by the coupling enzyme pyrophasphatase and the free orthophosphates (Pi) were measured in a colormetric assay using BIOMOL® Green reagent (Enzo Life Sciences). The production of AMP was quantified using the AMP-Glo™ Assay (Promega) method with a microplate luminometer (TECAN).
Co-immunoprecipitation analysis
HEK293 cells were co-transfected with constructs to express EGFP-tagged wild-type or His257Arg mutant TrpRS as well as Myc-tagged wild-type or His257Arg mutant TrpRS. Cell extracts were prepared 48 h post-transfection using Pierce IP Lysis Buffer (Thermo Scientific). Except for the 10% of sample volume loaded for input, the lysates were incubated with anti-Myc antibody-conjugated Dynabeads (Invitrogen) at 4°C overnight with rotation. After washing with RIPA buffer, bound proteins were eluted in SDS loading buffer at 95°C for 5 min and the eluates were subjected to western blotting with antibodies against TrpRS. Antibodies used in this study are summarized in Supplementary Table 4.
Cell viability assay
At 24 h after transfection, the HEK293 cells transfected with different TrpRS constructs were treated with WARS siRNAs (25 nM) specifically targeting the 3’UTR (Supplementary Table 2). After incubation for another 24 h, cell viability of the transfected cells was examined using the Cell Counting Kit-8 (CCK-8) assay (Sigma-Aldrich).
Neurite outgrowth assays and immunocytochemistry
Cells were grown on coverslips precoated with poly-l-lysine (100 μg/ml) and laminin (50 μg/ml) and transfected with wild-type or His257Arg TrpRS expression construct. Sixteen hours after transfection, the N2a cells or SH-SY5Y cells were induced to differentiate for 48 h and the neurite outgrowth phenotypes were observed under a phase-contrast light microscope (PrimoVert, Zeiss) prior to immunostaining. The differentiation step was omitted for rat primary neuron cells but culture medium was changed to remove residual Lipofectamine®. For immunocytochemistry, cells were fixed in 4% paraformaldehyde, permeabilized with 0.2% Tween-20 and blocked with 5% bovine serum albumin before incubation in primary antibody overnight at 4°C (Supplementary Table 4). Bound primary antibodies were detected using Alexa-conjugated secondary antibodies. Images were taken with an Olympus FluoView FV10i confocal laser scanning fluorescence microscopy system. Transfected cells bearing neurite-like extensions with length of at least one cell body were counted and scored as the percentage of total transfected cells. The determination of the average lengths of primary neurites was performed through the analysis of microscopic images using the Image-Pro PLUS (Meyer Instruments) measurement tool.
Yeast complementation assays
For details on yeast complementation assays, see the online Supplementary material.
In vitro pull-down assay
Protein pull-down experiments were performed using Pierce Pull-Down PolyHis Protein:Protein Interaction Kit (Thermo Scientific) according to the manufacturer’s instructions. In brief, His-tagged TrpRS was expressed in E. coli M15 and purified by Ni-NTA affinity chromatography. Bead-bound His-tagged TrpRS (40 nM) was mixed with equal amounts of lysates (2 mg) of the transfected HEK293 cells expressing Flag-tagged VE-cadherin, and then incubated for 2 h with 5 mM CaCl2. Pull-down mixtures and inputs were analysed by western blotting.
In vitro angiogenesis assays
HUVEC migration assays were performed using 24-well Millicell inserts (8 μm pore size; Merck Millipore) with fibronectin coating. HUVECs were seeded on the upper chamber with 105 cells per well, while vascular endothelial growth factor165 (VEGF165) (10 ng/ml) was added in the lower chamber. For inhibition assays, recombinant TrpRS (500 nM) was added to both upper and lower chambers and HUVECs were allowed to migrate at 37°C, 5% CO2 for 16 h. Migrant cells were fixed, stained with crystal violet, photographed at × 100 magnification, and counted in three random views. For Matrigel® angiogenesis assay, HUVECs were seeded in Matrigel® (BD Biosciences)-coated 24 well plates at a density of 8.5 × 104 cells/well and treated with VEGF165 (10 ng/ml). After incubation with recombinant TrpRS (500 nM) for 16 h, HUVECs were microscopically evaluated for tube formation under 40 × magnification. An average of vessel coverage (the total microscopic area covered by HUVEC vasculature) from four random fields in each well was calculated by ImageJ software (NIH).
Results
Exome sequencing and in silico analyses
We initially studied a three-generation Taiwanese family of Han Chinese origin afflicted with autosomal dominant dHMN, of which four affected individuals and two unaffected ones were enrolled (Pedigree A in Fig. 1A). Whole exome sequencing was performed in two affected patients (Patients A-II-1 and A-III-2) and two unaffected individuals (Subjects A-II-7 and A-III-1). On average, 23 Gb of sequence data were generated from each individual, resulting in an average coverage depth of 206× per targeted base (Supplementary Table 1). After analysis and filtering, four heterozygous coding variants were found to be shared by both affected individuals but not by either of the unaffected individuals and were not present in dbSNP Build 144, 1000 Genome Project, ExAC version 0.3, or the 24 in-house exome controls. Sanger sequencing of the four variants in all six individuals of Pedigree A showed that only one variant, c.770A > G (p.His257Arg) in WARS (RefSeq NM_004184.3) (Fig. 1B), perfectly segregates with the dHMN phenotype.
To ascertain the pathogenicity of WARS c.770A > G (p.His257Arg), this mutation was screened and not found in 1112 Taiwanese healthy controls (2224 chromosomes). This mutation is also absent in the genome Aggregation Database (gnomAD; www://gnomad.broadinstitute.org). In addition, it was predicted to be a disease-causing mutation by two bioinformatics programs, MutationTaster and CADD. The CADD v1.2 Phred score for this mutation was 22.3, ranking WARS p.His257Arg in the top 0.59% most deleterious variants in the genome (Kircher et al., 2014). Mutation Taster showed that this variant seemed to be disease-causing with a strong probability value, 0.99999998 (Schwarz et al., 2014). The His257 residue of TrpRS protein is evolutionarily conserved from human to zebrafish (Fig. 1C).
Human TrpRS consists of 471 amino acids, containing an N-terminal domain (residues 1–153), two catalytic domains (residues 154–362 and 453–471) and a C-terminal anticodon-binding domain (residues 363–452) (Fig. 1C) (Shen et al., 2006; Yang et al., 2006). From a structural viewpoint, human TrpRS functions as a homodimer and the His257 residue is situated within the connective polypeptide 1 (CP1) insertion (residues 247–292) of the catalytic domain (Fig. 1C). CP1 is known to play an essential role in dimer interface interaction and forms the substrate-binding pocket to grasp tRNA acceptor arm (Shen et al., 2006). The His257 is very close to residues Thr259 and Asn261, which are directly involved in the recognition and binding of the tRNA acceptor arm (Fig. 1D) (Shen et al., 2006; Yang et al., 2006). These bioinformatic approaches support that WARS p.His257Arg may compromise the enzymatic activity of human TrpRS.
WARS p.His257Arg is a recurrent mutation in dHMN pedigrees
To further investigate the pathogenic role of WARS mutations in inherited neuropathies, we screened additional 79 unrelated Taiwanese individuals, including 73 with molecularly unassigned axonal CMT and six with dHMN, and the same c.770A > G mutation accompanied with WARS c.1327G > T (p.Ala443Ser) in cis was found in one family with dHMN (Pedigree B; Fig. 1A). Among the five members of Pedigree B, WARS (c.770A > G; c.1327G > T) was present in all three affected individuals. One unaffected individual of Pedigree B carried only WARS c.1327G > T without WARS c.770A > G (Subject B-II-4; Fig. 1A). WARS c.1327G > T variant, denoted as rs139914390, may be a benign polymorphism because it had been previously identified in 1 of 104 southern Han Chinese samples and 1 of 103 Japanese samples in the 1000 Genome Project. Sanger sequencing demonstrated that WARS c.1327G > T variant was not present in Pedigree A (Fig. 1A), suggesting that WARS c.[770A>G; 1327G>T] mutation in Pedigrees A and B originated from different ancestors. Haplotype analysis using seven SNP markers flanking the p.His257Arg mutation revealed two distinct haplotypes (Fig. 1A). These findings strongly suggested that this identical mutation has independent origins in the two pedigrees and provided further evidence for the pathogenic role of WARS c.770A > G (p.His257Arg) mutation in dHMN.
We further checked other dHMN cohorts from different populations to affirm that a WARS mutation is indeed a cause of dHMN. In 163 unrelated index cases from Australia, Korea and the GENESIS collection in the USA, we identified one additional family of European ancestry with the same WARS c.770A > G (p.His257Arg) mutation in Antwerp, Belgium (Pedigree C; Fig. 1A).
Clinical features
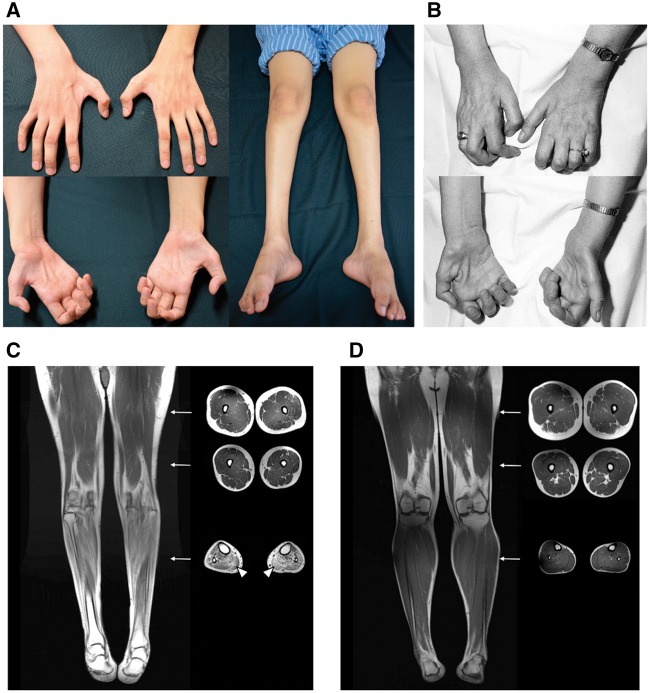
Among the two Taiwanese dHMN pedigrees, Subjects B-I-1 and B-I-2 both passed away at octogenarian age without distal limb muscle atrophy or weakness during their lifetime. Patient A-II-1 has an overt dHMN phenotype with disease onset at 12 years of age but refused to have a clinical evaluation. The clinical manifestations and electrophysiological features of the other six affected individuals in the two pedigrees harbouring WARS c.770A > G (p.His257Arg) mutation are similar (Table 1). All affected individuals have a juvenile-onset, slowly progressive, distal predominant, pure motor neuropathy. The average age of disease onset was 12.2 ± 1.2 years (range: 10–13). The affected individuals had normal developmental milestones at early childhood but developed muscle weakness and atrophy in the feet/legs followed by hand muscle involvement. The symptoms were mainly restricted to distal limbs and there was no subjective sensory complaint. Except for Patient A-II-3, who was wheelchair-bound by his fifties, the other five affected individuals could walk without assistance. All the affected individuals had high arches, a stork leg appearance and a variable degree of hand muscle atrophy and weakness (Fig. 2A). Paralysis and profound atrophy of intrinsic hand muscles were noted in the two older affected individuals (Patients A-II-3 and B-II-2). Nerve conduction studies showed a pure motor axonal neuropathy with preserved conduction velocities and no sensory involvement. Skeletal muscle MRI demonstrated that the affected Patient A-III-3 had widespread muscle atrophy with fatty infiltration in the leg muscles, but the muscles in the thighs were spared (Fig. 2C). This finding is concordant with the clinical observation of predominantly distal limb involvement in all the affected individuals. See Supplementary Video 1 for a clinical video of Patient A-III-3.
Table 1.
Clinical manifestations of the affected individuals carrying WARS p.His257Arg
| Pedigree A | Pedigree B | Pedigree C | |||||
|---|---|---|---|---|---|---|---|
| Subject | A-II-3 | A-III-2 | A-III-3 | B-II-2 | B-III-1 | B-III-2 | C-II-1 |
| Age at onset (years) | 12 | 10 | 13 | 12 | 13 | 13 | 9 |
| Age at clinical evaluation (years) | 64 | 25 | 30 | 58 | 22 | 20 | 45 |
| Gender | Male | Female | Male | Male | Male | Male | Female |
| Limb weakness UL (MRC) | Distal: 0 | Distal: 4 | Distal: 3 | Distal: 0 | Distal: 3 | Distal: 3 | Distal: 0 |
| Limb weakness LL (MRC) | Distal: 0 | Distal: 1 | Distal: 3 | Distal: 0 | Distal: 3 | Distal: 3 | Distal: 0 |
| Muscle atrophy UL | Distal: severe | Distal: mild | Distal: moderate | Distal: severe | Distal: mild | Distal: mild | Distal: severe |
| Muscle atrophy LL | Distal: severe | Distal: moderate | Distal: moderate | Distal: severe | Distal: moderate | Distal: moderate | Distal: severe |
| Pinprick sensation | Normal | Normal | Normal | Normal | Normal | Normal | Normal |
| Knee/ankle DTRs | Absent/absent | +/absent | Absent/absent | Absent/absent | Absent/absent | +/absent | Weak/absent |
| Age at NCS (years) | 64 | 25 | 13 | 50 | 14 | 13 | 45 |
| Ulnar nerve MNCV (m/s) | 46 | 55.6 | 57.9 | NR | 51.8 | 56.8 | 63 |
| Ulnar nerve cMAP (mV) | 0.1 | 5.3 | 5.2 | NR | 5.6 | 5.9 | 0.9 |
| Sural nerve SNAP (µV) | 17.0 | 46.0 | 34.0 | 8.9 | 13.2 | 11.9 | 11.0 |
cMAP = compound motor action potential; DTR = deep tendon reflex; LL = lower limbs; NCS = nerve conduction studies; MNCV = motor nerve conduction velocity; MRC = Medical Research Council scale; ND = not done; NR = not recordable; SNAP = sensory nerve action potential; UL = upper limbs.
Figure 2.
Clinical and muscle MRI features in patients with WARS p.His257Arg mutation. (A) Atrophy of the intrinsic hand muscles and muscles in the feet and legs seen in Patient A-III-3. (B) Atrophy of the intrinsic hand muscles seen in Patient C-II-1. (C) T1-weighted MRI of Patient A-III-3 revealing distal lower limb muscle atrophy with fatty infiltration (arrowheads). (D) T1-weighted lower limb MRI of a 30-year-old healthy male for comparison. Arrow: corresponding cross-section level of the axial view images.
The female index patient (Patient C-II-1) of the Belgian dHMN family has a disease onset at 9 years with distal weakness in the lower limbs, markedly progressive from the age of 20 years onwards, also with weakness in the hands. At the age of 45 years, there was a severe distal weakness in all limbs and no sensory involvement (Fig. 2B). Nerve conduction studies revealed a pure motor axonal neuropathy (Table 1). Sural nerve biopsy at the age of 32 years revealed normal pattern and muscle biopsy at the same time was compatible with neurogenic atrophy. The proband’s father and younger brother (both deceased at the time of evaluation) were reported to have a similar clinical presentation. Additional family members were not available for segregation studies. The patient died at the age of 66 years due to illness unrelated to the dHMN phenotype.
His257Arg TrpRS compromises protein synthesis in a dominant-negative manner
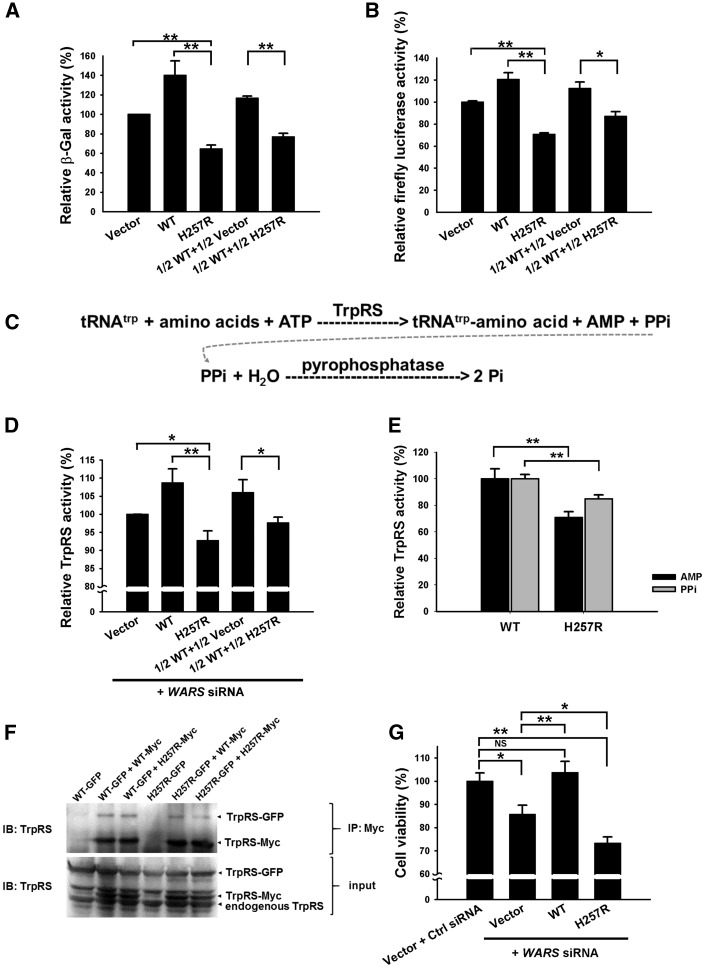
As TrpRS catalyses tRNA aminoacylation, which is an essential step of the translation process, we constructed wild-type and His257Arg mutant TrpRS expression plasmids to assess the effect of this mutation on protein synthesis. Two distinct reporter assays were used to examine the influence of this mutation on each reporter protein. We first analysed the β-Gal activities in HEK293 cells co-transfected with β-Gal reporter plasmids along with wild-type or mutant TrpRS expression construct. As shown in Fig. 3A, the transfectants expressing His257Arg TrpRS had a decreased β-Gal activity in comparison to those expressing wild-type TrpRS. Interestingly, the β-Gal activity in cells expressing mutant TrpRS was noticeably lower than that in cells without exogenous TrpRS (i.e. cells transfected with empty vector). Moreover, the β-Gal activity was lower in HEK293 cells transfected with half-dose of wild-type and half-dose of mutant TrpRS expression plasmids than those transfected with half-dose of wild-type TrpRS expression plasmid only (Fig. 3A). These findings suggested that the exogenous His257Arg mutant TrpRS had a dominant-negative effect to disturb the functions of both endogenous and exogenous wild-type TrpRSs. Similar results were obtained when experiments were performed using firefly luciferase as a reporter (Fig. 3B). The differences of the reporter activities between the studied cell groups appeared to be more evident in β-Gal assay than those in firefly luciferase assay given that β-Gal protein contains a higher proportion of tryptophan than firefly luciferase dose (3.8% versus 0.36%, respectively). Both assays supported that WARS p.His257Arg mutation possesses a dominant-negative effect on protein synthesis.
Figure 3.
In vitro characterization of wild-type and His257Arg (H257R) mutant TrpRS proteins. (A) β-Gal reporter assay and (B) firefly luciferase reporter assay demonstrating that H257R TrpRS has a dominant-negative effect on protein synthesis. HEK293 cells co-transfected with β-Gal or firefly luciferase reporter plasmids, along with different ratios of wild-type and H257R TrpRS expression plasmids were lysed and assayed for β-Gal or firefly luciferase activities at 48 h after transfection. The error bars indicate standard errors of the mean (n = 3) and asterisks indicate statistically significant differences (**P < 0.01, *P < 0.05). Vector: empty vector control. (C) Aminoacylation of tRNAtrp mediated by TrpRS generating adenosine monophosphate (AMP) and pyrophosphate (PPi), which can be used to assess aminoacylation activities. (D) Analyses of aminoacylation activities of TrpRS in HEK293 cells. Cells with different ratios of wild-type and mutant TrpRS constructs as well as the siRNAs specifically depleting endogenous WARS were lysed at 48 h post-transfection, and assayed for aminoacylation activities by assessing the amount of generated pyrophosphate. The data were displayed relative to the values detected in the control vector-transfected cells (n = 4). (E) Analyses of aminoacylation activities of purified TrpRSs. His-tagged wild-type and H257R TrpRS proteins were produced in E. coli and purified by Ni-NTA affinity chromatography. Both pyrophosphate and AMP generated through aminoacylation were quantified to estimate the aminoacylation activities (n = 3). (F) Investigating the dimerization properties of wild-type and H257R TrpRSs by co-immunoprecipitation (Co-IP) assay. HEK293 cells co-transfected with Myc- or/and GFP-tagged wild-type or/and H257R TrpRS expression plasmids were lysed at 48 h post-transfection. Dimerization was demonstrated via co-immunoprecipitation pull-downs of GFP-tagged wild-type or H257R TrpRS with the Myc-tagged TrpRS using anti-Myc antibody. These data showed that the H257R TrpRS could form dimers with wild-type TrpRS in cells. (G) Cell viability assays. Knockdown of endogenous WARS in HEK293 cells resulted in a significantly compromised cell viability, which could be rescued by expressing exogenous wild-type TrpRS but worsened by H257R TrpRS. Percentage of viable cells was displayed relative to cells transfected with empty vector along with non-targeting siRNA control (Ctrl siRNA) (n = 7). WT = wild-type.
His257Arg TrpRS has an impaired aminoacylation activity
The negative effect on protein synthesis might be attributed to the defective aminoacylation activity of mutant TrpRS. To test this hypothesis, the aminoacylation activities were analysed in cell lysates of HEK293 cells co-transfected with different combinations of wild-type and mutant TrpRS expression plasmids along with 25 nM of the WARS siRNAs knocking down∼70% of endogenous TrpRS in HEK293 cells (data not shown). The aminoacylation reaction was simulated in vitro using bovine tRNA as substrate, and the TrpRS activity was estimated by measuring the production of reaction byproducts—AMP and pyrophosphate (the schematic equation is shown in Fig. 3C) (Jordanova et al., 2006; Cestari and Stuart et al., 2013).
We first tested the TrpRS aminoacylation activity via measuring pyrophosphate concentration from lysates of the transfected HEK293 cells. Similar to the results obtained from the β-Gal or firefly luciferase experiments, the TrpRS aminoacylation activities were lowest in the cells transfected with His257Arg mutant construct only, followed by those transfected with half-dose of wild-type and half-dose of mutant constructs, and then those transfected with half-dose of wild-type construct only (Fig. 3D). The highest enzyme activity was observed in the cells over-expressing wild-type TrpRS.
Meanwhile, to specifically assess the enzymatic activity of mutant TrpRS, we purified recombinant His-tagged TrpRS proteins from E. coli. This cell-free assay also demonstrated that His257Arg mutant TrpRS has a significantly lower aminoacylation activity than wild-type TrpRS, no matter which measurement of aminoacylation activity (AMP or pyrophosphate) was used (Fig. 3E). These findings indicated that WARS p.His257Arg mutation has a direct and dominant-negative effect to compromise the aminoacylation activity of TrpRS, which subsequently perturbs protein synthesis.
His257Arg TrpRS retains the ability to form dimers with wild-type TrpRS
Since TrpRS functions as a homodimer (Fig. 1D), it would be intriguing to know whether the His257Arg mutation alters the dimerization properties of TrpRS. Co-immunoprecipitation experiments performed with HEK293 cells expressing EGFP-tagged wild-type or His257Arg mutant TrpRS as well as Myc-tagged wild-type or His257Arg mutant TrpRS revealed that the mutant TrpRS retained the ability to form dimers with wild-type proteins (Fig. 3F). The heterophilic association between the mutant and wild-type molecules might be a key precondition for the dominant-negative effect of WARS p.His257Arg mutation.
His257Arg TrpRS leads to attenuated cell viability
The mutant TrpRS may have a deleterious effect on cell viability due to insufficient protein synthesis. Therefore, we investigated the viability of HEK293 cells transfected with different TrpRS constructs and WARS siRNAs against endogenous TrpRS expression. Compared to cells transfected with empty vector and non-targeting siRNA control, knockdown of the endogenous TrpRS expression by the siRNAs significantly attenuated the cell viability, which could be rescued by expressing exogenous wild-type TrpRS but not His257Arg mutant TrpRS (Fig. 3G). Notably, cells expressing exogenous mutant TrpRS had a poorer viability than those transfected with empty vector, indicating that this mutation has a toxic effect rather than simply haplo-insufficiency. These cellular experiments clearly demonstrated a dominant-negative effect of WARS p.His257Arg mutation on cell viability.
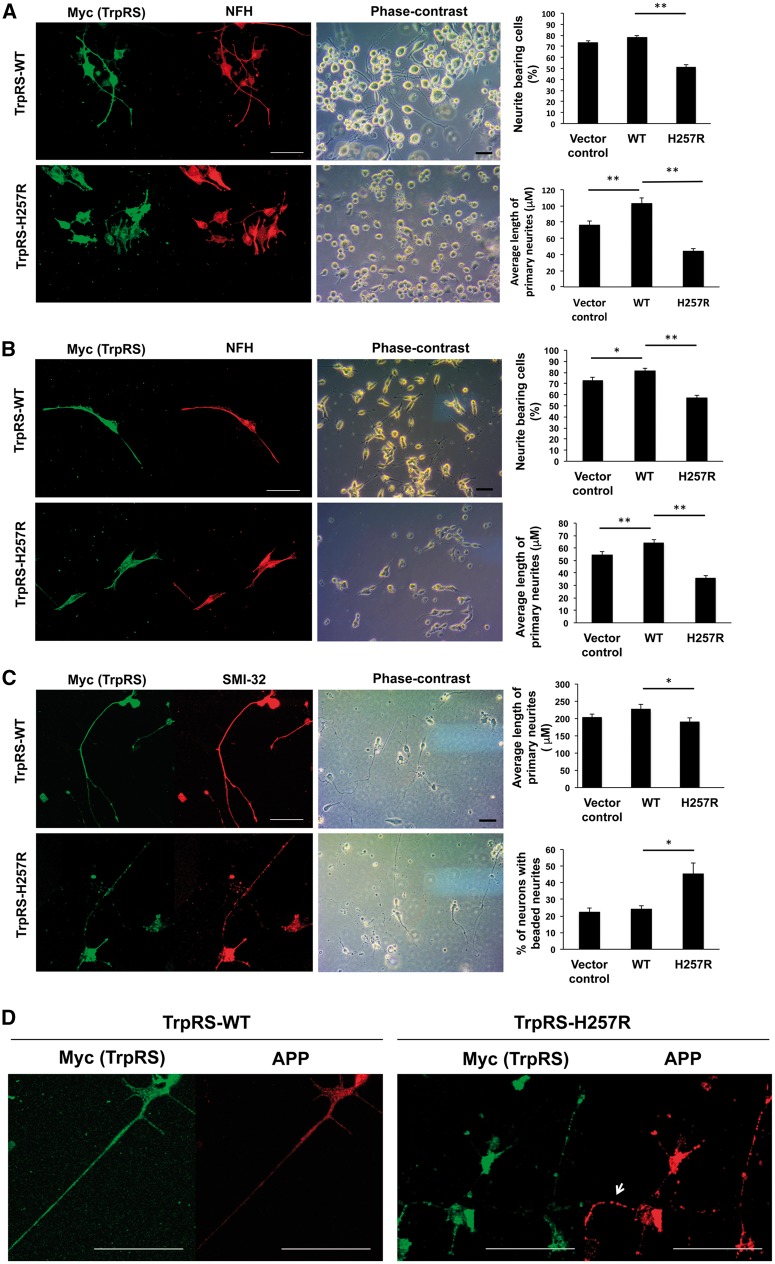
His257Arg TrpRS inhibits neurite outgrowth and leads to neurite degeneration
To investigate the effects of mutant TrpRS on neuronal cell differentiation, N2a cells were transiently transfected to overexpress wild-type or His257Arg TrpRS under retinoic acid-induced differentiated conditions. At 72 h after transfection, cells expressing wild-type TrpRS or those transfected with empty vectors (control cells) both had extensive neurite formation, and wild-type TrpRS overexpression further enhanced neurite outgrowth (Fig. 4A). Conversely, in cells expressing His257Arg mutant TrpRS, we observed a noticeable reduction in both the number of cells bearing neurite and neurite length, compared with cells expressing wild-type TrpRS (Fig. 4A). Similar results were obtained when TrpRS constructs were expressed in SH-SY5Y cells (Fig. 4B).
Figure 4.
His257Arg (H257R) TrpRS inhibits neurite outgrowth and leads to neurite degeneration. Neuro-2a (N2a) (A) or SH-SY5Y (B) cells were transfected with expression vector containing wild-type (WT) or H257R TrpRS or empty vector (vector control), grown under differentiation conditions, and immunostained against Myc (exogenous TrpRS staining) and neurofilament heavy polypeptide (NFH; neurite staining) at 72 h post-transfection. The representative fluorescence-immunostaining and phase-contrast images were shown, along with statistical results of per cent of cells bearing neurites and average primary neurite length. Scale bar = 50 μm. At least 100 cells from three independent experiments were measured for each preparation and data were expressed as the mean ± standard error of the mean (SEM). *P-value < 0.05; **P-value < 0.01 when compared to wild-type. (C) Rat motor neurons (embryonic Day 14) were transfected with expression vector containing wild-type or H257R TrpRS or empty vector, and processed for immunostaining using antibodies against Myc and SMI-32 (staining for motor neurons and their neurites) at 72 h post transfection. The representative fluorescence-immunostaining and phase-contrast images were shown. Neurites of motor neurons were traced and measured, and the average primary neurite length and per cent of cells bearing beaded neurites were estimated. Error bars represent the mean ± SEM of three independent experiments with at least 50 cells. (D) Rat motor neurons expressing above TrpRS constructs were processed for immunofluorescence analysis using antibody against amyloid precursor protein (APP; neurodegeneration staining) at 72 h post-transfection. Motor neurons expressing wild-type TrpRS showed only a faint or punctate staining of APP in the neurites, while motor neurons expressing TrpRS appeared to accumulate APP at neurites (arrow).
We then examined the effect of His257Arg TrpRS on neurite extension utilizing primary culture of rat embryonic motor neurons. Concordant to the results from the neuronal cell lines, motor neurons transfected with mutant TrpRS construct had shorter neurites than those expressing wild-type TrpRS or those transfected with empty vectors (control cells) (Fig. 4C). Interestingly, as judged by SMI-32 neurofilament immunostainings, we found that motor neurons expressing mutant TrpRS had prominent degenerative changes in the form of irregular swelling and beading of neurites, while this was not the case in cells expressing wild-type TrpRS or control cells (Fig. 4C). To further determine whether these abnormal phenotypes could be a sign of axonal degeneration, wild-type or mutant TrpRS-transfected motor neurons were stained with antibodies against amyloid precursor protein (APP), a marker widely used to denote neurodegeneration (Irobi et al., 2010). Motor neurons expressing wild-type TrpRS showed a faint or punctate APP staining (Fig. 4D), indicative of normal and active axonal transport, comparable to that observed in control cells. For mutant TrpRS-expressing motor neurons, there was abundant accumulation of APP in the neurites, which might occur as a consequence of neurite disruption and impaired axonal transport (Fig. 4D). These data suggested that mutant TrpRS compromises neurite growth in primary motor neurons and leads to neurite degeneration.
Yeast complementation assays and aminoacylation activities with yeast tRNA
Human full-length wild-type TrpRS failed to complement wrs1 deficiency in yeast cells and had a defective aminoacylation activity using yeast tRNA as substrates, whereas His257Arg TrpRS could partially complement wrs1 deficiency in yeast cells and had a significantly higher aminoacylation activity with yeast tRNA (Supplementary Fig. 1B–D). These findings support that WARS His257Arg mutation may significantly change the structure of human TrpRS so that the mutant TrpRS acquires exceptional catalytic ability toward yeast tRNATrp and can partially complement wrs1 deficiency in yeast cells (see Supplementary material for details).
His257Arg TrpRS acquires an enhanced angiostatic effect
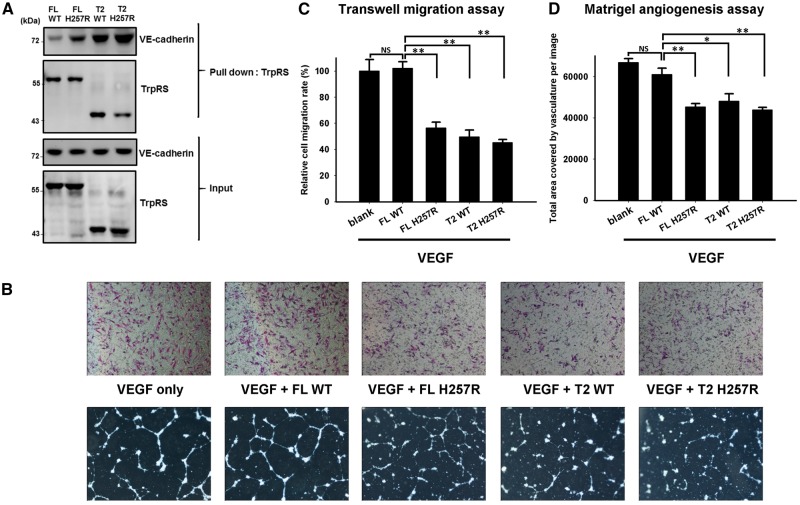
T2-TrpRS, generated by proteolysis of full-length TrpRS in physiological condition, has a potent angiostatic activity through binding to VE-cadherin (Tzima et al., 2005; Zhou et al., 2010) and inhibiting vascular endothelial growth factor (VEGF)-induced angiogenesis (Wakasugi et al., 2002; Yang et al., 2004). A previous study showed that the binding affinity between VE-cadherin and full-length TrpRS was only 5% of that between VE-cadherin and T2-TrpRS (Zhou et al., 2010). However, removal of the N-terminal WHEP domain exposes the essential residues for cell signalling and gives short forms of TrpRS (T2-, T1-, and mini-TrpRSs) an additional angiostatic function (Kise et al., 2004; Yang et al., 2004). As His257Arg mutant TrpRS showed a similar, even augmented effect akin to mini- and T1-TrpRSs in yeast complementation experiments (Supplementary material), we performed pull-down assays to investigate whether WARS p.His257Arg mutation altered the binding affinity between TrpRS and VE-cadherin. As shown in Fig. 5A, the p.His257Arg mutation significantly enhanced the interaction between VE-cadherin and full-length TrpRS as well as the bindings between VE-cadherin and T2-TrpRS. To examine whether the mutant TrpRS had an augmented angiostatic activity than wild-type TrpRS, transwell migration and Matrigel® angiogenesis assays were performed using HUVECs under VEGF165 stimulation (Wakasugi et al., 2002; Zhou et al., 2010). The VEGF-induced migration of HUVECs was significantly reduced in cells treated with mutant full-length TrpRS, mutant T2-TrpRS, and wild-type T2-TrpRS proteins, but not by wild-type full-length TrpRS (Fig. 5B and C). Matrigel® angiogenesis assay showed a similar pattern of tube formation between HUVECs treated with VEGF165 only and those treated with VEGF165 plus wild-type full-length TrpRS protein, supporting that full-length TrpRS had no or minimal angiostatic effect (Fig. 5B and D). However, mutant full-length TrpRS, mutant T2-TrpRS, and wild-type T2-TrpRS significantly inhibited angiogenesis of HUVECs in comparison to those treated with wild-type full-length TrpRS. These findings indicated that WARS p.His257Arg mutation had an enhanced angiostatic effect.
Figure 5.
Pull-down assay and in vitro angiogenesis assays showing that human His257Arg (H257R) mutant TrpRS has an augmented angiostatic effect through binding to VE-cadherin. (A) An in vitro pull-down assay used to assess the ability of His-tagged TrpRS to bind to Flag-tagged VE-cadherin. The immunoblots showed that both the full-length (FL) and the T2-H257R TrpRSs had a substantial increase in affinity to VE-cadherin compared to wild-type (WT) TrpRS. (B) Effect of the p.His257Arg mutation on the angiostatic activity of TrpRS evaluated by transwell migration assay and Matrigel angiogenesis assay using HUVECs under 16 h of VEGF165 stimulation with or without TrpRS treatment. Top: Representative microscopic images of the underside of the transwell assay membranes; Bottom: representative microscopic images of tube formation on Matrigel®. (C) Quantification of migration of HUVECs after 16 h treatment with VEGF165 and the indicated conditions of three independent experiments. Treatment with full-length H257R TrpRS, T2-H257R TrpRS, or T2-wild-type TrpRS significantly reduced in vitro cell mobility, while full-length wild-type TrpRS application did not further inhibit the cell migration compared to a control treatment of VEGF165 alone. The error bars represent standard errors of the mean (n = 3) and asterisks indicate statistically significant differences (**P < 0.01, *P < 0.05, NS = non-significant). (D) Quantification of vessel coverage (the total microscopic area covered by HUVEC vasculature) after 16 h treatment with VEGF165 and the indicated conditions of three independent experiments. The data revealed that full-length H257R TrpRS, T2-wild-type or T2-H257R TrpRS could significantly inhibit angiogenesis. Treatment of full-length wild-type TrpRS did not show inhibitory effect on tube formation.
Discussion
We identified a novel heterozygous mutation, WARS p.His257Arg, in seven individuals with dHMN from two Taiwanese families of Han-Chinese extraction but with different ancestries demonstrated by haplotype analysis. In addition, the same mutation was also identified in a Belgian kinship of Caucasian origin through screening 163 unrelated dHMN cases from Korea, Australia and the GENESIS collection in the USA. The WARS c.770A > G (p.His257Arg) mutation is not present in 1122 Taiwanese control subjects as well as in the gnomAD, which contains ∼126 000 exome sequences and 15 000 whole genomes from unrelated individuals (Lek et al., 2016). The recurrence of WARS c.770A > G (p.His257Arg) in dHMN patients with different genetic backgrounds and its absence from a large number of controls provide compelling evidence for the pathogenic role of WARS mutations in dHMN. Moreover, this study reveals that the WARS c.770A > G (p.His257Arg) mutation does not affect protein dimerization but possesses a damaging and dominant-negative effect on aminoacylation activity of TrpRS, which subsequently disturbs protein synthesis and reduces cell viability. The expression of mutant His257Arg TrpRS also inhibited neurite outgrowth and led to neurite degeneration in neuronal cell lines and rat motor neurons. Furthermore, we showed that the p.His257Arg mutation significantly changes the properties of TrpRS to facilitate normal function of yeast translational machinery and acquires an enhanced angiostatic activity. This study clearly demonstrates that WARS c.770A > G (p.His257Arg) mutation causes dHMN and alters the canonical and non-canonical functions of TrpRS.
The alteration of canonical and non-canonical TrpRS functions may both contribute to dHMN pathology. His257Arg TrpRS has a reduced aminoacylation activity, which results in disturbed protein synthesis and defective cell viability. The impaired aminoacylation activity may cause secondary neurotoxicity since a large volume of uncharged tRNAs would interfere with the function of eukaryotic initiation factor 2 (eIF2) and subsequently lead to a general suppression of protein translation (Dong et al., 2000; Wallen and Antonellis, 2013). Analogously, an impaired translation efficacy that fails to meet the high metabolic demand of protein synthesis in long axons has also been implicated in inherited neuropathies caused by mutations in other ARS genes (Jordanova et al., 2006; McLaughlin et al., 2012; Griffin et al., 2014). Multiple pathogenic mechanisms that are not mutually exclusive could jointly contribute to axonal degeneration. A loss-of-function effect has been documented in majority of disease-associated ARS mutations. However, haplo-insufficiency solely does not warrant the development of neuropathy. For example, mice heterozygous for a complete null allele caused by a gene-trap insertion in the second intron of Gars (GarsXM256/+) did not develop neuropathy (Seburn et al., 2006). Our study shows a dominant-negative toxicity of the WARS His257Arg mutation. Similar findings have been reported in neuropathies related to mutations in GARS and HARS (Brozkova et al., 2015; Malissovas et al., 2016). The ability of mutant His257Arg TrpRS to form dimers with wild-type TrpRS could be an important prerequisite for its dominant-negative effect. Recently, a zebrafish model illustrated that dimerization is required for the dominant GARS mutations to manifest a neuromuscular phenotype. Zebrafishes heterozygous for the Gars p.Thr209Lys mutation, which resulted in loss of dimerization, did not have any phenotype (Malissovas et al., 2016).
In addition, the enhanced angiostatic effects of His257Arg mutant full-length and T2-TrpRSs may interfere with the original neurotrophic effects of angiogenic factors, including regulating the growth, migration and survival of neurons, as well as guidance of the axonal outgrowth (Greenberg and Jin, 2005; Mackenzie and Ruhrberg, 2012). Under physiological conditions, short TrpRSs are the major forms that bind to VE-cadherin and inhibit the expression of downstream pro-angiogenic genes (Tzima et al., 2005; Zhou et al., 2010). However, the WARS c.770A > G (p.His257Arg) mutation enables and enhances the bindings of both full-length and T2-TrpRSs to VE-cadherin leading to inhibition of angiogenesis and migration of HUVECs. Similarly, GARS mutations which antagonize the physiological signalling between VEGF and neuropilin-1 were found to play a pathogenic role in CMT type 2D (CMT2D) and over-expression of VEGF improved the motor deficit of a CMT2D mouse model (He et al., 2015). These findings collectively support a link between derangement of angiogenesis pathway and axonal neuropathy.
The molecular epidemiology of dHMN in Taiwan remains obscure, and there was only one previous dHMN study in Taiwan, in which a dHMN patient was reported with a BSCL2 c.287G > A (p.Arg96His) mutation (Hsiao et al., 2016). In contrast, dHMN has been well studied in Caucasian populations and clinically categorized into seven subgroups (Harding, 2005). The patients harbouring the WARS c.770A > G (p.His257Arg) mutation manifest a slowly progressive, lower limb onset, length-dependent motor neuropathy starting at age 9–13 years. Such clinical features conform to dHMN type I, which presents as juvenile onset, autosomal-dominantly inherited, lower limbs predominant and pure motor neuropathy (Harding, 2005). Among the other ARS genes implicated in inherited neuropathy, AARS, HARS and GARS may also result in dHMN type I when mutated (Dubourg et al., 2006; Zhao et al., 2012; Brozkova et al., 2015). In addition to dHMN type I, the clinical phenotypes associated with these ARS genes include axonal CMT, intermediate CMT, and dHMN type V, which features an upper-limb predominant motor syndrome (Antonellis et al., 2003; Jordanova et al., 2006; Latour et al., 2010; Brozkova et al., 2015). Why different alleles of these functionally similar genes result in variable phenotypes is still unclear. Mutations in AARS, DYNC1H1, GARS, HARS, HSPB1 and HSPB8 have been identified to cause dHMN type I (Evgrafov et al., 2004; Rossor et al., 2012), and WARS mutations should also be considered in dHMN type I patients with unknown genetic diagnosis.
In conclusion, this study demonstrates a recurrent WARS mutation causing autosomal dominant dHMN and altering the canonical aminoacylation activity and non-canonical angiostatic functions of TrpRS. These findings establish WARS as a gene whose mutations can cause dHMN and extend our knowledge of the pathogenic role of tRNA-synthetases in inherited neuropathies.
Supplementary Material
Acknowledgements
We would like to thank the patients who participated in this study. We thank the High-throughput Genome and Big Data Analysis Core Facility of National Core Facility Program for Biotechnology, the Ministry of Science and Technology, Taiwan (MOST104-2319-B-010-001), for the NGS service and bioinformatics support.
Glossary
Abbreviations
- ARS
aminoacyl-tRNA synthetase
- β-Gal
beta-galactosidase
- CMT
Charcot-Marie-Tooth disease
- dHMN
distal hereditary motor neuropathy
- HUVEC
human umbilical vein endothelial cell
- siRNA
small interfering RNA
- TrpRS
tryptophanyl-tRNA synthetase
- VE-cadherin
vascular endothelial-cadherin
Funding
This work was supported by the grants from Ministry of Science and Technology, Taiwan, ROC (MOST102-2628-B-075-006-MY3 and MOST105-2628-B-075-002-MY3), and Taipei Veterans General Hospital (V104C-041 and V105C-027), the grants by the Association Belge contre les Maladies Neuromusculaire (ABMM) - Aide à la Recherche ASBL and the EU FP7/2007-2013 under grant agreement number 2012-305121 (NEUROMICS), and National Health and Medical Research Council Project Grant APP104668 awarded to M.L.K. and G.A.N. I.M. is supported by a PhD fellowship of the agency for Innovation by Science and Technology (IWT). J.B. is supported by a Senior Clinical Researcher mandate of the Research Fund - Flanders (FWO).
Supplementary material
Supplementary material is available at Brain online.
References
- 1000 Genomes Project Consortium; Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ. et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 2003; 72: 1293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brozkova SD, Deconinck T, Griffin LB, Ferbert A, Haberlova J, Mazanec R. et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain 2015; 138: 2161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari I, Stuart K. A spectrophotometric assay for quantitative measurement of aminoacyl-tRNA synthetase activity. J Biomol Screen 2013; 18: 490–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M. Aminoacyl-tRNA synthestases. Curr Opin Struct Biol 1995; 5: 48–55. [DOI] [PubMed] [Google Scholar]
- Dierick I, Baets J, Irobi J, Jacobs A, De Vriendt E, Deconinck T. et al. Relative contribution of mutations in genes for autosomal dominant distal hereditary motor neuropathies: a genotype-phenotype correlation study. Brain 2008; 131: 1217–27. [DOI] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged tRNA activates GCN2 by displacing the protein kinase moiety from a bipartite tRNA-binding domain. Mol Cell 2000; 6: 269–79. [DOI] [PubMed] [Google Scholar]
- Dubourg O, Azzedine H, Yaou RB, Pouget J, Barois A, Meininger V. et al. The G526R glycyl-tRNA synthetase gene mutation in distal hereditary motor neuropathy type V. Neurology 2006; 66: 1721–6. [DOI] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, Leung CL. et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet 2004; 36: 602–6. [DOI] [PubMed] [Google Scholar]
- Gonzalez M, McLaughlin H, Houlden H, Guo M, Liu YT, Hadjivassilious M. et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J Neurol Neurosurg Psychiatry 2013; 84: 1247–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Falk MJ, Gai X, Postrel R, Schüle R, Zuchner S. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat 2015; 36: 950–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DA, Jin K. From angiogenesis to neuropathology. Nature 2005; 438: 954–9. [DOI] [PubMed] [Google Scholar]
- Griffin LB, Sakaguchi R, McGuigan D, Gonzalez MA, Searby C, Zuchner S. et al. Impaired function is a common feature of neuropathy-associated glycyl-tRNA synthetase mutations. Hum Mutat 2014; 35: 1363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE. Inherited neuronal atrophy and degeneration predominantly of lower motor neurons. In: Dyck PJ, Thomas PK, editors. Peripheral neuropathy. 4th edn Philadelphia: Elsevier Saunders; 2005. p. 1603–21. [Google Scholar]
- He W, Bai G, Zhou H, Wei N, White NM, Lauer J. et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 2015; 526: 710–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CT, Tsai PC, Lin CC, Liu YT, Huang YH, Liao YC. et al. Clinical and molecular characterization of BSCL2 mutations in a Taiwanese cohort with hereditary neuropathy. PLoS One 2016; 11: e0147677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem 2000; 69: 617–50. [DOI] [PubMed] [Google Scholar]
- Irobi J, Almeida-Souza L, Asselbergh B, De Winter V, Goethals S, Dierick I. et al. Mutant HSPB8 causes motor neuron-specific neurite degeneration. Hum Mol Genet 2010; 19: 3254–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M. et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet 2006; 38: 197–202. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014; 46: 310–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise Y, Lee SW, Park SG, Fukai S, Sengoku T, Ishii R. et al. A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol 2004; 11: 149–56. [DOI] [PubMed] [Google Scholar]
- Latour P, Thauvin-Robinet C, Baudelet-Mery C, Soichot P, Cusin V, Faivre L. et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2010; 86: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T. et al. Analysis of protein-coding genetic variation in 60, 706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie F, Ruhrberg C. Diverse roles for VEGF-A in the nervous system. Development 2012; 139: 1371–80. [DOI] [PubMed] [Google Scholar]
- Malissovas N, Griffin LB, Antonellis A, Beis D. Dimerization is required for GARS-mediated neurotoxicity in dominant CMT disease. Hum Mol Genet 2016; 25: 1528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Liu C, Igarashi T, Pehlivan D, Chu K. et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am J Hum Genet 2010; 87: 560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Giblin W, Program NCS, Wilson TE, Biesecker L. et al. A recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with Charcot-Marie-Tooth disease type 2N (CMT2N). Hum Mutat 2012; 33: 244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Laura M, Fawcett K, Pandraud A, Liu YT, Davidson GL. et al. Charcot-Marie-Tooth disease: frequency of genetic subtypes and guidelines for genetic testing. J Neurol Neurosurg Psychiatry 2012; 83: 706–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossor AM, Kalmar B, Greensmith L, Reilly MM. The distal hereditary motor neuropathies. J Neurol Neurosurg Psychiatry 2012; 83: 6–14. [DOI] [PubMed] [Google Scholar]
- Rossor AM, Polke JM, Houlden H, Reilly MM. Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat Rev Neurol 2013; 9: 562–71. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–2. [DOI] [PubMed] [Google Scholar]
- Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 2006; 51: 715–26. [DOI] [PubMed] [Google Scholar]
- Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res 2006; 34: 3246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzima E, Reader JS, Irani-Tehrani M, Ewalt KL, Schwartz MA, Schimmel P. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J Biol Chem 2005; 280: 2405–8. [DOI] [PubMed] [Google Scholar]
- Vester A, Velez-Ruiz G, McLaughlin HM, Program NCS, Lupski JR, Talbot K. et al. A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum Mutat 2013; 34: 191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M. et al. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci USA 2002; 99: 173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallen RC, Antonellis A. To charge or not to charge: mechanistic insights into neuropathy-associated tRNA synthetase mutations. Curr Opin Genet Dev 2013; 23: 302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Schimmel P, Ewalt KL. Relationship of two human tRNA synthetases used in cell signaling. Trends Biochem Sci 2004; 29: 250–6. [DOI] [PubMed] [Google Scholar]
- Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C. et al. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J 2006; 25: 2919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Hashiguchi A, Hu J, Sakiyama Y, Okamoto Y, Tokunaga S. et al. Alanyl-tRNA synthetase mutation in a family with dominant distal hereditary motor neuropathy. Neurology 2012; 78: 1644–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Kapoor M, Guo M, Belani R, Xu X, Kiosses WB. et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol 2010; 17: 57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.